Abstract
Due to the current Coronavirus (COVID-19) pandemic, the rapid discovery of a safe and effective vaccine is an essential issue. Consequently, this study aims to predict a potential COVID-19 peptide-based vaccine utilizing the Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the Immunoinformatics approach. To achieve this goal, several Immune Epitope Database (IEDB) tools, molecular docking, and safety prediction servers were used. According to the results, The Spike peptide SQCVNLTTRTQLPPAYTNSFTRGVY is predicted to have the highest binding affinity to the B-Cells. The Spike peptide FTISVTTEI has the highest binding affinity to the Major Histocompatibility Complex class 1 (MHC I) Human Leukocyte Allele HLA-B*1503 (according to the MDockPeP and HPEPDOCK servers, docking scores were −153.9 and −229.356, respectively). The Nucleocapsid peptides KTFPPTEPK and RWYFYYLGTGPEAGL have the highest binding affinity to the MHC I HLA-A0202 allele and the three the Major Histocompatibility Complex class 2 (MHC II) Human Leukocyte Allele HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1-*06:02, HLA-DRB1, respectively. Docking scores of peptide KTFPPTEPK were −153.9 and −220.876. In contrast, docking scores of peptide RWYFYYLGTGPEAGL were ranged from 218 to 318. Furthermore, those peptides were predicted as non-toxic and non-allergen. Therefore, the combination of those peptides is predicted to stimulate better immunological responses with respectable safety.
Keywords: COVID-19, Multiple peptides vaccine, Nucleocapsid, Spike, And immunoinformatics
1. Introduction
The Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, COVID-19) is an enveloped, positive-sense, single-stranded polyadenylated RNA virus belongs to the Coronaviridae family [1].
COVID-19 is a novel strain detected firstly in the city of Wuhan, the Republic of China in December 2019 [2]. It causes fever, cough, dyspnea, bilateral pulmonary infiltrates that may progress to Pneumonia [3]. It is characterized by rapid spreading; “As the 27 Feb, it is reported in 47 countries, causing over 82,294 infections with 2804 deaths” [4] and till the 5th May, more than 3.5 million positive cases and 0.25 million deaths have been identified globally [5]. Unfortunately, until now COVID-19 has no effective antiviral drug for the treatment or vaccine for the prevention. Hence extensive researches should be conducted on the development of safe and effective vaccines and drugs [4].
S-protein in SARS-CoV-2 contains (S1 and S2 subunits); and receptor-binding domain (RBD) existing at the C-terminus on S1 subunit It plays an essential role with virus attachment, membrane fusion, and subsequent infection. Some studies have reported a multiple-mutations on this protein as main mutation of SARS-CoV-2 pathogen, the conserved deposits are placed mainly on S2 subunit - up until now, more than 1800 mutations are deposit in the S protein of SARS-CoV-2 [6]. Hence, the subunit vaccine protein as best candidate vaccine of multiple Peptides designated from different proteins can be helpful in the critical stages of the highly mutated pathogen.
A subunit vaccine must be devoid of allergenicity and also immunogenic; some of the experimental estimation of the designed vaccine were made in an animal model and may confer defense against F. gigantica infection [7].
The designing vaccine for Fascioliasis includes an investigation of the potential of glutathione s-transferase (GST) antigen as a subunit vaccine candidate via immunoinformatics [8].
To develop a safe and effective COVID-19 vaccine rapidly, the WHO recommended that “we must test all candidate vaccines until they fail to ensure that all of them have the chance of being tested at the initial stage of development”. Ensuing this point, recently, there are over 120 proposed vaccines. Six of them are in the clinical evaluation stage and 70 are in pre-clinical evaluation [9]. The vaccine development is achieved by multiple approaches including the Inactivated, Live-attenuated, Non-replicating viral vector, DNA, RNA, Recombinant proteins, and Peptide-based vaccines. “As of April 8, 2020, the global COVID-19 vaccine R&D landscape confirmed 78 active candidate vaccine” [10].
Consistent with global efforts, this study aims to predict potential COVID-19 Peptide-based vaccine utilizing the Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the Immunoinformatics approach. Due to the respectable antigenicity of the Nucleocapsid and Spike Glycoprotein, they are appropriate targets for vaccine design [11]. The peptide vaccines are sufficient to stimulate cellular and humoral immunity without allergic responses [12]. They are” safe, simply produced, stable, reproducible, cost-effective” [13], and permit a broad spectrum of immunity [12]; consequently, they are the targets for this study as well as being utilized in multiple studies concerning the COVID-19 vaccine [[14], [15], [16], [17], [18], [19], [20]].
2. Materials and methods
2.1. Protein sequence retrieval
A total of 100 sequences of Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) were retrieved from the National Center for Biotechnology Information (NCBI) database [20] as FASTA format in March 2020. The sequences with their accession numbers are listed in the Supplementary Data (supplementary table S1). The protein sequences of MHC I alleles HLA-A*02:01, HLA-B15:03, HLA-C*12:03, and MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DRB1 were obtained from the Immuno Polymorphism Database (IPD-IMGT/HLA) [22].
2.2. Multiple sequences alignment
The retrieved sequences of Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) were aligned using the ClustalW algorithm [25] on the BioEdit software version 7.2.5 [24] to identify the conserved regions within sequences.
2.3. B-Cells Peptides prediction
The B-Cells peptides were predicted from the conserved regions using the linear Epitope Prediction tool “BepiPred-test” on the Immune Epitope Database (IEDB) [25]. The default threshold value (−0.500) was used. To predict the epitopes accurately, a combination between the hidden (Parker and Levitt) method and the Markov model (HMM) [26] was used.
2.4. The surface accessibility prediction
The Surface Accessibility of B-Cells Peptides was predicted via the Emini Surface Accessibility tool [27] on the IEDB [25]. The default threshold holding value (1.000) was used.
2.5. The antigenic sites prediction
To identify the antigenic sites within the Nucleocapsid phosphoprotein and Spike Glycoprotein, the Kolaskar and Tongaonker method's on the IEDB [25] was used. The default threshold values were 0.988 and 1.041, respectively.
2.6. T-cell peptides prediction
To predict the interaction with different Major Histocompatibility Complex class protein 1 (MHC I) alleles, the MHC I binding prediction tool on the IEDB [25] was used. All peptide length was set as 9 amino acids. To predict the binding affinity, the Artificial Neural Network (ANN) prediction method was selected with a half-maximal inhibitory concentration (IC50) value of less than 100.
In contrast, to predict the interaction with different Major Histocompatibility Complex protein class II (MHC II) alleles, The Major Histocompatibility Complex class II (MHC II) binding prediction was used. To predict the binding affinity, the NN align algorithm was selected with an IC50 value of less than 500. The Human allele reference sets (HLA DR, DP, and DQ) were included in the prediction.
2.7. The population coverage prediction
To predict the percentage of peptides binding with various MHC I and MHC II alleles that cover the world population, the population coverage tool on the IEDB [25] was used.
2.8. Allergenicity and toxicity prediction
To predict the peptides' allergenicity, the AllergenFP v.1.0 [28] and AllerCatPro v. 1.7 [29] servers were used. In contrast, to predict the peptides’ toxicity, the ToxinPred server [30] was used.
2.9. 3D structure modeling and visualization
To model the 3D structure of the Nucleocapsid, Spike, and MHC molecules, the SWISS-MODEL server [31], and the Phyre2 web portal [32] were used. To visualize the modeled structures, the USCF Chimera 1.8 software [33] was used.
2.10. Molecular docking study
The predicted peptides were docked with MHC I alleles HLA-A*02:01, HLA-B15:03, HLA-C*12:03, and MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DRB1. The modeled MHC structures were prepared for the docking via Cresset Flare software [34] at the normal type calculation method. To dock the predicted peptides, the MDockPeP [35] and HPEPDOCK [[36], [37], [38], [39], [40]] servers were used. The predicted peptides were submitted as amino acid sequences. The 2D and 3D interactions were visualized using the PoseView [41] at the ProteinPlus web portal [42] and Cresset Flare viewer [34], respectively.
2.11. Molecular dynamics and binding energy calculations
Molecular dynamics and simulation (MDS) studies were carried out in order to determine stability and convergence of vaccine candidate peptide with the target MHC complexes. To set up the simulation, initially, the systems were built for peptides complexes derived from spike and nucleocapsid protein as ligands with MHC I and MHC II, respectively, in the system builder. For this purpose Desmond 2018–4 was used to set up the initial parameters within an explicit SPC water model within orthorhombic box 5.0 × 5.0 × 5.0 Å.Three sets of peptide MHC complex (FTISVTTEI–MHC I, KTFPPTEPK-MHC I and RWYFYYLGTGPEAGL-MHC II) were investigated - those displayed significant binding in docking studies.
