Highlights
-
•
The level of SARS-CoV-2 in wastewater varied over time during the outbreak in Gothenburg.
-
•
A peak of SARS-CoV-2 in wastewater preceded an increase in newly hospitalized patients with COVID-19 by 19-21 days.
-
•
The level of SARS-CoV-2 in wastewater varied when comparing different parts of Gothenburg.
-
•
SARS-CoV-2 was released to the recipient water from the wastewater treatment plant with a 4-log10 reduction.
Keywords: COVID-19, Surveillance, Sewage
Abstract
SARS-CoV-2 was discovered among humans in Wuhan, China in late 2019, and then spread rapidly, causing a global pandemic. The virus was found to be transmitted mainly by respiratory droplets from infected persons or by direct contact. It was also shown to be excreted in feces, why we investigated whether the virus could be detected in wastewater and if so, to which extent its levels reflects its spread in society. Samples of wastewater from the city of Gothenburg, and surrounding municipalities in Sweden were collected daily from mid-February until June 2020 at the Rya wastewater treatment plant. Flow proportional samples of wastewater were collected to ensure that comparable amounts were obtained for analysis. Daily samples were pooled into weekly samples. Virus was concentrated on a filter and analyzed by RT-qPCR. The amount of SARS-CoV-2 varied with peaks approximately every four week, preceding variations in number of newly hospitalized patients by 19-21 days. At that time virus testing for COVID-19 was limited to patients with severe symptoms. Local differences in viral spread was shown by analyzing weekly composite samples of wastewater from five sampling sites for four weeks. The highest amount of virus was found from the central, eastern, and northern parts of the city. SARS-CoV-2 was also found in the treated effluent wastewater from the WWTP discharged into the recipient, the Göta River, although with a reduction of 4-log10. The viral peaks with regular temporal intervals indicated that SARS-CoV-2 may have a cluster spread, probably reflecting that the majority of infected persons only spread the disease during a few days. Our results are important for both the planning of hospital care and to rapidly identify and intervene against local spread of the virus.
Graphical abstract
1. Introduction
The SARS-CoV-2 virus responsible for outbreaks of severe acute respiratory disease COVID-19 in Wuhan, China in late 2019 is a single stranded RNA virus of 30kb. It was found to be genetically similar to the coronavirus that caused severe acute respiratory syndrome (SARS) in 2002–2003, and to two coronaviruses infecting bats (Chan et al., 2020; Lu et al., 2020). Since its detection it has caused a large global pandemic with many deaths. Virus transmission is mainly through respiratory droplets, but may also spread by contact with contaminated surfaces (Biryukov et al., 2020). SARS-CoV-2 does not only infect the upper respiratory tract and the lungs, but also ilium and colon, where cells express the virus receptor angiotensin converting enzyme 2 (ACE2) in comparable levels to lung cells (Hamming et al., 2004; Zhang et al., 2020).
Although SARS-CoV-2 RNA has been detected in stool samples from infected individuals (Bwire et al., 2020; Wu et al., 2020c; Xu et al., 2020), and may cause diarrhea (Chen et al., 2020; Cholankeril et al., 2020) transmission by ingestion of contaminated food or the fecal-oral route has not been proven. Shedding of SARS-CoV-2 in feces may be common from patients with few symptoms as proposed from a study in the USA, showing a higher amount of viruses in wastewater than would be expected from the number of diagnosed patients (Wu et al., 2020). Since SARS-CoV-2 is excreted in feces it is also found in wastewater as has been shown in studies from several countries affected by the pandemic (Ahmed et al., 2020; Ahmed et al., 2020b; La Rosa et al., 2020; Nemudryi et al., 2020; Orive et al., 2020; Randazzo et al., 2020; Rimoldi et al., 2020; Venugopal et al., 2020). Wastewater based surveillance has been used to identify the spread of wild and vaccine derived polioviruses (Asghar et al., 2014; Hovi et al., 2012; Shulman et al., 2014) . It may also provide early warnings on outbreaks of norovirus and hepatitis A in society (Farkas et al., 2018; Hellmér et al., 2014). The excretion of SARS-CoV-2 in wastewater has so far, however, not been used to prospectively follow outbreaks by quantifying SARS-CoV-2 in consecutive wastewater samples for a longer period. This study was conducted to investigate if detection and quantification of SARS-CoV-2 in weekly samples can provide insight into whether there are both regional and quantitative variations of virus during a longer follow-up during the outbreak. Such an analysis may be used as an early warning for future outbreaks since it reflects the number of people who become ill with COVID-19, a portion of whom may require hospital care.
