Abstract
Calcium/calmodulin–dependent protein kinase II (CaMKII) plays a central role in Ca2+ signaling throughout the body. In the hippocampus, CaMKII is required for learning and memory. Vertebrate genomes encode four CaMKII homologs: CaMKIIα, CaMKIIβ, CaMKIIγ, and CaMKIIδ. All CaMKII homologs consist of a kinase domain, a regulatory segment, a variable linker region, and a hub domain, which is responsible for oligomerization. The four proteins differ primarily in linker length and composition due to extensive alternative splicing. Here, we report the heterogeneity of CaMKII transcripts in three complex samples of human hippocampus using deep sequencing. We showed that hippocampal cells contain a diverse collection of 70 CaMKII transcripts from all four CaMKII-encoding genes. We characterized the Ca2+/CaM sensitivity of hippocampal CaMKII variants spanning a broad range of linker lengths and compositions. The effect of the variable linker on Ca2+/CaM sensitivity depended on the kinase and hub domains. Moreover, we revealed a previously uncharacterized role for the hub domain as an allosteric regulator of kinase activity, which may provide a pharmacological target for modulating CaMKII activity. Using small-angle x-ray scattering and single-particle cryo–electron microscopy (cryo-EM), we present evidence for extensive interactions between the kinase and the hub domains, even in the presence of a 30-residue linker. Together, these data suggest that Ca2+/CaM sensitivity in CaMKII is homolog-dependent and includes substantial contributions from the hub domain. Our sequencing approach, combined with biochemistry, provides insights into understanding the complex pool of endogenous CaMKII homologs.
Introduction
Calcium/calmodulin-dependent protein kinase II (CaMKII) is a crucial oligomeric serine and threonine kinase involved in long-term memory formation (1), egg activation in fertilization (2,3), and cardiac pacemaking (4). CaMKII is expressed throughout the human body, which facilitates its role in these disparate functions. Four homologous CaMKIIs are encoded in the human genome: CaMKIIα (encoded by CAMK2A) and CaMKIIβ (encoded by CAMK2B), which are predominantly found in the brain, CaMKIIδ (encoded by CAMK2D), predominant in the heart, and CaMKIIγ (encoded by CAMK2G), found in multiple organ systems, including eggs and sperm (5–7). Each gene product is one CaMKII subunit that consists of a kinase domain, a regulatory segment, a variable linker region, and a hub domain (Fig. 1A). The kinase and hub domains are conserved across the four homologs, with a minimum of 90 and 75% pairwise amino acid identity, respectively. The linker that connects the kinase and hub domains is highly variable in length and composition due to alternative splicing. Herein, we focused on CaMKII in the hippocampus, the primary memory center in the brain (8). CaMKII is the most abundant enzyme in neuronal dendrites and is implicated functionally in memory (1). Transgenic mice deficient in neuronal CaMKIIα or CaMKIIβ have limited long-term memory and display specific learning impairments (9,10). More specifically, mice with mutations at critical phosphorylation sites in CaMKIIα display defects in higher-order memory and adaptive learning (11).
Fig. 1. CaMKII sequence divergence and structural organization.