3. Results
According to the IEDB [25] prediction, the average binding score for the Nucleocapsid phosphoprotein and Spike Glycoprotein were 0.558, 0.470, respectively. All values equal to or greater than the default threshold were predicted as potential B-cell binders.
The Emini surface accessibility tool predicts the average binding score of the Nucleocapsid phosphoprotein and Spike Glycoprotein as 1.00. All values equal to or greater than the default threshold were predicted to have good surface accessibility. The Kolaskar and Tongaonkar antigenicity prediction tool predicts the average threshold value for the Nucleocapsid phosphoprotein and Spike Glycoprotein as 0.988 and 1.04, respectively. Peptides with values equal to or greater than the average score are considered as antigenic peptides. The minimum and maximum values are listed in Table 1 . The Nucleocapsid peptide (DAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDD) had the highest antigenicity, surface accessibility, hydrophilicity, and flexibility. In contrast, the Spike peptides (LGKY) and (SQCVNLTTRTQLPPAYTNSFTRGVY) had the highest scores (Table 2 ). The location of those peptides within the 3D structure is shown in Fig. 1, Fig. 2 .
Table 1.
The average, minimum, and maximum Bepiprd epitopes, surface accessibility, Antigenicity, hydrophilicity, and Flexibility, prediction values.
| Test | Protein | Average Score | Minimum Score | Maximum Score |
|---|---|---|---|---|
| Bepiprd linear epitope prediction | Nucleocapsid | 0.558 | 0.297 | 0.764 |
| Spike Glycoprotein | 0.470 | 0.183 | 0.695 | |
| Emini surface accessibility prediction | Nucleocapsid | 1.000 | 0.050 | 7.006 |
| Spike Glycoprotein | 1.000 | 0.042 | 6.051 | |
| Kolaskar & Tongaonkar Antigenicity | Nucleocapsid | 0.988 | 0.874 | 1.197 |
| Spike Glycoprotein | 1.041 | 0.866 | 1.261 | |
| Parker Hydrophilicity Prediction | Nucleocapsid | 2.800 | −5.971 | 6.871 |
| Spike Glycoprotein | 1.238 | 7.629 | 7.743 | |
| Karplus & Schulz Flexibility Prediction | Nucleocapsid | 1.035 | 0.885 | 1.161 |
Table 2.
Result of the predicted B cell epitopes.
| Protein | Peptides | Position | Surface accessibility score | Antigenicity score | Hydrophobicity score |
|---|---|---|---|---|---|
| Nucleocapsid | DAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDD | 358–420 | 10.375 | 1.001 | 3.478 |
| Spike Glycoprotein | LGKY | 1203–1206 | 1.037 | 1.045 | – |
| SQCVNLTTRTQLPPAYTNSFTRGVY | 13–37 | 1.513 | 1.042 | – |
The Hydrophobicity scores of the Spike Glycoprotein peptides were not available.
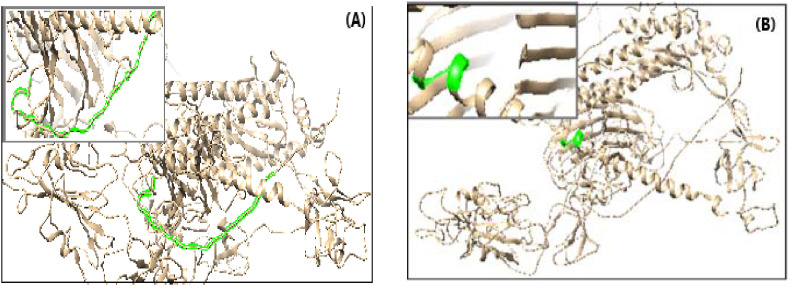
Fig. 1.
The location of predicted B cell peptide (DAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDD) (pink color) within 3D structure of the Nucleocapsid protein. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
The location of predicted B cell peptides (green color) within the 3D structure of the Spike Glycoprotein
a) The peptide (SQCVNLTTRTQLPPAYTNSFTRGVY)
b) The peptide ((LGKY). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
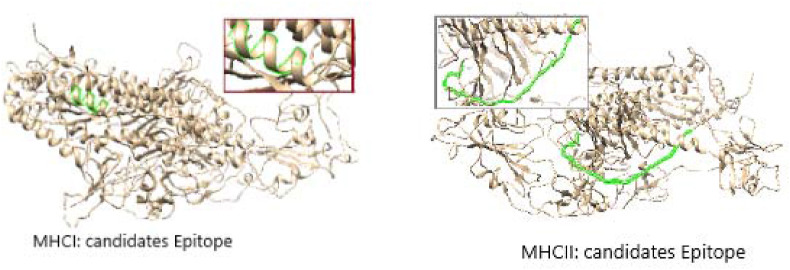
Regarding the cytotoxic T-lymphocyte peptides, the MHC I binding prediction tool predicts 46 peptides from the Nucleocapsid and 192 peptides from the Spike Glycoprotein could interact with the different MHC I alleles. The most promising peptides with their total population coverage were listed in Table 3 . In contrast, the MHC II binding prediction tool predicts 774 peptides from the Nucleocapsid and 1111 peptides from the Spike Glycoprotein could interact with the different MHC II alleles. The most promising peptides with their total population coverage were listed in Table 4 . The top predicted MHC I and MHC II peptides are listed in Table 6, Table 7 , respectively. The location of representative peptides from them within the 3D structure is shown in Fig. 3, Fig. 4 .
Table 3.
The most promising MHC I peptides with their corresponding alleles.
| Peptide | MHC 1 allele | Total population Coverage |
|---|---|---|
|
Nucleocapsid protein TPSGTWLTY FAPSASAFF KTFPPTEPK SPRWYFYYL NTASWFTAL KTFPPTEPK KAYNVTQAF FTALTQHGK LPAADLDDF LLLDRLNQL MEVTPSGTW KAYNVTQAF KAYNVTQAF LLNKHIDAY |
HLA-B*35:01 HLA-B*35:01 HLA-C*03:3 HLA-A*11:01 HLA-B*07:02 HLA-A*68:02 HLA-A*30:01 HLA-A*32:01 HLA-A*68:01 HLA-B*35:01 HLA-A*02:06 HLA-B*44:02 HLA-C*14:0 HLA-B*15:01 |
98.2% |
|
Spike glycoprotein FIAGLIAIV FVFLVLLPL MIAQYTSAL FTISVTTEI VVFLHVTYV |
HLA-A*68:02, HLA-A*02:01, HLA-A*02:06, HLA-C*12:03 HLA-A*02:06, HLA-A*02:01, HLA-A*68:02, HLA-C*12:03 HLA-C*14:02, HLA-B*08:01, HLA-B*35:01, HLA-B*07:02, HLA-A*68:02, HLA-B*39:01, HLA-A*02:06, HLA-C*03:03, HLA-A*02:01, HLA-B*15:01 HLA-A*68:02, HLA-A*02:06, HLA-A*02:01, HLA-B*58:01, HLA-C*15:02, HLA-C*12:03 HLA-A*02:06, HLA-A*02:01, HLA-A*68:02, HLA-C*12:03 |
99.73% |
Table 4.