2. Materials and methods
2.1. Wastewater sampling
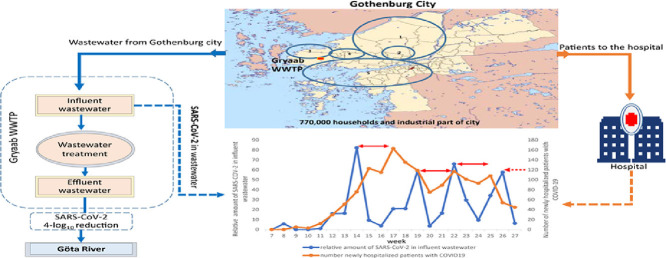
Influent and effluent wastewater samples were collected from the Rya wastewater treatment plant (WWTP), in Gothenburg, Sweden between February 10 and July 5, 2020. The Rya WWTP treats wastewater from nearly 800 000 inhabitants in the Gothenburg region on Sweden's west coast. The WWTP and tunnel system is owned and operated by Gryaab AB, a company fully owned by the municipalities it serves. The WWTP receives wastewater also from industry and from infiltration/inflow from older parts of the city. Flow proportional daily samples of influent wastewater were taken to ensure that the amount of wastewater from each household was comparable between sampling periods, as variation in influent flow are largely dependent on the amount of precipitation. The weekly variations of the influent flow during the sampling period is shown in Supplementary Fig. B1. The treated effluent from the WWTP, it is discharged into the Göta River estuary.
The influent wastewater was sampled over 24 hours using a fixed-site sampler, which was routinely used at the WWTP for monitoring the wastewater. The volume collected was proportional to the amount of influent wastewater. A composite daily sample contained 30mL wastewater per 1000 m3 influent wastewater taken over a 24-hour period between 07:00 day 1 until 07:00 day 2. These samples were combined into weekly samples by pooling the daily samples obtained during a given week. For virus analysis, 1.93% of the total weekly samples were used for analysis week 7 to 18, 3.85% of the total weekly samples from week 19 to 22 and from week 23 and onward 7.72% of the sampled volume was used for virus analysis. The virus sample volume was successively increased to simplify sample processing of the wastewater though a Nano-Ceram filter (Argonite Cooperation, Florida, USA), since larger amounts of water are easier to filter when using devices with 10L canisters and N2-gas to press the water through the filter. In total, 21 weekly samples were analyzed and designated week 7- 27. The volume for the weekly virus samples ranged from 0.975 to 5.79 liters and the samples were kept frozen at -20°C until processed.
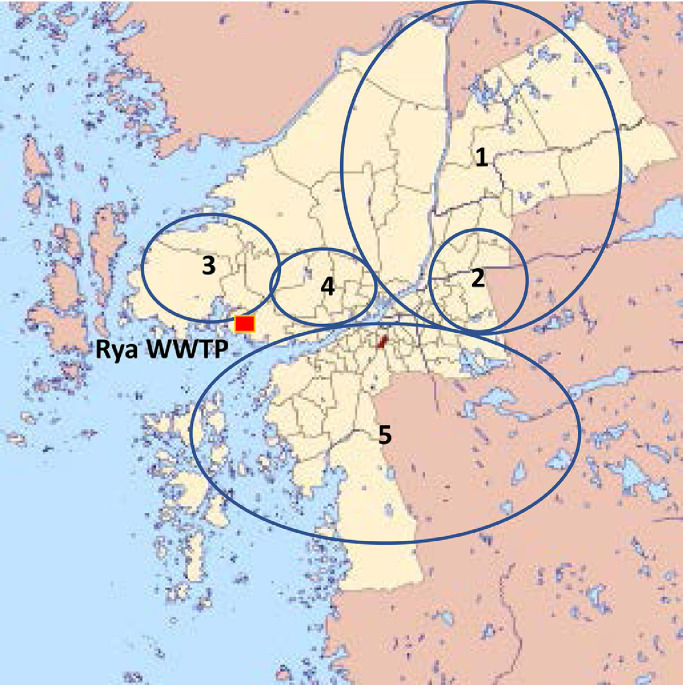
Wastewater samples from five samplings stations, receiving wastewater from different regions of Gothenburg (Fig. 1 ) were collected for three weeks (week 21-23). Samples from sampling points 2 and 3 were also collected week 20. The sampling stations collected 0.5L wastewater per day for 4 days (time proportional). On two occasions the sampler clogged and a grab samples were taken instead (Supplemental Table A1). Preparation of all influent virus samples was performed at the Clinical Microbiology Laboratory (CML) at Sahlgrenska University Hospital in Gothenburg.
Fig. 1.
Area covered by local collection points from which wastewater was sampled in this study. The wastewater from collection point 2 also passes through collection point 1.
For analysis of the treated effluent wastewater from the WWTP, a Nano-Ceram filter was connected to the flow pipeline before the treated water was discharged into the Göta River. The flow rate through the filter was set at 8 L/min and the sampling lasted for 24 hours resulting in about 11,500L of treated wastewater passing through the filter. In total 14 samples were collected by each week taking a daily sample between Monday to Tuesday week 14 until week 26 (March 31 until June 30 in 2020). After on-site sampling, the Nano-Ceram filter was kept moist in a plastic bag and shipped to CML for analysis.
Sampling date, flow and volume of influent and effluent samples are given in Table 1 .
Table 1.