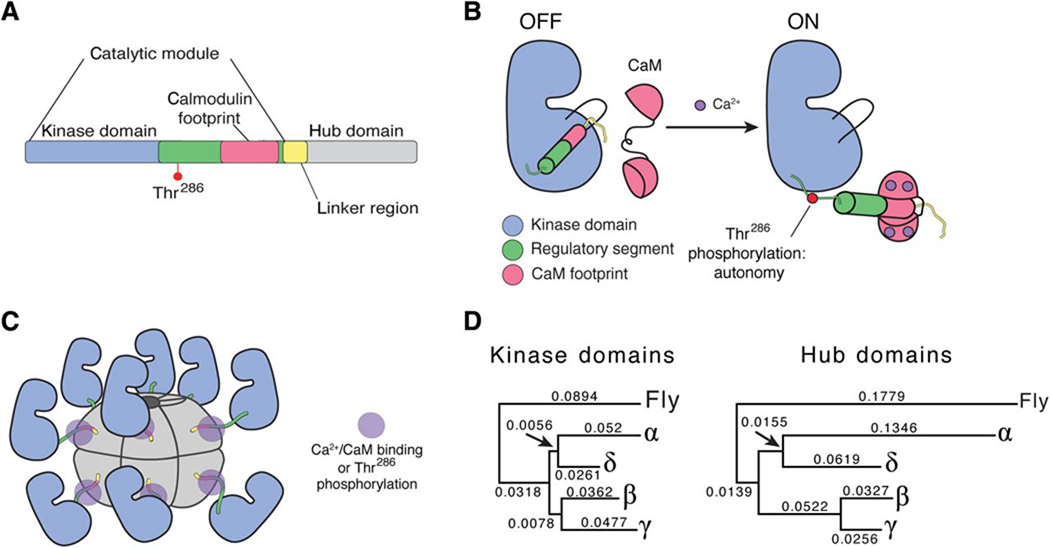
(A) Each subunit of CaMKII consists of a kinase domain, a regulatory segment, a variable linker region, and a hub domain. The regulatory segment houses the CaM-binding region as well as a regulatory autophosphorylation site, Thr286. (B) The regulatory segment acts as an autoinhibitory domain by blocking the substrate-binding pocket. In the presence of Ca2+, Ca2+/CaM binds to the regulatory segment, exposing the substrate-binding pocket and activating the enzyme. (C) The hub domain oligomerizes CaMKII into two stacked hexameric rings. Shown is a cartoon of an active structure with Ca2+/CaM bound to the regulatory segment. (D) Neighbor Joining reconstructions of kinase and hub domain divergence for the four human CaMKII genes, rooted with D. melanogaster CaMKII outgroups.
The structure and function of both the kinase and hub domains of CaMKII have been well studied. Kinase activity is regulated by the autoinhibitory regulatory segment, which blocks the substrate-binding site in the absence of Ca2+ (12). When the local cytoplasmic Ca2+ concentration increases, Ca2+-bound calmodulin (Ca2+/CaM) competitively binds to the regulatory segment and relieves inhibition by exposing the substrate-binding site (Fig. 1B) (13). The regulatory segment also contains three sites of autophosphorylation: Thr286, Thr305, and Thr306 (according to the numbering of residues in CaMKIIα) (14). Phosphorylation at Thr286 results in autonomous activation or sustained activity in the absence of Ca2+ (15). The hub domain oligomerizes the holoenzyme into both dodecameric and tetradecameric assemblies (Fig. 1C) (16–18).
The variable linker region has been difficult to study for several reasons. The sequence of the linker is predicted to be disordered. It is likely for this reason that high-resolution structures of the CaMKII holoenzyme including the linker region have not been solved. Indeed, the only atomic resolution structure of a CaMKII holoenzyme is one in which the variable linker is completely deleted (19). In that structure, the regulatory segment makes extensive contacts with the hub domain, thereby completely sequestering the CaM-binding segment from Ca2+/CaM in this compact conformation. This structure, combined with biochemical data, led to the hypothesis that linker length tunes the autoinhibition within the holoenzyme by either stabilizing a compact conformation (short linker forms) or populating an extended conformation in which the CaM-binding segment is more accessible (longer linker forms). The variable linker also varies in composition between the four CaMKIIs. Further complexity is generated by alternative splicing of up to 9 exons (exons 13 to 21) in the regions encoding each linker, generating a total of more than 80 splice variants possibly expressed from four genes (54). The combination of high identity within the conserved kinase and hub domains and high variability in the linker region makes it difficult to specifically identify CaMKII splice variants in cellular experiments.
There have been many studies that outline specific roles for exons in the variable linker region, which provide some clues as to why exons may be spliced in or out. Exon 15, which is encoded in all genes with the exception of CaMKIIβ has been documented to encode a nuclear localization sequence (NLS) (20–22). This NLS (KKRK) is a common motif identified as a sufficient sequence to achieve nuclear localization (23). Exon 13, which is found in CaMKIIβ and γ, but not α or δ, encodes a region that mediates an important interaction with F-actin (24,25). In dendritic spines, the interaction between CaMKII and actin is crucial for the maintenance of spine structure during long-term potentiation (26). Perhaps the most extensive form of alternative splicing results in the formation of alpha CaMKII associated protein (alpha KAP) where CaMKIIα is spliced to a form that includes only the hub domain (27). This splice variant plays an important role in membrane anchoring using its hydrophobic N-terminal domain (28). Finally, alternative splicing within the hub might regulate oligomerization (5). Roles for the remaining exons have yet to be delineated.