The most promising MHC II peptides with their corresponding alleles.
| Peptide | MHC II allele | Total Population Coverage |
|---|---|---|
| Nucleocapsid protein | 99.97% | |
| QIGYYRRATRRIRGG | HLA-DRB1*11:01 | |
| IGYYRRATRRIRGGD | HLA-DRB1*11:01 | |
| GYYRRATRRIRGGDG | HLA-DRB1*11:01 | |
| TPSGTWLTYTGAIKL | HLA-DRB1*07:01 | |
| DQIGYYRRATRRIRG | HLA-DRB1*11:01 | |
| PQIAQFAPSASAFFG | HLA-DRB1*09:01 | |
| WPQIAQFAPSASAFF | HLA-DRB1*09:01 | |
| QIAQFAPSASAFFGM | HLA-DRB1*09:01 | |
| IAQFAPSASAFFGMS | HLA-DRB1*09:01 | |
| AALALLLLDRLNQLE | HLA-DRB4*01:01, HLA-DPA1 03:01/DPB1*04, HLA-DRB3*01:0, HLA-DRB1*13:02, HLA-DRB1*11:0, HLA-DRB1*04:04, HLA-DRB1*01:01, HLA-DRB1*04, HLA-DPA1*02:01/DPB1*01:01, HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*04:05, HLA-DRB1*03:01, HLA-DRB1*08:02, HLA-DRB1*15:01, HLA DQA1*01:01/DQB1*05:01 | |
| ALALLLLDRLNQLES | HLA-DRB4*01:01, HLA-DPA1*03:01/DPB1*04:02, HLA-DRB3*01:01, HLA-DRB1*13:02, HLA-DRB1*11:01, HLA-DRB1*04:04, HLA-DRB1*04:01, HLA-DRB1*01:01, HLA-DRB1*03:01, HLA-DRB1*04:05, HLA-DPA1*02:01/DPB1*01:01, HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*08:02, HLA-DRB1*15:01, HLA-DQA1*01:01/DQB1*05:01 | |
| PRWYFYYLGTGPEAG | HLA-DRB1*07:01 | |
| RWYFYYLGTGPEAGL | HLA-DRB1*01:01 | |
| Spikes glycoprotein | 99.98% | |
| AAEIRASANLAATKM | HLA-DQA1*05:01/DQB1*03:01 | |
| NAQALNTLVKQLSSN | HLA-DRB1*11:01 | |
| EVFNATRFASVYAWN | HLA-DPB1*02:01, HLA DPB1*04:02, HLA-DPB1*05:01, HLA-DQA1*01:02, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQB1*06:02, HLA-DRB1*01:01, HLA-DRB1*04:04, HLA-DRB1*04:05, HLA-DRB1*07:01, HLA-DRB1*08:02, HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*15:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPA1*01:03, HLA-DPA1*02:01 | |
| VFRSSVLHSTQDLFL | HLA-DRB1*07:01, HLA-DRB1*01:01, HLA-DRB1*09:01, HLA-DRB1*04:05, HLA-DRB1*04:01, HLA-DRB1*03:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DPA1*03:01/DPB1*04:02, HLA-DRB1*13:02, HLA-DPA1*02:01/DPB1*01:01, HLA-DRB4*01:01, HLA-DQA1*05:01/DQB1*02:01, HLA-DRB1*04:04, HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*05:01/DQB1*03:01 |
The alleles HLA-DRB3*01:01, HLA-DRB4*01:01, HLA-DRB5*01:01 were not available, hence they were not included in the calculations.
Table 6.
Results of MHC II peptides Allergenicity and Toxicity prediction.
| Peptide | Allergicity |
Toxicity |
|
|---|---|---|---|
| AllergenFP v.1.0 | AllerCatPro v. 1.7 | ||
| EVFNATRFASVYAWN (Spike) | Allergen | No evidence | Non-Toxin |
| VFRSSVLHSTQDLFL (Spike) | Non-allergen | No evidence | Non-Toxin |
| AALALLLLDRLNQLE (Nucleocapsid) | Non-allergen | No evidence | Non-Toxin |
| ALALLLLDRLNQLES (Nucleocapsid) | Non-allergen | No evidence | Non-Toxin |
| PRWYFYYLGTGPEAG (Nucleocapsid) | Non-allergen | No evidence | Non-Toxin |
| RWYFYYLGTGPEAGL (Nucleocapsid) | Allergen | No evidence | Non-Toxin |
Table 7.
Docking scores of the predicted COVID-19 Spike Glycoprotein (S) and Nucleocapsid phosphoprotein (N) peptides with MHC I alleles HLA-A*02:02, HLA-B*15:03, and HLA-C*12:03.
| Peptide | HLA-A0202 |
HLA-B1503 |
HLA-C1203 |
|||
|---|---|---|---|---|---|---|
| MDockPeP | HPEPDOCK | MDockPeP | HPEPDOCK | MDockPeP | HPEPDOCK | |
| FTISVTTEI (Spike) | −148.1 | −250.625 | −153.9 | −229.356 | −134.2 | −215.735 |
| MIAQYTSAL (Spike) | −149.3 | −259.707 | −149.4 | −219.145 | −154.7 | −237.005 |
| KTFPPTEPK (Nucleocapsid) | −153.9 | −220.876 | −135.8 | −204.661 | −136.3 | −193.690 |
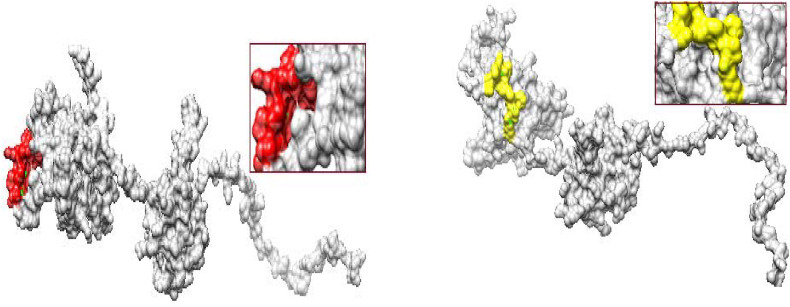
Fig. 3.
The location of a representative predicted MHC (red and yellow color) within the 3D structure of the Nucleocapsid
Upper: Red color indicates the peptide (AALALLLLDRLNQLE)
Lower: Yellow color indicates the peptide (KTFPPTEPK). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
The location of a representative predicted within the 3D structure of the Spike Glycoprotein
MHCI: MIAQYTSAL
MHCII: EVFNATRFASVYAWN.
According to the AllergenFP v.1.0 [15], AllerCatPro v. 1.7 [16], and ToxinPred servers [17], all the predicted peptides except the spike peptide (EVFNATRFASVYAWN) were Non-allergen and Non-Toxin (Table 5, Table 6).
Table 5.
Results of MHC I peptides Allergenicity and Toxicity prediction.
| Peptide | Allergicity |
Toxicity | |
|---|---|---|---|
| AllergenFP v.1.0 | AllerCatPro v. 1.7 | ||
| FTISVTTEI (Spike) | Non-allergen | No evidence | Non-Toxin |
| MIAQYTSAL (Spike) | Non-allergen | No evidence | Non-Toxin |
| KTFPPTEPK (Nucleocapsid) | Non-allergen | No evidence | Non-Toxin |
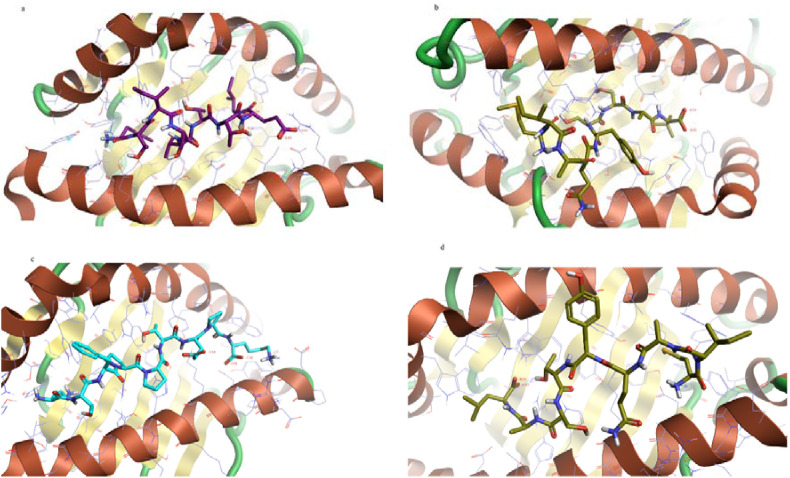
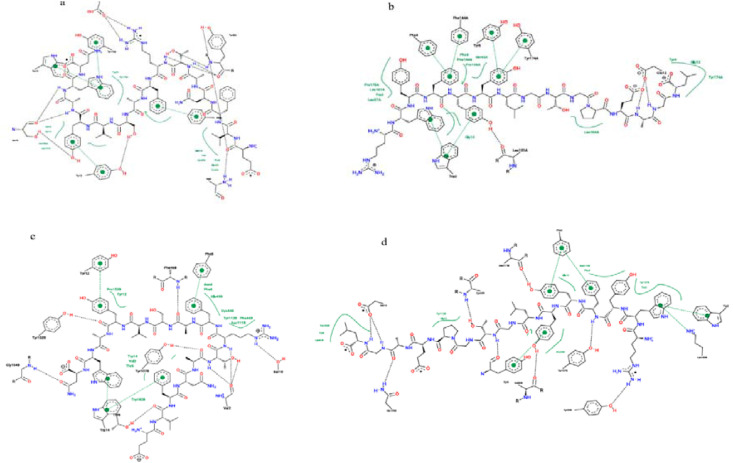
According to the MDockPeP [32] and HPEPDOCK [36] servers prediction, the spike peptide (FTISVTTEI) has the highest binding affinity to the MHC I HLA-B1503 allele and the spike peptide (MIAQYTSAL) has the highest binding affinity to the MHC I HLA-C1203 allele. The Nucleocapsid peptide (KTFPPTEPK) has the highest binding affinity to the MHC I HLA-A0202 allele (Table 7). Their 2D and 3D interactions are illustrated in Fig. 5, Fig. 6 .