Weekly determinations of amount/L and relative amount of SARS-CoV-2 genomes identified in weekly composite influent wastewater samples and daily effluent wastewater samples.
| Sampling week | Sampling date – time (Month Day hour:minute) | Mean flow m3/second | Volume analyzed (L) | Amount of SARS-CoV-2/L wastewater (log10) | Relative amount of SARS-CoV-2 |
|---|---|---|---|---|---|
| Incoming wastewater | |||||
| 7 | February 10-16 | ||||
| 7.75 | 2.65 | <3 | NA | ||
| 8 | 17-23 | ||||
| 8.46 | 2.91 | 4.23 | 5.7 | ||
| 9 | 24- March 02 | 6.12 | 2.09 | <3 | NA |
| 10 | March 03-09 | 4.78 | 1.65 | <3 | NA |
| 11 | 10-16 | 7.09 | 3.22 | 3.90 | 1 |
| 12 | 17-23 | ||||
| 5.12 | 1.90 | 4.90 | 16 | ||
| 13 | 24-30 | 3.37 | 1.00 | 5.61 | 16 |
| 14 | 31- April 06 | 3.22 | 1.13 | 6.27 | 82 |
| 15 | 07-13 | 3.17 | 1.12 | 5.33 | 9.5 |
| 16 | 14-20 | 2.96 | 1.02 | 4.96 | 3.7 |
| 17 | 21-27 | 2.75 | 0.98 | 5.74 | 21 |
| 18 | 28- May 04 | 3.74 | 2.6 | 5.31 | 21 |
| 19 | 05-11 | 3.00 | 2.12 | 6.15 | 59 |
| 20 | 12-18 | 2.78 | 1.95 | 4.67 | 3.6 |
| 21 | 19-25 | 3.13 | 3.30 | 5.10 | 16 |
| 22 | 26- June 01 | 2.70 | 2.84 | 5.77 | 66 |
| 23 | 02-08 | 3.03 | 4.21 | 5.25 | 30 |
| 24 | 09-15 | 2.60 | 3.65 | 4.82 | 9.5 |
| 25 | 16-22 | 2.96 | 4.17 | 5.32 | 34 |
| 26 | 23-29 | 3.06 | 4.50 | 5.51 | 58 |
| 27 | 29- July 06 | 4.14 | 5.79 | 4.44 | 6.3 |
| Treated wastewater | 8L/min | ||||
| 14 | March 31- April 1 9:35-9:45 | 11,600 | 1.80 | ||
| 15 | April 06- 07 8:35-8:25 | 11,510 | 1.42 | ||
| 16 | April 14- 15 8:15-8:14 | 11,512 | 0.84 | ||
| 17 | April 20- 21 8:15-7:45 | 11,280 | 1.82 | ||
| 18 | April 27- 28 8:22-8:18 | 11,504 | 1.87 | ||
| 19 | May 04 – 05 8:45-8:13 | 11,264 | 0.58 | ||
| 20 | May 11 - 12 8:38-8:14 | 11,328 | 0.14 | ||
| 21 | May 18- 19 8:33-8:08 | 11,336 | 0.52 | ||
| 22 | May 25- 26 8:56-8:21 | 11,248 | 1.38 | ||
| 23 | June 05- 06 7:40-7:50 | 11,600 | 1.10 | ||
| 24 | June 08 - 09 8:33-8:05 | 11,304 | 1.43 | ||
| 25 | June 15- 16 7:20-7:33 | 11,624 | 1.26 | ||
| 26 | June 22 –23 8:28-8:02 | 11,312 | 1.47 | ||
| 27 | June 29 - 30 7:45-7:44 | 11,520 | 1.80 | ||
2.2. Concentration of viruses in influent and effluent samples
Viruses in samples obtained from week 7 until week 11 were concentrated by milk powder as described previously (Hellmer et al., 2014). During week 19 and week 20 the samples were analyzed in parallel by adsorption to milk powder or to Nano-Ceram filter. The latter technique was found to be about 50 times more sensitive than adsorption to milk powder for SARS-CoV-2 detection. Stored samples from previous weeks (week 11 -18) were re-concentrated by adsorption on Nano-Ceram filters, which were subsequently used for the concentration assay (Supplementary Table A2).
Viruses concentrated from the influent wastewater samples by adsorption to Nano-Ceram filters were eluted from the filter followed by ultracentrifugation as described previously (Wang et al., 2018). For the treated effluent water samples, viruses were eluted from the Nano-Ceram filters by adding 5% Beef extract, 0.2 M phosphate buffer containing 0.05 M Glycine (pH 9.5), and thereafter further concentrated by ultracentrifugation as previously described (Wang et al., 2018). The resulting pellet was suspended in 2.4 mL 10 mM Tris-HCl (pH 8.0) buffer and stored at -80°C until analyzed.
2.3. Sensitivity assay
The amount of human coronavirus 229E (ATCC VR-740) in 100µL of a stock solution was determined by RT-qPCR of serial 10-step dilutions after nucleic acid extraction with DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacture's instruction. The RT-qPCR was performed with primers and probe as previously described (Lundin et al., 2014). To determine the sensitivity of the assay to concentrate coronaviruses 2.1 1011 genomic equivalents of human coronavirus 229E were added to 1L of wastewater and adsorbed to Nano-Ceram filter as performed for the sampled wastewater. One mL samples of the water were collected from the wastewater before and after addition of the virus, from the water that passed through the filter, from the buffer eluted from the filter, from the supernatant and from the dissolved pellet after ultracentrifugation. All samples were analyzed in triplicate by RT-qPCR in 10-step dilutions, undiluted to 10−3. Primers and probes used are given in Supplementary Table A3.
