Abstract
The COVID-19 pandemic has had a devastating impact worldwide and has brought clinical assays both for acute diagnosis and prior exposure determination to the forefront. Serological testing intended for point-of-care or laboratory use can be used to determine more accurate individual and population assessments of prior exposure to SARS-CoV-2; improve our understanding of the degree to which immunity is conveyed to subsequent exposures; and quantify immune response to future vaccines. In response to this pandemic, initially more than 90 companies deployed serology assays to the U.S. market, many of which made overstated claims for their accuracy, regulatory approval status, and utility for intended purpose. The U.S. Food and Drug Administration subsequently instituted an Emergency Use Authorization (EUA) procedure requiring that manufacturers submit validation data, but allowing newly developed serological tests to be marketed without the usual approval process during this crisis. Although this rapid deployment was intended to benefit public health, the incomplete understanding of immune response to the virus and lack of assay vetting resulted in quality issues with some of these tests, and thus many were withdrawn after submission. Common assay platforms include lateral flow assays which can serve an important niche of low cost, rapid turnaround, and increased accessibility whereas established laboratory-based platforms based on ELISAs and chemiluminescence expand existing technologies to SARS-CoV-2 and can provide throughput and quantification capabilities. While most of the currently EUA assays rely on these well-established platforms, despite their apparent technical simplicity, there are numerous practical challenges both for manufacturers in developing and for end-users in running and interpreting such assays. Within are discussed technical challenges to serology development for SARS-CoV-2, with an emphasis on lateral flow assay technology.
Keywords: SARS-CoV-2, COVID-19, Lateral flow assay, Serological testing, Rapid diagnostics, Immunoassays
Abbreviations: EUA, Emergency Use Authorization; LFA, Lateral flow assay; ELISA, Enzyme-linked immunosorbent assay; POC, Point-of-Care; PPV, Positive Predictive Value; NPV, Negative Predictive Value; CLIA, Clinical Laboratory Improvement Act
Graphical abstract
This work provides a thorough and technical view of COVID-19 serology assay development spanning considerations from engineering design of these assays to timing of antigen-specific antibody development. Within, we consider the regulatory aspects and correlations between analytical and clinical performance for such assays.
1. Introduction
The COVID-19 pandemic has been devastating worldwide, with known infections of COVID-19 exceeding 42.8 million and more than 1.15 million deaths as of October 25, 2020 [1]. In an attempt to contain the spread of this disease and limit the impact of high numbers of cases in short time spans on our healthcare systems, shutdowns of non-essential businesses and establishments and strict social distancing were instituted which had a destructive economic impact, such that in October 2020 the International Monetary Fund predicted the global GDP to be −4.4% in 2020 [2]. Collateral damage from stay-at-home orders and job losses ranging from increased domestic violence to delayed medical care exacerbated these challenges [3,4]. To mitigate further personal and economic hardships, an understanding of the epidemiology and naturally acquired immunity to COVID-19 through widespread serological testing is critical. Serology testing is not meant to diagnose an active infection but instead can answer important questions pertaining to public health, including determining accurate rates of infection, identifying people who have recovered from COVID-19 and can donate convalescent plasma, whether and for how long antibody presence conveys immunity and verifying future vaccine response. Estimates on secondary infections generated by an infected individual in the United States vary significantly with disease prevalence and population density, and are continuing to evolve as more accurate estimates of prevalence develop with more widespread testing [5]. The available data indicate that at its peak in New York state in late February 2020, the effective reproduction rate, or number of people who became infected by an infectious person, reached 4.17, but as of October 24, 2020, this number is estimated to be 1.02 [6]. Rates of antibody-development vary widely—as of mid-August 2020, tracked by a nationwide commercial laboratory seroprevalence study via the CDC, it was estimated that approximately 22.5% of the population in New York had positive serological results, while only 0.4% were antibody positive in Maine [7].
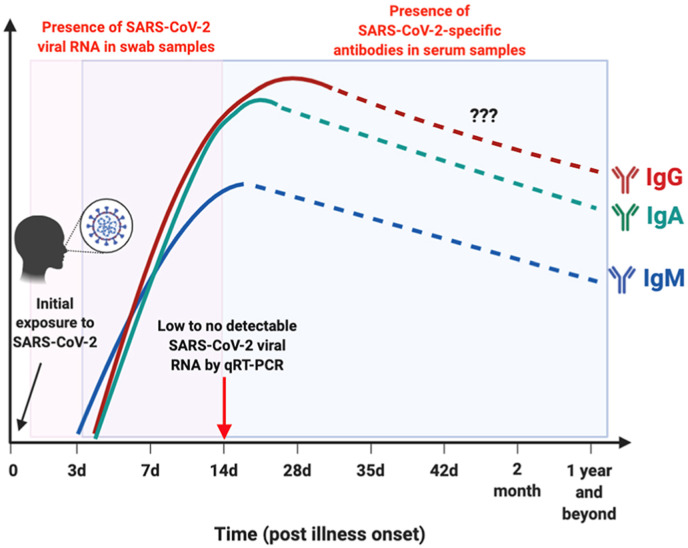
In serum, IgM is present at currently detectable levels in COVID-19 on average after approximately 8 days after onset of symptoms, while IgG can usually be detected after 11 days although some patients reportedly seroconvert starting between 2 and 6 days after symptom onset [[8], [9], [10], [11], [12]]. While the IgM/IgG time profile is often illustrated by a rise and fall of IgM prior to the rise of IgG, in both SARS and SARS-CoV-2, these time profiles have been shown to largely overlap (Fig. 1 ) [9,13]. By the peak of SARS-CoV-2 specific antibody production, little to no detectable viral RNA is present by qRT-PCR [14]. Serology tests offer a wider window of opportunity to detect prior exposure which may have been limited by inability to access RT-PCR testing at peak levels and may provide clarity when clinical presentation of symptoms suggests false negative RT-PCR results [15].
Fig. 1.
Time course of IgM, IgG, IgA development relative to exposure and presence of detectable levels of SARS-CoV-2 viral RNA [14]. Copyright © 2020 Lee, Lin, Renia and Ng. This image was sourced from an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY).
IgA develops at the same time as IgG and is important for immune response in the mucous membranes coating body surfaces [[16], [17], [18]]. IgM, IgG, and IgA comprise approximately 5%, 75%, and 15% of serum immunoglobulins, respectively [19,20]. Many of the serology tests developed to date detect both IgM as an early response and IgG as a more sustained response to disease exposure. The ability to detect these antibodies at early time points depends not only on a patient's immune response, but also on the limit of detection of the assay. Thus, it is important to assess both analytical sensitivity and specificity, which are measures of the performance of the assay itself, as well as diagnostic sensitivity and specificity which are measures of the clinical performance of the assay.
2. Questionable claims of SARS-CoV-2 serology assays
During the early stages of the COVID-19 pandemic in the early spring of 2020, more than 90 companies developed and marketed SARS-CoV-2 antibody tests in the U.S. to meet the demand and the market opportunity that it provided [21,22]. Unfortunately, many of these manufacturers made inaccurate claims for the performance, point of application, or FDA review status of their assays [23]. This included inaccurate claims that some of these products were FDA approved or had been granted FDA permission to distribute; claims by some that they were available for “home-use”; and overreaching suggestions in their documentation for the information that a positive antibody result could provide [[24], [25], [26], [27]]. Hundreds of thousands to multi-million dollar investments in rapid antibody tests were made by private hospital administrators and government officials which were later found to be too inaccurate for use [[28], [29], [30]]. The FDA has maintained a list of such products, issuing as of November 4, 2020, a list of 128 warning letters to sellers, including some for SARS-CoV-2 rapid diagnostic assays directly marketed to consumers [31].
Manufacturers who originally benefited from relaxed regulations amid the urgent need for testing in the early spring of 2020 were then required to submit proof of accuracy through validation to the FDA within 10 days to continue selling their tests [23]. The FDA since cleared some of these tests through their Emergency Use Authorization (EUA) guidance, while many others were removed from distribution [23,24,32]. EUAs can be issued to use unapproved medical products when there are no approved medical products to diagnose, treat, or prevent a life-threatening disease and there are no adequate alternatives [33]. EUAs have been used for other diseases posing a significant risk to public health or national security, including Zika virus, H7N9 Influenza, Ebola, and MERS-CoV [34]. As of 10/24/20, there were 56 FDA EUA cleared immunoassays, 16 of which were lateral flow assays (LFAs, similar to home pregnancy tests), and 40 of which were based on instrumentation-based assays, including enzyme-linked immunosorbent assays (ELISAs) and microparticle-based assays [35]. Per the worldwide list maintained by the Foundation for Innovative New Diagnostics (FIND), as of 10/24/20, 487 serology assays, 309 of which were rapid diagnostic tests, were in various stages of development or regulatory approval status [36]. The numbers of these assays increased markedly during a short span of time, with 24 serology assays with an EUA and 236 serology assays on the FIND list as of 7/12/20, indicative of both the diagnostic need and broad commercial market available during this unprecedented time [35,36].
Part of the demand to make serology tests available rapidly was due to the severity of the health and economic effects demonstrated during the pandemic, but there is a market-driven opportunity to develop these tests as well, given the potential importance of serology tests in broad economic reopening [21]. As of July 14, 2020, Abbott, who had two EUA-authorized COVID-19 serology tests in the spring, showed a year-to-date increase of stock price of 21.3% [37]. For a company that as of May 13th, 2019 had a net worth of 134.4 billion USD [38], this increase is significant. Although serology tests are not the sole driver in increases for most established manufacturers, some manufacturers may have incentive to provide such tests while an unprecedented demand exists. The global market for serology tests has been estimated to grow at a rate of 8.7%, reaching 4.8 billion USD by 2025 [39]. Many of the companies marketing SARS-CoV-2 serological tests are established diagnostic companies with a solid track record of success in assay development, but others with tests under development are from manufacturers without the same history in serological or lateral flow testing [36,40]. Within, we outline some of the technical considerations with respect to immunoassay development, many of which are not trivial to successful implementation.
3. SARS-CoV-2 serology assay platforms
LFAs and other similar rapid diagnostic tests have limited processing requirements for the end-user, can provide yes/no or semi-quantitative results in typically 10–15 min and are usually deployed in the point-of-care setting. They have particular utility for resource-limited settings [15]. Often used as screening assays, the demands for these assays to limit false positives or negatives (depending on the disease context) are high. With the implications of false results during COVID-19, to date SARS-CoV-2 serology tests irrespective of format have been carried out in centralized laboratories, though the first POC serology test received an EUA on 9/23/20 [41]. By contrast, assays such as ELISAs and microparticle-based immunoassays are designed to yield quantitative results in a laboratory setting [[42], [43], [44]]. Aside from submission to a centralized facility, sufficient sample numbers must be accumulated before these typically high-throughput analyses are run. In the current pandemic, this has been secondary to the backlog of samples awaiting testing, but nevertheless, these laboratory-based tests require significant infrastructure and equipment costs, operator skill, and take longer to return results to patients, typically on the order of days rather than minutes possible in a POC setting. With the supplies and resources of many laboratories straining to keep up with the demand for not only SARS-CoV-2 serology tests but also real-time PCR and antigen tests, along with their usual clinical analysis workloads for non-COVID-19 conditions, the availability of serology tests in the POC domain may signal an important shift.
3.1. Lateral-flow/immunochromatographic assays
LFA technology emerged in the 1980s, with patents filed by inventors at major companies including Becton Dickinson, Unilever, and Carter Wallace within a short span of time in the late 1980s for similar technologies [[45], [46], [47], [48]]. In 1988, the first lateral flow assay released commercially was the Clear view home pregnancy test by Unipath, Ltd, a subsidiary of Unilever [49]. The patents by Unilever and Carter Wallace were subsequently acquired by Inverness Medical Innovations, who became a significant stakeholder in the lateral flow market [45]. Patents covering lateral flow technology have been strongly protected historically and the LFA landscape has been marked with commercial acquisitions, with Inverness maintaining a dominant position [[50], [51], [52], [53], [54]]. This company was subsequently renamed Alere in 2010, which was acquired by Abbott in 2017 [55]. The global lateral flow assay market was reportedly worth 6.51 billion USD in 2019 and estimated to grow to 9.65 billion USD by 2024 [56]. LFAs have continued to advance in technology and application, with numerous patent filings and commercial and academic research on cassette designs, matrix improvements, processing methods, signaling species, and improved result readouts [57,58].
