Abstract
Introduction
Protein neddylation, one of the most important posttranslational modifications that tagging neuronal precursor cell‐expressed developmentally downregulated protein 8 onto substrate proteins, plays fundamental roles in the process of many cellular functions. A number of studies have demonstrated the critical roles of neddylation modification in multiple pathophysiological processes, but its regulatory role in the immune system has only been finitely unveiled.
Methods
In this review, the latest advances in the field of neddylation modification in regulating the immune responses are succinctly discussed.
Results
Neddylation modification acts as a crucial modulator of innate immune cells (neutrophils, macrophages, and dendritic cells) and lymphocytes. Dysregulation of neddylation alters characteristics and functions of those cells due to abnormal degradation of key signaling molecules involved in immunoregulation. Furthermore, the ectopic immune responses caused by the abnormal neddylation play pivotal roles in a variety of immune‐related diseases, such as infection, inflammation, and cancer.
Conclusions
The pivotal roles of neddylation pathway in immunoregulation are attracted more and more attention, which may provide new insights into the pathogenesis of a variety of immune‐related diseases and help to indicate new therapeutic targets and potential treatment strategies.
Keywords: immune cells, immune response, immune‐related disease, neddylation, signaling molecule
Abbreviations
- ATP, adenosine triphosphate; CCL2
C‐C motif chemokine ligand 2
- CRLs
Cullin‐RING E3 ligases
- CSN
COP9 signalosome
- DAMPs
damage associated molecular patterns
- DCN1
defective in Cullin neddylation 1
- DCs
dendritic cells
- Deptor
Dep domain containing mTOR interacting protein
- Erk
extracellular regulated protein kinases
- FoxO1
forkhead box O1
- HIF‐1
hypoxia‐inducible factor 1
- Hu‐antigen R
HuR
- HUVECs
human umbilical vein endothelial cells
- ICAM
intercellular cell adhesion molecule
- IFN‐β
interferon‐β
- IFN‐γ
interferon‐1γ
- IL‐10
interleukin‐10
- IL‐1β
interleukin‐1β
- IL‐6
interleukin‐6
- Itch
itchy E3 ubiquitin‐protein ligase
- IκB
inhibitor of nuclear factor kappa B
- LPS
lipopolysaccharide
- MDM2
murine double minute 2
- mTOR
mechanistic target of rapamycin kinase
- NAE1
NEDD8‐activating enzyme E1 subunit 1
- NEDD8
neuronal precursor cell‐expressed developmentally downregulated protein 8
- NF‐κB
nuclear factor kappa B
- OVA, ovalbumin; PAMPs
pathogen‐associated molecular patterns
- PD‐1
programmed cell death protein 1; PD‐L1
- RBX1/2
RING‐box protein ½
- RING
really interesting new gene
- SAG
sensitive to apoptosis gene
- SENP8
SUMO peptidase family member, NEDD8 specific
- SKP‐1
S‐phase kinase‐associated protein 1
- ssRNA
single‐stranded ribonucleic acid
- TAMs
tumor‐associated macrophages
- Tfh
follicular helper T
- Th
T helper
- TNF‐α
tumor necrosis factor‐α
- Tregs
regulatory T Cells
- UBA3
ubiquitin‐like modifier activating enzyme 3
- UBE2F
ubiquitin conjugating enzyme E2 F
- UBE2M
ubiquitin conjugating enzyme E2 M
- VHL
von Hippel‐Lindau protein
- βTrCP
β‐transducin repeat‐containing protein
1. INTRODUCTION
Protein neddylation is the posttranslational modification that conjugating an ubiquitin‐like molecule neuronal precursor cell‐expressed developmentally downregulated protein 8 (NEDD8) to the lysine residue in substrate proteins via a successive three‐step enzymatic reaction. 1 , 2 , 3 Analogous to ubiquitylation, the exposed C‐terminal glycine of NEDD8 is activated by NEDD8‐activating enzyme E1, a heterodimer composed of NEDD8‐activating enzyme E1 subunit 1 (also known as APPBP1) and ubiquitin‐like modifier activating enzyme 3 (UBA3) (also known as NAEβ), in an ATP‐dependent manner. 4 Then, activated NEDD8 is transferred to NEDD8‐conjugating enzyme E2, UBE2M (also known as UBC12) or UBE2F, via forming a thiolester linkage. 5 The substrate‐specific NEDD8‐E3 ligases subsequently transfer NEDD8 to substrate protein forming an isopeptide bond. Unlike E3 ubiquitin ligases, a limited number of NEDD8‐E3 ligases is identified. Most of them, such as really interesting new gene (RING)‐box protein 1 (RBX1) (also known as ROC1), RBX2 (also known as ROC2) and murine double minute 2, have the RING domain structure. 6 DCN1 is the sole exception that does not contain a RING domain for its catalytic activity. 7 , 8 Cullin‐RING E3 ligases (CRLs) represent the largest superfamily of multisubunit E3s as well as the best‐characterized substrates of neddylation. CRLs activation requires neddylation of Cullin proteins to facilitate conformational change of CRLs and subsequent substrate ubiquitination. 9 Besides of Cullins, several non‐Cullin proteins (eg, von Hippel‐Lindau protein [VHL], Hu‐antigen R) have also been identified as the substrates of neddylation. 10 , 11 Neddylation is a reversible modification of substrates and can be reversed by the action of deneddylating enzymes including COP9 Signalosome (CSN), SENP8 etc. 12 , 13 CSN mainly deneddylates Cullin proteins and ectopic CSN expression may affect the stability of CRLs. 14
Neddylation modification is of vital importance in biological processes and its dysregulation causes abnormal degradation of their crucial substrate proteins along with diseases. MLN4924 (known as pevonedistat) is a first‐in‐class small molecular inhibitor of UBA3 currently in phase I/II clinical trials for patients suffering from cancer, capable of blocking the entire neddylation modification. 15 MLN4924 covalently adducts with NEDD8 and competitively binds to the active site of NAEβ, effectively blocks CRLs activation. Numerous investigations have revealed that inactivation of CRLs triggers multiple biological events including cell cycle arrest, apoptosis, senescence, autophagy, angiogenesis, ciliogenesis, mitochondrial morphology etc. 16 , 17 , 18 , 19 , 20 , 21 , 22 All the above have been linked to cancer (reviewed by Zhou, 2018), 2 neurodegenerative disorders, 23 cardiac disease (reviewed by Kandala, 2014), 24 and others.