In addition to roles assigned to specific exons, the variable linker region also plays a critical role in CaMKII activation. A standard metric for CaMKII activity is the concentration of Ca2+/CaM required for half-maximal activation (EC50 value) (19,29). CaMKII activation is also cooperative (29). With regard to linker length, a CaMKIIα variant with a 30-residue linker requires substantially less Ca2+/CaM to achieve maximal activity compared to a CaMKIIα variant with a 0-residue linker (19). This general trend also holds in comparing a long-linker CaMKIIβ variant to a short-linker CaMKIIα variant (30). Here, we addressed the gaps in knowledge in three major areas. First, which specific CaMKII splice variants are expressed in human hippocampal cells. Second, how splicing affects CaMKII activation. Third, structural analysis of an autoinhibited CaMKII holoenzyme with a 30-residue linker.
Results
The exon architecture is conserved between CaMKII paralogs
The four CaMKIIs are highly homologous. Using the Neighbor Joining method (52,53), we generated reconstructions of the kinase and hub domain divergence for the four human CaMKII genes rooted with Drosophila melanogaster CaMKII kinase and hub outgroups (Fig. 1D). Because of the clear one-to-one correspondence of exons across genes, we use a common exon numbering system (Fig. 2A). Herein, we implemented a new naming scheme for CaMKII variants based on: (i) gene identity and (ii) incorporated exons. Moving forward, we propose this is the simplest way to efficiently catalog current CaMKII variants and include new variants as they are discovered (54).
Fig. 2. Illumina sequencing reveals a heterogeneous population of CaMKII variants in the human hippocampus.

(A) Human CaMKII genes are encoded by 24 exons with clear one-to-one correspondence across genes. Exons 1 to 10 encode the catalytic kinase domain, exons 11 and 12 encode the regulatory segment, exons 13 to 21 encode the variable-length linker, and exons 22 to 24 encode the association (hub) domain. Missing linker exons indicate that the exon is not encoded in the corresponding gene. Constitutively incorporated kinase, regulatory segment, and hub exons (gray) are spliced into every transcript. CaMKIIδ encodes two versions of kinase exon 6, with one or the other, but not both, incorporated into transcripts. Several exons contain alternative splice sites, enabling omission of a fraction of the exon (indicated by hatching). CaMKIIβ encodes three nearly identical proline-rich linker exons (19 to 21), which are homologous to CaMKIIγ exon 19. (B) Illumina library construction is illustrated with CaMKIIβ. Initial PCR amplification of a region encompassing all alternative splice sites (including the linker and hub exon 23) was performed with primers designed for specificity for the target CaMKII gene, to the exclusion of the other three genes (orange and red arrows). Initial amplicons served as templates for the amplification of library inserts in a second PCR reaction with a different primer pair (green and blue arrows). The resulting inserts were fully spanned by paired-end 251-bp reads for nearly all splice variants. For CaMKIIβ, this was true as long as exons 19, 20, and 21 were not incorporated. These exons are shown as semi-transparent because they were not incorporated in any detected hippocampal splice variant. (C) Summary of CaMKIIα and CaMKIIβ transcript variants for two replicates of donor 1 (r1, r2), donor 2, and donor 3 as detected by Illumina sequencing. All detected variants, including all previously annotated variants, are shown for CaMKIIα. For CaMKIIβ, the top eight read-mapping variants are shown above the black line and the remaining previously annotated variants are shown below the line. Variants are marked as ‘unreported’ if they have not been previously annotated or ‘undetected’ if we did not detect the transcript in our experiments. Underlined percentages indicate inconsistency between libraries. Variants highlighted in blue are those that were used in activity measurements.
