Abstract
Lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) are the two major subtypes of non–small-cell lung cancer (NSCLC). This study aimed to compare mRNA and circRNA expression patterns between LUSC and LUAD. Cancer tissues from 8 LUSC patients and 12 LUAD patients were collected to obtain mRNA and circRNA expression profiles. The differentially expressed mRNAs (DEmRNAs) and circRNAs (DE-circRNAs) between LUSC and LUAD were screened. Afterwards, miRNA–DEcircRNA pairs and miRNA–DEmRNA pairs were predicted to construct a competing endogenous RNAs (ceRNAs) network, followed by functional enrichment analysis and survival analysis. In total, 635 DEmRNAs and 245 DEcircRNAs were obtained. The ceRNA analysis revealed that genes, such as EPHA2, EPHA7, NTRK2, CDK6, hsa_circ_027570, hsa_circ_006089, and hsa-circ_035997, had distinct expression patterns between LUSC and LUAD. Also, functional enrichment analysis indicated that DEmRNAs were mainly enriched in ERK1 and ERK2 cascade. Survival analyses suggested that STXBP1 and PMEPA1 were associated the prognosis of with both LUAD and LUSC, whereas EPHA2 and CDK6 might serve as prognostic factors for LUSC and LUAD, respectively. In conclusion, genes such as EPHA2, EPHA7, NTRK2, and CDK6 had different patterns in the two major histological subtypes of NSCLC. Notably, EPHA2 and CDK6 might be considered as potential therapeutic targets for LUSC and LUAD, respectively.
Keywords: Non–small-cell lung cancer, circRNAs, lung adenocarcinoma, lung squamous cell carcinoma, ceRNA network, survival analysis
Introduction
Non–small-cell lung cancer (NSCLC) is a prevalent malignant tumor characterized by a considerably high incidence and mortality (Jacky and Baik, 2017). Despite some achievements in NSCLC treatment, the overall survival is still poor (Dreyer et al., 2018). Recently, with the identification of oncogenes and development of corresponding targeted therapies, molecular testing has become an important means for treating NSCLC (Nellesen et al., 2018). Thus, understanding the molecular basis of NSCLC progression is vital to improve the treatment and prognosis of patients. The most abundant subtypes of NSCLC are lung squamous cell carcinoma (LUSC), and lung adenocarcinoma (LUAD), which are distinct in their histological, molecular, and clinical characteristics; thus, accurate classification of NSCLC into LUAD and LUSC is essential for both clinical practice and lung cancer research (Coudray et al., 2018).
Accumulating evidence has reported the similarities and differences of gene expression patterns between LUAD and LUSC. Yang et al. (2019) investigated the differences in the gene expression and methylation patterns of LUAD and LUSC, and observed that cathepsin E (CTSE) and solute carrier family 5 member 7 (SLC5A7) could be considered as potential biomarkers in the personalized treatment for LUAD and LUSC. Liu et al. (2017) found that lncRNA DGCR5 was correlated with the prognosis of LUSC, whereas MIR31HG was associated with the overall survival of LUAD, indicating that these lncRNAs had diagnostic and therapeutic potential in for LUSC and LUAD treatment. Furthermore, Wang et al. (2019) demonstrated that hsa_circ_0001073 and hsa_circ_0001495 as diagnostic markers for LUAD and LUSC, respectively. Despite the above studies, the differences of mRNA and circRNA in two major subtypes of NSCLC have not been fully explored.
This study aimed to explore the differences in the gene expression of LUAD and LUSC by comparing the mRNA and circRNA expression profiles. In addition, the prognostic genes related to LUSC and LUAD were also respectively identified. Our study could provide more insights into the molecular mechanism of LUSC and LUAD, and provide potential prognostic and diagnostic biomarkers for NSCLC.
Material and Methods
Sample collection
All lung cancer tissues were collected from 20 patients, including 8 LUSC and 12 LUAD, who were admitted to Shanxi Provincial People's Hospital, China between October 2015 and March 2018. Histopathological diagnosis of all specimens was in accordance with the World Health Organization criteria for pathological classification of lung cancer and AJCC/UICC staging system for lung cancer. Furthermore, surgical resection complied with the standards from NCCN guideline staging and surgical indications. The excluding criteria of samples were as follows: 1) patients who refuse surgery; 2) patients who do not undergo pulmonary lobectomy and receive general anesthesia; 3) patients with contraindications to anesthesia; and 4) patients who have received adjuvant therapy. The clinical characteristics of included patients are shown in Table S1 (67.2KB, pdf) . This study was approved by the Ethics Committee for Shanxi Provincial People's Hospital and the University of Arkansas for Medical Science. Informed consent was obtained.
RNA extraction and sequencing
To meet the sequencing criteria, each sample required a certain amount of repeated measurement data. Therefore, we created three repetitions for each group. Three LUAD groups and three LUSC groups were then subjected to transcriptome sequencing. Total RNA of the collected samples was extracted using TRIzol kit following the manufacturer's procedure. The concentration and purity of RNA were detected using a NanoPhotometerN60 (IMPLEN, Munich, Germany). The integrity of RNA was examined using the Agilent 2100 Bioanalyzer (Agilent Corporation, CA, USA). Then, rRNA from the total RNA was depleted using Ribo-Zero GoldrRNARemova Kit (Illumina, San Diego, USA). The treated RNAs were interrupted randomly to 200–300 bp to construct cDNA library. The insert size and total concentration of library were detected using Agilent 2100 Bioanalyzer and qPCR. Finally, high-quality library was sequenced on the Illumina HiSeq platform.
Data processing and identification of RNAs
To ensure the accuracy of the information analysis, the quality control of the sequencing data was performed to obtain clean reads. Reference genome was acquired from Ensembl database (http://www.ensembl.org/org/). Then, the clean data were aligned to the reference genome to obtain the mapped data.