Notably, neddylation has emerged as a critical mechanism in regulating the immune system. Abnormal activation of neddylation pathway affects the proliferation, effector function, and signal transduction of diverse immune cells, leading to ectopic immune responses in vitro and in vivo. Accumulated evidence has proven that pharmacological intervention by MLN4924 and genetic deletion of key molecules in neddylation modification are inseparably related to alleviated immune‐mediated diseases, which include infection (reviewed by Han, 2018), 25 inflammation and tumor. Herein, we aim to deeply clarify the roles and underlying mechanisms of neddylation modification as a regulator in the immune system. We also summarize various immune‐related diseases associated with neddylation dysregulation.
1.1. Neddylation emerges as a regulator of immune cells
Posttranslational protein modification is an essential influencing factor on innate and adaptive immune cells in aspects of survival, differentiation, recruitment, and effector function (Figure 1). In addition, it served in an indirect manner by modulating the crosstalk between immune cells and others.
Figure 1.
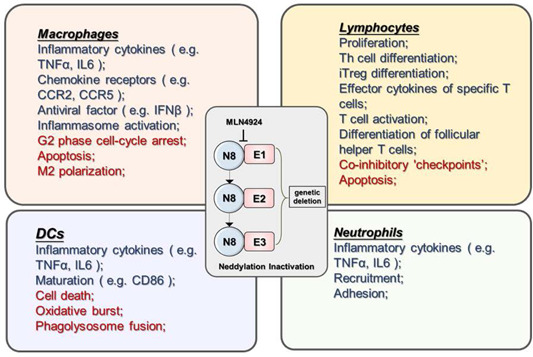
Neddylation inactivation by MLN4924 treatment or genetic deletion is involved in multiple regulatory responses within immune cells. Red, enhanced biological process by repressing neddylation modification; Blue, impaired biological process
1.2. Neddylation in innate immune cells
Neutrophils, a type of polymorphonuclear leukocyte, are recognized as the first line of defense in innate immune response who are able to be recruited to an inflammatory site to eliminate pathogens. 26 Multiple mechanisms are involved in modulating biological roles of neutrophils, such as producing multifarious cytokines with wide functional diversity. 27 Researches have shown that neddylation plays an important role in regulating neutrophils. Zhu et al 28 found that deficiency of Cullin‐5, which belongs to the Cullin family that can form CRLs, attenuated alveolar neutrophil recruitment in lipopolysaccharide (LPS)‐challenged mice. In vitro study indicated that pharmacological agent MLN4924 led to decreased production of tumor necrosis factor‐α (TNF‐α), interleukin (IL)‐6, and IL‐1β in response to LPS in a dose‐dependent manner. Meanwhile, the viability of neutrophils was marginally affected. 29 Intriguingly, Asare et al 30 showed that MLN4924 treatment in vivo elevated neutrophil counts in blood. This phenotype may be caused by overcompensation for the early‐stage anti‐inflammatory effects of MLN4924 on endothelium. To sum up, neddylation modification acts as a crucial modulator of the neutrophil recruitment and effector function.
The “mononuclear phagocyte system” is identified as a population of originally bone marrow‐derived myeloid cells that circulate in the blood as monocytes and migrate to tissues as macrophages in a steady‐state and during inflammation. 31 , 32 It was demonstrated that blocking neddylation with MLN4924 or siRNA abrogated LPS‐induced proinflammatory cytokines (TNF‐α and IL‐6) secreted from murine and human macrophages. 33 , 34 The production of interferon‐β (IFN‐β), which is important for the elimination of viral infection, was also repressed in macrophage with MLN4924 treatment, whereas NEDD8 knockdown had no effects. 35 Moreover, NEDD8 silencing or MLN4924 treatment led to diminished inflammasome activation and reduced IL‐1β maturation in macrophages. 36 In a mouse model of bileductligation‐ or carbon tetrachloride‐induced fibrosis, neddylation inhibition was reported to reduce the expression of chemokine receptors and cytokines in Kupffer cells, the liver‐resident macrophages. 37 Besides altered effector functions of macrophages, the viability of RAW264.7 was obviously impaired with persistent and severe inactivation of neddylation by MLN4924 while insignificant changed with partial inactivation. 34 Due to the properties of the functional polarization and plasticity, macrophages are polarized into classically activated macrophages (M1) and alternatively activated macrophages (M2) in response to different signals. 38 , 39 Research indicated that MLN4924 drove macrophage polarization toward an anti‐inflammatory M2 phenotype even in the absence of exogenous polarizing signals. 30 Collectively, neddylation modification is involved in controlling the polarization, the survival, and the inflammatory responses of macrophage.
Dendritic cells (DCs) are professional antigen‐presenting cells that bridge between innate and adaptive immunity. Beyond that, DCs also providing multiple soluble and surface‐bound signals so as to tailor distinct T‐cell differentiation programs. 40 , 41 Studies have proven that MLN4924 treatment, as well as siRNA‐mediated knockdown of RBX2/SAG in DCs, exhibited remarkable repression in the release of cytokines. 42 Another research reported that inhibition of neddylation suppressed the release of proinflammatory cytokines by DCs but which was is not due to the result of decreased cell viability or phenotypic change. 43 Notably, Mohamed El‐Mesery et al 44 found that MLN4924 inhibited DC maturation by sensitizing immature DCs to TNF‐ and LPS‐induced necroptosis. Functionally, RNAi mediated knockdown of NEDD8 promoted the oxidative burst and phagolysosome fusion in mycobacteria infected DCs, as well as higher expression of autophagy and apoptosis‐associated proteins. 45 However, current evidence demonstrated that the E3 ligase CRL4DCAF2 in DCs negatively regulated IL‐23 production. 46 Thus, the immunoregulation of different neddylation components in DC inflammatory response remains elusive.