2.4. Detection of SARS-CoV-2 and enteric viruses in wastewater by RT-qPCR
Total nucleic acids were extracted from 200 µL dissolved pellet after ultracentrifugation using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacture's instruction. The nucleic acids were eluted with 200 µL elution buffer (Qiagen). RT-qPCR was performed for detection of SARS-CoV-2. The qPCR was performed on a 7300 Fast Real-Time PCR system (Applied Biosystems), and all samples were tested in triplicate. The reaction was performed in a 50µL reaction mixture containing 10 µL RNA, 1x Reaction Mix (Invitrogen), 20 U RNaseOUTTM (Invitrogen), 1 µL SuperScript® III/platinum® Taq Mix (Invitrogen), and 0.3 µM of each primer, and 0.2 µM of probe. The qPCR was initiated with reverse transcription at 46°C for 30 min followed by one cycle of 95°C for 10 min and 45 cycles of 95°C for 15 sec and 56°C 1 min. Primers and probes used are listed in Supplementary Table A3. Four ten-fold serial dilutions (1/104 – 1/107) of a 2ug/mL pEX-A128-Wuhan nCov 2plasmid (Eurofinns genomics) containing the target regions was used as positive control in all RT-qPCR analyses and W4502 sterile water (Sigma Life Science) was used as negative control. Analysis of other fecal/oral transmitted viruses was used for control of the extraction and concentration technique (Wang et al., 2020)
The samples were also analyzed for norovirus GI, GII, and GIV, sapovirus, adenovirus and astrovirus by qPCR as previously described (Hellmer et al., 2014). The reaction was performed in a 25 µL reaction mixture containing 5 µL extracted nucleic acids, 1 × TaqMan Universal Master Mix (Applied Biosystems, Foster City, CA, USA), 6 µL of H2O, and 0.6 µM of each primer and 0.2 µM of probe. The RT-qPCR reactions were performed with initial one cycle at 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 55°C for 1 min for all viruses except for adenovirus, which had an extension at 60°C for 1 min. All qPCRs were performed on the 7500 Fast Real-Time PCR system (Applied Biosystems) and all samples were analyzed in duplicate.
2.5. Number of identified viral genomes
Serial dilutions of a plasmid containing all targeted viral regions inserted into the EcoRV site of a pUC157 plasmid (pUC57cl; GenScript HK, Ltd., Hong Kong) were used as positive controls in the RT-qPCR assays. The Ct values obtained were used to calculate the number of viral genomes in the wastewater samples by performing linear regression of the Ct value obtained from the serial dilutions of the pUC157 plasmid (Wang et al., 2018).
The estimated number of viruses in the influent wastewater per week, Yin, was calculated by the formula, Yin=(Xin*100)/(N*Z%), Xin is estimated number of viral genomes in wastewater based on the RT-qPCR assay (without consideration of 80% loss); X*100 is the total amount of viruses in the sample analyzed (2,400µL of diluted pellet after ultracentrifugation/25µL used for RT-qPCR), N is the fraction of wastewater obtained out of the total influent of waste water during the week. For weeks 7 to18 N= 5.79 × 10−10; for weeks 19 to 23 N=1.74 × 10−9 and for weeks 24 to 27 N=2.31 × 10−9. Z is the efficiency of virus concentration in wastewater by the technique used as determined previously to 15% for adenovirus and assumed for the other fecal transmitted viruses (Wang et al., 2018), and 6% for SARS-CoV-2 as determined in this study.
The number of SARS-CoV-2 genomes per L and day was calculated according by the formula for the influent wastewater Win=(Yin/7)/((F*60*60*24)*1000*6%), where F is the wastewater flow (m3/second; Table A2). For the treated effluent wastewater it was calculated according to the formula Wout=(Xout*100)/(Aout*6%), where Xout is calculated as for Xin, Aout is the amount of liters of purified wastewater that had passed through the filter (Table 1).
The relative amount of viral genomes per liter wastewater was calculated by dividing the amount of viral genomes in the sample with the amount of SARS-CoV-2 genomes in the influent wastewater during week 11, which was the first week where all subsequent weeks contained detectable SARS-CoV-2 genomes.
The amount of genomes per L and day at the five different sampling stations was calculated according to the formula Wx = ((Xreg/ L) *100)/Freg*6%), where x is the sampling station number. Xreg is the estimated number of viral genomes in wastewater based on the RT-qPCR assay, L is the volume analyzed, Freg is the fraction of collected wastewater from the total flow, 6% the recovery rate of viral genomes. Freg was calculated according to the formula Freg= (C/D)/(((F*60*60*24)*1000)*Q) where C is the number of collected liters of waste water. D is the number of collection days; Q is the percentage of the total flow into to the WWTP passing the collection point.