HTseq 0.6.1p2 (http://www huber.embl.de/users/anders/HTSeq) was applied to calculate the read count value of each mRNA and find_circ was used to identify the counts of circRNA. The raw counts of mRNA and circRNA were normalized using reads per kilo bases per million reads (RPKM).
Identification of differentially expressed mRNAs and circRNAs
After sequencing, raw expression levels were processed by log2 transformation. Differential expression analysis between LUSC and LUAD was performed using a paired t-test with Benjamini–Hoachberg method. The log2FC (fold change) > 1 and adjusted p value (adj. p value) < 0.05 were set as cut-off threshold of DEmRNA and DEcircRNA. The heatmap of DE-mRNAs and DE-circRNAs were constructed using pheatmap (version: 1.0.10, https://cran.r-project.org/web/packages/pheatmap/index.html) software in R.
Construction of miRNA regulatory network
We adopted the overrepresentation analysis (ORA) method in WebGestalt (http://www.webgestalt.org/option.php) (Zhang et al., 2005) to predict miRNAs associated with DEmRNAs. The p value < 0.05 was set as the significant level. Afterwards, miRNA–DEmRNA pairs were identified and visualized using Cytoscape software. In addition, miRanda (version 3.3a, https://omictools.com/miranda-tool) (Agarwal et al., 2018) was utilized to predict potential target miRNAs of the DEcircRNAs. The miRNA–DEcircRNA pairs with score > 150 and energy < −20 were selected to establish a regulatory network.
CeRNA network construction
The ceRNA network was established to explore and determine the interactive relationship between mRNAs, miRNAs, and circRNAs. Notably, mRNAs–DEcircRNAs positive correlation pairs were further extracted with the cutoffs of p value < 0.05 and r > 0.9. Following this, DEcircRNA–miRNA–DEmRNA ceRNA network was structured by combining miRNAs–DEmRNAs pairs and DEmRNAs–DEcircRNAs positive correlation pairs.
Functional enrichment analysis of DEmRNAs
To better understand the biological functions of genes, the functional enrichment analysis of DEmRNAs existed in ceRNA network was performed using DAVID (version 6.8, https://david-d.ncifcrf.gov/) (Huang et al., 2009). GO-biological process (BP) terms (Ashburner et al., 2000) or pathways (Kanehisa and Goto, 2000) with the threshold of p < 0.05 and gene count > 2 were considered as statistically significant.
Survival analysis
TCGA database provides various expression profiling and clinical data, which contributes to improve the accuracy of prevention and diagnosis of various cancers (Tomczak et al., 2015). The gene expression profiles and clinical information of LUAD and LUSC were obtained from TCGA database, including 501 LUSC samples and 517 LUAD samples. DEmRNAs in ceRNA network were divided into high expression group and low expression group according to the median of expression values. The Kaplan–Meier curves were acquired using survival package (version: 2.42-6, https://cran.r-project.org/web/packages/survival/index.html) in R. The p value < 0.05 was considered statistically significant.
Results
RNA sequencing overview
We obtained 427,794,228 raw data from LUSC samples and 412,360,880 raw data from LUAD samples. The LUSC and LUAD clean reads accounted for more than 99.5% and 99.6% of raw data, suggesting that our data were high quality and could be used for subsequent analysis. Ultimately, a total of 17284 mRNAs and 4997 circRNAs were screened.
Identification of DEmRNAs and DEcircRNAs
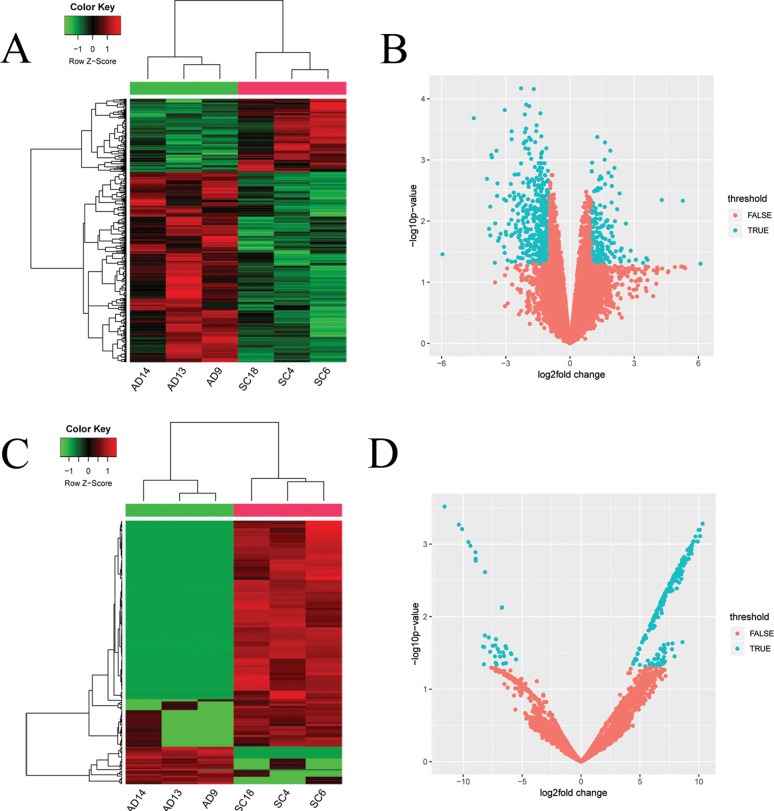
A total of 635 DE-mRNAs (176 up- and 459 down-regulated) and 245 DEcircRNA (210 up- and 35 down-regulated) were obtained in LUAD vs LUSC. The complete list of DEmRNAs and DEcircRNAs is shown in Table S2 (106KB, xls) and Table S3 (52.5KB, xls) . Hierarchical cluster analyses on DEmRNAs (Figure 1A) and DEcircRNAs (Figure 1C) indicated that LUSC clearly segregated from LUAD. In addition, the volcano diagrams of DEmRNAs (Figure 1B) and DEcircRNAs (Figure 1D) are displayed.