1.3. Neddylation in adaptive immune cells
T cells are central regulators of adaptive immune responses. Due to antigen stimulation, naïve T cells subsequently proliferate, differentiate and play their roles in connection to infection, cancer, autoimmunity disease, and alloreactivity. 47 Jin et al 48 have investigated the role of neddylation in regulating T‐cell function and demonstrated that deficiency of Ubc12 in CD4+ T cells resulted in diminished proliferation, Th1, and Th2 differentiation and cytokine production in vitro and in vivo. However, no obvious effect was observed on the development of CD4+, CD8+, or CD4+CD8+ thymocytes. 48 SAG genetic knockout showed phenotypically normal mature T‐cell development and decreased activation, proliferation, and T‐effector cytokine release as well. 49 The naïve T cells stimulated with various endogenous signals can polarize to different T‐cell subsets. Upon exposure to the NAE inhibitor, the proportion of Th1 cells moderately increased, whereas Th2 cells decreased. 50 Meanwhile, NAE inhibition downregulates the differentiation of inducible Tregs. 50 Apart from that, neddylation is also required for maintaining CD4+ T‐cell survival through repressed mitochondria‐dependent apoptosis. 51 Tfh cells are a unique CD4+ T‐cell subset and are essential for the germinal center formation and effective humoral immunity. 52 Evidence indicated that neddylation promotes Tfh cell differentiation via elevated forkhead box O1 (FoxO1) degradation in an Itch‐dependent manner. 51 Coinhibitory “checkpoints,” such as PD‐1 and its ligand PD‐L1, are considered to cause T‐cell exhaustion and terminally diminished adaptive immune response. 53 It was reported that inhibition of Cullin‐3 activity by MLN4924 attenuated T‐cell killing through blocked PD‐L1 protein degradation. 54 In view of intricate differentiation and function of adaptive immune cells, our knowledge of that of neddylation is far from being desired.
1.4. Neddylation in other cells
Neddylation is also considered to regulate substrate proteins in other types of cells such as endothelial cells or epithelial cells, and their interaction with immune cells is influenced as a consequence. The state‐of‐art research has proven that treatment of human umbilical vein endothelial cells with a COP9 signalosome inhibitor (pharmacological activation of CRLs) enhanced intercellular cell adhesion molecule (ICAM) expression and resulted in consequent adhesion of neutrophils to endothelial cells. 55 Moreover, the glioma cells after MLN4924 treatment were reported to have a stronger potential to induce T‐cell anergy. 56 Recently, our colleagues uncovered that neddylation inactivation in tumor cell increased C‐C motif chemokine ligand 2 (CCL2) secretion and then promoted tumor‐associated macrophage infiltration. 57
2. NEDDYLATION‐MODULATED SIGNALING MOLECULE IN THE IMMUNE RESPONSE
2.1. Neddylation and NF‐κB signal
Nuclear factor‐kappa B (NF‐κB) exists in most cell types and serves as a core regulatory molecular in the inducible expression of many proteins in innate and adaptive immune responses, including cytokines, cell adhesion molecules, and acute phase response proteins. 58 , 59 There are two types of NFκB signaling pathways: the classical and the alternative pathway. The classical pathway is rapidly and transiently activated by proinflammatory cytokines, pathogen‐associated molecular patterns, and damage‐associated molecular patterns. The translocation of NFκB dimers from the cytoplasm to the nucleus depends on the proteasomal degradation of the inhibitor of nuclear factor kappa B (IκB), following the phosphorylation by IKK complex. 60 , 61 Recent studies have highlighted our current knowledge of the posttranslational modifications in this signaling pathway. Studies have provided that the NEDD8 conjugation of Cullin‐1 has profound effects on the signal‐dependent degradation of IκBα. 62 , 63 Its polyubiquitination is triggered by the activation of SKP1‐CUL1/RBX1‐βTrCP CRL (SCFβTrCP), which is composed of cullin‐1 and RBX1 (the enzymatic core), SKP1 (an adaptor) and βTrCP (the substrate‐binding F‐box protein). 64 Deneddylation removes the small protein NEDD8 from CRLs to the accumulation of IκB, which inactivating NF‐κB. The pharmacological blockade of the entire neddylation pathway by MLN4924 has been proven to cause accumulated IκB and stabilized NF‐κB as well. In a consequence, typical targets for NF‐κB such as proinflammatory cytokines (TNF‐α, IL‐6), chemokines (CCL2/MCP1), antirival factor (IFN‐β) and adhesion molecules (ICAM) are downregulated. 25 , 55 , 57 , 65 However, what's puzzling is that neddylation inactivation usually promotes the accumulation of phosphorylated IκBα, rather than total IκBα, in some studies. 34 , 66 It may be interpreted that other protein degradation pathways are involved in the regulation.
Few physiologic regulatory factors have been demonstrated to function on neddylation components so as to modulate the NFκB pathway. The observations indicated that the interaction of nonpathogenic commensal bacteria with epithelial cells blocked the neddylation of the Culllin‐1 subunit of E3‐SCFβTrCP. Bacteria‐derived butyrate and other short‐chain fatty acids also rapidly induced the generation of reactive oxygen species and caused oxidative inactivation of Ubc12, resulting in attenuation of NFκB activation. 67 , 68 , 69 Besides, Glutamine administration could enhance Cullin‐1 neddylation and attenuate NEDD8 expression, which contributes to decreased NFκB activation. 70 It has been revealed that adenosine could modulate Cullin‐1 neddylation and then inhibit NF‐κB through modulating proteasomal degradation of IκB proteins. 71 Additionally, Nedd8 is identified as a novel TRIM40‐binding protein. TRIM40 enhances neddylation of IKKγ and inhibits the NFκB‐mediated transcription. 72 The funny thing is that MyD88, a novel substrate of NEDD8, was thought to antagonize its ubiquitination and dimerization, and negatively regulate MyD88‐dependent NF‐κB signaling. 73
2.2. Neddylation and HIF‐1α signal
Hypoxia‐inducible factor 1 (HIF‐1), a heterodimer composed of an oxygen‐regulated HIF‐1α subunit and a constitutively expressed HIF‐1β subunit, 74 usually acts as a signaling hub to coordinate the activities of many transcription factors and signaling molecules. 75 Under normoxic conditions, HIF‐1α is hydroxylated by prolyl hydroxylase domain proteins. Hydroxylated HIF‐1α can interact with the VHL protein, a substrate recognition subunit of E3 ubiquitin ligases, for proteasomal degradation. 76 It's worth noting that specific metabolic imbalances, such as bacterial lipopolysaccharide stimulation, are able to induce HIF‐1α accumulation in the case of normal oxygen levels. 77 Under hypoxic conditions, such as sites of inflammation and tumor, hydroxylation is repressed and HIF‐1α is stabilized and translocated to the nucleus for enhancing transcription of a number of inflammatory genes. 78
HIF‐1α is also tightly regulated in a posttranslational fashion. As early as 2003, it was reported that VHL was covalently conjugated by NEDD8, not requiring for the E3 activity to ubiquitylated degradation of HIF‐1α. However, expression of a cullin neddylation‐defective VHL protein mutant in renal clear‐cell carcinoma cells restored the regulation of HIF‐1α, 11 indicating mysterious neddylation modification of HIF‐1α. Sufan RI et al discovered that VHL triggered Rbx1‐mediated of Cullin‐2 neddylation, which promotes the engagement of UbcH5a to the ECV complex and subsequent recognition of HIF‐1α in an oxygen‐dependent manner. 79 Interestingly, HIF‐1α substrate binding to ECV also was involved in promoting Cullin‐2 neddylation and mutation of HIF‐1α residues of VHL binding resulted in reduced Cullin‐2 neddylation. 80 Additionally, NEDD8 covalently modified and stabilized HIF‐1 both in normoxia and hypoxia. Such an effect was diminished by antioxidants and mitochondrial respiratory chain blockers. 81 The greatest attraction was that stress‐inducible UBE2M was cis‐transactivated by HIF‐1 and MLN4924 upregulated UBE2M via blocking degradation of HIF‐1α, 82 indicating that HIF‐1α may also regulate transcription and degradation of neddylation components, not just as a substrate of neddylation.