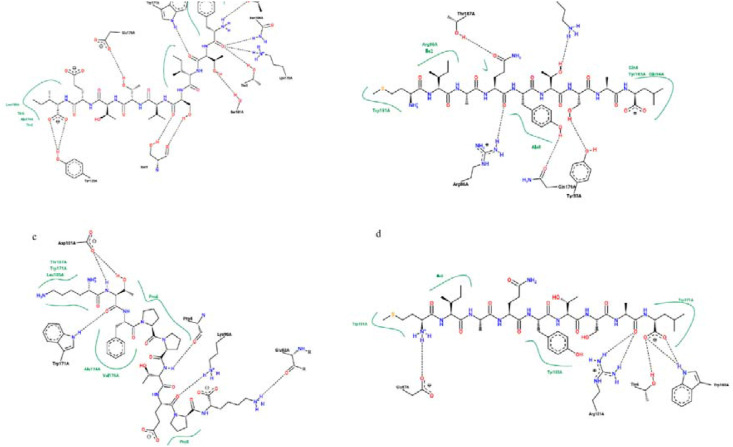
Fig. 5.
The 2D interaction of the predicted peptides with the MHC I alleles
a) Spike peptide (FTISVTTEI) with MHC I HLA-B1503 allele
b) Spike peptide (MIAQYTSAL) with MHC I HLA-C1203 allele
c) Nucleocapsid peptide (KTFPPTEPK) with MHC I HLA-A0202 allele
d) Spike peptide (MIAQYTSAL) with MHC I HLA-A0202 allele.
Fig. 6.
The 3D interaction of the predicted peptides with the MHC I alleles
a) Spike peptide (FTISVTTEI) with MHC I HLA-B1503 allele
b) Spike peptide (MIAQYTSAL) with MHC I HLA-C1203 allele
c) Nucleocapsid peptide (KTFPPTEPK) with MHC I HLA-A0202 allele
d) Spike peptide (MIAQYTSAL) with MHC I HLA-A0202 allele.
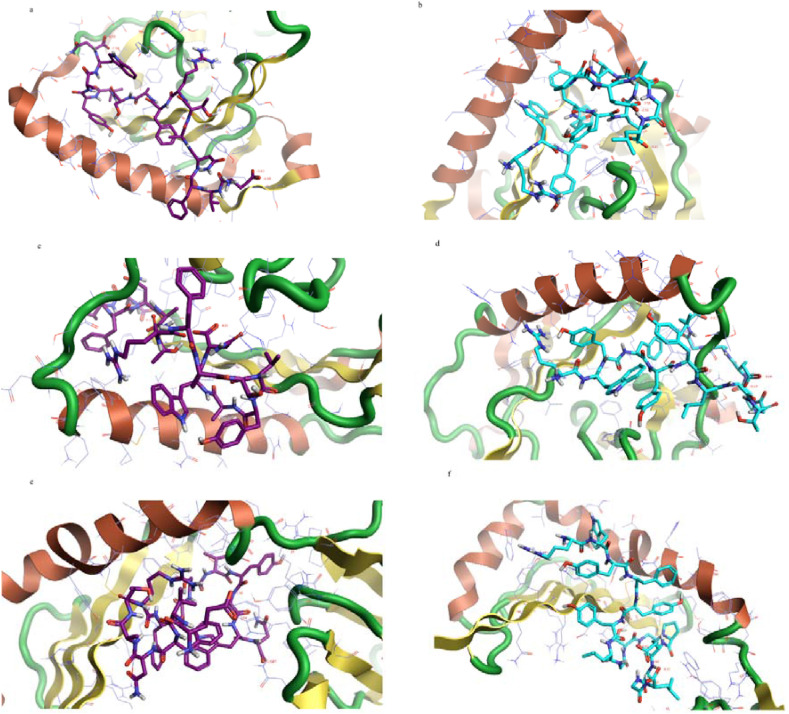
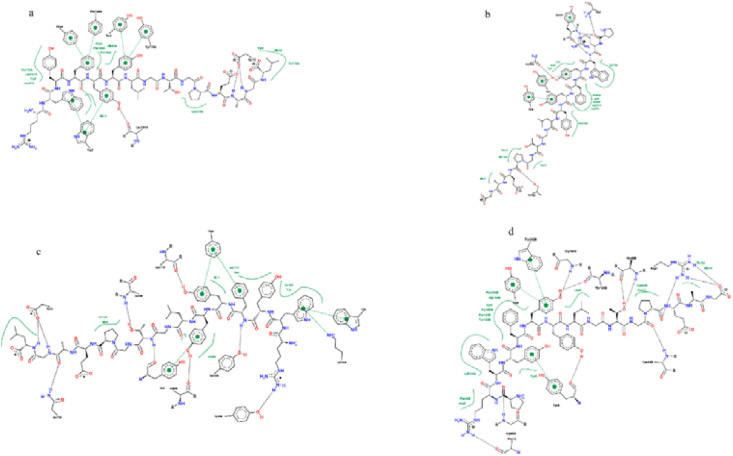
In contrast, the spike peptide (EVFNATRFASVYAWN) has the highest binding affinity to the MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, and HLA-DRB1. The Nucleocapsid peptide (RWYFYYLGTGPEAGL) has the highest binding affinity to the MHC II allele HLA-DQA1*01:02/DQB1*06:02 and the Nucleocapsid peptides (RWYFYYLGTGPEAGL) (PRWYFYYLGTGPEAG) and has a higher binding affinity to the MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB1 (Table 8 ). The 2D and 3D interactions are shown in Fig. 7, Fig. 8 .
Table 8.
Docking scores of the predicted COVID-19 Spike Glycoprotein (S) and Nucleocapsid phosphoprotein (N) peptides with MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, and HLA-DRB1.
| Peptide | HLA-DPA1*01:03/DPB1*02:01 |
HLA-DQA1*01:02/DQB1*06:02 |
HLA-DRB1 |
|||
|---|---|---|---|---|---|---|
| MDockPeP | HPEPDOCK | MDockPeP | HPEPDOCK | MDockPeP | HPEPDOCK | |
| EVFNATRFASVYAWN (Spike) | −233.5 | −284.222 | −253.0 | −251.477 | −235.9 | −213.801 |
| VFRSSVLHSTQDLFL (Spike) | −229.5 | −249.637 | −233.7 | −227.509 | −214.5 | −189.293 |
| AALALLLLDRLNQLE (Nucleocapsid) | −218.1 | −234.533 | −212.5 | −193.846 | −208.9 | −160.346 |
| ALALLLLDRLNQLES (Nucleocapsid) | −223.4 | −220.495 | −208.1 | −211.481 | −224.3 | −157.521 |
| PRWYFYYLGTGPEAG (Nucleocapsid) | −250.5 | −319.883 | −248.3 | −266.145 | −241.0 | −230.567 |
| RWYFYYLGTGPEAGL (Nucleocapsid) | −251.5 | −317.700 | −253.9 | −274.600 | −245.8 | −218.061 |
Fig. 7.
The 2D interaction of the predicted peptides with the MHC II alleles; a) and b) Spike peptide (EVFNATRFASVYAWN) with MHC II HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB, respctively. c) and d) Nuclcapsid peptide (RWYFYYLGTGPEAGL) with MHC II alleles HLA- 15 DPA1*01:03/DPB1*02:01 and HLA-DRB, respectively.
Fig. 8.
The 3D interaction of the predicted peptides with the MHC II alleles. a), c), and e) Spike peptide (EVFNATRFASVYAWN) with MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, and HLA-DRB, respectively
b), d), and f) Nucleocapsid peptide (RWYFYYLGTGPEAGL) with MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, and HLA-DRB, respectively.