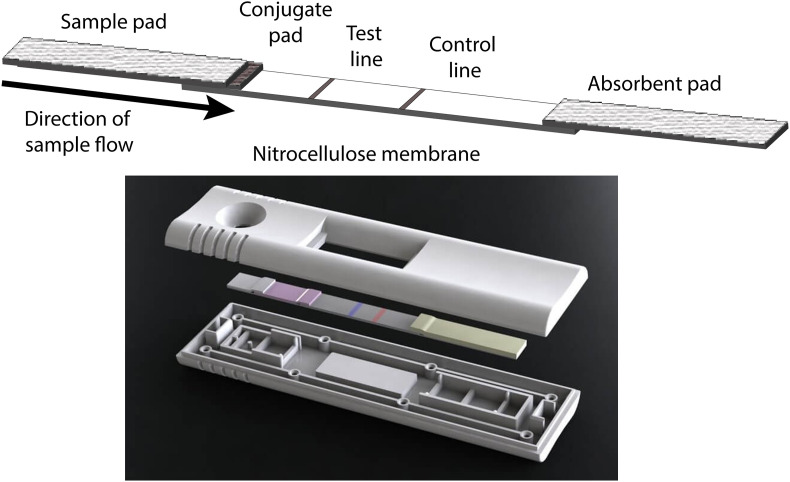
In most lateral flow assays, an antibody or antigen specific to the analyte is immobilized onto a nitrocellulose membrane by adsorption forming a test line (Fig. 2 ) [59,60]. A second zone distal to the point of origin is designated as a control line, where either unbound signaling materials or a second type of particle are captured to indicate that sufficient fluid was applied for successful particle release and capillary flow. Remaining sites on the membrane are blocked with a non-specific protein in the presence or absence of a low concentration of surfactant (such as bovine serum albumin and Tween-20, respectively.) A glass fiber conjugate pad is impregnated with colloidal gold or dyed latex spheres that are conjugated with either another antibody or antigen specific to the analyte of interest. Colloidal gold is available commercially in several sizes, with typical sizes of 20–60 nm used for signal generation in LFAs [61,62]. The small size of these particles allows for significant accumulation at test lines and the intense color of these particles aids in the detection of low level analytes, relative to larger latex beads (typically 100–300 μm). Functionalization of colloidal gold is usually through non-covalent adsorption or dative interactions with biomolecule thiol groups, versus latex beads which usually are covalently modified [61,63]. A cellulose fiber sample pad is assembled onto the conjugate pad and serves to reduce interferences such as red blood cells, white blood cells, and particulates that may discolor the membrane or alter binding of the analyte at the capture zone. An absorbent pad is assembled on the distal end of the membrane to serve as a wick for fluids passing through the assembly. The assemblies are compiled with a user-optimized overlap of components (in our hands, 2 mm) using an adhesive backing to support and retain components, then cut into strips typically approximately 5 mm wide [42]. A key component of these assays is the housing in which they are subsequently encased. Aside from providing physical protection to the sensitive components, the housings restrict evaporation from the membrane surface during running samples, thus ensuring more consistent flow. They are also designed with protrusions so that the LFA components remain in direct contact with each other, ensuring transfer of fluids from one material to the next (Fig. 2). They further provide the user with viewing windows and ports for sample and reagent addition as needed. This established technology allows ready access to both these standard components and a variety of alternate materials are commercially available while research and development on other strategies remains active.
Fig. 2.
Typical layout of lateral flow components. (Top) Lateral flow assay fluid path. The sample is applied to a sample pad through a port in the plastic housing. The sample pad is chosen to remove constituents that interfere with signal generation or cause unnecessary background. The fluid from the sample then passes through the conjugate pad where the dehydrated signaling species (most commonly colloidal gold) with an attached biorecognition molecule becomes rehydrated. The solution then passes onto the analytical membrane where capture and control molecules are immobilized, then lastly is wicked by the absorbent pad at the opposite end of the assembly. (Bottom) The lateral flow assembly is inserted into a plastic housing which protects the sensitive components from mechanical disruption, keeps the strip in place during sample application, and restricts evaporation while the sample is being run. The protrusions in the device parallel to the membrane components are spaced to provide physical contact of the sample pad with the conjugate pad, the conjugate pad with the nitrocellulose membrane, and the absorbent pad with the nitrocellulose membrane to promote and ensure consistent fluid flow. Image used with permission of DCN Diagnostics. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
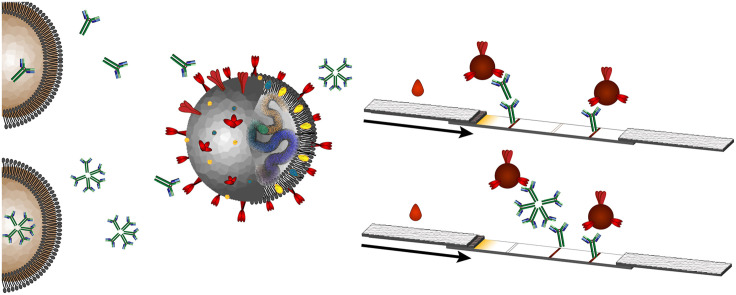
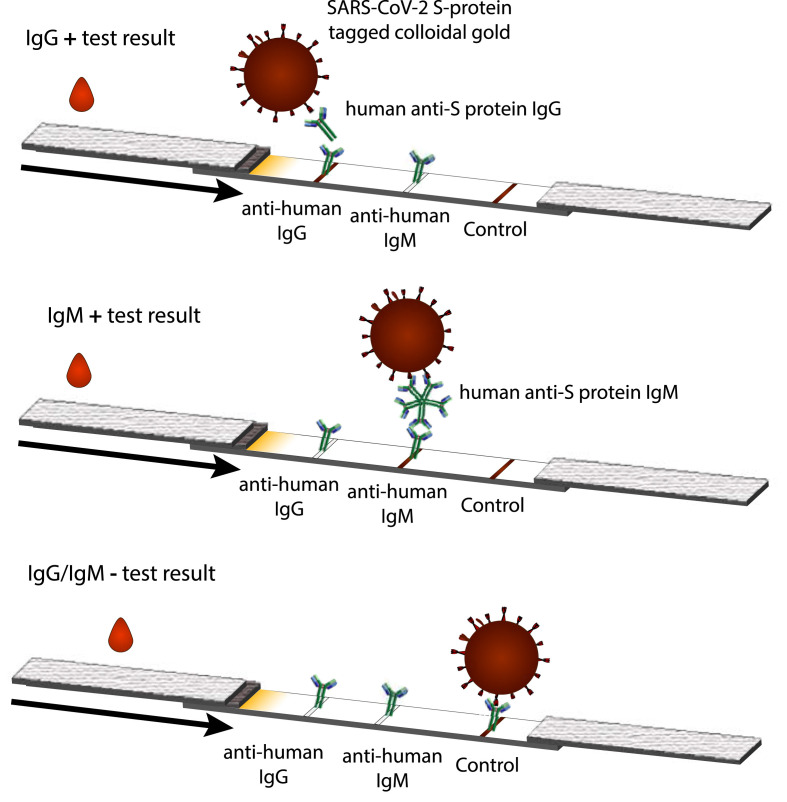
Most of the SARS-CoV-2 semi-quantitative serological tests rely on capture of anti-COVID IgG and/or IgM by immobilized anti-human IgG and/or anti-human IgM, respectively, subsequent to specific recognition of sample anti-SARS-CoV-2 antibodies using SARS-CoV-2 protein tagged colloidal gold for a visually-detectable signal (Fig. 3 ) [23]. The control lines in these assays are usually an immobilized antibody against the antigen labeled on the gold or secondary antibody against a non-specific antibody functionalized on the colloidal gold. This may also be against an antibody on a second control particle that does not participate in specific detection (e.g. goat anti-rabbit control line to capture rabbit IgG colloidal gold) [23,64].
Fig. 3.
For anti-human IgG/IgM detection: (Top) If SARS-CoV-2 IgG is present, sandwich complexes with the SARS-CoV-2 protein on the colloidal gold and the anti-human IgG immobilized on the nitrocellulose membrane can be formed. A visible signal where the anti-IgG is captured is observed as well as a visible signal where a control antibody is immobilized to indicate successful fluid flow and conjugate release. (Middle) As above, but detecting anti-SARS-CoV-2 IgM in a spatially-distinct zone (Bottom) If the antibody is not present or is at concentrations too low to yield an analyte-specific signal, a signal at the control line only is observed. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Although simple technology, lateral flow assays are capable of detecting analytes at low concentrations in complex matrices. The limits of detection possible with a lateral flow assay stem from the analyte being forced to pass the capture lines by capillary action with minimal mass transfer limitations, relative to solution-phase assays which rely on diffusion from the bulk solution volume to a surface immobilized biomolecule. Further, the high binding capacity of nitrocellulose allows significant levels of protein to be immobilized (mg/mL range, binding capacity between 50 and 200 μg IgG/cm2) relative to polystyrene microplates (μg/mL range, binding capacity: 600–650 ng/cm2) typical of higher-throughput ELISAs [65,66]. These properties can mitigate the rapid binding kinetics required of these assays relative to the extended incubation times (typically 1–2 h) in a standard microplate-based ELISA format.
3.2. ELISA, chemiluminescence, and microparticle-based platforms
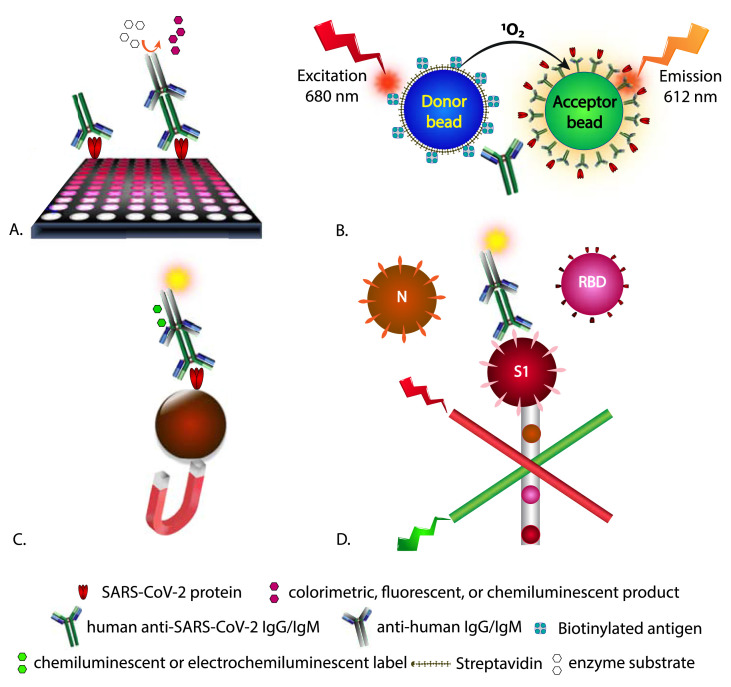
Laboratory-based heterogeneous serology methods for SARS-CoV-2 rely on similar assay designs, but typically use enzymatic or chemiluminescent signaling in lieu of colloidal gold for signal generation. Another variation on assay design is the formation of sandwich complexes between immobilized SARS-CoV-2 proteins, patient IgG, IgM, and IgA, and a second SARS-CoV-2 protein that is labeled, allowing for total antibody detection [67]. Such assays can provide quantitative results in a high-throughput manner (96-well or larger microplates), but also can be used for semi-quantitative or qualitative results. Available assays provide a read-out for total SARS-CoV-2 antibody (IgG, IgM, and IgA) or separate immunoglobulin classes. Commonly, such assays rely on capture of anti-SARS-CoV-2 IgG and/or IgM by immobilized SARS-CoV-2 proteins and detection by labeled anti-human IgG and/or IgM with intermediate wash steps. In such assays, the target antigen is typically coated on the surface of microplate wells (Fig. 4 A) or bead-based microparticles (Fig. 4B-D) by adsorption or covalent modification. Many of the high-throughput clinical analyzers rely on magnetic microparticles in lieu of the surface of a microwell plate which provide increased antigen coating surface area, improved diffusion throughout the sample matrix, and reduced incubation times [68,69]. In these assays, a static magnet or electromagnet is used to isolate the magnetic beads from the bulk volume. Anti-SARS-CoV-2 antibodies potentially present in patient samples are incubated with the immobilized SARS-CoV-2 antigen to allow binding. This is subsequently followed typically by adding a secondary anti-human IgG or IgM antibody conjugated with enzyme followed by a colorimetric, fluorometric, or chemiluminescent signal-producing substrate for ELISAs or chemiluminescent enhancement reagent for chemiluminescent immunoassays or application of voltage for electrochemiluminescent immunoassays, respectively. Specific examples include horseradish peroxidase with tetramethylbenzidine for colorimetric detection at 450 nm; alkaline phosphatase with 4-methylumbelliferyl phosphate for fluorescence detection; directly conjugated labels including isoluminol and acridinium derivatives with signal enhancement reagents for chemiluminescence and ruthenium-based labels for electrochemiluminescence [35]. An imaging microarray platform using colloidal-gold labeled antibodies similar to those employed in LFAs, but using silver-enhancement has also received an EUA [70].