Neddylation modulates various HIF‐1α‐transactivated effectors. MLN4924 was confirmed to repress transcriptional expression of ICAM‐1 and LPS‐induced endothelial permeability in mucosal inflammation. MLN4924 attenuated the transcription of a number of inflammatory cytokines, including IL‐1β, IL‐6, TNF‐α, IL‐12p70, and IFN‐γ. Additionally, an increase, albeit not statistically significant, in the anti‐inflammatory cytokine IL‐10 was also observed with MLN4924 treatment, which revealed an anti‐inflammatory effect of neddylation inactivation. 83 , 84 Targeting neddylation pathway contributed to a significant increase of HIF‐1α and PD‐L1 in gliomas and further resulted in T‐cell energy. 56 Interestingly, most of the HIF‐1α‐transactivated inflammatory genes or other effect genes can be regulated by NF‐κB and HIF hydroxylases also regulate NF‐κB. In hypoxia inflammatory conditions, these two central transcription factors display an intimate interdependence via several mechanistic ways. Hence, the effects of neddylation on HIF‐1α and NF‐κB signaling should be more complicated and further balanced. 85
2.3. Neddylation and other signals
Other than NF‐κB and HIF‐1α, little knowledge of neddylation‐targeted signals has been mentioned in regulating the immune response. Some CRL‐dependent and CRL‐independent substrates have already been identified, despite not being widely detected. Neddylation leads to repressed Deptor accumulation by the conjugation of Nedd8 to Cullin‐1 and sequentially promotes the mechanistic target of rapamycin kinase (mTOR) activity. 43 Neddylation inactivation also upregulates PD‐L1 expression on glioblastoma cell lines by stabilization of c‐Myc. 54 Adaptor protein Shc is another target for neddylation, which facilitating the formation of the ZAP70‐Shc‐Grb2 complex and downstream Erk activation. 48 NEDD8 modification of Itch, leading to degradation of FoxO1, promotes Tfh cell differentiation. 51 Treatment with MLN4924 contributes to increased Lipin‐2 protein stability by repressing interaction with Cullin‐1 and concomitantly decreased IL‐1β expression. 86
3. NEDDYLATION EMERGES AS A REGULATOR OF IMMUNE‐MEDIATED DISEASES
3.1. Neddylation in infection
Infection is referred to a process that bacteria, viruses, fungi, or other pathogens enter the body and the subsequent biological changes. By far, the immunomodulatory effects of neddylation inhibition against viral infections have been concerned (Figure 2). First, neddylation is considered to control the amplification and replication of viruses. It is reported that blocking or silencing neddylation dramatically increased spring viremia of carp virus (an ssRNA virus that causes an important disease affecting cyprinids) replication. 87 Zhang et al 88 reported that neddylation of the polymerase basic protein 2 of influenza A virus reduces its stability and blocks the replication of influenza A virus. On the other hand, neddylation inhibition restores the restriction of human immunodeficiency virus by repressing the degradation of host restriction factors, a kind of cellular proteins inhibiting the replication of viruses at various stages of their life cycle. 89 , 90 Intriguingly, activated neddylation modification pathway led to increased virus growth in influenza A virus‐infected A549 cells. 91 Hence, the modulatory effects of neddylation on amplification and replication may be disparate, depending on the virus species. Second, neddylation prevents viruses against immune‐evasive activities by controlling the production of type I interferon. It is demonstrated that neddylation inhibition with MLN4924 or upon UBA3 deficiency led to impaired IκBα degradation and NF‐κB‐promoted production of type I interferon in the early phase of herpes simplex virus type I (HSV‐1) infection, rather than the late phage, whereas no obvious effect of interferon regulatory factor (IRF) on type I interferon. 92 The transcription of type I interferon genes is also mediated by IRF family members. Neddylation inhibition also results in the stabilization of IRF3 and production of type I interferon following Sendai virus infection. 93 More interestingly, NEDD8 knockdown exhibits no effect on type I interferon production triggered by poly (I:C) or Sendai virus, which is reported to induce IRF3 degradation, but HSV‐1 does not. 93 Hence, the modulatory effects of neddylation on the production of type I interferon may be determined by the interplay between the IRF and NF‐κB signal. Besides viruses, Cheng et al 51 have described a crucial role of neddylation during the primary blood‐stage Plasmodium infection, in controlling the CD4+ T‐cell responses and subsequent humoral immunity.
Figure 2.