3. Results
3.1. Sensitivity assay
The titre of a stock solution of human coronavirus 229E was determined by RT-qPCR of tenfold dilutions to 3.6 × 108 genomes per 5µL stock solution. To 1L of wastewater 2.5mL of the stock solution 229E (2.1 × 1011 genome equivalents) was added. In the final pellet 1.24 × 1010 genome equivalents were recovered, which corresponds to 5.9 % of the number of genome equivalents in the original sample. Table 2 shows the recovery rate between each of the concentration steps.
Table 2.
Amount of coronavirus 229E detected during the different concentration steps.
| Volume | Total log10 viral genomes | % recovery | |
|---|---|---|---|
| wastewater without 229E virus addition | 1L | 0 | |
| Wastewater after 229E addition | 1L | 11.32 | 100% |
| In effluent 1 not bound to the Nano-Ceram Filter | 1L | 10.33 | 10.33% |
| In effluent 2 not bound to the Nano-Ceram Filter | 1L | 9.63 | 2.0% |
| Effluent from bound viruses to the Nano-Ceram filter | 300mL | 10.04 | 5.2% |
| Supernatant after ultracentrifugation | 300 mL | 9.20 | 0.8% |
| Dissolved pellet after ultracentrifugation | 2.4 mL | 10.10 | 5.9% |
3.2. Comparison of the assays
In the beginning of the study, viruses were concentrated by flocculation and adsorption to milk powder. To compare the two techniques available, viruses were concentrated both by adsorption to milk powder and to Nano-Ceram filters during weeks 19 and 20. The milk powder technique was about 50-times less sensitive than the Nano-Ceram technique for detection of SARS-CoV-2 (Table A2). However, it was equal or nearly equally sensitive for fecal transmitted viruses as norovirus GII, sapovirus and astrovirus and somewhat less sensitive for detection of adenovirus and more sensitive for detection of rotavirus (Table A1). The Nano-Ceram technique was subsequently used and stored previous wastewater samples from weeks 11 – 18 were reanalyzed by this technique.
3.3. Detection of SARS-CoV-2
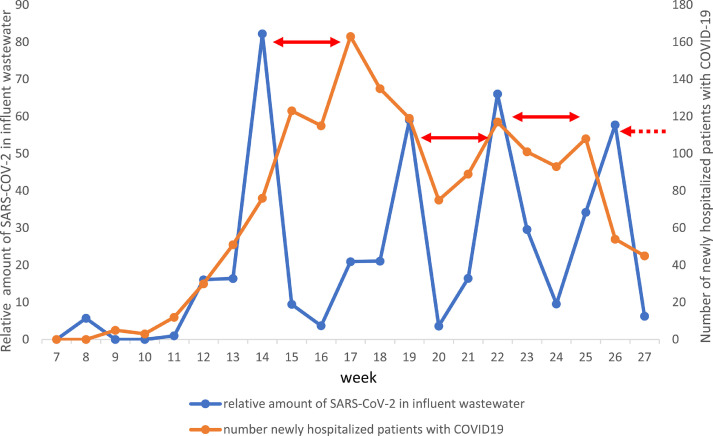
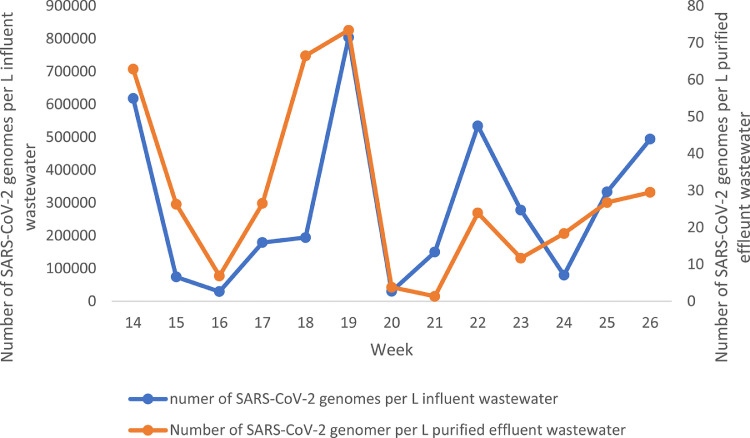
The wastewater sampling started in week 7 (February 11-17, 2020), which was the winter break week for schools in Gothenburg. The virus was detected in the sample from week 8 (February 18-24, 2020), but not in the samples from weeks 9-10, but reappeared again in week 11 (March 10 to 16) and was detected in all subsequent samples until the end of the study in week 27 (June 29 to July 6) (Table 1). The amount of detectable SARS-CoV-2 genomes in the total wastewater flow entering the treatment plant week 11 was used as a reference. The amount found in the subsequent samples were related to the amount found in this sample, which was set as 1 (Table 1). There were four peaks of SARS-CoV-2 during the sampling period, weeks 14,19, 22 and 26 (Fig. 2 ). These peaks coincided with the peaks of SARS-CoV-2 in treated effluent wastewater (Fig. 3 ), although in the treated effluent the concentration of viral genomes per L wastewater was approximately 4-log10 lower than in the influent wastewater. The SARS-CoV-2 peaks in wastewater preceded the peaks of number of hospitalized patients with COVID-19 at Sahlgrenska University Hospital by three to four weeks (Fig. 2).