Deep sequencing reveals at least 70 CaMKII transcript variants in the human hippocampus
We performed targeted deep sequencing of hippocampal CaMKII transcripts from three separate human donors (82, 26, and 66 year-old males) without neurological or neuropsychiatric diagnoses. To identify all of the alternatively spliced CaMKII transcripts expressed in a human total RNA sample, these transcripts must be substantially enriched. Similar to previous approaches (5,7,24), we reverse-transcribed human hippocampal RNA to generate cDNA and then used gene-specific primer pairs to amplify variable regions of CaMKII transcripts by PCR (Fig. 2B). Agarose gel separation of PCR amplicons followed by Sanger sequencing of excised gel bands was inadequate for detailed characterization of alternative CaMKII splicing, because many bands contained mixtures of variants (fig. S1). To circumvent this problem, we devised an Illumina next-generation sequencing approach. Given the locations at which CaMKII transcripts are variably spliced and the maximal lengths of variable regions when all optional exons are incorporated, strategic placement of primer pairs facilitates PCR-based construction of Illumina sequencing libraries such that 251-bp paired-end sequencing enabled unambiguous identification of virtually all possible CaMKII transcripts from all four genes (Fig. 2B, blue/green primer pair). Compared to traditional whole-transcriptome Illumina RNA-Seq library preparation, this approach emphasizes the sensitivity of unambiguous variant detection over accuracy in the quantification of relative transcript abundance, due to the greater potential for amplification bias resulting from additional PCR cycles. Therefore, we make no claims with respect to transcript abundance beyond classifying transcripts into several detection level categories based on the number of reads mapped to each transcript. However, spurious detection of transcripts is extremely unlikely, because we applied a stringent similarity cutoff of a minimum of 10 reads mapping to each transcript (see Materials and Methods).
We performed this experiment with three separate donor samples of human hippocampal tissue. We detected a large number of distinct transcript variants (Fig. 2C, tables S1 to S16, file S1), including all three possible CaMKIIα variants and more than 15 each of CaMKIIβ, CaMKIIγ, and CaMKIIδ. In addition to variants previously annotated in genomic databases (NCBI Consensus CDS collection, NCBI GenBank, Ensembl), we also identified numerous previously unobserved variants of each gene, which we refer here to as ‘unreported.’. One unreported variant of note was the CaMKIIα transcript that included no optional linker exons (0 aa linker). This is important because the crystal structure of the CaMKIIα no-linker variant is the only available high-resolution structure of full-length CaMKII [PDB code: 3SOA (19)], but this variant was previously not known to be biologically relevant.
Nearly every identified transcript incorporating optional linker exons contained exons 14 and 18, which are required for the protein to remain in frame when any linker exons are spliced in. Given the constraints on linker exon connectivity imposed by the requirement to remain in frame (54), we observed a large fraction of allowable linker sequences for each homolog. Two notable exceptions that were not detected in our experiment were CaMKIIβ variants incorporating exons 19, 20, or 21 (encoding proline-rich sequences) and CaMKIIγ variants incorporating exon 13 (encoding an F-actin interaction site).
We assessed the reproducibility of this experiment with two replicates of sample preparation and sequencing for donor 1, the highest quality tissue and RNA sample (Fig. 2C, tables S1 to S4). For these technical replicates, 99% of the reads mapped to the same collection of variants. Not surprisingly, variants on the edge of detection (cumulatively mapping to 1% of reads) differed between the replicates (tables S2, S6, S10, and S14). However, there was too much variability between the replicates to precisely quantify detection levels of variants and confidently order them based on detection level (see Materials and Methods, tables S17 to S19). Instead we binned variants into several detection-level categories.
Expressed variants were also quite consistent across donors, with at least 97.5% of all reads from each donor mapped to a collection of variants detected in all three donors (tables S1 to S16). Most, but not all, variants fell into the same broad expression category across all donors. Accordingly, we designated variants as ‘consistent’ and ‘inconsistent’ (tables S1, S5, S9, and S13). Large differences in detection levels between donors observed for inconsistent variants may reflect underlying physiological differences between donors.
The variable linker affects activation of CaMKIIα, but not CaMKIIβ
We focused on the activation properties of CaMKIIα and CaMKIIβ variants because of their well-documented involvement in learning and memory (9,11). First, we determined the EC50 values for Ca2+/CaM activation of all three CaMKIIα variants sequenced from the human hippocampus. For all EC50 measurements, the curve fit is shown for simplicity (Fig. 3), and individual fits with all data points used are shown in fig. S2. For brevity we refer to variants by linker length: α-41 (α14,15,18 in Fig. 2C), α-30 (α14,18), and α-0 (α-no-linker). Consistent with previous experiments comparing α-30 to α-0 (19), we also observed a large right-shift in the EC50 value (from 24 nM to 313 nM, respectively, Fig. 3A). The EC50 values of CaMKIIα-41 (25 nM, Fig. 3A) and CaMKIIα-30 were identical within error estimation.