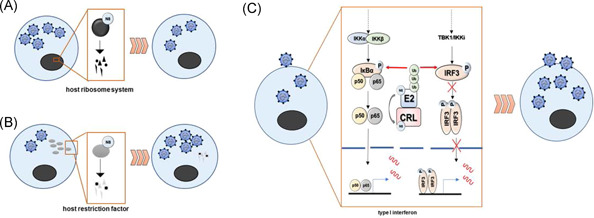
Mechanisms of neddylation modification against viral infection. A, Neddylation of proteins involved in the host ribosome system reduces its stability and blocks the replication of the virus. B, Neddylation modification contributes to the degradation of host restriction factors and releases suppression to the replication of the virus. C, Neddylation modulates the production of type I interferons mainly in NF‐κB‐ and IRF3‐dependent manner. Phosphorylated IRF3 or p50/p65 dimerizes and is translocated into the nuclear compartment to induce the expression of antiviral genes. Phosphorylated IRP‐3 and IκB are recognized by CRLs and induced the proteasomal degradation. CLR, Cullin‐RING E3 ligase; IRF, interferon regulatory factor; NF‐κB, nuclear factor kappa B
3.2. Neddylation in inflammatory disease
Inflammation is considered as a defense mechanism that triggered by damage to the body and neddylation inactivation by genetic silencing or the specific inhibitor MLN4924 usually exhibits beneficial effects on the disease control. For LPS‐induced inflammatory responses such as acute kidney injury and acute lung injury, inactivation of neddylation has been demonstrated to inhibit the CRL/NF‐κB signal mediated production of proinflammatory cytokines (eg, TNF‐α and IL‐6) so as to alleviate the inflammation. 28 , 65 Additionally, diminished neddylation activity via MLN4924 could be important for resolving liver fibrosis and pulmonary fibrosis through reduced expression of the chemokines from epithelial cells and subsequently impaired induction of chemokine receptors and cytokines in activated macrophages. 37 , 94 Mice transferred with Ubc12 knockdown CD4+ T cells exhibited a deficiency in the ability to develop Th2‐mediated allergic response in OVA‐driven airway inflammation. 48 The immunomodulation of neddylation inhibition on mucosal inflammation has also been described, indicating a potential therapeutic opportunity in inflammatory bowel diseases. Neddylation inhibition suppressed CRL/Deptor‐mediated mTOR signaling pathway in DCs. 43 Meanwhile, inhibition of the neddylation is capable of enhancing barrier function of intestinal epithelial cells in a CRL/HIF‐dependent way, which may be disturbed by DEN‐1. 84 However, neddylation inhibition by MLN4924 treatment combined with proinflammatory cytokines may lead to increased epithelial CRL/NF‐κB‐mediated apoptosis and barrier disruption, 95 indicating that MLN4924 treatment in inflammatory conditions may cause side effects as well as more aggravated diseases.
3.3. Neddylation in cancer
A blend of epithelial cells, endothelial cells, stromal cells (eg, immunocytes and fibroblast cells), and soluble factors interplay with each other and constitute the tumor immune microenvironment. 96 Neddylation inactivation, as well as MLN4924 (also known as pevonedistat, currently in phase I/II clinical trials for patients suffering from cancer), have been proven to exert anti‐tumor activity by modulating cellular processes in tumor cells, but their influences on the functions of immunocytes in the tumor microenvironment are rarely studied. Zhou L et al 57 discover for the first time to our knowledge that, neddylation inactivation exhibits its anti‐tumor activity by suppressing the recruitment of monocytes/tumor‐associated macrophages in the tumor microenvironment, in response to Cullin‐1/NF‐κB‐modulated CCL2 derived from tumor cells. 57 Another study indicated that neddylation inhibition significantly enhances Cullin‐1/c‐Myc‐modulated PD‐L1 expression in glioblastoma cancer cell lines, resulting in T‐cell exhaustion and attenuated T‐cell killing. 54 Besides of solid tumors, unique effects of neddylation inhibition on lymphoid malignancies have been discussed recently. NAE inhibition treated T cells derived from patients with chronic lymphocytic leukemia exert markedly differential expression of NF‐κB‐regulated genes and downregulated of interleukin‐2 signaling during T‐cell activation. In addition, NAE inhibition causes decreased Treg differentiation and a shift toward the Th1 phenotype, accompanied by increased interferon‐γ production. 50 Altogether, the neddylation pathway is capable of modulating the immune microenvironment in tumors via diverse ways, but much more remains to be concerned.
4. CONCLUDING REMARKS
Neddylation modification was originally identified as a posttranslational regulator that highly conserved exists in a variety of cell types and species. New insights into regulatory roles for neddylation in immune responses have come of interest but efforts will still be made to intensify the investigations in this field. The existing investigations highlight that neddylation pathway is engaged in many aspects of the immune cell biology such as survival, differentiation, and immune effector function. Abnormal neddylation alters the characteristics and functions of immune cells and further plays pivotal roles in different stages of immune‐related diseases. Additionally, the following aspects of the immunology of neddylation modification need to be concerned.
First, most of our knowledge about the role of neddylation in various processes of immune responses depends on MLN4924, a potent and selective inhibitor which blocks the first cascade of neddylation pathway by competitively binding to NAEβ. By far, few inhibitors targeting other components of neddylation pathway exist. It would be fascinating whether other targeting inhibitors of neddylation pathway would show effective modulation as well as disparate effects compared with MLN4924 on immune responses. More selective targeting inhibitors effective on immunoregulation will kindle a spark of hope in clinical application.
Second, neddylation modification is extremely well conserved and NEDD8 is ubiquitously expressed in all cell types in the body. 3 Considering that MLN4924 treatment gives rise to global inactivation of neddylation, it may be difficult to clarify the detailed mechanisms for intricate diseases in vivo. Hence, conditional knockout mice that lack specific components of neddylation pathway are in demand. Previous research has directly elucidated the effects of neddylation on T‐cell mediated parasite control and host survival against Plasmodium infection by crossing the Uba3 fl/fl mice with the Lck‐Cre transgeneic strain to generate Uba3 fl/fl Lck‐Cre + mice, in which neddylation inhibition is confined to the T‐cell compartment. 89 Likewise, despite of numerous researches of neddylation acting on tumor cells, the regulation of other components in the microenvironment altered by neddylation modification could be investigated by crossing flox mice with LysM‐Cre transgenic strain, Fsp1‐Cre transgenic strain and so on.
Third, little is known about the immunoregulation of neddylation on chronic inflammation and inflammation‐associated tumorigenesis which undergo a long time of progression. It is reported that p38α negatively regulates the initiation of colitis‐associated colon tumors, whereas transformed intestinal epithelial cells rely on p38α signaling for survival and proliferation, 97 indicating a dual function of p38α signaling in colitis‐associated cancer. Then, dose neddylation inhibition induced in different stages of the disease progression exhibit disparate function? An in vivo research demonstrated that MLN4924 treatment decreased the progression of early atherosclerotic lesions in mice, but had no net effect on the progression of more advanced lesions. In contrast to inflammation elimination in the early stage, MLN4924 treatment exhibited an increased level in neutrophil and monocyte counts in blood in the advanced stage, 30 suggesting discrepant and complicated roles of neddylation in chronic (ongoing) diseases. Notably, the inducible CreERT2 transgenic mice, in which the Cre activity is only detected in the presence of tamoxifen, 98 are considered to be appropriate for such investigations.