In MD and binding energy calculation, all the protein peptide complexes were neutralized with NaCl salt by adding 0.15 M Na+ ions. The prepared systems were relaxed using the Desmond default protocol of relaxation [55]. MDS run of 20 ns was set up at constant temperature and constant pressure (NPT) for the final production run. The NPT ensemble was set up using Nosé–Hoover chain coupling scheme [56] at temperature 300 K for final production and throughout the dynamics with relaxation time 1 ps RESPA integrator was used to calculate the bonding interactions for a time step 2 fs [57]. All other parameters were associated in the settings followed as described elsewhere [58]. After the final production run, the simulation trajectories of apo protein and complexed with xylotetraose were analyzed for final outcome of RMSD and RMSF, ligand RMSF, derived from simulation. Binding energies of the complexes were calculated using MM-GBSA for every 5 ns trajectories up to 20 ns, and the average binding energies with standard deviations were measured for accurate binding approximation and stability.
4. Discussion
Due to the current COVID-19 pandemic, the rapid discovery of a safe and effective vaccine is an essential issue [43].
Since the successful vaccine relies on the selection of the most antigenic parts and the best approaches [44], COVID-19 Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) were selected to design a peptide vaccine. The antigenicity of the Nucleocapsid and Spike is well predicted [11], and the advantages of the peptide vaccines are well established [12,13].
The peptide design via the Immunoinformatics approach is achieved through multiple steps including the prediction of B-Cells and T-cell Peptides, the surface accessibility, antigenic sites, and the Population coverage. After the selection of the candidate peptides, their interaction with the MHC molecules is simulated and their safety is predicted [45].
Regarding the B-Cells peptides prediction, the successful candidates must pass the threshold scores in the Bepipred, Parker hydrophilicity, Kolaskar, and Tongaonkar antigenicity, as well as Emini surface accessibility tests [46] (Table 1). The IEDB Bepipred test [25] on the Nucleocapsid showed that eleven peptides were predicted, however, the peptide (DAYKTFPPTEPKKDK-KKKADETQALPQRQKKQQTVTLLPAADLDD) was the only one that passed all the tests. In contrast, the IEDB Bepipred test [21] on the Spike showed that forty-two peptides were predicted, but the peptides (SQCVNLTTRTQLPPAYTNSFTRGVY) and (LGKY) were the only two that passed all the tests. As the length of effective B-cell peptides varies from 5 to 30 amino acids [47], the peptide (LGKY) is too short and the peptide (DAYKTFPPTEPKKDK-KKKADETQALPQRQKKQQTVTLLPAADLDD) is too long. Consequently, the Spike peptide peptides (SQCVNLTTRTQLPPAYTNSFTRGVY) is predicted to have the highest binding affinity to the B-Cells (Table 2, and Fig. 1, Fig. 2).
Concerning the T-Cells peptides prediction, the test measures the peptides’ binding affinity to the MHC molecules [25]. The available MHC I alleles HLA A, HLA B, HLA C, HLA E, and MHC II alleles HLA-DR, HLA-DQ, and HLA-DP were used. The MHC I IEDB tests [25] on the Spike glycoprotein predicted 192 peptides could interact with 2–4 MH C I alleles. The top five are (FIAGLIAIV), (FTISVTTEI), (FVFLVLLPL), (MIAQYTSAL), and (VVFLHVTYV) with population coverage of 99.73%. The tests on the Nucleocapsid protein predicted 46 peptides could interact with 2–4 MH C II alleles. The top three are (KAYNVTQAF), (KTFPPTEPK), and (LLNKHIDAY) with total population coverage of 98.20% (Table 3, and Fig. 3, Fig. 4).
In contrast, the MHC II test on the Spike glycoprotein predicted 1111 peptides could interact with the main MHC II alleles. The top two are (EVFNATRFASVYAWN) and (VFRSSVLHSTQDLFL) with total population coverage of 99.55%. The tests on the Nucleocapsid protein predicted 177 could interact with the main MHC II alleles. The top six are (AALALLLLDRLNQLE), (ALALLLLDRLNQLES), (ALLLLDRLNQLESKM), (LALLLL-DRLNQLESK), (PRWYFYYLGTGPEAG), and (RWYFYYLGTGPEAGL) with total population coverage up to 98.6% (Table 4, and Fig. 3, Fig. 4).
The results of collective IEDB tests [25] revealed that the Spike glycoprotein peptides (FTISVTTEI), (MIAQYTSAL), and the Nucleocapsid peptide (KTFPPTEPK) are the most promising MHC1 peptides. On the other hand, the Spike peptides (EVFNATRFASVYAWN), (VFRSSVLHSTQDLFL), and the Nucleocapsid peptides (AALALLLLDRLNQLE), (ALALL-LLDRLNQLES), (PRWYFYYLGTGPEAG), (RWYFYYLGTGPEAGL) are the most promising MHC II peptides.
Besides the binding with the MHC molecules, the predicted peptide must be non-toxic and non-allergen, hence, their safety was predicted using the AllergenFP v.1.0 [28], AllerCatPro [29] v. 1.7, ToxinPred [30] servers. The result showed that all the peptides were non-toxic. The AllerCatPro v. 1.7 [26] server results showed there is no evidence about the allergenicity of all peptides, however, the AllergenFP v.1.0 [28] server predicts Spike peptide (EVFNATRFASVYAWN) as an allergen (Table 5, Table 6).
To stimulate better immunological responses by the predicted peptides, they must interact and bind effectively with the MHC1 and MHC II molecules [48]; therefore, we must study their interaction with the MHC molecules.
The simulation and prediction of the interaction between the predicted peptides and the MHC molecules are conducted using molecular docking studies that rely on the calculation of the binding free energy. The lowest binding energy scores of the MHC-Peptide complex will indicate the best interaction and the highest stability [49].
To validate the results of molecular docking, MDockPeP [35] and HPEPDOCK [[36], [37], [38], [39], [40]] servers were used. The MDockPeP server predicts the MHC-Peptides interaction by “docking the peptides onto the whole surface of protein independently and flexibly using a novel the conformation restriction in its novel iterative approach. It ranks the docked Peptides via the ITScorePeP scoring function that uses the known protein-peptide complex structures in the calculations” [35]. In contrast, HPEPDOCK uses “a hierarchical flexible peptide docking approach” to predict the MHC-Peptides interaction [36]. The MHC I, HLA-A0202, HLA-B1503, HLA-C1203 was predicted to present the highly conserved SARS-CoV-2 peptides more effectively [50]; hence, they were used in the molecular docking study.
The molecular docking results showed that the Spike peptide (FTISVTTEI) has the lowest docking energy score with the MHC I HLA-B1503 allele; thus it is predicted to have the highest binding affinity. The Spike peptide (MIAQYTSAL) showed the lowest docking energy score with the MHC I HLA-C1203; consequently, it is predicted to have the highest binding affinity to the MHC I HLA-C1203 allele. In contrast, regarding the MHC I HLA-A0202, the results of the MDockPeP [35] server showed that the Nucleocapsid peptide (KTFPPTEPK) has the lowest docking energy score, but the results of the HPEPDOCK [36] server showed the Spike peptide (MIAQYTSAL) has the lowest docking energy score (Table 7).
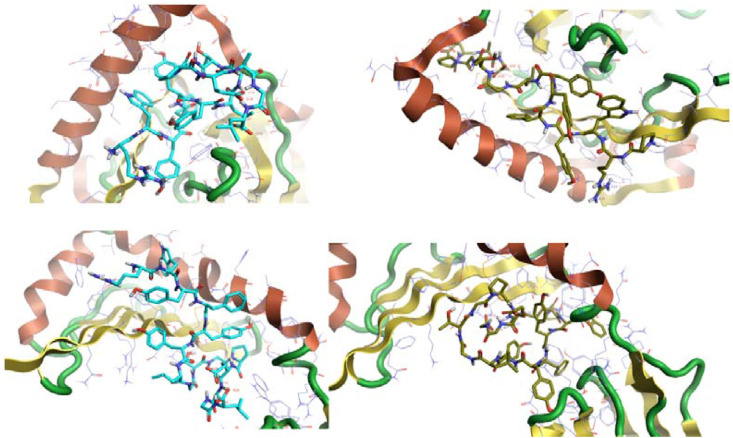
To illustrate the MHC-Peptide interaction, the PoseView [38] at the ProteinPlus web portal [42] that illustrate the 2D interactions, and Cresset Flare viewer [11] that illustrate the 3D interaction were used.
The Spike peptide (FTISVTTEI) interacts with the MHC I allele HLA-B1503 by forming nine hydrogen bonds with amino acids; threonine (two molecules), serine (three molecules), asparagine, tyrosine, lysine, tryptophan, and glutamic acid. Also, it forms seven hydrophobic bonds with amino acids; threonine (two molecules), tryptophan (two molecules), serine, alanine, and leucine). (Fig. 5, Fig. 6).