Fig. 4.
Example assay formats for anti-SARS-CoV-2 IgG/IgM detection designed for a laboratory setting. These formats commonly use an immobilized recombinant SARS-CoV-2 protein for class-specific antibody isolation or total antibodies developed against SARS-CoV-2, followed by detection with a labeled secondary anti-human antibody A.) ELISA formats using visible, fluorescent, or chemiluminescent substrates in microtiter plates, B.) homogeneous assays reliant on proximity of binding entities and energy transfer prior to light emission. C.) magnetic bead-based isolation of immunocomplexes reliant on luminescence, or electrochemiluminescence detection and D.) coding of binding events using the Luminex® platform where beads with specific dye formulations are conjugated to a single SARS-CoV-2 protein and form a sandwich complex with a fluorescently labeled antibody detection species with separation of beads/complexes in a flow-based channel.
Additions in these multi-step assays are separated by appropriate wash steps to remove unbound materials. The signal generated is proportional to the amount of antibody-enzyme conjugate bound to the underlying complex. Commercially available clinical analyzers carry out these steps in an automated manner allowing for processing of up to hundreds of samples per hour (e.g., 400 samples per hour using the Abbott Architect i4000SR analyzer [71] and 18 min per sample using the Elecsys® anti-SARS-CoV-2 assay on the Roche cobas® e immunoassay analyzer) [72]. Some of the other platforms with currently issued EUAs include microfluidic devices with spatially distinct anti-IgG/IgM immobilization zones and analysis using fluorescent, antigen-coated beads and a benchtop reader [73]; a Luminex® bead-based multiplex platform for IgG using the N protein and RBD and S1 subunits of the S protein [74] (Fig. 4); and a multiplex assay based on ring-resonators reliant on a change in resonance wavelength upon binding of patient antibodies to immobilized viral antigens on a silicon chip, preceding goat anti-human IgG and IgM [75].
Aside from heterogeneous assays requiring wash steps, homogeneous assay platforms reliant on proximity-based signaling resulting from complex formation are available for SARS-CoV-2 serology assays. The Luminescent Oxygen Channeling Immunoassay (LOCI) technology from Siemens relies on anti-fluorescein isothiocyanate (FITC) coated, chemiluminescent microbeads (Chemibeads®) bound to a fluorescein-labeled RBD S1 antigen which in the presence of patient anti-SARS-CoV-2 antibodies forms a complex. Following a 1 min incubation, a biotinylated antigen is then added followed by streptavidin coated microbeads (Sensibeads®). The bead proximity due to the antigen-antibody complex formation allows for a signaling event without the requirement of wash steps [76]. Upon excitation at 680 nm, singlet oxygen is released from the Sensibead which can diffuse into the nearby Chemibead and triggers a chemiluminescent signal which can then be measured at 612 nm (Fig. 4).
4. Challenges with serology assay development and implementation
Full consideration of clinical sample matrices and assay formats is beyond the scope of this article, but included here are technical considerations that are notable to SARS-CoV-2 serological assay development. Readers are directed to references such as [77,78] which provide a thorough treatment of possible interferences in immunoassays and evaluation of clinical considerations.
4.1. Sample matrix and sampling
SARS-CoV-2 serodiagnostic assays currently use either serum, plasma, venous whole blood, or capillary blood [64,67,73,79]. Laboratory studies typically use serum or plasma as the test matrix, though many EUA LFAs state all three samples can be used without apparent preference [23]. The FDA has recommended 95% agreement across matrices to be considered equivalent [80]. In serology tests developed previously for other infectious diseases, there was no evidence of a difference in capillary versus venous blood for antibody concentrations or profiles that impacted testing capabilities [[81], [82], [83]]. Thus, finger-stick blood obtained through a lancet, as is common with home glucometers, rather than venous draw which necessitates trained phlebotomists and a clinical setting is an option and is currently used in some EUA tests. Although at present there is only one assay designed for the point-of-care setting with an EUA, ultimately, self-sampling would reduce the risk exposure of healthcare providers to COVID-19 and other communicable diseases and allow for decentralized testing [84]. The low collection volumes further can facilitate testing in infants and the elderly, yet as most studies reference serum or plasma, the comparison to whole blood does warrant further investigation [15,78]. A high rate of false negatives was noted in a SARS-CoV-2 LFA when capillary blood rather than serum was used [85].
Heat-inactivation of serum samples for 30 min at 56 °C has been suggested to reduce virus infectivity and exposure concerns and can minimize background through inactivation of the complement system, but can also yield denaturation and aggregation of IgG and IgM thus interfering with serological detection [[86], [87], [88]]. The limited available information to date indicates mixed results on the subsequent impact of heat inactivation on immunological detection for SARS-CoV-2, indicating increased false negatives in an IgM immunochromatographic assay and no significant impact on an ELISA for detection of IgG and IgM [89,90]. Dried blood spot collections provide an opportunity for greater access to testing through easier sample collection and submission, though have similarly had mixed results with one study showing a 30-fold higher limit of detection using dried blood spots than for plasma but another showing comparable results [91,92]. Assays using saliva as a matrix offer less invasive sampling and recent studies have shown good correlation of saliva IgG and IgM to serum in COVID-19 patients [93,94]. Saliva-based antigen and molecular diagnostic assays have received an EUA, but no serological saliva tests have received this authorization as of 10/24/20 [35,94,95].
Non-specific binding by plasma constituents in such complex matrices can yield false positive or false negative results [96]. Variable hematocrit levels can impact the rate of blood flow on the assay as a higher percentage of red blood cells can result in faster coagulation and therefore less fluid movement [97,98]. For example, blood samples from people with anemia or who are on anticoagulants would migrate faster than those with more viscous blood which may change the binding characteristics and assay sensitivity. Additionally, red blood cells that either are inadequately separated by the sample membrane or become lysed during processing can contribute background color that can mask the reaction of the target analyte. Colloidal gold usually offers a lower limit of detection, but the reddish-brown color may be difficult to discern against the background imparted by colored sample matrices such as whole blood, hence dyed latex microspheres which are available in various colors are sometimes employed in LFAs [61,97].
At peak levels, specific SARS-CoV-2 IgA, IgG, and IgM concentrations of 8.84 μg/mL, 16.6 μg/mL, and 7.25 μg/mL were observed in patient serum samples, respectively [19]. This concentration range is well above the typical pg/mL and ng/mL limit of detection of most ELISAs and lateral flow assays, respectively. For LFAs, the samples are usually applied neat to a sample well in the LFA housing followed by introduction of buffer to a separate well downstream of the sample [64,99]. The concentrated application can allow these assays to achieve detection of relatively low-level analytes, but also can make them prone to endogenous or exogenous interferences as other components in the sample matrix are also not diluted and may increase flow challenges depending on sample viscosity. For SARS-CoV-2 assays, a typical volume ratio of sample to buffer is 1:5 to 1:10. By contrast, with ELISAs, chemiluminescence immunoassays, or electrochemiluminescence immunoassays, the samples are typically diluted more significantly and prior to incubation with the surface-bound antigen, reducing the potential impact of the sample matrix and interpatient variability thereof. Dilutions of 1:50 to 1:101 are common for screening purposes and antibody titers may be obtained with a series of dilutions which are useful for quantitative characterization of convalescent plasma or understanding of seroconversion [100]. In validation panels run by the CDC, serum or plasma samples were diluted 1:100–1:6400 and any sample yielding a positive signal in an ELISA at the highest dilution was assigned a titer of 1:6400 [101].
4.2. Affinity and kinetics
As is the case with any assay reliant on biomolecular interactions, binding and thus limits of detection are a function of the kinetics and affinity of the antigen-antibody interactions [102]. This can be a greater challenge in non-equilibrium-based assays such as LFAs than for assays that rely on prolonged incubation steps and externally applied mixing. Nitrocellulose membranes used for LFAs are produced commercially that can yield varying wicking rates and adsorption capacities. As nitrocellulose is formed as a fibrous matrix, rather than defined particles, membranes are characterized in terms of capillary rise rate, defined as the time required for a fluid to transverse 4 cm. This characterization is more appropriate than traditional flow rate since fluid flow is not constant over the length of a LFA membrane [103]. For most commercially available membranes, this time varies between 60 and 180 s [42]. There is typically a trade-off of increasing assay speed with an increase in the limit of detection [104]. As the width of the test line is usually only on the order of 1 mm and fluid flows only in one direction, there is limited time for the antibody-antigen binding event to occur. If the capillary rise rate were to double, the limit of detection of the assay would be estimated to decrease four-fold [66]. The kinetics of the on-rate for the interaction are critical given the transient time that an antibody-antigen interaction can take place [105]. LFAs intended as SARS-CoV-2 serology assays often test for both IgG and IgM antibodies immobilized in spatially distinct zones. Aside from the choice in nitrocellulose membrane as a function of capillary flow rate, there are design considerations to placement of the LFA lines, with respect to the altered binding kinetics that occur over the length of the nitrocellulose membrane [106,107]. When immobilized adjacent to the sample pad, a relatively large amount of fluid is available and hence fluid flow through the test line is at its greatest. As fluid passes further up the membrane towards the absorbent pad, evaporation from the surface and depletion of fluid from the reservoir causes fluid flow to slow [[106], [107], [108], [109]]. These altered kinetics affect the duration of the interaction with the immobilized species, having an impact on the limit of detection of these assays as well as the increasing the potential for non-specific interactions as the fluid flow is reduced [106].
4.3. Assay engineering
Technical challenges with manufacturing and storage of LFAs also need to be considered [42,66,103,105]. For LFAs, the nitrocellulose membrane is made through precipitating a polymer of a defined viscosity onto a backing for support [103]. These polymers can be difficult to generate consistently as multiple polymer grades are often blended to create the correct viscosity, but this mixing can lead to molecular weight variation between lots, which along with storage conditions of the membranes, can impact wicking rate [61,102,103,110]. Aggregation and poor release from the conjugate pad or migration through the nitrocellulose can occur, especially with latex beads used for signal generation. Additionally, inconsistent conjugate release may be caused by either a lack of contact or excessive compression by the housing leading to poor fluid transfer from the conjugate pad to the nitrocellulose membrane (Fig. 3). In an independent evaluation of SARS-CoV-2 LFAs, instances of assays not yielding a control line were noted, requiring the addition of another portion of buffer for the sample migration to be complete [96]. Such user interventions can alter the performance characteristics of the assay and are usually not accounted for by the manufacturers. These tests typically use low sample volumes (on the order of 10–20 μL) which can be restrictive as far as the lower end of detectable analyte concentrations, including antibodies present at low titer [19,61,103]. Some of this may be compensated for by increasing immobilized biomolecule concentrations, but there is a binding saturation limit for the surface. Lacking sufficient adsorption sites, biomolecule concentrations that are too high may leach from the immobilization site on the nitrocellulose yielding a more diffuse line and increased limits of detection. Labeled particles typically undergo multiple washing steps prior to dehydration on the conjugate pads, but loosely adsorbed biorecognition molecules can also leach from the particles, yielding unlabeled entities that still capable of participating in the antibody-antigen complex [111].