In a word, the investigation we have summarized here deepens our understanding of the role of neddylation pathway in fundamental immunology which help to unveil the pathogenesis of immune‐related diseases, and provides a foundation for neddylation‐based therapies in disease treatment.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
This research is supported by grants from the National Natural Science Foundation of China (81972627, 81900591), Shanghai Sailing Program (19YF1449500).
Lu Y, Yang X. The pivotal roles of neddylation pathway in immunoregulation. Immun Inflamm Dis. 2020;8:782–792. 10.1002/iid3.335
DATA AVAILABILITY STATEMENT
All data are shown within the manuscript and figures.
REFERENCES
- 1. Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin‐RING ligases. Nat Rev Mol Cell Biol. 2015;16(1):30‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou L, Zhang W, Sun Y, Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal. 2018;44:92‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down‐regulated ubiquitin‐like protein. J Biol Chem. 1997;272(45):28557‐28562. [DOI] [PubMed] [Google Scholar]
- 4. Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23(11):1985‐1997. [DOI] [PubMed] [Google Scholar]
- 5. Huang DT, Ayrault O, Hunt HW, et al. E2‐RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33(4):483‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011;19(2):168‐176. [DOI] [PubMed] [Google Scholar]
- 7. Kurz T, Chou YC, WillemS AR, et al. Dcn1 functions as a scaffold‐type E3 ligase for cullin neddylation. Mol Cell. 2008;29(1):23‐35. [DOI] [PubMed] [Google Scholar]
- 8. Kurz T, Ozlü N, Rudolf F, et al. The conserved protein DCN‐1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435(7046):1257‐1261. [DOI] [PubMed] [Google Scholar]
- 9. Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of Cullin‐RING ligases: conformational control of conjugation. Cell. 2008;134(6):995‐1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Embade N, Fernández‐Ramos D, Varela‐Rey M, et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 2012;55(4):1237‐1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24(8):3251‐3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xirodimas DP. Novel substrates and functions for the ubiquitin‐like molecule NEDD8. Biochem Soc Trans. 2008;36(Pt 5):802‐806. [DOI] [PubMed] [Google Scholar]
- 13. Lyapina S, Cope G, Shevchenko A, et al. Promotion of NEDD‐CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292(5520):1382‐1385. [DOI] [PubMed] [Google Scholar]
- 14. Ying J, Zhang MM, Qiu XY, Lu Y. Targeting the neddylation pathway in cells as a potential therapeutic approach for diseases. Cancer Chemoth Pharm. 2018;81(5):797‐808. [DOI] [PubMed] [Google Scholar]
- 15. Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8‐activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732‐U67. [DOI] [PubMed] [Google Scholar]
- 16. Mao H, Tang Z, Li H, et al. Neddylation inhibitor MLN4924 suppresses cilia formation by modulating AKT1. Protein Cell. 2019;10:726‐744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo Z, Yu G, Lee HW, et al. The Nedd8‐activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72(13):3360‐3371. [DOI] [PubMed] [Google Scholar]
- 18. Jia L, Li H, Sun Y. Induction of p21‐dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia. 2011;13(6):561‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen P, Hu T, Liang Y, et al. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4‐CHOP‐DR5 axis in human esophageal cancer cells. Clin Cancer Res. 2016;22(16):4145‐4157. [DOI] [PubMed] [Google Scholar]
- 20. Yao WT, Wu JF, Yu GY, et al. Suppression of tumor angiogenesis by targeting the protein neddylation pathway. Cell Death Dis. 2014;5:e1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mackintosh C, García‐Domínguez DJ, Ordóñez JL, et al. WEE1 accumulation and deregulation of S‐phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene. 2013;32(11):1441‐1451. [DOI] [PubMed] [Google Scholar]
- 22. Zhou Q, Li H, Li Y, et al. Inhibiting neddylation modification alters mitochondrial morphology and reprograms energy metabolism in cancer cells. JCI Insight. 2019;4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, Neve RL, Liu H. Neddylation dysfunction in Alzheimer's disease. J Cell Mol Med. 2012;16(11):2583‐2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kandala S, Kim IM, Su H. Neddylation and deneddylation in cardiac biology. Am J Cardiovasc Dis. 2014;4(4):140‐158. [PMC free article] [PubMed] [Google Scholar]
- 25. Han K, Zhang J. Roles of neddylation against viral infections. Cell Mol Immunol. 2018;15(3):292‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13(3):159‐175. [DOI] [PubMed] [Google Scholar]
- 27. Cassatella MA, Ostberg NK, Tamassia N, Soehnlein O. Biological roles of neutrophil‐derived granule proteins and cytokines. Trends Immunol. 2019;40:648‐664. [DOI] [PubMed] [Google Scholar]
- 28. Zhu Z, Sun L, Hao R, Jiang H, Qian F, Ye RD. Nedd8 modification of Cullin‐5 regulates lipopolysaccharide‐induced acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2017;313(1):L104‐L114. [DOI] [PubMed] [Google Scholar]
- 29. Jin J, Jing Z, Ye Z, et al. MLN4924 suppresses lipopolysaccharide‐induced proinflammatory cytokine production in neutrophils in a dose‐dependent manner. Oncol Lett. 2018;15(5):8039‐8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asare Y, Ommer M, Azombo FA, et al. Inhibition of atherogenesis by the COP9 signalosome subunit 5 in vivo. Proc Natl Acad Sci USA. 2017;114(13):E2766‐E2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity. 2018;49(4):595‐613. [DOI] [PubMed] [Google Scholar]
- 32. Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656‐661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang FM, Reyna SM, Granados JC, et al. Inhibition of neddylation represses lipopolysaccharide‐induced proinflammatory cytokine production in macrophage cells. J Biol Chem. 2012;287(42):35756‐35767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li L, Liu B, Dong T, et al. Neddylation pathway regulates the proliferation and survival of macrophages. Biochem Biophys Res Commun. 2013;432(3):494‐498. [DOI] [PubMed] [Google Scholar]