The Spike peptide (MIAQYTSAL) interacts with the MHC I allele HLA-C1203 by forming five hydrogen bonds with amino acids; tyrosine, arginine, lysine, glutamine, and threonine. Also, it forms seven hydrophobic bonds amino acids; isoleucine, glutamine (two molecules), alanine, arginine, tyrosine, and tryptophan. In comparison, it interacts with the MHC I allele HLA-A0202 by forming four hydrogen bonds with amino acids threonine, glutamine, arginine, and tryptophan, and four hydrophobic bonds with amino acids; isoleucine, tryptophan (two molecules), and tyrosine). Since it forms five hydrogen bonds and seven hydrophobic bonds with the MHC I allele HLA-C1203, it indicates the higher binding affinity to the MHC I allele HLA-C1203 and supports the MDockPeP [3] server score (Table 7 and Fig. 5, Fig. 6).
The Nucleocapsid peptide (KTFPPTEPK) interacts with the MHC I allele HLA-A0202 by forming five hydrogen bonds with amino acids; proline, glutamine, lysine, aspartic acid, and tryptophan, and seven hydrophobic bonds with amino acids (two Proline molecules, Leucine, Threonine, Trp1, Alanine, and Valine) (Fig. 5, Fig. 6).
Among the reported MHC I alleles, the allele HLA-B1503 was predicted to have “the greatest ability to present the highly conserved SARS-CoV-2 peptides” [50]. Therefore, the Spike peptide (FTISVTTEI) is predicted to make the highest response, since the binding with the MHC I stimulates the natural killer and the cytotoxic T cells [51].
Regarding the interaction with the MHC II molecule, the Spike peptide (EVFNATRFASVYAWN) showed the lowest docking energy score with the three MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, and HLA-DRB1; therefore it is predicted to have the highest binding affinity to the three alleles. Hence, it is predicted to stimulate the CD4+ (helper) T cells more effectively, since the MHC II molecule presents the antigenic peptides to the CD4+ (helper) T cells [52].
On the contrary, the Nucleocapsid peptide (RWYFYYLGTGPEAGL) showed lower docking energy scores with the MHC II allele HLA-DQA1*01:02/DQB1*06:02. The results of the MDockPeP [35] server showed that the peptide (PRWYFYYLGTGPEAG) has the lowest docking energy score; however, the results of the HPEPDOCK [36] server differed from it in the MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB1 (Table 8).
The Spike peptide (EVFNATRFASVYAWN) interacts with the allele HLA-DPA1*01:03/DPB1*02:01 by forming five hydrogen bonds with the amino acids; valine, serine, tyrosine, tryptophan, and glutamic acid, ten hydrophobic bonds with the amino acids; valine, phenylalanine, tyrosine, alanine (two molecules), asparagine, leucine (two molecules), tryptophan, tyrosine, and five pi-pi bonds with amino acids; phenylalanine, serine, tyrosine (two molecules), and tryptophan. It interacts with the allele HLA-DRB1 by forming seven hydrogen bonds with the amino acids valine, threonine, serine, phenylalanine, tyrosine, tryptophan, and glycine, thirteen hydrophobic bonds with the amino acids; valine, aspargine (two molecules), threonine, phenylalanine (two molecules), tyrosine (two molecules), tryptophan (two molecules), glutamic acid, cysteine, and proline, and three pi-pi bonds with the aromatic amino acid; phenylalanine, tyrosine, and tryptophan. Its interaction with the allele HLA-DQA1*01:02/DQB1*06:02 was not obtained from the server.
The Nucleocapsid peptide (RWYFYYLGTGPEAGL) interacts with the allele HLA-DPA1*01:03/DPB1*02:01 by forming two hydrogen bonds with the amino acids; glutamic acid and leucine, thirteen hydrophobic bonds with the amino acids; tryptophan, phenylalanine (two molecules), tyrosine (two molecules), glycine, glutamic acid, glutamine, leucine (three molecules), and proline (two molecules), and five pi-pi bonds with the aromatic amino acid Tryptophan, phenylalanine (two molecules), and tyrosine (two molecules). It interacts with the allele HLA-DQA1*01:02/DQB1*06:02 by forming six hydrogen bonds with the amino acids; glutamic acid, asparagine (three molecules), leucine, and threonine, thirteen hydrophobic bonds with the amino acids; tryptophan, phenylalanine, tyrosine, glycine (two molecules), glutamic acid, glutamine, phenylalanine, alanine (three molecules), tyrosine, and threonine, and pi-pi bonds with the aromatic amino acid tryptophan, phenylalanine, tyrosine (two molecules). It interacts with the allele HLA-DRB1 by seven forming hydrogen bonds with the amino acids; glutamic acid, valine, cysteine, tyrosine (two molecules), asparagine, and glutamine, twelve hydrophobic bonds with the amino acids; tryptophan (two molecules), phenylalanine, tyrosine (two molecules), glycine, glutamic acid, lysine, histidine, tyrosine, asparagine, and three pi-pi bonds with the aromatic amino acids; tryptophan, phenylalanine, and tyrosine. The Nucleocapsid peptide (RWYFYYLGTGPEAGL) interacts most effectively with the allele HLA-DQA1*01:02/DQB1*06:02; thus it showed the highest binding affinity to it (Fig. 7, Fig. 8).
Concerning the two Nucleocapsid peptides (RWYFYYLGTGPEAGL) and (PRWYFYYLGTGPEAG) in the interaction with the MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB1, the 2D and 3D interaction results showed that the peptide (PRWYFYYLGTGPEAG) more effectively with the allele HLA-DPA1*01:03/DPB1*02:01 and the peptide (RWYFYYLGTGPEAGL) more effectively with the HLA-DRB1 allele; however, they differ only in the first and last amino acid (Fig. 9, Fig. 10 ).
Fig. 9.
The 2D interaction of the predicted Nucleocapsid peptides (RWYFYYLGTGPEAGL) and (PRWYFYYLGTGPEAG) with the MHC II alleles. a) and b) (RWYFYYLGTGPEAGL) with MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB, respectively. b) and d) Nucleocapsid peptide (PRWYFYYLGTGPEAG) with MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB, respectively.
Fig. 10.
The 3D interaction of the predicted Nucleocapsid peptides (RWYFYYLGTGPEAGL) and (PRWYFYYLGTGPEAG) with the MHC II alleles a) and b) (RWYFYYLGTGPEAGL) with MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB, respectively
b) and d) Nucleocapsid peptide (PRWYFYYLGTGPEAG) with MHC II alleles HLA-DPA1*01:03/DPB1*02:01 and HLA-DRB, respectively.
In comparison between the Spike and Nucleocapsid peptides, the Spike peptide (FTISVTTEI) showed a higher binding affinity to the MHC I allele HLA-B1503. The Nucleocapsid peptides (KTFPPTEPK) and (RWYFYYLGTGPEAGL) showed a higher binding affinity to the MHC I allele HLA-A0202 and the three MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DRB1, respectively. However, the total population coverage of the peptides (FTISVTTEI) and (KTFPPTEPK) is not high (Table 9 ).
Table 9.
The total population coverage of the best predicted MHC peptides.
| Peptide | Total Population Coverage |
|---|---|
| FTISVTTEI | 52.54% |
| KTFPPTEPK | 43.03% |
| RWYFYYLGTGPEAGL | 97.60% |
Joshi A et al. in their predictive study found that the ORF-7A protein's peptide (ITLCFTLKR) binds most effectively with the MHC I alleles HLA-A*11:01, HLA-A*68:01 [16]. Enayatkhani M et al. in their predictive study included Nucleocapsid N, but they studied its interaction with the MHC I HLA-A*11:01 allele [16]. Kalita P et al. included the Nucleocapsid protein and Spike glycoprotein. They used the predicted peptides from Nucleocapsid, Spike, and Membrane glycoprotein to design a subunit vaccine [15]. Singh A et al. [17] used the Nucleocapsid protein to design a multi-Peptides vaccine. Also, M. Bhattacharya et al. [16] used the Spike glycoprotein and P. Kalita et al. [20] used the Spike glycoprotein (S), Membrane glycoprotein (M), and Nucleocapsid protein (N) to design a subunit vaccine. Besides the previous three proteins, B. Sarkar et al. [18] used ORF3a protein to design a subunit vaccine. Since we did not include the two alleles HLA-A*11:01, HLA-A*68:01 in our study and did not design a subunit vaccine, the logical comparison will not be applied. We aim to predict the most antigenic peptides in the Spike glycoprotein (S) and Nucleocapsid protein (N) to be a multi-peptide vaccine candidate. Since peptides could be chemically synthesized readily [53], we propose chemical synthesis as a production method to deliver large doses from the candidate vaccine after approval.