Fig. 3. The hub domain plays a crucial role in Ca2+/CaM sensitivity.

(A to D) CaMKII activity against a peptide substrate (syntide) was measured as a function of Ca2+/CaM concentration. To simplify comparisons, fitted curves were normalized by the corresponding Vmax fit parameter and data points are not shown (see fig. S3). The EC50 value for each fit is indicated by a single box, whereas dashed gray lines in (B) to (D) indicate reference EC50 values for CaMKIIα-30 and CaMKIIα-0 (B) or CaMKIIβ-0 and CaMKIIα-0 (C and D), as labeled on the x-axis. Activity measurements and EC50 and nH (Hill coefficient) fit parameters are shown for all three CaMKIIα splice variants (A), eight selected CaMKIIβ variants (B), chimeras of CaMKIIα/β with no linker (C), and CaMKIIγ and CaMKIIδ with no linker (D). EC50 and Hill coefficient (nH) fit parameters with 95% confidence intervals (see Materials and Methods) are listed for each tested variant, N=3 titrations for each protein sample. Cartoon representations of CaMKII holoenzymes in their autoinhibited states are shown in (A). For (C) and (D), each domain is labeled according to the CaMKII gene.
Next, we examined CaMKIIβ variants with the broadest possible range of variable linker lengths: previously annotated and detected variants β-93, β-69, β-68, β-54, β-30, and β-0, as well as the annotated, but undetected, variant β-217 (Fig. 3B). We observed no major differences in EC50 across the entire range of linker lengths, with the EC50 values of all CaMKIIβ variants falling in the narrow range between 14 and 31 nM. This result suggests that the variable linker does not tune activation the same way in different CaMKII homologs. The Hill coefficients (nH) from these measurements of CaMKIIα and CaMKIIβ variants were all quite similar, ranging from 1.5 to 2.5, which was consistent with previous measurements (19). Note that the cooperativity in CaMKIIβ variants was consistently higher compared to that in CaMKIIα variants but did not correlate with EC50 value differences.
We observed a 15-fold difference in the EC50 for Ca2+/CaM between α-0 and β-0 (313 nM vs. 21 nM). The difference in the activation properties of these two variants may be attributed to sequence differences outside of the variable linker: in the kinase domain, the hub domain, or a combination of the two. Indeed, the only CaMKIIβ variant that we tested with an EC50 outside of the narrow range around 20 nM was the novel variant β-93-short-hub, which was slightly right-shifted from the other CaMKIIβ variants (Fig. 3B). The only difference between β-93 (EC50 = 25 nM) and β-93-short-hub (EC50 = 43 nM) is due to the lack of 26 residues in the hub domain, indicating that hub alone may affect activation properties. These results motivated our next experiments.
Hub identity plays a regulatory role in holoenzyme activation
We directly assessed the regulatory contributions of the kinase and hub domains of CaMKIIα and CaMKIIβ (90% and 77% identity, respectively). To do this, we generated two chimeras with 0aa linkers. The first chimera was composed of the CaMKIIα kinase domain and regulatory segment fused to the CaMKIIβ hub domain, and the second chimera was composed of the CaMKIIβ kinase domain and regulatory segment fused to the CaMKIIα hub domain. The EC50 value of the β kinase/α hub chimera was substantially right-shifted compared to that of the α kinase/β hub chimera (Fig. 3C). Both CaMKIIβ-0 and the chimera with the β hub had approximately the same EC50 values (24 nM and 21 nM, respectively). On the other hand, CaMKIIα-0 and the chimera with the α hub were both right-shifted. However, the EC50 value of the chimera was still three-fold lower than that of wild-type CaMKIIα-0 (102 nM vs. 313 nM). This suggests that the role of the hub domain in determining activation properties is dominant with respect to the kinase domain, with no contribution from the kinase in the context of the β hub, but a substantial contribution in the context of the α hub.