In both rapid diagnostic and laboratory-based platforms, immobilization onto solid supports or linkage to signal-generating species can yield inactivation of some potentially significant fraction of the biomolecule by way of denaturation, obscuring binding sites, deactivation of binding sites, or steric hindrance. Higher concentrations of immobilized biomolecule do not necessarily yield corresponding returns in assay performance. This is especially the case when biorecognition is reliant on a conformational change [112,113]. Hydrophilic spacers, selective covalent conjugation chemistry, or oriented immobilization strategies can be used to mitigate these effects and also reduce the concentrations of biorecognition element needed. In some cases, increasing the labeled particle or labeled antibody concentration can be useful to increase the signal intensity, but this may result in increased assay costs and potential for increased background. In sandwich LFAs, as there typically is either no wash step or at most only one buffer addition step after sample addition, assay developers need to be concerned with the formation of incomplete sandwich complexes. In this so-called ‘Hook effect’, the analyte complexes with one, but not both biomolecules used for recognition, hence the actual analyte concentration is lower than would appear based on the signal [114]. While this effect can be tested for by running the sample at greater dilutions, this increases per sample cost and reduces throughput. For LFAs, if the same signaling particle is used both for recognition of the analyte as well as the control line, high analyte concentrations may deplete the particles, yielding a reduction in the control signal.
The assay dynamic range and limit of detection can be altered by the placement of the immobilized lines, biomolecule concentrations and conjugation chemistry, but are also influenced by blocking reagents and stabilizers added. As the surface area available in a given nitrocellulose membrane increases, the protein immobilization capacity increases [66]. The breadth of the immobilization line impacts its signal intensity, with greater signal intensity and lower limits of detection expected with narrower immobilization lines. This parameter is impacted by the capillary rise time of a membrane and the diluent used, which can both influence the opportunity for the biomolecule to spread during deposition. Reagent optimization and handling conditions during immobilization, dehydration, and storage are further critical to ensure sufficiently active capture reagent and conjugate upon rehydration during the analysis. Some of this may be tempered by correct pH, salt, buffer, and stabilizer levels during the manufacturing of LFAs [115]. Optimal buffers, immobilization conditions, signaling species, housing designs and standards vary across assays, often necessitating analyte-dependent development. As proprietary information gained through their development or challenges within is often not reported, optimization of a new LFA often occurs from the ground up, without the benefit of a priori information [116,117].
4.4. Interpretation of results
Errors in running and interpreting some assays may occur due to improper training of operators, failure to follow protocols, or poorly written protocols [116,118]. In a survey of 436 laboratories performing point-of-care testing before the emergence of SARS-CoV-2, 19% of testing personnel had not been trained or evaluated in running assays, and 25% did not follow the assay's procedure [118]. The subjective nature of visually interpreted lateral flow assays is an inherent weakness in their implementation, as can be the lack of a quantitative read-out. The cut-off refers to the level of analyte that indicates whether a result is deemed diagnostically positive or negative. In a qualitative assay such as an LFA, this is usually the presence of a visually discernible signal over the background, with semi-quantitative measurements sometimes addressed against a color intensity chart or densitometry measurement. During evaluation of LFAs for SARS-CoV-2 early in the pandemic, a relatively high rate of false positives found in pre-COVID-19 pandemic samples. Of the 39 false positives observed, 22 were attributed to weak signals visually scored as positive results [96]. To reduce the rate of false positives, the cut-off for assigning a positive or negative result can be increased to assume a more intense signal indicating higher concentrations of antibody. However, adjusting the cut-off results in a trade-off. While increasing the criteria for visually assigning a sample as positive improved the specificity through reducing false positives, it also decreased the sensitivity through yielding more false negatives for these point-of-care assays [96].
Similarly, considerations for instrument-based assays include the selection of an appropriate threshold signal whereby signals below this value are deemed non-reactive and those above this value are deemed reactive. For example, in a colorimetric SARS-CoV-2 IgG ELISA, signals above an OD490 = 0.15 were reported as a presumptive positive result [119]. Results are also expressed as a signal/cut-off ratio where the signal from the patient sample is reported with respect to that of a calibrator. For example, in the Euroimmun colorimetric SARS-CoV-2 IgG ELISA, a ratio of <0.8 would be deemed negative, ≥1.1 positive, and 0.8 to 1.1 borderline [100]. The selection of this level has implications for the rate of false negatives and false positives. Assay performance becomes more variable for assays if the peak window for antibody development in the course of the infection is missed, hence multiplex assay designs targeting multiple immunoglobulin classes have a sensitivity advantage. The time needed until detection is possible in patient samples depends on the performance of the individual assay and has been defined by variable criteria making comparisons difficult [23]. By adjusting the threshold and time post symptom onset for samples, reported performance of commercial assays could be adjusted to meet a desired specificity level [120].
5. SARS-CoV-2 antigen selection and cross-reactivity
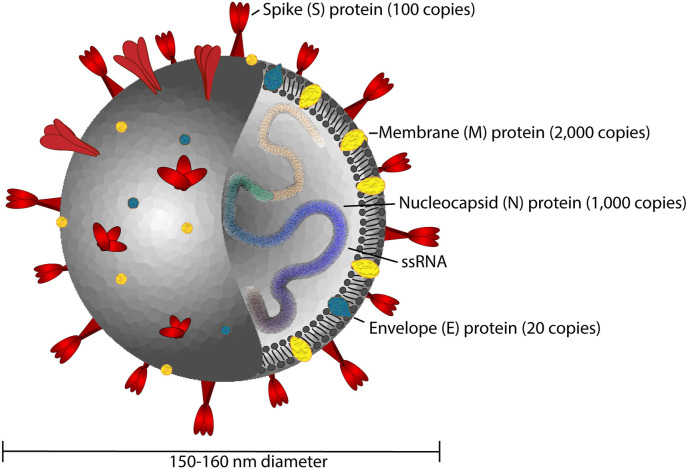
The structure of SARS-CoV-2 is typical of coronaviruses, consisting of nucleocapsid (N), membrane (M), envelope (E) and spike (S) proteins (Fig. 5 ) [121,122]. This is an enveloped, positive sense RNA virus approximately 150–160 nm in diameter [123]. The most commonly used antigens for testing SARS-CoV-2 immune response have been the N and S proteins. The phosphorylated N-protein binds to the viral RNA genome and is an approximately 50–60 kDa non-glycosylated species present in high abundance [124]. The S proteins are ~150 kDa glycoproteins with significant N-linked glycosylation which form trimeric species responsible for interaction with host cell receptors [121]. An estimated 74 S proteins and 1000 N proteins are present per virus particle, respectively [121,125]. S proteins are composed of two subunits, S1 and S2. The receptor binding domain (RBD) formed from the distal tips of the S1 subunit, binds to receptors on the host cell, whereas the S2 domain mediates fusion with the cell membrane [126,127]. The known coronavirus types that infect humans (OC43, HKU1, NL63, and 229E) have variable modes of entry into host cells. OC43 and HKU1 bind to 5-N-acetyl-9-O-acetyl-sialosides on host cell surface glycoproteins; 229E binds to CD13, whereas NL63, SARS, and SARS-CoV-2 target the angiotensin-converting enzyme 2 (ACE-2) receptor [126,128,129]. The affinity of ACE-2 for the S1 subunit constituting the RBD of SARS-CoV-2 is reportedly at least 10-fold greater than for SARS which may contribute to its greater infectivity [130].
Fig. 5.
Structure of SARS-CoV-2 consisting of envelope (E), membrane (M), spike (S), and nucleocapsid (N) proteins, the latter of which is complexed with single stranded RNA in the interior of the virus particle (Approximate protein copy numbers sourced from Refs. [122].).
Given the high prevalence of endemic coronaviruses, cross-reactivities with conserved viral antigens are a concern for assay development. Specificity towards SARS-CoV-2 in these tests is conferred by either S or N proteins, both of which have been shown to prompt an immune response and yield neutralizing antibodies [12,131]. The N protein is expressed at high levels and is responsible for early immune response in SARS [132,133]. As it is not glycosylated, it can readily be expressed in high yield and comparatively low cost in bacterial expression systems and its recombinant form would mimic the antibody binding to the natural virus [133,134]. However, there is approximately 90% shared sequence identity of the SARS-CoV-2 N-protein with SARS, and ~49% and 20–36% sequence identity with MERS-CoV and less pathogenic coronaviruses (HCoV-OC43, 229E, NL63, HKU1), respectively [12,123,135,136]. A pilot study examining cross-reactivity of the S and N proteins of SARS-CoV-2 versus the less pathogenic viruses showed that the N protein had more cross-reactivity, reducing its appeal as a focus for serological tests [137]. With SARS, cross-reactivity against the N protein from mild CoV strains such as OC43 and 229E was believed to contribute to false positives in samples tested from various patient populations [132,138]. This may be tempered through selection of unique epitopes of the N-protein, though given the homology, the use of the N-protein as an antigen would be unlikely to confer specificity between exposure to SARS or SARS-CoV-2 [123]. However, given the low prevalence of SARS in most populations, with no new reported cases worldwide since 2004 [139], this cross-reactivity would seem an insignificant source of error. The ubiquity of strains responsible for symptoms of the common cold is a larger concern, despite lower homology. While sera obtained from patients before the start of the pandemic can be used as negative controls, the limited availability of positive control sera from patients without prior immunity to these strains can be a challenge for assay development [140,141].
The S-protein is more integral to the viruses' interaction with the cell and the SARS-CoV-2 S-protein shares only 75% homology with the SARS spike protein, with 28–33% amino acid sequence identity with other human coronavirus strains [12,142]. The ELISA developed by the CDC uses the S-protein and showed significant cross-reactivity with MERS and SARS, though recognition by sera from NL63, OC43, 229E, and HKU1 was below the limit of detection [143]. Within the S protein, there was ~90% sequence identity with SARS for the S2 domain and this region was 35–43% homologous to other coronaviruses [12]. The differences across human coronaviruses primarily lie in the S1 domain, with 66% homology to SARS and only 21–25% amino acid identity shared with other human coronaviruses [12]. Hence reliance on it for detection would be expected to yield less cross-reactivity not only with SARS, but also with other less pathogenic and more common strains. The N-terminus of the S1 domain of this protein has even less homology between strains, with 51% homology between SARS and SARS-CoV-2 [142]. Though the receptor binding domain (RBD) on the C-terminus portion of S1 has 74% homology with the RBD of SARS [142], it has also been shown to provide high specificity towards SARS-CoV-2 versus HKU-1, NL63, OC43, and 229E, (19–21% amino acid identity) [12,131]. Thus the N-terminus of S1 and the RBD are both critical determinants for neutralizing antibodies [131]. In SARS, antibodies against the S-protein developed later in the infection, whereas antibodies against the N-protein were more readily detectable early in the infection and in mildly infected patients [12,133,144]. This can be a concern for serological diagnostics as a differential expression of IgG/IgM against N and S-proteins has been observed in patients with varying disease severity and time-course [145]. Some assays are designed to monitor antibodies to multiple SARS-CoV-2 proteins, providing a more comprehensive understanding of immune response [74]. Potential cross-reactivity towards not only other coronaviruses, but also viruses such as HIV, hepatitis B virus, and Epstein-Barr virus should also be evaluated, including an assessment of assays deployed in regions with more endemic infectious agents [15].
6. The push for regulation of SARS-CoV-2 serology assays
On February 4, 2020, the Secretary of the Department of Health and Human Services (HHS) declared that SARS-CoV-2 was a significant threat to the health and security of United States citizens abroad which warranted authorization of diagnostics, PPE, and alternative medical treatments for emergency use [146]. The FDA issued a guidance document for EUAs for In Vitro Diagnostics 25 days after the announcement from HHS, with non-binding recommendations for serology tests subsequently being released in early May 4, 2020 [12,80]. The goal of issuing this guidance document was to expedite the availability of tests developed by both laboratories and commercial manufacturers [80]. Until an assay received an EUA, it was not authorized for use. For serological tests, the FDA suggests cross-reactivity or analytical specificity, class specificity, and clinical agreement studies to establish performance characteristics. Cross reactivity studies carried out by test manufacturers commonly assess response from antibodies that bind to multiple strains of the coronavirus, other common human pathogens, including hepatitis B virus and influenza virus, and autoimmune markers, including anti-nuclear antibodies and rheumatoid factors [100,147]. Class specificity refers to the ability of an assay to discern between different immunoglobulins, typically IgM and IgG in SARS-CoV-2 testing [80]. If testing for total antibodies in clinical specimens, the test should demonstrate a minimum sensitivity of 90% and a minimum specificity of 95%. If the test is antibody specific, the test should demonstrate a minimum sensitivity for IgG of 90%, a minimum sensitivity for IgM of 70%, and a minimum specificity of 95% [80].