Figure 1. Heatmaps and volcano plots of DEmiRNAs differentially expressed mRNAs and DEcircRNA, miRNAs and lncRNAs. (A) heatmap for DEmRNAs, (B) volcano plot for DEmRNAs, (C) heatmap for DEcircRNA, and (D) heatmap for DEcircRNA. DEmRNAs: differentially expressed mRNAs; DEcircRNA: differentially expressed circRNAs.
Construction of miRNA-mRNA and circRNA-miRNA networks
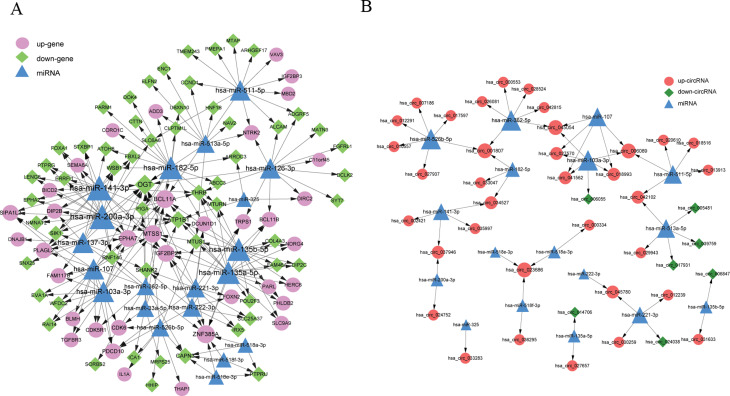
According to the prediction analysis with WebGestalt, 229 miRNA–DEmRNAs pairs were obtained to construct the miRNA–DEmRNA network, which contained 94 DEmRNAs and 20 miRNAs (Figure 2A). Among these, the top five miRNAs that had more co-expressive relationship with DEmRNAs included hsa-miR-200a-3p, hsa-miR-141-3p, hsa-miR-135b-5p, hsa-miR-135a-5p, and hsa-miR-182-5p. Moreover, the top five DEmRNAs with higher degrees in this network included OGT, ZNF385A, BCL11A, MTSS1, and ATP1B1 (Table 1).
Figure 2. The miRNA–mRNA and miRNA–circRNA regulatory networks. (A) miRNA–mRNA network and (B) miRNA–circRNA regulatory network. Blue triangles indicate miRNA, pink circles represent down-regulated genes, light green rhombuses indicate down-regulated genes, red circles denote up-regulated circRNA, and dark green rhombuses represent down-regulated circRNA.
Table 1. The top 5 DEGs and miRNAs in miRNA-DEG regulatory network.
| Genes | Description | Degree | miRNAs | Degree |
|---|---|---|---|---|
| OGT | down | 12 | hsa-miR-200a-3p | 22 |
| ZNF385A | up | 8 | hsa-miR-141-3p | 22 |
| BCL11A | up | 8 | hsa-miR-135b-5p | 18 |
| MTSS1 | up | 7 | hsa-miR-135a-5p | 18 |
| ATP1B1 | down | 7 | hsa-miR-182-5p | 18 |
Moreover, we further used miRanda software to predict the combination of these 20 miRNAs and DEcircRNAs, total 57 miRNA–DEcircRNA regulatory interactions were selected, and miRNA–circRNA network included 42 DEcircRNAs and 17 miRNAs (Figure 2B). The top five miRNAs (hsa-miR-362-5p, hsa-miR-103a-3p, hsa-miR-526b-5p, hsa-miR-513a-5p, and hsa-miR-511-5p) and circRNAs (hsa_circ_023686, hsa_circ_045054, hsa_circ_001807, hsa_circ_006089, and hsa_circ_018993EPG5) identified based on the degree ranking are listed in Table 2.
Table 2. The top 5 DEcircRNAs and miRNAs in miRNA-DEcircRNA network.
| circRNAs | Description | Degree | miRNAs | Degrees |
|---|---|---|---|---|
| hsa_circ_023686 | up | 3 | hsa-miR-362-5p | 6 |
| hsa_circ_045054 | up | 3 | hsa-miR-103a-3p | 6 |
| hsa_circ_001807 | up | 3 | hsa-miR-526b-5p | 6 |
| hsa_circ_006089 | up | 3 | hsa-miR-513a-5p | 5 |
| hsa_circ_018993 | up | 2 | hsa-miR-511-5p | 5 |
Construction of ceRNA network
To investigate the ceRNA regulation in LUSC and LUAD, the miRNA–DEmRNA pairs and miRNA–DEcircRNA pairs were integrated to establish the ceRNA network. The ceRNA network comprised 86 DEmRNAs, 42 DEcircRNAs, and 17 miRNAs (Figure 3A). Top 5 DEmRNAs, DEcircRNAs, and miRNAs are shown in Table 3. Additionally, the functional enrichment analysis of DEmRNAs in ceRNA network was performed. As showed in Figure 3B and Table 4, GO analysis revealed that DEGs were significantly enriched in the regulation of ERK1 and ERK2 cascade (GO:0070372), transmembrane receptor protein tyrosine kinase signaling pathway (GO:0007169), and negative regulation of epithelial cell proliferation (GO:0050680). These genes, including EPHA2, EPHA7, NTRK2, and CDK6, had different patterns in two major histological subtypes of NSCLC.
Figure 3. The ceRNA regulatory network and functional enrichment analysis of DEmRNAs in ceRNA network. (A) The ceRNA network. Pink circles represent up-regulated genes, blue triangles represent miRNAs, light green rhombuses indicate up-regulated cicrRNAs, deep green rhombuses represent down-regulated cicrRNAs, blue solid lines depict the regulatory relations among different RNA transcripts, and dotted orange line indicates the co-expression relationships. (B) Functional enrichment analysis of DEmiRNAs in ceRNA network. The dot size shows the gene ratio and color ranging from blue to red indicates the increasing significance.