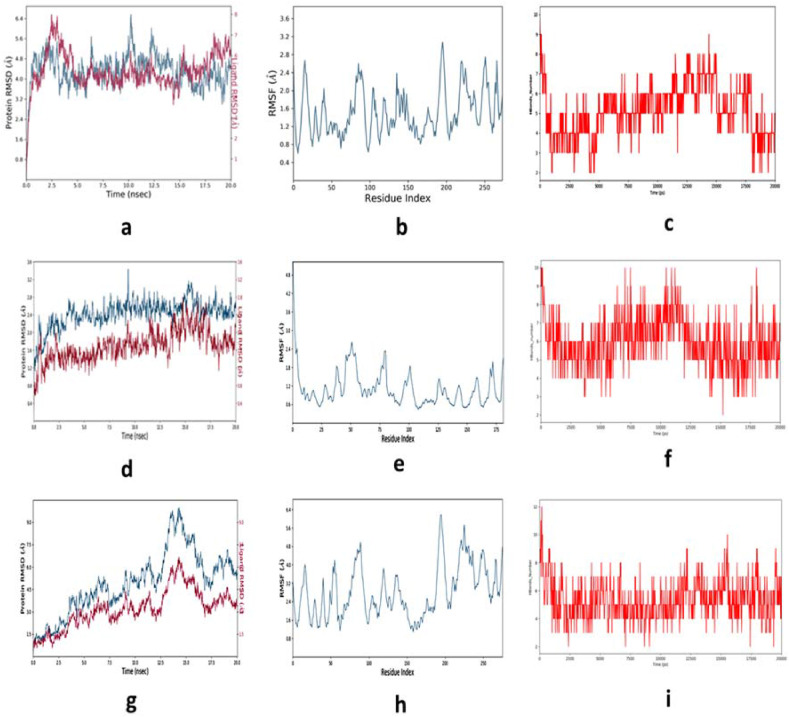
Molecular dynamics and simulation for the 20 ns convergence study revealed the conformational properties of peptide bound MHCs (Fig. 11 ). Simulation interaction displayed the significant stability of Cα-backbone of MHC I in presence of spike protein derived peptide FTISVTTEI (Fig. 11 a) in the RMSD plot. The total conformational deviation was observed to be 1.6 Å. The root mean square fluctuations (RMSF) displayed less deflections of each amino acid residues during the convergence to highly stable conformation of MHC I in the peptide bound state from the mean (Fig. 11 b).
Fig. 11.
Depiction of simulation events RMSD, RMSF and number of H-bonds formed during the entire course, respectively for FTISVTTEI-MHC I, RWYFYYLGTGPEAGL-MHC II and KTFPPTEPK- MHC I complexes. (a), (d), (g) are the RMSD plots, (b), (e), (h) RMSF plots and (c), (f), (i) H-bonds number plots.
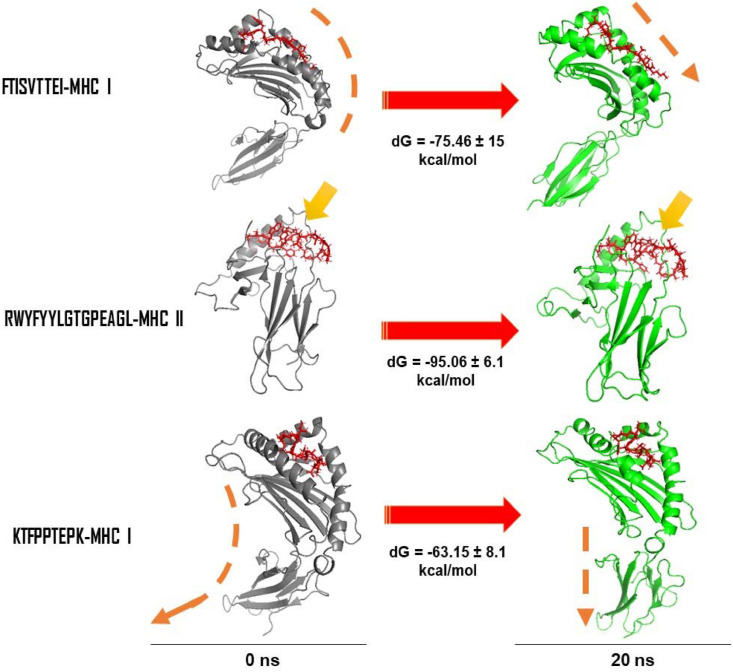
H-bond formation is an important parameter to deduce the stability of the conformation through the simulation period in the protein ligand complex. It was observed that the significant number was on an average 5 H-bonds created within the MHC I and peptide complex throughout the simulation period. In the final convergence period at 20 ns the number of H-bonds was maintained to be 5 numbers (Fig. 11 c). On the other hand, for complex RWYFYYLGTGPEAGL-MHC II displayed RMS deviation 1.4 Å (Fig. 11 d), fluctuations at each residue position were found to be least (Fig. 11 e) and the average H-bonds were created to be 6 throughout the simulation (Fig. 11 f). The overall complex displayed good stability and highly converged structure. The KTFPPTEPK-MHC I complex displayed higher RMS deviation as compared to the other complexes as discussed above. The RMS deviation was found to be high ~4.5 Å before the complex reached to convergence. This might be due to high arrangement of the Cα-backbone of MHC I during the course of simulation. The RMS fluctuation also seemed to be high from the mean point at the Ala114, Asp220 andGln250 residues, and later converged (20 ns) with an acceptable deviation of 1.6 Å. The significant average number of H-bonds was depicted throughout simulation and significantly explained the converged stability of the simulation complex (Fig. 11 i). Binding free energy calculations using molecular mechanics and generalized born model and solvent accessibility displayed significant energies between peptide and MHCs (Fig. 12 ). Interestingly it was observed that FTISVTTEI with MHC I at the beginning of simulation was bound in the binding pocket with low binding energies and later after convergence of the complex at 20 ns the peptide seemed to be rearranged in a much straighter orientation. This rearrangement gave much stable conformation of binding with dG = −75.46 ± 15 kcal/mol (Fig. 12). On the other hand RWYFYYLGTGPEAGL-MHC II complex displayed small rearranged binding site loop for the peptide and therefore conforming highly stable structure from the beginning to end of the simulation. The average binding energy was calculated to be dG = −95.06 ± 6.1 kcal/mol (Fig. 12). Significant changes of the conformation of KTFPPTEPK-MHC I complex was observed after MM/GBSA calculations. The final step of the simulation complex displayed straightening of the foot of MHC I away from peptide binding site (Fig. 12). This conformational change provided significant rearrangement space for the peptide as to bind with binding energies dG = −63.15 ± 8.1 (Fig. 12) with MHC I. Therefore, from the simulation event analysis and binding free energy calculation, it could be suggested that the peptides analyzed in this study would be potent candidates for vaccine against COVID-19.
Fig. 12.
Structural conformation of receptor (MHC I and MHC II) at 0th ns and at final 20th ns. Binding free energies (MM/GBSA) with standard deviation averaging from each frame is displayed as (dG). Arrows are indicating changes observed in the peptide binding to the MHC receptors or conformational changes observed.
5. Conclusion
A potential COVID-19 Peptide-based vaccine was predicted from the Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the Immunoinformatics approach. The Spike peptide peptides SQCVNLTTRTQLPPAYTNSFTRGVY is predicted to have the highest binding affinity to the B-Cells. The Spike peptide FTISVTTEI has the highest binding affinity to the MHC I HLA-B1503 allele. The Nucleocapsid peptides KTFPPTEPK and RWYFYYLGTGPEAGL have the highest binding affinity to the MHC I HLA-A0202 allele and the three MHC II alleles HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1-*06:02, HLA-DRB1, respectively. Furthermore, those peptides were predicted as non-toxic and non-allergen. Therefore, the combination of those peptides is predicted to stimulate better immunological responses.
Since the study is an in silico predictive work, further experimental studies are recommended to validate the obtained results.
Data availability
The data that support the findings of this study are available within the article. Additional data are published in the online open-access repository Figshare [54] under the DOI: (https://doi.org/10.6084/m9.figshare.13011878.v3).
Declaration of competing interest
There’s no direct or indirect to any actual or potential conflict of interest relationship including any financial, personal or other relationships with other people or organizations between the author's manuscript and any outside contacts.