The Clinical Laboratory Improvement Act (CLIA) was passed in 1988 to ensure a high quality of laboratory testing on human specimens used for diagnosing, preventing, or treating disease [148]. Tests are categorized as either low, medium, or high complexity depending on criteria including technical knowledge and extent of reagent or material preparation [148]. Test manufacturers can apply for a CLIA waiver if the particular test is simple to carry with a low rate of error and risk of incorrect results [149]. Laboratory-based assays for SARS-CoV-2 are deemed moderate to high complexity and hence are limited to CLIA certified sites with this clearance [35,100]. During the pandemic, the FDA deemed any EUA point-of-care test to be CLIA-waived and hence could be carried out in facilities that have a CLIA waiver [26]. However, until the test received an EUA, was deemed high complexity by default and as of 10/24/20, all serology tests with the exception of the one point-of-care assay remain moderate to high complexity assays [35,150]. Additional considerations would apply for potential home use [80].
To facilitate the deployment of serology assays, on April 28, 2020, an umbrella EUA was issued by the FDA to centralize validation and streamline review. Assays covered under this umbrella EUA were only those intended for use in moderate to high complexity CLIA labs. Tests eligible for the umbrella EUA were required to undergo an independent validation by the National Cancer Institute (NCI) or another agency designated by the FDA [35]. In some cases, tests that had been independently validated by commercial laboratories were also directed to the NCI to confirm their studies. The NCI independently assessed SARS-CoV-2 serology tests from commercial manufacturers against a panel of known positive and known negative samples. Thirty samples collected from nucleic acid positive patients were used in the panel along with 80 negative samples collected before the 2020 pandemic, including 10 HIV positive samples. The presence of antibodies (IgG, IgM and RBD) in the panels in these samples were confirmed using well-established ELISAs [101]. Out of the 73 serology tests assessed in this study to date, 26 assays yielded performance consistent with their validation and were granted EUA status [151]. Others were voluntarily withdrawn by the manufacturers or removed from the EUA notification list and not issued an EUA. Removal from the notification list occurred due to some manufacturers not submitting an EUA request within the requisite time frame, significant quality control or performance issues that could not or had not been addressed in a reasonable time period, or voluntary withdrawal of the notification to distribute by the manufacturer [23]. Two EUAs were revoked for tests after they were found through further testing to have very poor diagnostic sensitivity despite promising early pre-authorization data [[151], [152], [153]]. However, some caveats with the NCI assay validation process apply, including a lack of understanding of whether the sample panel employed was representative of the general population; that only serum or plasma samples were included, not whole blood; and that information on anticoagulants used in these samples was not known [151]. The process to obtain an EUA evolved during the course of the pandemic along with submission expectations. Without clear benefit to streamlining the EUA process, the umbrella EUA was subsequently revoked as of July 21, 2020 in lieu of issuing individual EUAs to address assay specific considerations and applications [35]. As of October 7, 2020, per the FDA, the current priority of EUA review requests is for assays that can be employed in the point-of-care setting, those for home use or home sample collection, those that reduce dependence on testing supplies, and those that are high-throughput and widely distributed [32].
7. Clinical performance and utility of serology assays for SARS-CoV-2
The technical considerations behind assay design and corresponding analytical performance underlie the ability of an assay to assess patients clinically. Analytical sensitivity and limit of detection are often used interchangeably, but while an assay with greater sensitivity, defined as the slope of the relationship between response versus concentration, can yield a lower limit of detection, the noise and reproducibility of the assay must also be considered [154]. The FDA defines sensitivity as the lowest analyte concentration in the matrix that can be measured with acceptable accuracy and precision (i.e., lower limit of quantification, LLOQ) [155]. An assay that has a low limit of detection will be able to detect IgG/IgM at earlier time points after symptom onset and would be more likely to detect positive patients that may have a lower antibody titer, yielding fewer false negatives (improved clinical sensitivity). Assays with greater analytical specificity are less likely to yield false positive results (improved clinical specificity) due to reduced non-specific interactions and cross-reactivity to other viruses. The value of a positive or negative result in a population is encompassed in its positive or negative predictive value. In a population where the disease is prevalent, a positive result would be more likely to indicate a true positive case and the PPV would be greater, and vice versa for the NPV [156,157].
Numerous studies have surveyed the clinical performance of specific SARS-CoV-2 serology assays to date, many of which are compiled elsewhere [15,23,78,96,117,142,151,[158], [159], [160], [161], [162]]. Some of these tests indicated an insufficient diagnostic reliability for patient testing while others demonstrated clinically solid performance. ELISA and chemiluminescence immunoassays tended to yield improved diagnostic sensitivities in various surveys versus LFAs, with the latter showing moderate rates of both false positives and false negatives in studies using multiple LFAs [96,99,161,162]. The majority of the assays voluntarily withdrawn or not issued an EUA were LFAs [151]. The engineering design of these assays likely varied (e.g., signaling species concentration, particle diameter, capillary flow rate, antigen, immobilized concentrations, and analyte) impacting their analytical sensitivity and specificity. Such technical parameters are often not detailed in clinical performance evaluations. Further, the review status by regulatory bodies of assays used in peer-reviewed publications is often not known. Thus, it is important to consider the technical design and validation status given the challenges with assays released without full vetting especially early in the pandemic. Some of these assays had good clinical accuracy and with limited access to tests, it is also important to consider that currently available assessments of test performance review only a sampling of available assays.
Evaluating comparative assay performance in the context of disease-development timeline and patient populations is also critical to assessment of the accuracy of reported results from serology assays. Assays reliant on total antibody detection or a combination of IgG, IgA, and IgM have yielded improved sensitivity versus IgG or IgM alone [15,159]. With IgG antibody levels reaching a maximum titer approximately two to three weeks after symptom onset, improvements in diagnostic sensitivity would be expected over this period [121,122]. IgG tests offer more consistency and potentially pertinent information on long-term immunity. However, the duration of IgG presence in the body in response to the virus is not yet known [163]. Neutralizing antibodies against SARS peaked four months after disease presentation and could be detected in serum through 16 months, but declined to undetectable levels by 24 months [164]. Available studies show antibody levels dropping amongst patients that elicited an immune response to SARS-CoV-2 with one study showing a median decrease in IgG antibodies from the acute phase to the convalescent phase of infection of 71.1% in asymptomatic patients and 76.2% in symptomatic patients [165]. At least one study found confirmed positive patients, especially those who had been asymptomatic, to become seronegative for IgG in the convalescent phase, although the universality of this finding requires further understanding [117,166]. Such studies may need to be taken into context with the antigen used in the assay employed. Recent studies indicated stability of SARS-CoV-2 IgG for at least three months using a spike trimer antigen whereas one using a linear segment of the C-terminus region suggested an earlier decline [93,165,167]. Further, it has been shown that viral RNA can persist for more than 20 days after symptom onset, hence it is not clear whether the presence of antibodies indicates lack of infectivity [146,168].
The comparator for assay performance is also important. Some assays are compared for their clinical performance against RT-PCR results rather than a vetted antibody assay which has significant timing considerations. RT-PCR assays have high specificity, indicating few false positives, however, for an assay to yield an accurate negative result and thus high sensitivity, the virus needs to be present at detectable levels in the given sample, leading to concern over sampling technique, timing of collection, and virus migration to other parts of the body [169]. Other assays have been compared to viral neutralization assays, which may provide a better understanding of possible protective immunity [99]. While our understanding of the protection afforded by SARS-CoV-2 antibody development continues to evolve, such assays may be able to provide a surrogate to such viral neutralization assays when addressing a universal need for convalescent plasma [99,119]. As such assays typically require the use of the actual pathogen to assess prevention of infectivity in cell culture, most laboratories are not equipped for the necessary Biosafety Level 3 (BSL3) conditions. The availability of this testing is thus limited and assays that allow quantification of antibodies against epitopes of SARS-CoV-2 known to yield neutralization of infectivity will be critical. For vaccine development, a need for serological assays that can distinguish between immunity induced by natural infection and that induced by vaccination will be key.
8. Conclusions and future outlook
The limited availability, but high demand for, diagnostic or serological testing for SARS-CoV-2 led to an influx of tests without the usual FDA-mandated vetting process. In vitro diagnostic companies were under pressure to both increase testing capacity while at the same time facing scrutiny of politicians, the media, and general public. This led to instances of premature marketing of some assays that showed poor sensitivity, specificity, or both. While most of these assays were and are based on well-established lateral-flow assay and ELISA platforms, the technical considerations to developing such assays are not trivial despite apparent simplicity for the end-user. Many of these assays were subsequently withdrawn from regulatory review due to sub-par performance. Regulatory and commercial efforts evolved to validate the many assays being produced thus limiting the impact of poor performing assays on clinical use and epidemiology. However, there is a sizeable market for COVID-19 serological assays and numerous assays based on the same well established platforms remain awaiting potential clearance for distribution. With the number of assays that submitted notification, but did not advance to receive an EUA to date, one question is whether these additional assays provide technical improvements or only put a strain on resources for validation and regulatory consideration. Focusing efforts on developing and distributing a few, well-vetted assay platforms rather than spreading available resources and regulatory efforts on redundant technologies would seem prudent. It is critical to have sound serology assays to allow health care professionals to better track the extent of the spread of SARS-CoV-2 in populations, understand the implications of the presence of antibodies for immunity to reinfection and the effectiveness of future vaccination campaigns, and help develop strategies for containment of future COVID-19 outbreaks.
From a broader analytical perspective, some of the issues within should perhaps give scientists pause for reagents or kits marketed for ‘research use only’ that do not face the same scrutiny and validation that was observed during the pandemic. The costs of such assays are often prohibitive for most laboratories to carry out in-house validation, yet important decisions and research directions are based upon their output. While not used for clinical decision making, the importance of qualified reagents and assays to basic science cannot be overlooked.
Funding
This work was supported in part by the State University of New York (SUNY) Research Seed Grant Program RFP #20-03-COVID, proposal #COVID202059.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge Peter McEntee for his preliminary literature review and writing efforts towards this project.