Fig. 2.
Relative amount of SARS-CoV-2 genomes in influent wastewater per week in relation to the number of newly hospitalized patients in Gothenburg. The time difference between peaks of viral genomes and newly hospitalized patients are indicated with red arrows.
Fig. 3.
The number of SARS-CoV-2 genome equivalents per L incoming wastewater in relation to those in treated effluent wastewater between weeks 14 and 26.
3.4. Local differences in prevalence of SARS-CoV-2
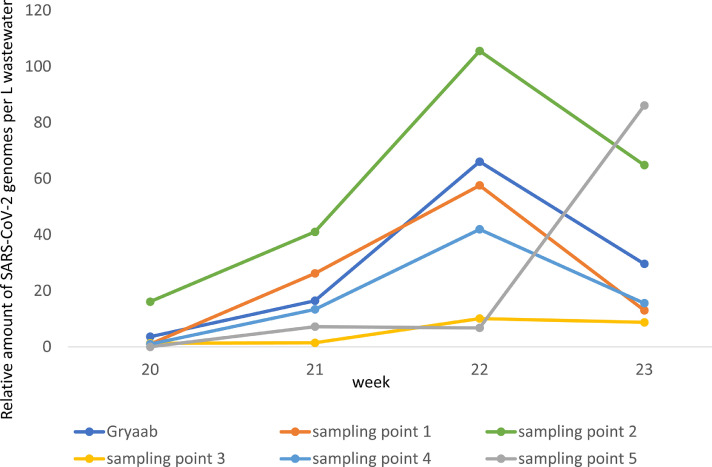
Since there were four distinct peaks of SARS-CoV-2 genome equivalents with regular intervals of three to four weeks, possible local differences in prevalence within Gothenburg were investigated during a four-week period (weeks 20 to 23). Five different sampling points were selected, each covering one part of the city (Fig. 1). In samples from three of the collection points 1, 2 and 4, covering the north-east and central part of the city, there was a peak of viral genomes week 22, which coincided with the peak observed in the influent wastewater at the WWTP that week. The relative amount of SARS-CoV-2 increased from 0 – 5 times to 42-105 times of the amount found in week 11 in the wastewater. At collection point 5, covering the southern part of the city, there was an increase of 86 times of the amount found in week 11 in the wastewater in week 23, while the wastewater from collection point 3, covering the western part of the city, had low level of SARS-CoV-2 genomes throughout the four weeks analyzed (Fig. 4 ).
Fig. 4.
Relative amount of SARS-CoV-2 genome equivalents per L wastewater samples from five different collection points and from the total amount of wastewater collected at the Rya WWTP between weeks 20 and 23.
3.5. Enterically transmitted viruses in wastewater
All wastewater samples were also analyzed for caliciviruses such as norovirus GII and GIV and sapovirus, and for rotavirus, astrovirus and adenovirus. The level of rotavirus and norovirus GII were relatively high the first 8 weeks, and declined thereafter, while the levels for the other viruses was rather constant during the study period with some elevations of adenoviruses (Fig. S2). There was no apparent covariation in genome levels of these viruses and SARS-CoV-2.
4. Discussion
There were weekly variations in the concentration of SARS-CoV-2 genomes in wastewater with distinct peaks of viruses occurring in three to four-week intervals. This indicated cluster transmission of COVID-19 in Gothenburg during the study period, and that the period of transmission may be rather short. Few studies have reported longitudinal monitoring of SARS-CoV-2 in wastewater over several months. One study from Massachusetts followed the amount of virus in wastewater between March and May 2020 and could show some fluctuations with an increase of viral genomes until mid-April and thereafter a declining trend (Wu et al., 2020a). In this study we found a correlation between the increase in concentrations of viral genomes in wastewater and the number of newly hospitalized patients with COVID-19 with the former preceding the latter by three to four weeks. The transmission of virus usually precedes the hospitalization by 2-3 weeks corresponding to an incubation period of 2-14 days followed by hospitalization approximately one week after the onset of symptoms (Bi et al., 2020). The discovery of repeated peaks observed in this study and the only peak in Massachusetts may be due to the profound differences in strategies to mitigate the outbreak between Sweden and the United States. In Massachusetts, there was a state school closure on March 17 and advice for everyone to stay at home from March 24, while in Sweden there has been no lockdown of society. Residents have instead been advised to adhere to good personal hygiene, physical distancing, working from home, and minimize travelling as much as possible (The-Public-Health-Agency-of-Sweden, 2020). Day care and primary schools have been kept open during the pandemic.