Table 3. The top 5 DEGs, DEcircRNAs, and miRNAs in ceRNA network.
| DEGs | Description | Degree | DEcircRNAs | Description | Degree | miRNAs | Degree |
|---|---|---|---|---|---|---|---|
| ZNF385A | up | 33 | hsa_circ_027570 | up | 23 | hsa-miR-141-3p | 26 |
| TRPS1 | up | 29 | hsa_circ_010557 | up | 15 | hsa-miR-200a-3p | 24 |
| SLC9A9 | up | 21 | hsa_circ_000553 | up | 12 | hsa-miR-182-5p | 21 |
| THAP1 | up | 19 | hsa_circ_006089 | up | 10 | hsa-miR-103a-3p | 20 |
| DIRC2 | up | 17 | hsa_circ_014706 | down | 10 | hsa-miR-135a-5p | 20 |
Table 4. The functional enrichment analysis of DEmRNAs in ceRNA network.
| Category | Term | Count | P Value | Genes |
|---|---|---|---|---|
| KEGG | hsa05222:Small cell lung cancer | 3 | 0.034643 | COL4A3, CCND1, CDK6 |
| GO_BP | GO:0070372~regulation of ERK1 and ERK2 cascade | 3 | 0.006112 | EPHA7, TGFBR3, EPHA2 |
| GO_BP | GO:0048013~ephrin receptor signaling pathway | 4 | 0.007751 | EPHA7, CDK5R1, VAV3, EPHA2 |
| GO_BP | GO:0010976~positive regulation of neuron projection development | 4 | 0.008515 | NDRG4, BCL11A, ENC1, NTRK2 |
| GO_BP | GO:0007169~transmembrane receptor protein tyrosine kinase signaling pathway | 4 | 0.010466 | MTSS1, DOK4, PTPRG, NTRK2 |
| GOT_BP | GO:0006915~apoptotic process | 8 | 0.017216 | CLPTM1L, SEMA6A, EVA1A, EPHA7, OGT, CAPN3, ZNF385A, IL1A |
| GO_BP | GO:0050680~negative regulation of epithelial cell proliferation | 3 | 0.02857 | MTSS1, TGFBR3, CDK6 |
| GO_BP | GO:0017148~negative regulation of translation | 3 | 0.030483 | ENC1, IGF2BP2, IGF2BP3 |
| GO_BP | GO:0042035~regulation of cytokine biosynthetic process | 2 | 0.032477 | IGF2BP2, IGF2BP3 |
| GO_BP | GO:0048671~negative regulation of collateral sprouting | 2 | 0.037031 | EPHA7, BCL11A |
| GO_BP | GO:0033598~mammary gland epithelial cell proliferation | 2 | 0.037031 | CCND1, EPHA2 |
| GO_BP | GO:0045944~positive regulation of transcription from RNA polymerase II promoter | 10 | 0.040381 | HNF1B, THRB, BCL11B, POU2F3, TRPS1, FOXA1, BCL11A, OGT, IL1A, PLAGL2 |
| GO_BP | GO:0007399~nervous system development | 5 | 0.046348 | SEMA6A, DOK4, NAV2, ENC1, ATOH8 |
Survival analysis of DEmRNA
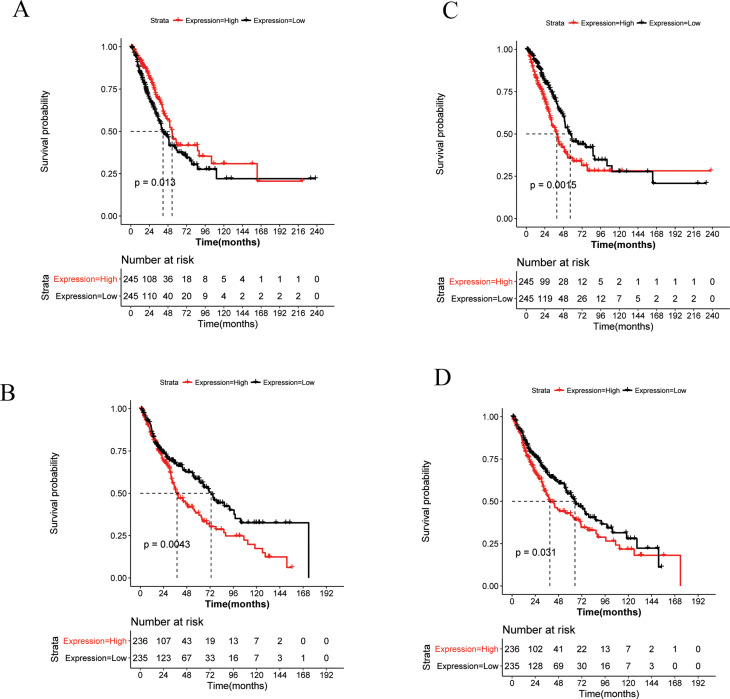
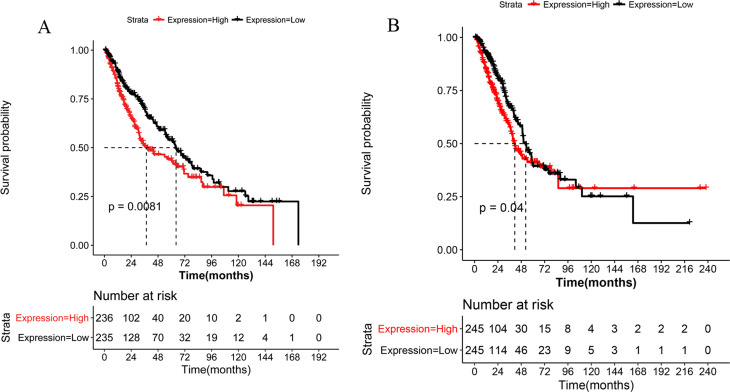
Based on the expression profile data and clinical information of TCGA-LUAD and TCGA-LUSC, the survival analysis of 86 DEmRNAs in ceRNA network was performed. Among these, 22 genes exhibited remarkable correlation with the prognosis of LUAD, whereas 9 genes were closely associated with LUSC clinical outcomes (Table S4 (57.9KB, pdf) ). Interestingly, both STXBP1 and PMEPA1 were significantly related to the prognosis of LUAD and LUSC. Furthermore, decreased expression of STXBP1 predicted a relatively worse prognosis for LUAD (Figure 4A), whereas elevated STXBP1 expression level represented a poor LUSC clinical outcome (Figure 4C). Additionally, the overexpression of PMEPA1 was associated with LUAD and LUSC inferior prognosis (Figure 4B and 4D). We also noted that a high expression level of EPHA2 represented a poor prognosis for LUSC (Figure 5A) and high level of CDK6 indicated a poor prognosis for LUAD (Figure 5B).