References
- 1.Coronavirus Resource Center 2020. https://coronavirus.jhu.edu/
- 2.World Economic Outlook . 2020. October 2020: A Long and Difficult Ascent.https://www.imf.org/en/Publications/WEO [Google Scholar]
- 3.Campbell A.M. An increasing risk of family violence during the Covid-19 pandemic: strengthening community collaborations to save lives. Forensic Sci. Int.: Report. 2020;2:100089. doi: 10.1016/j.fsir.2020.100089. 100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfefferbaum B., North C.S. Mental health and the covid-19 pandemic. N. Engl. J. Med. 2020;383:510–512. doi: 10.1056/NEJMp2008017. [DOI] [PubMed] [Google Scholar]
- 5.Ives A.R., Bozzuto C. State-by-State estimates of R0 at the start of COVID-19 outbreaks in the USA. medRxiv. 2020 2020.05.17.20104653. [Google Scholar]
- 6.Rt COVID-19. 2020. https://rt.live/ [Google Scholar]
- 7.Commercial laboratory seroprevalence survey data. 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/commercial-lab-surveys.html
- 8.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H., Wei X., Cai P., Ma W.-L. Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020:1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L., Liu W., Zheng Y., Jiang X., Kou G., Ding J., Wang Q., Huang Q., Ding Y., Ni W., Wu W., Tang S., Tan L., Hu Z., Xu W., Zhang Y., Zhang B., Tang Z., Zhang X., Li H., Rao Z., Jiang H., Ren X., Wang S., Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microb. Infect. 2020;22(4–5):206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okba N.M.A., Müller M., Li W., Wang C., GeurtsvanKessel C., Corman V., Lamers M., Sikkema R., de Bruin E., Chandler F., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B. Severe acute respiratory syndrome coronavirus 2−Specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349(5):508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 14.Lee C.Y.-P., Lin R.T.P., Renia L., Ng L.F.P. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front. Immunol. 2020;11(879) doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cota G., Freire M.L., de Souza C.S., Pedras M.J., Saliba J.W., Faria V., Alves L.L., Rabello A., Avelar D.M. Diagnostic performance of commercially available COVID-19 serology tests in Brazil. Int. J. Infect. Dis. 2020;S1201–9712(20):32211–32216. doi: 10.1016/j.ijid.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10(12):1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.South M.A., Copper M.D., Wollheim F.A., Good R.A. The IgA system. II. The clinical significance of IgA deficiency: studies in patients with agammaglobulinemia and ataxia-telangiectasia. Am. J. Med. 1968;44(2):168–178. doi: 10.1016/0002-9343(68)90148-4. [DOI] [PubMed] [Google Scholar]
- 18.Goldblum R.M. The role of IgA in local immune protection. J. Clin. Immunol. 1990;10(6):64S–71S. doi: 10.1007/BF00918693. [DOI] [PubMed] [Google Scholar]
- 19.Ma H., Zeng W., He H., Zhao D., Yang Y., Jiang D., Zhou P., Qi Y., He W., Zhao C., Yi R., Wang X., Wang B., Xu Y., Yang Y., Kombe Kombe A.J., Ding C., Xie J., Gao Y., Cheng L., Li Y., Ma X., Jin T. 2020. COVID-19 Diagnosis and Study of Serum SARS-CoV-2 Specific IgA, IgM and IgG by Chemiluminescence Immunoanalysis, medRxiv. 2020.04.17.20064907. [Google Scholar]
- 20.Immunoglobulin structure and classes. https://www.thermofisher.com/us/en/home/life-science/antibodies/antibodies-learning-center/antibodies-resource-library/antibody-methods/immunoglobulin-structure-classes.html
- 21.Eder S., Twohey M., Mandavilli A. New York Times; New York, N.Y.: 2020. Antibody Test, Seen as Key to Reopening Country, Does Not yet Deliver [With Graphic(s)] [Google Scholar]
- 22.Antibody Tests Go to Market Largely Unregulated. Warns House Subcommittee Chair; 2020. https://www.npr.org/sections/coronavirus-live-updates/2020/04/26/845164212/antibody-tests-go-to-market-largely-unregulated-warns-house-subcommittee-chair [Google Scholar]
- 23.EUA Authorized Serology Test Performance. 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance [Google Scholar]
- 24.Certain COVID-19 Serology/Antibody Tests Should Not Be Used - Letter to Clinical Laboratory Staff and Health Care Providers. 2020. https://www.fda.gov/medical-devices/letters-health-care-providers/certain-covid-19-serologyantibody-tests-should-not-be-used-letter-clinical-laboratory-staff-and [Google Scholar]
- 25.Warning Letter-Modern Allergy Management LLC Dba Direct Med Solutions LLC. 2020. https://www.fda.gov/inspections-compliance-enforcement-and-criminal-investigations/warning-letters/modern-allergy-management-llc-dba-direct-med-solutions-llc-607769-06172020 [Google Scholar]
- 26.Krishnamoorthi R., editor. C. United States, House, Committee on Oversight and Reform, to ARCpoint Labs Franchise Group, LLC [Letter] 2020. [Google Scholar]
- 27.Warning Letter-Medakit Ltd MARCS-CMS 607603. 2020. https://www.fda.gov/inspections-compliance-enforcement-and-criminal-investigations/warning-letters/medakit-ltd-607603-06102020
- 28.U.K. Paid $20 . They Didn’t Work; 2020. Million for New Coronavirus Tests.https://www.nytimes.com/2020/04/16/world/europe/coronavirus-antibody-test-uk.html [Google Scholar]
- 29.Dudik A., Tomek R. Faulty virus tests cloud China's European outreach over COVID-19. 2020. https://www.bloomberg.com/news/articles/2020-04-01/faulty-virus-tests-cloud-china-s-european-outreach-over-covid-19?srnd=premium
- 30.A Laredo ER spent $500,000 on coronavirus tests. Health officials say they’re unreliable. 2020. https://www.texastribune.org/2020/04/10/laredo-texas-coronavirus-tests-did-not-work/
- 31.Fraudulent Coronavirus Disease 2019 (COVID-19) Products. 2020. https://www.fda.gov/consumers/health-fraud-scams/fraudulent-coronavirus-disease-2019-covid-19-products [Google Scholar]
- 32.FAQs on Testing for SARS-CoV-2. 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-testing-sars-cov-2#nolonger-ivd [Google Scholar]
- 33.FAQs on Emergency Use Authorizations (EUAs) for Medical Devices during the COVID-19 Pandemic. 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/faqs-emergency-use-authorizations-euas-medical-devices-during-covid-19-pandemic [Google Scholar]
- 34.Emergency Use Authorizations for Medical Devices. 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations [Google Scholar]
- 35.In Vitro Diagnostics EUAs. 2020. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas#individual-serological [Google Scholar]
- 36.SARS-CoV-2 Diagnostic Pipeline - FIND. 2020. https://www.finddx.org/covid-19/pipeline/ [Google Scholar]
- 37.Abbott Laboratories. 2020. https://www.marketwatch.com/investing/stock/abt/charts [Google Scholar]
- 38.Abbott Laboratories Net Worth 2006-2020. ABT; 2020. https://www.macrotrends.net/stocks/charts/ABT/abbott-laboratories/net-worth [Google Scholar]
- 39.Covid-19 serology testing market global opportunity and growth analysis 2020. https://www.marketwatch.com/press-release/covid-19-serology-testing-market-global-opportunity-and-growth-analysis-2020-2020-10-09?mod=mw_quote_news
- 40.Coronavirus Test Tracker . 2020. Commercially Available COVID-19 Diagnostic Tests.https://www.360dx.com/coronavirus-test-tracker-launched-covid-19-tests [Google Scholar]
- 41.Coronavirus (COVID-19) Update . 2020. FDA Authorizes First Point-of-Care Antibody Test for COVID-19.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-point-care-antibody-test-covid-19?utm_medium=email&utm_source=govdelivery [Google Scholar]
- 42.Edwards K.A., Korff R., Baeumner A.J. Liposome-enhanced lateral-flow assays for clinical analyses. Methods Mol. Biol. 2017;1571:407–434. doi: 10.1007/978-1-4939-6848-0_25. [DOI] [PubMed] [Google Scholar]
- 43.Nakatomi Y., Sugiyama J. A rapid latex agglutination assay for the detection of penicillin-binding protein 2'. Microbiol. Immunol. 1998;42(11):739–743. doi: 10.1111/j.1348-0421.1998.tb02347.x. [DOI] [PubMed] [Google Scholar]
- 44.Serology-based Tests for COVID-19. 2020. https://www.centerforhealthsecurity.org/resources/COVID-19/serology/Serology-based-tests-for-COVID-19.html#sec2 [Google Scholar]
- 45.Rosen S. Market trends in lateral flow immunoassays. Lateral Flow Immunoassay. 2008:1–15. [Google Scholar]
- 46.Campbell R.L., Wagner D.B., O'Connell J.P. 1984. Solid Phase Assay with Visual Readout [Patent]https://patents.google.com/patent/US4703017A/en [Google Scholar]
- 47.May K.P., Evans Michael, Richards Ian Evans. 1988. Analytical Test Device for Performing Specific Union Tests [Patent]https://patents.google.com/patent/ES2184734T3/en [Google Scholar]
- 48.Rosenstein R.W., Bloomster T.G. 1987. Solid Phase Assay Employing Capillary Flow [Patent]https://patents.google.com/patent/US4855240A/en [Google Scholar]
- 49.Pregnancy Test Kit. 1988. https://collection.sciencemuseumgroup.org.uk/objects/co96440/pregnancy-test-kit-england-1988-pregnancy-test England. [Google Scholar]
- 50.Quidel Affirms Confidence in Patent Litigation. 2004. https://www.sec.gov/Archives/edgar/data/353569/000110465904037120/a04-14020_1ex99d1.htm [Google Scholar]
- 51.Legal Fight between Inverness and Acon Labs Ends with a Purchase. 2006. [Google Scholar]
- 52.Becton, Dickinson Co v. 2001. Inverness Medical Tech., 176 F. Supp. 2d 258 | Casetext Search + Citator.https://casetext.com/case/becton [Google Scholar]
- 53.law . 2020. HIV-testing Companies Settle Patent Suit - Law360.https://www.law360.com/articles/11298/hiv-testing-companies-settle-patent-suit [Google Scholar]
- 54.Inverness Medical Innovations Settles FTC Charges that it Stifled Future Competition in U.S. Market for Consumer Pregnancy Tests. 2008. https://www.ftc.gov/news-events/press-releases/2008/12/inverness-medical-innovations-settles-ftc-charges-it-stifled [Google Scholar]
- 55.Abbott acquisition of Alere set to close on tuesday. https://abbott.mediaroom.com/2017-09-29-Abbott-Acquisition-of-Alere-Set-to-Close-on-Tuesday-October-3-2017 October 3, 2017, 2020.