Many different techniques have been used for concentrating SARS-CoV-2 from wastewater, for example flocculation to beef extract or aluminium hydroxid adsorption (Mlejnkova et al., 2020; Randazzo et al., 2020), direct ultracentrifugation of the waste water (Prado et al., 2020), polyethylene glycol precipitation (Wu et al., 2020a) or filtration through 0.22µm Millipore filters (Rimoldi et al., 2020). When two techniques for detection of viruses were applied in this study, flocculation and adsorption to milk powder and adsorption to Nano-Ceram filter, it was evident that the Nano-Ceram filter was more sensitive for SARS-CoV-2 but both techniques had equal sensitivity for detection of fecal-oral transmitted viruses. The reason for this may be that the milk powder did not adsorb enveloped viruses and only the non-enveloped SARS-CoV-2 nucleocapsids were adsorbed, while both encapsidated and non-encapsidated were adsorbed to the Nano-Ceram filter. There is a need for a reference technique with which different studies could be compared. By using all these different techniques, the relative amounts of viruses in wastewater in the investigated region could be determined, but the amount of virus could be difficult to compare between studies.
The variations of genome levels of SARS-CoV-2 in the influent wastewater in this study correlated well with the concentration of genomes found in the treated effluent wastewater, with a 4-log10 reduction in the amount of genomes in the effluent compared to the influent wastewater. This finding supports the sharp variation in virus concentration found in the influent wastewater, and probably reflects the variation of infected individuals in the community as was stated for Massachusetts (Wu et al., 2020a). If only 60-70 % of the infected humans excrete the virus as has been shown in China (Wu et al., 2020c; Zheng et al., 2020) there must be a substantial number of infected individuals not requiring hospitalization, most of whom will likely remain undiagnosed. The sharp peak of virus genomes in only one week followed by a rapid decline, indicates both that there may be cluster spread and that excretion of higher amounts of viruses only last for a short period, thus contrasting to the findings from China (Wu et al., 2020c). The reason for this difference may either be a genetic difference in virus strains prevalent in China and Europe, since according to the open
-source project Nextstrain, five larger clades of SARS-CoV-2 have been identified so far (Hadfield et al., 2018). Another reason may be that many persons with subclinical infection more often excrete the virus in feces than those with clinical infections, who were investigated in the Chinese studies.
Analysis of wastewater from five different sampling points, covering different parts of Gothenburg showed differences in local incidence of SARS-CoV-2 in the city. During the four weeks of sampling there was a high concentration of viruses in the wastewater from the northern and north-western part of the city during one week, while the amount of genomes seemed to increase in the wastewater from the southern part at the end of the study period. This clearly indicates a patten of cluster spread and that analyses of wastewater originating from different regions enables the detection of local outbreaks rapidly before the occurrence of more severe cases. Such monitoring would be beneficial for both public health to track and mitigate the spread and for hospital authorities planning the availability of hospital beds and care needs.
The genome levels of five fecally transmitted viruses in the analyzed wastewaters were similar or lower than during the same period in 2014 and 2017 in the same region (Hellmer et al., 2014; Wang et al., 2020) and did not covariate with the levels of SARS-CoV-2. There were normal numbers of reported cases with norovirus infection (Norwalk infection, winter vomiting disease) during weeks 7 and 9, while it declined during week 11 when the outbreak usually peaks. Thereafter the number of reported cases rapidly declined to very few cases (Swedish-Public-Health-Agency, 2020). This pattern was reflected in our study by the concentration of these viruses in wastewater and may indicate that the advice from the authorities to the public to practice good hand hygiene, keep physical distance, work from home, etc. have had effect also on the transmission of these viruses with fecal-oral spread.
The fourth peak of SARS-CoV-2 genomes found in the wastewater from Gothenburg during week 22 (May 26 to Junie 1), was not followed by an increase of newly hospitalized patients. The reason for this remains unclear, but may in part be explained by that people were outdoors more with less risk for transmission and that other coronavirus infections in general get milder during summer months, or that the COVID-19 infected were younger, who often get mild illness, than older persons, who more often need hospital care (Brittain-Long et al., 2012; Gaunt et al., 2010). There may also be genetic changes of the viral genome during the spread between many hosts, thereby inducing lower virulence, or lower induction of the host immune response, which often is the reason for the manifestations of COVID-19.
This study showed that detection and quantification of SARS-CoV-2 in wastewater may be used to identify the prevalence of the virus in society, as well as to often, but not always, predict imminent hospitalization needs, especially when availability of patient testing is limited to those with severe symptoms. The disease dynamics in a community can thus be quantifiably monitored and used as an early warning of outbreaks. If wastewater could be collected from different regions of a city, as done in this study, an observed increase in a specific region could facilitate contact tracing for public health representatives. It can be especially important to monitor such fluctuations throughout the year, since it mirrors the number of infected individuals in the population. An increase may not always reflect an immediate need for increased burden for the health care but reflects that the virus is prevalent in society. This is important if there are seasonal differences in the manifestations of the disease with more severe cases during autumn and winter.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are grateful to Annette Roth for assisting in handling the wastewater samples. This study was supported by Swedish Research Council for Sustainable Development, Formas, grant number 942-2015-306.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2020.116620.