Figure 4. The overall survival curve of key genes. (A) STXBP1 for LUAD, (B) PMEPA1 for LUAD, (C) STXBP1 for LUSC, and (D) PMEPA1 for LUSC.
Figure 5. The survival curve of EPHA2 (A) and CDK6 (B). Red line represents high risk group and black line denotes low risk group.
Discussion
In this study, we performed a systematic bioinformatics analysis to compare the gene expression patterns of LUSC and LUAD. A total of 635 DEmRNAs and 245 DEcircRNAs were identified in the LUSC vs. LUAD group. The ceRNA analysis identified several key nodes with different patterns in LUAD and LUSC, such as EPHA2, EPHA7, NTRK2, CDK6, hsa-circ_035997, hsa_circ_027570, and hsa_circ_006089. Additionally, functional enrichment analysis revealed that these genes were significantly enriched in the regulation of ERK1 and ERK2 cascade, transmembrane receptor protein tyrosine kinase signaling pathway, and negative regulation of epithelial cell proliferation pathway. Finally, we found that STXBP1 and PMEPA1 could be prognostic factors for both LUAD and LUSC, whereas EPHA2 and CDK6 could be diagnostic marker for LUSC and LUAD, respectively.
Eph receptor A7 (EPHA7) belongs to the receptor subfamily of the protein-tyrosine kinase family, which inhibits cancer survival and migration via a ligand and tyrosine kinase dependent signaling (Oricchio et al., 2011). Giaginis et al. (2014) indicated that EPHA7 was up-regulated in lung cancer tissues and positively associated with the proliferation of lung cancer cells. Subsequently, Liu et al. (2016) confirmed that knockdown of EPHA7 could suppress the growth of NSCLC cells, suggesting that silencing EPHA7 might provide a novel approach for the treatment of NSCLC. Moreover, the functional enrichment analysis implied that EPHA7 was significantly enriched in the regulation of ERK1 and ERK2 cascade. ERK1/ERK2 signaling cascades have been widely reported to serve vital roles in various diseases, such as cancer and chronic inflammation (Lu and Malemud, 2019). Lim et al. (2017) indicated that coumestrol (a novel chemotherapeutic agent) might play a therapeutic role in prostate cancer by mediating ERK1/2 signaling pathway. Thus, we speculated that EPHA7 played an important role in NSCLC via affecting the regulation of ERK1 and ERK2 cascade. In this study, we observed that the expression level of EPHA7 was different in LUSC and LUAD. EPHA7 expression was significantly correlated with the overall survival (OS) of LUAD patients and not with the OS of patients with LUSC (Giaginis et al., 2014), indicating that the secretory form of EPHA7 might have the potential to distinguish the type of tumor histopathology in lung cancer. However, further research based on large cohorts need to be performed to explore the function of EPHA7 in LUAD and LUSC.
Another gene, neurotrophic receptor tyrosine kinase 2 (NTRK2), also presented different expression level in LUAD and LUSC. NTRK2 encodes a member of the neurotrophic tyrosine kinase (NTRK) family, which phosphorylates itself and the members of the MAPK pathway by binding to neurotrophins (Park et al., 2017). Gomez et al. (2018) compared potential therapeutic targets in LUSC and LUAD using proteomic analysis, they observed that the expression level of NTRK2 was significantly increased in LUSC as compared to LUAD, and the combination of NTRK2 with tyrosine kinase inhibitors could be considered as a promising approach for NSCLC treatment. Similarly, NTRK2 was found highly specific in LUSC (96.4%) compared with other carcinoma subtypes (including LUAD), suggesting that NTRK2 was a potential immunohistochemical marker that might be particularly helpful in separating LUSC from LUAD (Terry et al., 2011). Therefore, NTRK2 might be a target for personalized treatment of NSCLC.
In addition to mRNA, our study also revealed that several circRNAs showed distinct expression patterns between LUSC and LUAD, such as hsa_circ_027570, hsa_circ_006089, and hsa-circ_035997. Unfortunately, the biological function of these circRNA in the pathogenesis of NSCLC has not been reported. The ceRNA analysis indicated that hsa_circ_006089 and hsa_circ_027570 bound to hsa-miR-103a-3p. Fan et al. (2019) confirmed that miR-103a-3p played an anti-oncogenic role in cancer, suggesting that it might serve a novel potential therapeutic target for NSCLC. Moreover, hsa-circ_035997 was bound to miR-141-3p. The expression level of miR-141-3p was significantly down-regulated in NSCLC tissues, suggesting that miR-141-3p might be a prognostic tumor suppressor involved in the NSCLC progression (Li et al., 2019). Thus, we speculated that these circRNA might play roles by targeting miRNAs. However, the roles of these circRNAs in distinguishing LUAD from LUSC need further investigate.