- 56.Global Lateral Flow Assay Market Analysis: Forecast 2019-2024. 2020. https://www.marketdataforecast.com/market-reports/lateral-flow-assay-market [Google Scholar]
- 57.Beesley N.E., Brewster B.S., Day S.C., Walker A.L. 2002. Test Strips Containing Control Agents and Timing Agents [Patent]https://patents.google.com/patent/DE60233107D1/en [Google Scholar]
- 58.Tang Z., Xiong D., Wu S., Guan Z. 2005. Devices and Methods for Analyte Assays with Built-In Result Reporting Using Recognizable Symbols [Patent.https://patents.google.com/patent/US7704753B2/en).52 [Google Scholar]
- 59.Nakamura K., Tanaka T., Takeo K. Characterization of protein binding to a nitrocellulose membrane. Seibutsu Butsuri Kagaku. 1989;33:293–303. [Google Scholar]
- 60.Pristoupil T.I., Kramlová M., Stĕrbíková J. On the mechanism of adsorption of proteins to nitrocellulose in membrane chromatography. J. Chromatogr. 1969;42(3):367–375. doi: 10.1016/s0021-9673(01)80636-1. [DOI] [PubMed] [Google Scholar]
- 61.O'Farrell B. In: Lateral Flow Immunoassay. Wong R T.H., editor. Humana Press; 2009. Evolution in lateral flow–based immunoassay systems; pp. 1–33. [Google Scholar]
- 62.Sajid M., Kawde A.-N., Daud M. Designs, formats and applications of lateral flow assay: a literature review. J. Saud. Chem. Soc. 2015;19(6):689–705. [Google Scholar]
- 63.Jazayeri M.H., Amani H., Pourfatollah A.A., Pazoki-Toroudi H., Sedighimoghaddam B. Various methods of gold nanoparticles (GNPs) conjugation to antibodies. Sensing and Bio-Sensing Research. 2016;9:17–22. [Google Scholar]
- 64.RightSign COVID-19 IgM/IgG Rapid Test Cassette. 2020. https://www.fda.gov/media/138660/download [Google Scholar]
- 65.Nunc MaxiSorp™ Flat-Bottom. 2020. https://www.thermofisher.com/order/catalog/product/44-2404-21#/44-2404-21 [Google Scholar]
- 66.Rapid Lateral Flow Strips . 2013. Considerations for Product Development.https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma-Aldrich/General_Information/1/tb500en00em-rapid-lateral-flow-test-strips.pdf [Google Scholar]
- 67.Platelia SARS-CoV-2 Total Ab. 2020. https://www.fda.gov/media/137493/download [Google Scholar]
- 68.Yakub G.P., Stadterman-Knauer K.L. Immunomagnetic separation of pathogenic organisms from environmental matrices. Methods Mol. Biol. 2004;268:189–197. doi: 10.1385/1-59259-766-1:189. [DOI] [PubMed] [Google Scholar]
- 69.Safarík I., Safaríková M. Use of magnetic techniques for the isolation of cells. J. Chromatogr. B Biomed. Sci. Appl. 1999;722(1–2):33–53. [PubMed] [Google Scholar]
- 70.Martinaud C., Hejl C., Igert A., Bigaillon C., Bonnet C., Mérens A., Wolf A., Foissaud V., Leparc-Goffart I. Evaluation of the Quotient® MosaiQTM COVID-19 antibody microarray for the detection of IgG and IgM antibodies to SARS-CoV-2 virus in humans. J. Clin. Virol. 2020;130:104571. doi: 10.1016/j.jcv.2020.104571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chew K.L., Tan S.S., Saw S., Pajarillaga A., Zaine S., Khoo C., Wang W., Tambyah P., Jureen R., Sethi S.K. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020;26(9):1256. doi: 10.1016/j.cmi.2020.05.036. e9-1256.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roche Develops New Serology Test to Detect COVID-19 Antibodies. 2020. https://www.roche.com/media/releases/med-cor-2020-04-17.htm [Google Scholar]
- 73.FREND™ COVID-19 Total Ab. 2020. https://www.fda.gov/media/142557/download [Google Scholar]
- 74.xMAP® SARS-CoV-2 Multi-Antigen IgG Assay Package Insert. 2020. https://www.fda.gov/media/140256/download [Google Scholar]
- 75.Maverick™ SARS-CoV-2 Multi-Antigen Serology Panel V2 01030ART-01. 2020. https://www.fda.gov/media/142915/download [Google Scholar]
- 76.SARS-CoV-2 Total Antibody Assay (CV2T) 2020. https://www.fda.gov/media/138757/download [Google Scholar]
- 77.Tate J., Ward G. Interferences in immunoassay. Clin. Biochem. Rev. 2004;25(2):105–120. [PMC free article] [PubMed] [Google Scholar]
- 78.Hanson K., Caliendo A., Cesar A., Englund J., Hayden M., Lee M., Loeb M., Patel R., Altayar O., Alayli A., Sultan S., Falck-Ytter Y., Lavergne V., Morgan R., Murad M.H., Bhimraj A., Mustafa R. Serologic Testing; 2020. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19.https://www.idsociety.org/COVID19guidelines/serology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.SARS-CoV-2 Immunoassays. 2020. https://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease/sars-cov-2 [Google Scholar]
- 80.Policy for Coronavirus Disease-2019 Tests during the Public Health Emergency (Revised) 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised [Google Scholar]
- 81.Vejtorp M., Leerhoy J. Rubella IgG antibody detection by ELISA using capillary blood samples collected on filter paper and in microtainer tubes. Acta Pathol. Microbiol. Scand. B. 1981;89(5):369–370. doi: 10.1111/j.1699-0463.1981.tb00202_89b.x. [DOI] [PubMed] [Google Scholar]
- 82.Novello F., Ridolfi B., Fiore L., Buttinelli G., Medda E., Favero A., Marchetti D., Gaglioppa F. Comparison of capillary blood versus venous blood samples in the assessment of immunity to measles. J. Virol. Methods. 1996;61(1–2):73–77. doi: 10.1016/0166-0934(96)02071-x. [DOI] [PubMed] [Google Scholar]
- 83.Chen T.S., Chang F.Y., Lee S.D. No difference of accuracy between capillary and venous blood in rapid whole blood test for diagnosis of Helicobacter pylori infection. Dig. Dis. Sci. 2002;47(11):2519–2522. doi: 10.1023/a:1020516211030. [DOI] [PubMed] [Google Scholar]
- 84.Miočević O., Cole C.R., Laughlin M.J., Buck R.L., Slowey P.D., Shirtcliff E.A. Quantitative lateral flow assays for salivary biomarker assessment: a review. Front. Public Health. 2017;5(133) doi: 10.3389/fpubh.2017.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Santos V.A.D., Rafael M.M., Sabino E.C., Duarte A.J.d.S. Sensitivity of the Wondfo One Step COVID-19 test using serum samples. Clinics. 2020;75 doi: 10.6061/clinics/2020/e2013. e2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van den Akker R., Hekker A.C., Osterhaus A.D. Heat inactivation of serum may interfere with HTLV-III/LAV serology. Lancet. 1985;2(8456):672. doi: 10.1016/s0140-6736(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 87.Fipps D.R., Damato J.J., Burke D.S. Effects of heat inactivation on HIV antibody screening and confirmatory test systems. Diagn. Microbiol. Infect. Dis. 1988;10(2):103–107. doi: 10.1016/0732-8893(88)90047-8. [DOI] [PubMed] [Google Scholar]
- 88.Soltis R.D., Hasz D., Morris M.J., Wilson I.D. The effect of heat inactivation of serum on aggregation of immunoglobulins. Immunology. 1979;36(1):37–45. [PMC free article] [PubMed] [Google Scholar]
- 89.Hu X., Zhang R., An T., Li Q., Situ B., Ou Z., Wu C., Yang B., Tian P., Hu Y., Ping B., Wang Q., Zheng L. Impact of heat-inactivation on the detection of SARS-CoV-2 IgM and IgG antibody by ELISA. Clin. Chim. Acta. 2020;509:288–292. doi: 10.1016/j.cca.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu X., An T., Situ B., Hu Y., Ou Z., Li Q., He X., Zhang Y., Tian P., Sun D., Rui Y., Wang Q., Ding D., Zheng L. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. J. Clin. Lab. Anal. 2020;34(9) doi: 10.1002/jcla.23411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mulchandani R., Brown B., Brooks T., Semper A., Machin N., Linley E., Borrow R., Wyllie D. 2020. Use of Dried Blood Spot Samples for SARS-CoV-2 Antibody Detection Using the Roche Elecsys ® High Throughput Immunoassay, medRxiv. 2020.10.19.20215228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morley GL T.S., Jossi S., Perez-Toledo M., Faustini S.E., Marcial-Juarez E., et al. Sensitive detection of SARS-CoV-2–specific antibodies in dried blood spot samples. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2612.203309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., Pu A., Christie-Holmes N., Gervais C., Ceccarelli D., Samavarchi-Tehrani P., Guvenc F., Budylowski P., Li A., Paterson A., Yue F.Y., Marin L.M., Caldwell L., Wrana J.L., Colwill K., Sicheri F., Mubareka S., Gray-Owen S.D., Drews S.J., Siqueira W.L., Barrios-Rodiles M., Ostrowski M., Rini J.M., Durocher Y., McGeer A.J., Gommerman J.L., Gingras A.-C. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5(52) doi: 10.1126/sciimmunol.abe5511. eabe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Randad P.R., Pisanic N., Kruczynski K., Manabe Y.C., Thomas D., Pekosz A., Klein S., Betenbaugh M.J., Clarke W.A., Laeyendecker O., Caturegli P.P., Larman H.B., Detrick B., Fairley J.K., Sherman A.C., Rouphael N., Edupuganti S., Granger D.A., Granger S.W., Collins M., Heaney C.D. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. medRxiv. 2020 doi: 10.1128/JCM.02204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sri Santosh T., Parmar R., Anand H., Srikanth K., Saritha M. A review of salivary diagnostics and its potential implication in detection of Covid-19. Cureus. 2020;12(4):e7708. doi: 10.7759/cureus.7708. e7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., Rathore U., Goldgof G.M., Whitty C., Woo J.M., Gallman A.E., Miller T.E., Levine A.G., Nguyen D.N., Bapat S.P., Balcerek J., Bylsma S.A., Lyons A.M., Li S., Wong A.W.-y., Gillis-Buck E.M., Steinhart Z.B., Lee Y., Apathy R., Lipke M.J., Smith J.A., Zheng T., Boothby I.C., Isaza E., Chan J., Acenas D.D., Lee J., Macrae T.A., Kyaw T.S., Wu D., Ng D.L., Gu W., York V.A., Eskandarian H.A., Callaway P.C., Warrier L., Moreno M.E., Levan J., Torres L., Farrington L.A., Loudermilk R.P., Koshal K., Zorn K.C., Garcia-Beltran W.F., Yang D., Astudillo M.G., Bernstein B.E., Gelfand J.A., Ryan E.T., Charles R.C., Iafrate A.J., Lennerz J.K., Miller S., Chiu C.Y., Stramer S.L., Wilson M.R., Manglik A., Ye C.J., Krogan N.J., Anderson M.S., Cyster J.G., Ernst J.D., Wu A.H.B., Lynch K.L., Bern C., Hsu P.D., Marson A. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 2020;38:1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hegener M.A., Li H., Han D., Steckl A.J., Pauletti G.M. Point-of-care coagulation monitoring: first clinical experience using a paper-based lateral flow diagnostic device, Biomed. Microdevices. 2017;19(3):64. doi: 10.1007/s10544-017-0206-z. [DOI] [PubMed] [Google Scholar]
- 98.Li H., Han D., Pauletti G.M., Hegener M.A., Steckl A.J. Correcting the effect of hematocrit in whole blood coagulation analysis on paper-based lateral flow device. Anal. Methods. 2018;10(24):2869–2874. [Google Scholar]
- 99.GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J., van den Akker J.P.C., van Kampen J.J.A., van der Eijk A.A., van Binnendijk R.S., Haagmans B., Koopmans M. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 2020;11(1):3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anti-SARS-CoV-2 ELISA (IgG) 2020. https://www.fda.gov/media/137609/download
- 101.Serology Test Evaluation Report for “Architect I1000 SARS-CoV-2 IgG” from Abbott. 2020. https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3305-a001.pdf [Google Scholar]
- 102.Hsieh H.V., Dantzler J.L., Weigl B.H. Analytical tools to improve optimization procedures for lateral flow assays. Diagnostics. 2017;7(2) doi: 10.3390/diagnostics7020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mansfield M.A. In: Lateral Flow Immunoassay. Wong R T.H., editor. Humana Press; 2009. Nitrocellulose membranes for lateral flow immunoassays: a technical treatise; pp. 1–19. [Google Scholar]
- 104.Hi-Flow™ Plus Membranes and SureWick® Pad Materials. EMD Millipore; 2015. [Google Scholar]
- 105.Brown M.C. Lateral Flow Immunoassay, Humana. 2009. Antibodies: key to a robust lateral flow immunoassay; pp. 1–16. W. R., T. H. (Eds.) [Google Scholar]
- 106.Bartosh A.V., Sotnikov D.V., Hendrickson O.D., Zherdev A.V., Dzantiev B.B. Design of multiplex lateral flow tests: a case study for simultaneous detection of three antibiotics. Biosensors. 2020;10(3):17. doi: 10.3390/bios10030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berli C.L.A., Kler P.A. A quantitative model for lateral flow assays. Microfluid. Nanofluidics. 2016;20(7):104. [Google Scholar]
- 108.Fenton E.M., Mascarenas M.R., López G.P., Sibbett S.S. Multiplex lateral-flow test strips fabricated by two-dimensional shaping. ACS Appl. Mater. Interfaces. 2009;1(1):124–129. doi: 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- 109.Moghadam B.Y., Connelly K.T., Posner J.D. Two orders of magnitude improvement in detection limit of lateral flow assays using isotachophoresis. Anal. Chem. 2015;87(2):1009–1017. doi: 10.1021/ac504552r. [DOI] [PubMed] [Google Scholar]
- 110.Pierce H.F. Nitrocellulose membranes of graded permeability. J. Biol. Chem. 1927;75:795–815. [Google Scholar]