Appendix. Supplementary materials
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas Dsc K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travelers. J. Travel Med. 2020 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L., Akande A.O., Al Maamoun E., Zaidi S., Adeniji A.J., Burns C.C., Deshpande J., Oberste M.S., Lowther S.A. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 2014;210(Suppl 1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Q., Wu Y., Mei S., Ye C., Zou X., Zhang Z., Liu X., Wei L., Truelove S.A., Zhang T., Gao W., Cheng C., Tang X., Wu X., Wu Y., Sun B., Huang S., Sun Y., Zhang J., Ma T., Lessler J., Feng T. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect. Dis. 2020;20(8):911–919. doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukov J., Boydston J.A., Dunning R.A., Yeager J.J., Wood S., Reese A.L., Ferris A., Miller D., Weaver W., Zeitouni N.E., Phillips A., Freeburger D., Hooper I., Ratnesar-Shumate S., Yolitz J., Krause M., Williams G., Dawson D.G., Herzog A., Dabisch P., Wahl V., Hevey M.C., Altamura L.A. Increasing temperature and relative humidity accelerates inactivation of SARS-CoV-2 on surfaces. mSphere. 2020;5(4):e00441–20. doi: 10.1128/mSphere.00441-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; DOI: 10.1128
- Brittain-Long R., Andersson L.M., Olofsson S., Lindh M., Westin J. Seasonal variations of 15 respiratory agents illustrated by the application of a multiplex polymerase chain reaction assay. Scand. J. Infect. Dis. 2012;44(1):9–17. doi: 10.3109/00365548.2011.598876. [DOI] [PubMed] [Google Scholar]
- Bwire G.M., Majigo M.V., Njiro B.J., Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: a systematic review and meta-analysis. J. Med. Virol. Epub ahead of print. 2020 doi: 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]; doi:10.1002.
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholankeril G., Podboy A., Aivaliotis V.I., Tarlow B., Pham E.A., Spencer S., Kim D., Hsing A., Ahmed A. High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology. 2020;159(2):775–777. doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; doi:0.1053/j.gastro.2020.04.008
- Farkas K., Marshall M., Cooper D., McDonald J.E., Malham S.K., Peters D.E., Maloney J.D., Jones D.L. Seasonal and diurnal surveillance of treated and untreated wastewater for human enteric viruses. Environ. Sci. Pollut. Res. Int. 2018;25(33):33391–33401. doi: 10.1007/s11356-018-3261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48(8):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmer M., Paxeus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergstrom T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T., Shulman L.M., van der Avoort H., Deshpande J., Roivainen M., DE Gourville E.M. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140(1):1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., Hannoun C., Kindler E., Jónsdóttir H.R., Muth D., Kint J., Forlenza M., Müller M.A., Drosten C., Thiel V., Trybala E. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Preliminary study of SARS-CoV-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health. 2020;17(15):5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep Med. 2020;1(6):100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020;732 doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Maranhão A.G., Siqueira M.M., Miagostovich M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman L.M., Mendelson E., Anis E., Bassal R., Gdalevich M., Hindiyeh M., Kaliner E., Kopel E., Manor Y., Moran-Gilad J., Ram D., Sofer D., Somekh E., Tasher D., Weil M., Gamzu R., Grotto I. Laboratory challenges in response to silent introduction and sustained transmission of wild poliovirus type 1 in Israel during 2013. J. Infect. Dis. 2014;210(Suppl 1):S304–S314. doi: 10.1093/infdis/jiu294. [DOI] [PubMed] [Google Scholar]
- Swedish-Public-Health-Agency 2020 Calicivirus reports 2019-2020.
- The-Public-Health-Agency-of-Sweden 2020. Protect yourself and others from spread of infection. https://www.folkhalsomyndigheten.se/the-public-health-agency-of-sweden/communicable-disease-control/protect-yourself-and-others-from-spread-of-infection/.
- Wang H., Neyvaldt J., Enache L., Sikora P., Mattsson A., Johansson A., Lindh M., Bergstedt O., Norder H. One year variations of viruses in influent and effluent water at a wastewater plant–assessed by qPCR and metagenomics. Appl. Environ. Microbiol. 2020 doi: 10.1128/AEM.02073-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sikora P., Rutgersson C., Lindh M., Brodin T., Bjorlenius B., Larsson D.G.J., Norder H. Differential removal of human pathogenic viruses from sewage by conventional and ozone treatments. Int. J. Hyg. Environ. Health. 2018;221(3):479–488. doi: 10.1016/j.ijheh.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A., Ganesan H., Sudalaimuthu Raja S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin. Environ. Sci. Health. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Xiao, A., Zhang, J., Moniz, K., Endo, N., Armas, F., Bonneau, R., Brown, M.A., Bushman, M., Chai, P.R., Duvallet, C., Erickson, T.B., Foppe, K., Ghaeli, N., Gu, X., Hanage, W.P., Huang, K.H., Lee, W.L., Matus, M., McElroy, K.A., Nagler, J., Rhode, S.F., Santillana, M., Tucker, J.A., Wuertz, S., Zhao, S., Thompson, J. and Alm, E.J. 2020a. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. medRxiv 10.1101/2020.06.15.20117747. [DOI]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4):e00614–20.. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; DOI: 10.1128/mSystems.00614-20
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Li Z., Cui X., Xiao J., Zhan J., Meng T., Zhou W., Liu J., Xu H. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69(6):1010–1018. [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.