- 35. Song H, Huai W, Yu Z, et al. MLN4924, a first‐in‐class NEDD8‐activating enzyme inhibitor, attenuates IFN‐beta production. J Immunol. 2016;196(7):3117‐3123. [DOI] [PubMed] [Google Scholar]
- 36. Segovia JA, Tsai SY, Chang TH, et al. Nedd8 regulates inflammasome‐dependent caspase‐1 activation. Mol Cell Biol. 2015;35(3):582‐597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zubiete‐Franco I, Fernández‐Tussy P, Barbier‐Torres L, et al. Deregulated neddylation in liver fibrosis. Hepatology. 2017;65(2):694‐709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677‐686. [DOI] [PubMed] [Google Scholar]
- 40. Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol. 2019;19(2):89‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Segura E, Touzot M, Bohineust A, et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013;38(2):336‐348. [DOI] [PubMed] [Google Scholar]
- 42. Mathewson N, Toubai T, Kapeles S, et al. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122(12):2062‐2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng M, Hu S, Wang Z, et al. Inhibition of neddylation regulates dendritic cell functions via deptor accumulation driven mTOR inactivation. Oncotarget. 2016;7(24):35643‐35654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. El‐Mesery M, Seher A, Stuhmer T, Siegmund D, Wajant H. MLN4924 sensitizes monocytes and maturing dendritic cells for TNF‐dependent and ‐independent necroptosis. Brit J Pharmacol. 2015;172(5):1222‐1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chadha A, Mehto S, Selvakumar A, et al. Suppressive role of neddylation in dendritic cells during mycobacterium tuberculosis infection. Tuberculosis (Edinb). 2015;95(5):599‐607. [DOI] [PubMed] [Google Scholar]
- 46. Huang T, Gao Z, Zhang Y, et al. CRL4(DCAF2) negatively regulates IL‐23 production in dendritic cells and limits the development of psoriasis. J Exp Med. 2018;215(8):1999‐2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Masopust D, Schenkel JM. The integration of T cell migration, differentiation and function. Nat Rev Immunol. 2013;13(5):309‐320. [DOI] [PubMed] [Google Scholar]
- 48. Jin HS, Liao L, Park Y, Liu YC. Neddylation pathway regulates T‐cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc Natl Acad Sci USA. 2013;110(2):624‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mathewson ND, Fujiwara H, Wu SR, et al. SAG/Rbx2‐dependent neddylation regulates T‐cell responses. Am J Pathol. 2016;186(10):2679‐2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Best S, Lam V, Liu T, et al. Immunomodulatory effects of pevonedistat, a NEDD8‐activating enzyme inhibitor, in chronic lymphocytic leukemia‐derived T cells [published online ahead of print March 16, 2020]. Leukemia. 2020. 10.1038/s41375-020-0794-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng Q, Liu J, Pei Y, et al. Neddylation contributes to CD4+ T cell‐mediated protective immunity against blood‐stage Plasmodium infection. PLoS Pathog. 2018;14(11):e1007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu Y, Zhao Y, Zou L, Zhang D, Aki D, Liu YC. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic‐epigenetic control. J Exp Med. 2019;216:1664‐1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18(3):153‐167. [DOI] [PubMed] [Google Scholar]
- 54. Zhou S, Zhao X, Yang Z, et al. Neddylation inhibition upregulates PD‐L1 expression and enhances the efficacy of immune checkpoint blockade in glioblastoma. Int J Cancer. 2019;145(3):763‐774. [DOI] [PubMed] [Google Scholar]
- 55. Majolée J, Pronk MCA, Jim KK, et al. CSN5 inhibition triggers inflammatory signaling and Rho/ROCK‐dependent loss of endothelial integrity. Sci Rep. 2019;9(1):8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Filippova N, Yang X, An Z, Nabors LB, Pereboeva L. Blocking PD1/PDL1 interactions together with MLN4924 therapy is a potential strategy for glioma treatment. J Cancer Sci Ther. 2018;10(8):190‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou L, Jiang Y, Liu X, et al. Promotion of tumor‐associated macrophages infiltration by elevated neddylation pathway via NF‐kappaB‐CCL2 signaling in lung cancer. Oncogene. 2019;38(29):5792‐5804. [DOI] [PubMed] [Google Scholar]
- 58. Ghosh S, May MJ, Kopp EB. NF‐kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225‐260. [DOI] [PubMed] [Google Scholar]
- 59. Taniguchi K, Karin M, NF‐kappa B. inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309‐324. [DOI] [PubMed] [Google Scholar]
- 60. Bonizzi G, Karin M. The two NF‐kappa B activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25(6):280‐288. [DOI] [PubMed] [Google Scholar]
- 61. Karin M, Greten FR. NF‐kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749‐759. [DOI] [PubMed] [Google Scholar]
- 62. Read MA, Brownell JE, Gladysheva TB, et al. Nedd8 modification of cul‐1 activates SCF(beta(TrCP))‐dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20(7):2326‐2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Amir RE, Iwai K, Ciechanover A. The NEDD8 pathway is essential for SCF beta‐TrCp‐mediated ubiquitination and processing of the NF‐kappa B precursor p105. J Biol Chem. 2002;277(26):23253‐23259. [DOI] [PubMed] [Google Scholar]
- 64. Karin M, Ben‐Neriah Y. Phosphorylation meets ubiquitination: the control of NF‐kappa B activity. Annu Rev Immunol. 2000;18:621‐663. [DOI] [PubMed] [Google Scholar]
- 65. Fu Z, Liao W, Ma H, et al. Inhibition of neddylation plays protective role in lipopolysaccharide‐induced kidney damage through CRL‐mediated NF‐kappaB pathways. Am J Transl Res. 2019;11(5):2830‐2842. [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Y, Luo Z, Pan Y, et al. Targeting protein neddylation with an NEDD8‐activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer Biol Ther. 2015;16(3):420‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Collier‐Hyams LS, Sloane V, Batten BC, Neish AS. Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of cullin‐1. J Immunol. 2005;175(7):4194‐4198. [DOI] [PubMed] [Google Scholar]