Notably, our survival analyses suggested that two down-regulated genes (STXBP1 and PMEPA1) were responsible for both LUAD and LUSC. Syntaxin binding protein 1 (STXBP1) encodes a syntaxin-binding protein, which plays a role in release of neurotransmitters via regulation of syntaxin (a transmembrane attachment protein receptor) (Rezazadeh et al., 2019). A recent article reported the relationship between STXBP1 and prognosis of LUAD (Wang et al., 2020). They observed that STXBP1 expression phenotypes could be categorized as membrane, cytoplasm, and mixed expression (both membrane and cytoplasm), revealing that entire STXBP1 phenotypes or membrane phenotypes were closely connected with poor prognosis of LUAD and considered as independent prognostic factors of LUAD. Additionally, the detection of STXBP1 helped in screening the patients with poor prognosis and more accurately strengthen adjuvant therapy. Consistent with previous study, our study also revealed the correlation between STXBP1 expression and LUAD prognosis. However, we did not considered the relationship between STXBP1's subcellular localization and LUAD prognosis, and this will would be a focus of our further research. Notably, we also observed an association between STXBP1 and LUSC prognosis, whereas the expression pattern of STXBP1 in LUSC was different from that in LUAD. Thus, the function of STXBP1 and its role in LUSC still need to be clarified. Prostate transmembrane protein, androgen induced 1 (PMEPA1) encodes transmembrane protein containing the Smad interaction motif (SIM), which could inhibit the transforming growth factor signaling pathways and play a role in multiple types of cancer (Fournier et al., 2015; Zhang et al., 2019). PMEPA1 was found highly expressed in the lung cancer cell lines, whereas knockdown of PMEPA1 could significantly inhibit the proliferation of cancer cells (Vo Nguyen et al., 2014). Overexpression of PMEPA1 was associated with poor prognosis of lung cancer, which was in line with our findings. Taken together, STXBP1 and PMEPA1 might be prognostic and diagnostic markers of NSCLC.
We observed that EPHA2 was a prognostic factor for LUSC. EPHA2 mutation was present in LUSC, and it could increase tumor invasion and survival by activating the focal adhesions and actin cytoskeletal regulatory proteins, indicating EPHA2 as a potential therapeutic target of LUSC (Faoro et al., 2010), which was consistent with our findings. In addition, the expression level of CDK6 was significantly associated with the prognosis of LUAD. Cyclin-dependent kinase 6 (CDK6) was reported to be a vital regulator of cell cycle progression and correlated with various cancers including NSCLC. The expression level of CDK6 was negatively associated with the overall survival of patients with LUAD (Li et al., 2017), which was further confirmed in our results. Taken together, we speculated that EPHA2 and CDK6 were potential biomarkers in the personalized treatment for LUSC and LUAD, respectively.
There were some limitations in our study. Our results only indicated that the obtained DEmRNAs and DEcircRNAs had different expression patterns in the LUSC and LUAD, whether they served roles in the development of NSCLC needs to be explored. In addition, our study was performed based on bioinformatics analysis, further experimental studies were demanded to validate our results.
In summary, we compared two major histological subtypes of NSCLC (LUAD and LUSC) based on mRNA and circRNA expression patterns. We identified several genes, such as EPHA2, EPHA7, NTRK2, CDK6, hsa_circ_027570, and hsa_circ_035997, might be used to distinguish LUAD and LUSC. Notably, STXBP1 and PMEPA1 could predict clinical outcomes of both LUAD and LUSC. Meanwhile, EPHA2 and CDK6 might be considered as prognostic biomarkers for LUSC and LUAD, respectively.
Acknowledgments
This work was supported by Shanxi Province Natural Science Research Project (Program No.2018JM7044), Key Projects of Social Development Research Programs of Shanxi Province (Program No.2017SF-199, 2019-SF-185), Research foundation of Shanxi Provincial Administration of traditional Chinese Medicine(Program No.JCPT039), Xi'an Science and Technology Bureau of social development projects[Program No.2016047SF/YX03(4); 201805093YX1SF27(9)], Xi'an Medical College Advanced Subjects Project Funded (Program No.16SXK) and Shanxi Province Health Family Planning Commission research projects (Program No. 2016D036) and National Key R&D Program of China (2018YFC2002100, 2018YFC2002103).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Ricardo G. Correa
Reference
- Agarwal V, Subtelny A, Thiru P, Ulitsky I, Bartel D. Predicting microRNA targeting efficacy in Drosophila. Genome Biol. 2018;19:152. doi: 10.1186/s13059-018-1504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Agarwal V, Subtelny A, Thiru P, Ulitsky I and Bartel D (2018) Predicting microRNA targeting efficacy in Drosophila. Genome Biol 19:152. [DOI] [PMC free article] [PubMed]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25-29. [DOI] [PMC free article] [PubMed]
- Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL and Razavian N (2018) Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24:1559-1567. [DOI] [PMC free article] [PubMed]
- Dreyer J, Bremer M, Henkenberens C. Comorbidity indexing for prediction of the clinical outcome after stereotactic body radiation therapy in non-small cell lung cancer. Radiat Oncol. 2018;13:213. doi: 10.1186/s13014-018-1156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dreyer J, Bremer M and Henkenberens C (2018) Comorbidity indexing for prediction of the clinical outcome after stereotactic body radiation therapy in non-small cell lung cancer. Radiat Oncol 13:213. [DOI] [PMC free article] [PubMed]