Acknowledgements
I would like to express my appreciation and thanks to my supervisor Prof. Mohammed A salih whose supported me and my colleagues with valuable help and encouragement throughout the period of this study, all authors read and approved the final version of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100476.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gonzalez J., Gomez-Puertas P., Cavanagh D., Gorbalenya A., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148(11):2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization W.H. Novel Coronavirus ( 2019-nCoV): situation report. 2020;3 [Google Scholar]
- 3.McIntosh K. Uptodate; 2020. Coronavirus disease 2019 (COVID-19): epidemiology, virology, clinical features, diagnosis, and prevention; p. 2020. [Google Scholar]
- 4.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Centre for Disease Prevention and Control COVID-19 situation update worldwide. https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases as of 5 May 2020. accessed 6/5/2020, 2020.
- 6.Aditya K. Padhi and timir tripathi "can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacology & Translational Science. 2020;3(5):1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalita P. Development of multi-epitope driven subunit vaccine against Fasciola gigantica using immunoinformatics approach. Int J Biol Macromol. 2019;138:224–233. doi: 10.1016/j.ijbiomac.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Kalita J., Padhi A.K., Tripathi T. Designing a vaccine for fascioliasis using immunogenic 24 kDa mu-class glutathione s-transferase"infection. Genetics and Evolution. 2020;83:104352. doi: 10.1016/j.meegid.2020.104352. [DOI] [PubMed] [Google Scholar]
- 9.(Who) T.W.H.O. WHO Solidarity Trial – accelerating a safe and effective COVID-19 vaccine. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-trial-accelerating-a-safe-and-effective-covid-19-vaccine accessed 6/5/2020, 2020.
- 10.Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020 May;19(5):305–306. doi: 10.1038/d41573-020-00073-5. PMID: 32273591. [DOI] [PubMed] [Google Scholar]
- 11.Chan J.F.-W. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Joshi M.D., Singhania S., Ramsey K.H., Murthy A.K. Peptide vaccine: progress and challenges. Vaccines. 2014;2(3):515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petitdidier E. Peptide-based vaccine successfully induces protective immunity against canine visceral leishmaniasis. npj Vaccines. 2019;4(1):49. doi: 10.1038/s41541-019-0144-2. 2019/11/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enayatkhani M. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalita P., Padhi A.K., Zhang K., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020;145:104236. doi: 10.1016/j.micpath.2020.104236. 2020/08/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi A., Joshi B.C., Mannan M.A.-u., Kaushik V. Epitope based vaccine prediction for SARS-COV-2 by deploying immuno-informatics approach. Informatics in Medicine Unlocked. 2020;19:100338. doi: 10.1016/j.imu.2020.100338. 2020/01/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A., Thakur M., Sharma L.K., Chandra K. Designing a multi-epitope peptide-based vaccine against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.15.040618. 04.15.040618, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar B., Ullah M.A., Johora F.T., Taniya M.A., Araf Y. Immunoinformatics-guided designing of epitope-based subunit vaccines against the SARS Coronavirus-2 (SARS-CoV-2) Immunobiology. 2020;225(3):151955. doi: 10.1016/j.imbio.2020.151955. 2020/05/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya M. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol. 2020;92(6):618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalita P., Padhi A., Zhang K.Y., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020:104236. doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017;45(D12) doi: 10.1093/nar/gkw1071. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E. IPD-IMGT/HLA database. Nucleic Acids Res. 2019;48(D1):D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. citeulike-article-id:691774. [Google Scholar]
- 25.Vita R. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen J.E.P., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2(1):2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emini E.A., Hughes J.V., Perlow D., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55(3):836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivan Dimitrov L.N. Irini Doytchinova, and Ivan Bangov, "AllergenFP: allergenicity prediction y descriptor fingerprints. Bioinformatics. 2013;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 29.Maurer-Stroh S. AllerCatPro—prediction of protein allergenicity potential from the protein sequence. Bioinformatics. 2019;35(17):3020–3027. doi: 10.1093/bioinformatics/btz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta S. In silico approach for predicting toxicity of peptides and proteins. PloS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. e73957-e73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torsten Schwede J.K., Guex Nicolas, Peitsch Manuel C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.al K.e. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen Ef G.T., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Flare v. A. Molecular field extrema as descriptors of biological Activity: definition and validation. Cheeseright T., Mackey M., Rose S., Vinter, editors. J Chem Inf Model. 2006;46(2):665–676. doi: 10.1021/ci050357s. http://www.cresset-group.com/flare/ Cresset®, Litlington, Cambridgeshire, UK. [DOI] [PubMed] [Google Scholar]
- 35.Xu X., Yan C., Zou X. MDockPeP: an ab-initio protein–peptide docking server. J Comput Chem. 2018;39(28):2409–2413. doi: 10.1002/jcc.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P., Jin B., Li H., Huang S.-Y. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. eng, editor. Nucleic Acids Res. 2018;46(W1):W443–W450. doi: 10.1093/nar/gky357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9(2):173–175. doi: 10.1038/nmeth.1818. 2012/02/01. [DOI] [PubMed] [Google Scholar]
- 38.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., and F.M., Šali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29(1):291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 40.Berman H.M. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rarey K.S.a.M. PoseView-- molecular interaction patterns at galance. J Cheminf. 2010;2(1):50. [Google Scholar]
- 42.Fährrolfes R. ProteinsPlus: a web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017;45(W1):W337–W343. doi: 10.1093/nar/gkx333%J. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lurie N., Saville M., Hatchett R., Halton J. Developing covid-19 vaccines at pandemic speed. N Engl J Med. 2020 doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 44.Rueckert C., Guzmán C.A. Vaccines: from empirical development to rational design. PLoS Pathog. 2012;8(11) doi: 10.1371/journal.ppat.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adhikari U.K., Tayebi M., Rahman M.M. Immunoinformatics approach for epitope-based peptide vaccine design and active site prediction against polyprotein of emerging oropouche virus. Journal of Immunology Research. 2018:6718083. doi: 10.1155/2018/6718083. 2018/10/08 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Resource I.A. Antibody epitope prediction - tutorial. http://tools.iedb.org/bcell/help/ accessed 5/9/2020, 2020.
- 47.Flaherty D.K., editor. Immunology for pharmacy. Mosby; Saint Louis: 2012. Chapter 3 - immunogenicity and antigenicity; pp. 23–30. [Google Scholar]
- 48.Rock K.L., Reits E., Neefjes J. Present yourself! By MHC class I and MHC class II molecules. eng, editor. Trends Immunol. 2016;37(11):724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cournia Z., Allen B., Sherman W. Relative binding free energy calculations in drug discovery: recent advances and practical considerations. J Chem Inf Model. 2017;57(12):2911–2937. doi: 10.1021/acs.jcim.7b00564. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen A. 2020. Human leukocyte antigen susceptibility map for SARS-CoV-2. medRxiv, p. 2020.03.22.20040600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wieczorek M. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. (in English), Frontiers in Immunology, Review. 2017;8(292) doi: 10.3389/fimmu.2017.00292. March-17 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holling T.M., Schooten E., van Den Elsen P.J. Function and regulation of MHC class II molecules in T-lymphocytes: of mice and men. Hum Immunol. 2004;65(4):282–290. doi: 10.1016/j.humimm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Hou W., Zhang X., Liu C.-F. vol. 23. Transactions of Tianjin University; 2017. pp. 401–419. (Progress in chemical synthesis of peptides and proteins). 5. 2017/09/01. [DOI] [Google Scholar]
- 54.Singh J. FigShare. J Pharmacol Pharmacother. 2011;2(2):138. doi: 10.4103/0976-500X.81919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J Biomol Struct Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dayer M.R., Taleb-Gassabi S., Dayer M.S. Lopinavir; a potent drug against coronavirus infection: insight from molecular docking study. Arch Clin Infect Dis. 2017;12(4) doi: 10.5812/archcid.13823. [DOI] [Google Scholar]
- 57.Kaiser & Jocelyn Chemists want NIH to curtail database. Science. 2005;308(5723):774. doi: 10.1126/science.308.5723.774a. [DOI] [PubMed] [Google Scholar]
- 58.Chowdhury P. Insilico investigation of phytoconstituents from Indian medicinal herb ‘Tinosporacordifolia (giloy)’against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J Biomol Struct Dynam. 2020:1–18. doi: 10.1080/07391102.2020.1803968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the article. Additional data are published in the online open-access repository Figshare [54] under the DOI: (https://doi.org/10.6084/m9.figshare.13011878.v3).