- 68. Kumar A, Wu H, Collier‐Hyams LS, et al. Commensal bacteria modulate cullin‐dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26(21):4457‐4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kumar A, Wu HX, Collier‐Hyams LS, Kwon YM, Hanson JM, Neish AS. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species‐mediated changes in Cullin‐1 neddylation. J Immunol. 2009;182(1):538‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Singleton KD, Wischmeyer PE. Glutamine attenuates inflammation and NF‐kappa B activation via Cullin‐1 deneddylation. Biochem Bioph Res Co. 2008;373(3):445‐449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Khoury J, Ibla JC, Neish AS, Colgan SR. Antiinflammatory adaptation to hypoxia through adenosine‐mediated cullin‐1 deneddylation. J Clin Invest. 2007;117(3):703‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Noguchi K, Okumura F, Takahashi N, et al. TRIM40 promotes neddylation of IKKgamma and is downregulated in gastrointestinal cancers. Carcinogenesis. 2011;32(7):995‐1004. [DOI] [PubMed] [Google Scholar]
- 73. Yan FX, Guan JH, Peng YY, Zheng XF. MyD88 NEDDylation negatively regulates MyD88‐dependent NF‐kappa B signaling through antagonizing its ubiquitination. Biochem Bioph Res Co. 2017;482(4):632‐637. [DOI] [PubMed] [Google Scholar]
- 74. Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia‐inducible factor‐1 Is a basic‐helix‐loop‐helix‐pas heterodimer regulated by cellular O‐2 tension. Proc Natl Acad Sci USA. 1995;92(12):5510‐5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Balamurugan K. HIF‐1 at the crossroads of hypoxia, inflammation, and cancer. Int J Cancer. 2016;138(5):1058‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365(6):537‐547. [DOI] [PubMed] [Google Scholar]
- 77. Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF‐1 activation in human monocytes via p44/42 MAPK and NF‐kappaB. Biochem J. 2006;396(3):517‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Grenz A, Clambey E, Eltzschig HK. Hypoxia signaling during intestinal ischemia and inflammation. Curr Opin Crit Care. 2012;18(2):178‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sufan RI, Ohh M. Role of the NEDD8 modification of Cul2 in the sequential activation of ECV complex. Neoplasia. 2006;8(11):956‐963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chew EH, Hagen T. Substrate‐mediated regulation of cullin neddylation. J Biol Chem. 2007;282(23):17032‐17040. [DOI] [PubMed] [Google Scholar]
- 81. Ryu JH, Li SH, Park HS, Park JW, Lee B, Chun YS. Hypoxia‐inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species‐dependent. J Biol Chem. 2011;286(9):6963‐6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhou W, Xu J, Tan M, et al. UBE2M is a stress‐inducible dual E2 for neddylation and ubiquitylation that promotes targeted degradation of UBE2F. Mol Cell. 2018;70(6):1008‐24 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ehrentraut SF, Kominsky DJ, Glover LE, et al. Central role for endothelial human deneddylase‐1/SENP8 in fine‐tuning the vascular inflammatory response. J Immunol. 2013;190(1):392‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Curtis VF, Ehrentraut SF, Campbell EL, et al. Stabilization of HIF through inhibition of Cullin‐2 neddylation is protective in mucosal inflammatory responses. FASEB J. 2015;29(1):208‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Taylor CT. Interdependent roles for hypoxia inducible factor and nuclear factor‐kappaB in hypoxic inflammation. J Physiol. 2008;586(17):4055‐4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Treatment of locally unresectable carcinoma of the pancreas: comparison of combined‐modality therapy (chemotherapy plus radiotherapy) to chemotherapy alone . Gastrointestinal tumor study group. J Natl Cancer Inst. 1988;80(10):751‐755. [PubMed] [Google Scholar]
- 87. Yu G, Liu X, Tang J, Xu C, Ouyang G, Xiao W. Neddylation facilitates the antiviral response in Zebrafish. Front Immunol. 2019;10:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang T, Ye Z, Yang X, et al. NEDDylation of PB2 reduces its stability and blocks the replication of influenza A virus. Sci Rep. 2017;7:43691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hughes DJ, Wood JJ, Jackson BR, Baquero‐Perez B, Whitehouse A. NEDDylation is essential for Kaposi's sarcoma‐associated herpesvirus latency and lytic reactivation and represents a novel anti‐KSHV target. PLoS Pathog. 2015;11(3):e1004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nekorchuk MD, Sharifi HJ, Furuya AK, Jellinger R, de Noronha CM. HIV relies on neddylation for ubiquitin ligase‐mediated functions. Retrovirology. 2013;10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sun H, Yao W, Wang K, Qian Y, Chen H, Jung YS. Inhibition of neddylation pathway represses influenza virus replication and pro‐inflammatory responses. Virology. 2018;514:230‐239. [DOI] [PubMed] [Google Scholar]
- 92. Zhang X, Ye Z, Pei Y, et al. Neddylation is required for herpes simplex virus type I (HSV‐1)‐induced early phase interferon‐beta production. Cell Mol Immunol. 2016;13(5):578‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bibeau‐Poirier A, Gravel SP, Clément JF, et al. Involvement of the IkappaB kinase (IKK)‐related kinases tank‐binding kinase 1/IKKi and cullin‐based ubiquitin ligases in IFN regulatory factor‐3 degradation. J Immunol. 2006;177(8):5059‐5067. [DOI] [PubMed] [Google Scholar]
- 94. Deng Q, Zhang J, Gao Y, et al. MLN4924 protects against bleomycin‐induced pulmonary fibrosis by inhibiting the early inflammatory process. Am J Transl Res. 2017;9(4):1810‐1821. [PMC free article] [PubMed] [Google Scholar]
- 95. Ehrentraut SF, Curtis VF, Wang RX, et al. Perturbation of neddylation‐dependent NF‐kappaB responses in the intestinal epithelium drives apoptosis and inhibits resolution of mucosal inflammation. Mol Biol Cell. 201627;(23):3687–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity. 2020;52(1):55‐81. [DOI] [PubMed] [Google Scholar]
- 97. Gupta J, del Barco Barrantes I, Igea A, et al. Dual function of p38alpha MAPK in colon cancer: suppression of colitis‐associated tumor initiation but requirement for cancer cell survival. Cancer Cell. 2014;25(4):484‐500. [DOI] [PubMed] [Google Scholar]
- 98. Ichise H, Hori A, Shiozawa S, et al. Establishment of a tamoxifen‐inducible Cre‐driver mouse strain for widespread and temporal genetic modification in adult mice. Exp Anim. 2016;65(3):231‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are shown within the manuscript and figures.