- 111.Kramarcy N.R., Sealock R. Commercial preparations of colloidal gold-antibody complexes frequently contain free active antibody. J. Histochem. Cytochem. 1991;39(1):37–39. doi: 10.1177/39.1.1983872. [DOI] [PubMed] [Google Scholar]
- 112.Edwards K.A., Baeumner A.J. Aptamer sandwich assays: label-free and fluorescence investigations of heterogeneous binding events. Anal. Bioanal. Chem. 2010;398(6):2635–2644. doi: 10.1007/s00216-010-3765-x. [DOI] [PubMed] [Google Scholar]
- 113.Edwards K.A., Wang Y., Baeumner A.J. Aptamer sandwich assays: human alpha-thrombin detection using liposome enhancement. Anal. Bioanal. Chem. 2010;398(6):2645–2654. doi: 10.1007/s00216-010-3920-4. [DOI] [PubMed] [Google Scholar]
- 114.Fernando S.A., Wilson G.S. Studies of the 'hook' effect in the one-step sandwich immunoassay. J. Immunol. Methods. 1992;151(1–2):47–66. doi: 10.1016/0022-1759(92)90104-2. [DOI] [PubMed] [Google Scholar]
- 115.Mansfield M.A. In: Troubleshooting Problems with Lateral Flow Assays. Millipore E., editor. EMD Millipore; 2015. [Google Scholar]
- 116.Biehl M., Velten T. Gaps and challenges of point-of-care technology. IEEE Sensor. J. 2008;8(5):593–600. [Google Scholar]
- 117.Adams E.R., Ainsworth M., Anand R., Andersson M.I., Auckland K., Baillie J.K., Barnes E., Beer S., Bell J., Berry T., Bibi S., Carroll M., Chinnakannan S., Clutterbuck E., Cornall R.J., Crook D.W., De Silva T., Dejnirattisai W., Dingle K.E., Dold C., Espinosa A., Eyre D.W., Farmer H., Fernandez Mendoza M., Georgiou D., Hoosdally S.J., Hunter A., Jeffrey K., Klenerman P., Knight J., Knowles C., Kwok A.J., Leuschner U., Levin R., Liu C., Lopez-Camacho C., Martinez Garrido J.C., Matthews P.C., McGivern H., Mentzer A.J., Milton J., Mongkolsapaya J., Moore S.C., Oliveira M.S., Pereira F., Perez Lopez E., Peto T., Ploeg R.J., Pollard A., Prince T., Roberts D.J., Rudkin J.K., Sanchez V., Screaton G.R., Semple M.G., Skelly D.T., Slon-Campos J., Smith E.N., Sobrino Diaz A.J., Staves J., Stuart D., Supasa P., Surik T., Thraves H., Tsang P., Turtle L., Walker A.S., Wang B., Washington C., Watkins N., Whitehouse J. Antibody testing for COVID-19: a report from the national COVID scientific advisory panel. Wellcome Open Res. 2020;5(139) doi: 10.12688/wellcomeopenres.15927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meier F.A., Jones B.A. Point-of-care testing error: sources and amplifiers, taxonomy, prevention strategies, and detection monitors. Arch. Pathol. Lab Med. 2005;129(10):1262–1267. doi: 10.5858/2005-129-1262-PTESAA. [DOI] [PubMed] [Google Scholar]
- 119.Salazar E., Kuchipudi S.V., Christensen P.A., Eagar T.N., Yi X., Zhao P., Jin Z., Long S.W., Olsen R.J., Chen J., Castillo B., Leveque C., Towers D.M., Lavinder J., Gollihar J.D., Cardona J., Ippolito G.C., Nissly R.H., Bird I.M., Greenawalt D., Rossi R.M., Gontu A., Srinivasan S., Poojary I.B., Cattadori I.M., Hudson P.J., Joselyn N., Prugar L., Huie K., Herbert A., Bernard D.W., Dye J., Kapur V., Musser J.M. 2020. Relationship between Anti-spike Protein Antibody Titers and SARS-CoV-2 in Vitro Virus Neutralization in Convalescent Plasma, bioRxiv : the Preprint Server for Biology. 2020.06.08.138990. [Google Scholar]
- 120.Ainsworth M., Andersson M., Auckland K., Baillie J.K., Barnes E., Beer S., Beveridge A., Bibi S., Blackwell L., Borak M., Bown A., Brooks T., Burgess-Brown N.A., Camara S., Catton M., Chau K.K., Christott T., Clutterbuck E., Coker J., Cornall R.J., Cox S., Crawford-Jones D., Crook D.W., D’Arcangelo S., Dejnirattsai W., Dequaire J.M.M., Dimitriadis S., Dingle K.E., Doherty G., Dold C., Dong T., Dunachie S.J., Ebner D., Emmenegger M., Espinosa A., Eyre D.W., Fairhead R., Fassih S., Feehily C., Felle S., Fernandez-Cid A., Fernandez Mendoza M., Foord T.H., Fordwoh T., Fox McKee D., Frater J., Gallardo Sanchez V., Gent N., Georgiou D., Groves C.J., Hallis B., Hammond P.M., Hatch S.B., Harvala H.J., Hill J., Hoosdally S.J., Horsington B., Howarth A., James T., Jeffery K., Jones E., Justice A., Karpe F., Kavanagh J., Kim D.S., Kirton R., Klenerman P., Knight J.C., Koukouflis L., Kwok A., Leuschner U., Levin R., Linder A., Lockett T., Lumley S.F., Marinou S., Marsden B.D., Martinez J., Martins Ferreira L., Mason L., Matthews P.C., Mentzer A.J., Mobbs A., Mongkolsapaya J., Morrow J., Mukhopadhyay S.M.M., Neville M.J., Oakley S., Oliveira M., Otter A., Paddon K., Pascoe J., Peng Y., Perez E., Perumal P.K., Peto T.E.A., Pickford H., Ploeg R.J., Pollard A.J., Richardson A., Ritter T.G., Roberts D.J., Rodger G., Rollier C.S., Rowe C., Rudkin J.K., Screaton G., Semple M.G., Sienkiewicz A., Silva-Reyes L., Skelly D.T., Sobrino Diaz A., Stafford L., Stockdale L., Stoesser N., Street T., Stuart D.I., Sweed A., Taylor A., Thraves H., Tsang H.P., Verheul M.K., Vipond R., Walker T.M., Wareing S., Warren Y., Wells C., Wilson C., Withycombe K., Young R.K. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect. Dis. 2020;0(0) doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bar-On Y., Flamholz A., Phillips R., Milo R. eLife; 2020. Science Forum: SARS-CoV-2 (COVID-19) by the Numbers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kannan S., Shaik Syed Ali P., Sheeza A., Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020;24(4):2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- 124.Malik Y.A. Properties of coronavirus and SARS-CoV-2, malays. J. Pathol. 2020;42(1):3–11. [PubMed] [Google Scholar]
- 125.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hofmann H., Simmons G., Rennekamp A.J., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., Pöhlmann S. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80(17):8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A.J., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maache M., Komurian-Pradel F., Rajoharison A., Perret M., Berland J.L., Pouzol S., Bagnaud A., Duverger B., Xu J., Osuna A., Paranhos-Baccalà G. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid-based western blot assay were rectified by the use of two subunits (S1 and S2) of spike for detection of antibody to SARS-CoV. Clin. Vaccine Immunol. 2006;13(3):409–414. doi: 10.1128/CVI.13.3.409-414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang L.R., Chiu C.M., Yeh S.H., Huang W.H., Hsueh P.R., Yang W.Z., Yang J.Y., Su I.J., Chang S.C., Chen P.J. Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J. Med. Virol. 2004;73(3):338–346. doi: 10.1002/jmv.20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Q., Li C., Zhang Q., Wang T., Li J., Guan W., Yu J., Liang M., Li D. Interactions of SARS coronavirus nucleocapsid protein with the host cell proteasome subunit p42. Virol. J. 2010;7:99. doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.-W., Xu S.-Y., Zhu H.-D., Xu Y.-C., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Khan S., Nakajima R., Jain A., de Assis R.R., Jasinskas A., Obiero J.M., Adenaiye O., Tai S., Hong F., Milton D.K., Davies H., Felgner P.L. 2020. Analysis of Serologic Cross-Reactivity between Common Human Coronaviruses and SARS-CoV-2 Using Coronavirus Antigen Microarray. bioRxiv. [Google Scholar]
- 138.Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J., Macdonald D., David S.T., Srour L.F., Parker R., Andonov A., Isaac-Renton J., Loewen N., McNabb G., McNabb A., Goh S.-H., Henwick S., Astell C., Guo J.P., Drebot M., Tellier R., Plummer F., Brunham R.C. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can. J. Infect Dis. Med. Microbiol. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Severe Acute Respiratory Syndrome (SARS) 2017. https://www.cdc.gov/sars/index.html [Google Scholar]
- 140.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., Meade P., Brito R.N., Teo C., McMahon M., Simon V., Krammer F. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Prot. Microbiol. 2020;57(1) doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368(6495):1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 142.Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432(10):3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Freeman B., Lester S., Mills L., Rasheed M.A.U., Moye S., Abiona O., Hutchinson G.B., Morales-Betoulle M., Krapinunaya I., Gibbons A., Chiang C.-F., Cannon D., Klena J., Johnson J.A., Owen S.M., Graham B.S., Corbett K.S., Thornburg N.J. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and sero-surveillance. bioRxiv. 2020 2020.04.24. [Google Scholar]
- 144.Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P., Peng P., Liu X., Chen Z., Huang H., Zhang F., Luo W., Niu X., Hu P., Wang L., Peng H., Huang Z., Feng L., Li F., Zhang F., Li F., Zhong N., Chen L. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microb. Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Emergency Use Authorization. 2020. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization [Google Scholar]
- 147.Elecsys® Anti-SARS-CoV-2. 2020. https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html [Google Scholar]
- 148.CLIA Law and Regulations. 2018. https://www.cdc.gov/clia/law-regulations.html [Google Scholar]
- 149.How to Obtain a CLIA Certificate of Waiver. 2019. https://www.cms.gov/regulations-and-guidance/legislation/clia/downloads/howobtaincertificateofwaiver.pdf [Google Scholar]
- 150.Assure COVID-19 IgG/IgM Rapid Test Device. 2020. https://www.fda.gov/media/139789/download [Google Scholar]
- 151.Independent Evaluations of COVID-19 Serological Tests. 2020. https://open.fda.gov/apis/device/covid19serology/ [Google Scholar]
- 152.Revocation of EUA200179. 2020. https://www.fda.gov/media/139109/download [Google Scholar]
- 153.Revocation of EUA200349. 2020. https://www.fda.gov/media/140908/download [Google Scholar]
- 154.Saah A.J., Hoover D.R. “Sensitivity” and “specificity” reconsidered: the meaning of these terms in analytical and diagnostic settings. Ann. Intern. Med. 1997;126(1):91–94. doi: 10.7326/0003-4819-126-1-199701010-00026. [DOI] [PubMed] [Google Scholar]
- 155.Bioanalytical Method Validation Guidance for Industry. 2018. https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf [Google Scholar]
- 156.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. 108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Parikh R., Mathai A., Parikh S., Chandra Sekhar G., Thomas R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008;56(1):45–50. doi: 10.4103/0301-4738.37595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.-P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Ahmad Khan F. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wechselberger C., Süßner S., Doppler S., Bernhard D. Performance evaluation of serological assays to determine the immunoglobulin status in SARS-CoV-2 infected patients. J. Clin. Virol. 2020;131:104589. doi: 10.1016/j.jcv.2020.104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang H., Ai J., Loeffelholz M.J., Tang Y.-W., Zhang W. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg. Microb. Infect. 2020;9(1):2200–2211. doi: 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ong D.S.Y., de Man S.J., Lindeboom F.A., Koeleman J.G.M. Comparison of diagnostic accuracies of rapid serological tests and ELISA to molecular diagnostics in patients with suspected coronavirus disease 2019 presenting to the hospital. Clin. Microbiol. Infect. 2020;26(8):1094. doi: 10.1016/j.cmi.2020.05.028. e7-1094.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., Bruno R., Mojoli F., Baldanti F., Members of the San Matteo Pavia C.-T.F. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020 doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu T., Wu S., Tao H., Zeng G., Zhou F., Guo F., Wang X. 2020. Prevalence of IgG Antibodies to SARS-CoV-2 in Wuhan - Implications for the Ability to Produce Long-Lasting Protective Antibodies against SARS-CoV-2, medRxiv. 2020.06.13.20130252. [Google Scholar]
- 164.Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L., Tang F., Lv H., Cao W.C. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193(6):792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 166.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wajnberg A., Amanat F., Firpo A., Altman D., Bailey M., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., Stadlbauer D., Stone K., Strohmeier S., Aberg J., Reich D., Krammer F., Cordon-Cardo C. SARS-CoV-2 infection induces robust, neutralizing antibody responses that are stable for at least three months. medRxiv. 2020 doi: 10.1126/science.abd7728. [DOI] [Google Scholar]
- 168.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.-C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.-F., Lina B., van-der-Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:S1473–S3099. doi: 10.1016/S1473-3099(20)30200-0. 30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]