- Fan Z, Yang J, Zhang D, Zhang X, Ma X, Kang L, Liu Y, Yan X, Ji Q, Wang J, et al. The risk variant rs884225 within EGFR impairs miR-103a-3p's anti-tumourigenic function in non-small cell lung cancer. Oncogene. 2019;38:2291–2304. doi: 10.1038/s41388-018-0576-6. [DOI] [PubMed] [Google Scholar]; Fan Z, Yang J, Zhang D, Zhang X, Ma X, Kang L, Liu Y, Yan X, Ji Q, Wang J et al. (2019) The risk variant rs884225 within EGFR impairs miR-103a-3p's anti-tumourigenic function in non-small cell lung cancer. Oncogene 38:2291-2304. [DOI] [PubMed]
- Faoro L, Singleton PA, Cervantes GM, Lennon FE, Choong NW, Kanteti R, Ferguson BD, Husain AN, Tretiakova MS, Ramnath N, et al. EphA2 mutation in lung squamous cell carcinoma promotes increased cell survival, cell invasion, focal adhesions, and mammalian target of rapamycin activation. J Biol Chem. 2010;285:18575–18585. doi: 10.1074/jbc.M109.075085. [DOI] [PMC free article] [PubMed] [Google Scholar]; Faoro L, Singleton PA, Cervantes GM, Lennon FE, Choong NW, Kanteti R, Ferguson BD, Husain AN, Tretiakova MS, Ramnath N et al. (2010) EphA2 mutation in lung squamous cell carcinoma promotes increased cell survival, cell invasion, focal adhesions, and mammalian target of rapamycin activation. J Biol Chem 285:18575-18585. [DOI] [PMC free article] [PubMed]
- Fournier PG, Juárez P, Jiang G, Clines GA, Niewolna M, Kim HS, Walton HW, Peng XH, Liu Y, Mohammad KS, et al. The TGF-β Signaling Regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell. 2015;27:809–821. doi: 10.1016/j.ccell.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fournier PG, Juárez P, Jiang G, Clines GA, Niewolna M, Kim HS, Walton HW, Peng XH, Liu Y, Mohammad KS et al. (2015) The TGF-β Signaling Regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell 27:809-821. [DOI] [PMC free article] [PubMed]
- Giaginis C, Tsoukalas N, Bournakis E, Alexandrou P, Kavantzas N, Patsouris E, Theocharis S. Ephrin (Eph) receptor A1, A4, A5 and A7 expression in human non-small cell lung carcinoma: associations with clinicopathological parameters, tumor proliferative capacity and patients’ survival. BMC Clin Pathol. 2014;14:8. doi: 10.1186/1472-6890-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Giaginis C, Tsoukalas N, Bournakis E, Alexandrou P, Kavantzas N, Patsouris E and Theocharis S (2014) Ephrin (Eph) receptor A1, A4, A5 and A7 expression in human non-small cell lung carcinoma: associations with clinicopathological parameters, tumor proliferative capacity and patients’ survival. BMC Clin Pathol 14:8. [DOI] [PMC free article] [PubMed]
- Gomez DR, Byers LA, Nilsson M, Diao L, Wang J, Li L, Tong P, Hofstad M, Saigal B, Wistuba I, et al. Integrative proteomic and transcriptomic analysis provides evidence for TrkB (NTRK2) as a therapeutic target in combination with tyrosine kinase inhibitors for non-small cell lung cancer. Oncotarget. 2018;9:14268–14284. doi: 10.18632/oncotarget.24361. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gomez DR, Byers LA, Nilsson M, Diao L, Wang J, Li L, Tong P, Hofstad M, Saigal B, Wistuba I et al. (2018) Integrative proteomic and transcriptomic analysis provides evidence for TrkB (NTRK2) as a therapeutic target in combination with tyrosine kinase inhibitors for non-small cell lung cancer. Oncotarget 9:14268-14284. [DOI] [PMC free article] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]; Huang DW, Sherman BT and Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44-57. [DOI] [PubMed]
- Jacky J, Baik C. Symptom management strategies for patients receiving anaplastic lymphoma kinase inhibitors for non-small cell lung cancer. J Adv Pract Oncol. 2017;8:729–735. [PMC free article] [PubMed] [Google Scholar]; Jacky J and Baik C (2017) Symptom management strategies for patients receiving anaplastic lymphoma kinase inhibitors for non-small cell lung cancer. J Adv Pract Oncol 8:729-735. [PMC free article] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kanehisa M and Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27-30. [DOI] [PMC free article] [PubMed]
- Li H, Zhang J, Xie Y. Elevated nuclear CDK6 is associated with an unfavorable prognosis in lung adenocarcinoma patients. Int J Clin Exp Pathol. 2017;10:9614–9620. [PMC free article] [PubMed] [Google Scholar]; Li H, Zhang J and Xie Y (2017) Elevated nuclear CDK6 is associated with an unfavorable prognosis in lung adenocarcinoma patients. Int J Clin Exp Pathol 10:9614–9620. [PMC free article] [PubMed]
- Li W, Cui Y, Wang D, Wang Y, Wang L. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;509:647–656. doi: 10.1016/j.bbrc.2018.12.089. [DOI] [PubMed] [Google Scholar]; Li W, Cui Y, Wang D, Wang Y and Wang L (2019) MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun 509:647-656. [DOI] [PubMed]
- Lim W, Jeong M, Bazer FW, Song G. Coumestrol inhibits proliferation and migration of prostate cancer cells by regulating AKT, ERK1/2, and JNK MAPK Cell Signaling Cascades. J Cell Physiol. 2017;232:862–871. doi: 10.1002/jcp.25494. [DOI] [PubMed] [Google Scholar]; Lim W, Jeong M, Bazer FW and Song G (2017) Coumestrol inhibits proliferation and migration of prostate cancer cells by regulating AKT, ERK1/2, and JNK MAPK Cell Signaling Cascades. J Cell Physiol 232:862-871. [DOI] [PubMed]
- Liu B, Chen Y, Yang J. LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget. 2017;8:24275–24291. doi: 10.18632/oncotarget.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu B, Chen Y and Yang J (2017) LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget 8:24275-24291. [DOI] [PMC free article] [PubMed]
- Liu M, Zhou K, Cao Y. MicroRNA-944 Affects Cell growth by targeting EPHA7 in Non-Small Cell Lung Cancer. Int J Mol Sci. 2016;17:1493. doi: 10.3390/ijms17101493. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu M, Zhou K and Cao Y (2016) MicroRNA-944 Affects Cell growth by targeting EPHA7 in Non-Small Cell Lung Cancer. Int J Mol Sci 17:1493. [DOI] [PMC free article] [PubMed]
- Lu N, Malemud CJ. Extracellular signal-regulated kinase: a regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated. Int J Mol Sci. 2019;20:3792. doi: 10.3390/ijms20153792. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu N and Malemud CJ (2019) Extracellular signal-regulated kinase: a regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated. Int J Mol Sci 20:3792. [DOI] [PMC free article] [PubMed]
- Nellesen D, Dea K, Guerin A, Culver KW, Mutebi A, Dalal A. Reimbursement landscape for molecular testing in non-small cell lung cancer (NSCLC) Am J Manag Care. 2018;24:SP37-SP42. [PubMed] [Google Scholar]; Nellesen D, Dea K, Guerin A, Culver KW, Mutebi A and Dalal A (2018) Reimbursement landscape for molecular testing in non-small cell lung cancer (NSCLC). Am J Manag Care 24:SP37-SP42. [PubMed]
- Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, Liu X, Bruno J, Heguy A, Olshen AB, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–564. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, Liu X, Bruno J, Heguy A, Olshen AB et al. (2011) The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell 147:554-564. [DOI] [PMC free article] [PubMed]
- Park SJ, More S, Murtuza A, Woodward BD, Husain H. New targets in Non-Small Cell Lung Cancer. Hematol Oncol Clin North Am. 2017;31:113–129. doi: 10.1016/j.hoc.2016.08.010. [DOI] [PubMed] [Google Scholar]; Park SJ, More S, Murtuza A, Woodward BD and Husain H (2017) New targets in Non-Small Cell Lung Cancer. Hematol Oncol Clin North Am 31:113-129. [DOI] [PubMed]
- Rezazadeh A, Uddin M, Snead OC, Lira V, Silberberg A, Weiss S, Donner EJ, Zak M, Bradbury L, Scherer SW, et al. STXBP1 encephalopathy is associated with awake bruxism. Epilepsy Behav. 2019;92:121–124. doi: 10.1016/j.yebeh.2018.12.018. [DOI] [PubMed] [Google Scholar]; Rezazadeh A, Uddin M, Snead OC, Lira V, Silberberg A, Weiss S, Donner EJ, Zak M, Bradbury L, Scherer SW et al. (2019) STXBP1 encephalopathy is associated with awake bruxism. Epilepsy Behav 92:121-124. [DOI] [PubMed]
- Terry J, De Luca A, Leung S, Peacock G, Wang Y, Elliot WM, Huntsman D. Immunohistochemical expression of neurotrophic tyrosine kinase receptors 1 and 2 in lung carcinoma: potential discriminators between squamous and nonsquamous subtypes. Arch Pathol Lab Med. 2011;135:433–439. doi: 10.5858/2010-0038-OA.1. [DOI] [PubMed] [Google Scholar]; Terry J, De Luca A, Leung S, Peacock G, Wang Y, Elliot WM and Huntsman D (2011) Immunohistochemical expression of neurotrophic tyrosine kinase receptors 1 and 2 in lung carcinoma: potential discriminators between squamous and nonsquamous subtypes. Arch Pathol Lab Med 135:433-439. [DOI] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tomczak K, Czerwinska P and Wiznerowicz M (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19:A68-77. [DOI] [PMC free article] [PubMed]
- Vo Nguyen TT, Watanabe Y, Shiba A, Noguchi M, Itoh S, Kato M. TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci. 2014;105:334–341. doi: 10.1111/cas.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vo Nguyen TT, Watanabe Y, Shiba A, Noguchi M, Itoh S and Kato M (2014) TMEPAI/PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci 105:334-341. [DOI] [PMC free article] [PubMed]
- Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu Y, Liu X, Cheng W, Wei YQ, Peng Y, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18:134. doi: 10.1186/s12943-019-1061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu Y, Liu X, Cheng W, Wei YQ, Peng Y et al. (2019) RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer 18:134. [DOI] [PMC free article] [PubMed]
- Wang X, Fu G, Wen J, Chen H, Zhang B, Zhu D. Membrane location of Syntaxin-Binding Protein 1 is correlated with poor prognosis of lung adenocarcinoma. Tohoku J Exp Med. 2020;250:263–270. doi: 10.1620/tjem.250.263. [DOI] [PubMed] [Google Scholar]; Wang X, Fu G, Wen J, Chen H, Zhang B and Zhu D (2020) Membrane location of Syntaxin-Binding Protein 1 is correlated with poor prognosis of lung adenocarcinoma. Tohoku J Exp Med 250:263-270. [DOI] [PubMed]
- Yang Y, Wang M, Liu B. Exploring and comparing of the gene expression and methylation differences between lung adenocarcinoma and squamous cell carcinoma. J Cell Physiol. 2019;234:4454–4459. doi: 10.1002/jcp.27240. [DOI] [PubMed] [Google Scholar]; Yang Y, Wang M and Liu B (2019) Exploring and comparing of the gene expression and methylation differences between lung adenocarcinoma and squamous cell carcinoma. J Cell Physiol 234:4454-4459. [DOI] [PubMed]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741-748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang B, Kirov S and Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741-748. [DOI] [PMC free article] [PubMed]
- Zhang L, Wang X, Lai C, Zhang H, Lai M. PMEPA1 induces EMT via a non-canonical TGF-β signalling in colorectal cancer. J Cell Mol Med. 2019;23:3603–3615. doi: 10.1111/jcmm.14261. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang L, Wang X, Lai C, Zhang H and Lai M (2019) PMEPA1 induces EMT via a non-canonical TGF-β signalling in colorectal cancer. J Cell Mol Med 23:3603-3615. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.