Activation properties track with evolutionary divergence
We compared the divergence of the human CaMKII kinase and hub domains using the corresponding domains from the single CaMKII homolog in fly as an outgroup to root the trees (Fig. 1D). The kinase and hub trees have consistent topologies in which the common ancestor of CaMKIIα and CaMKIIδ diverged from the common ancestor of CaMKIIβ and CaMKIIγ. However, whereas the four kinase domains diverged in almost a star topology (the segments separating the α/δ and β/γ ancestors from their common ancestor are much shorter than any other segment), hub divergence was not nearly as uniform. CaMKIIα and CaMKIIδ are much more diverged from each other and from CaMKIIβ/γ. The CaMKIIβ and CaMKIIγ hubs have diverged by far the least, with the fewest substitutions between them. Based on this similarity between CaMKIIβ and CaMKIIγ, and the apparent involvement of the hub domain in activation, we hypothesized that CaMKIIγ-0 would behave similarly to CaMKIIβ-0. We measured the activation of γ-0 and δ-0 (Fig. 3D), which confirmed our hypothesis about γ-0 (EC50 = 28 nM). Consistent with the order in which α, β/γ, and δ split, as well as with the greater degree of divergence between the α and δ hubs, δ-0 required an intermediate amount of Ca2+/CaM for activation (EC50 = 167 nM), which was approximately 7-fold greater than that of β-0, but 2-fold less than that of α-0.
Insight into the structural mechanism of activity regulation
The difference that we observed in the activation of CaMKIIα-0 compared to that of CaMKIIβ-0 (EC50 = 313 nM and 21 nM, respectively) led us to investigate whether there was a structural explanation. The crystal structure of CaMKIIα-0 shows a compact conformation with a radius of gyration (Rg) of 61 Å, as determined using small angle X-ray scattering (SAXS) (19). We performed SAXS measurements to compare the overall shapes of CaMKIIα-0 and CaMKIIβ-0 in the unactivated conformation (without the addition of Ca2+/CaM). Guinier and pair distribution analyses revealed a substantial difference in the Rg and Dmax values of CaMKIIα-0 [~59 Å and 155 Å, consistent with previous data (19)] compared to those of CaMKIIβ-0 (~66.4 Å and 250 Å) (fig S3). These results showed that CaMKIIα-0 had a more compact conformation than that of CaMKIIβ-0.
Our current understanding of the CaMKII holoenzyme state (compact versus extended) has emerged from a combination of negative stain EM, SAXS, and X-ray crystallography, as reviewed previously (31). The central hub is consistently observed as a dodecameric or tetradecameric assembly. The kinase domains have been observed in multiple conformations, including above and below the hub domain plane in EM studies (32), in compact positions docked against the hub in an X-ray crystal structure (19), or in the same plane as the central hub at extended positions, from EM and SAXS modeling (33,34). Another negative stain EM study of a CaMKIIα-30 construct showed the kinases adopting positions at extended distances from the central hub (35). A general challenge for 3D EM reconstructions of CaMKII has been the strong preferred orientation of the sample under both cryo and negative stain conditions. In addition, the average size of negative stain particles and the linker distances present in CaMKII isoforms have motivated careful comparison of negative stain and cryo-conditions when interpreting kinase domain position (32).
To revisit questions surrounding the CaMKII holoenzyme conformational state, we analyzed samples of CaMKIIα-30 using single-particle cryo–electron microscopy (cryo-EM). Two-dimensional (2D) class averages of the sample, frozen at 0.5 to 1 mg/ml, showed clearly distinguishable six- and seven-fold versions of the central hub with visible secondary structure elements (Fig. 4A). Of ~800,000 obvious top views, ~500,000 (63%) showed clear six-fold features, whereas 290,000 (36%) showed clear seven-fold features. Obvious side views were visible in the 2D classes, unlike in previous negative stain analyses (Fig. 4B and D) (17,35,36). 2D classes also showed additional, less ordered density, proximal of the hub (presumably from the kinase domains) (Fig. 4C, D). This density was still present when the mask diameter was expanded during 2D classification (Fig. 4E). No additional ordered density, at more extended positions, appeared to be excluded due to masking. In top views of the six- and seven-fold hub assemblies, additional kinase domain density was observed as a strong signal positioned between hub subunits (Fig. 4C) and was observed in side-views above and below the mid plane of the hub (Fig. 4D, E).
Fig. 4. 2D classes of single-particle cryo-EM analysis of CaMKIIα-30.
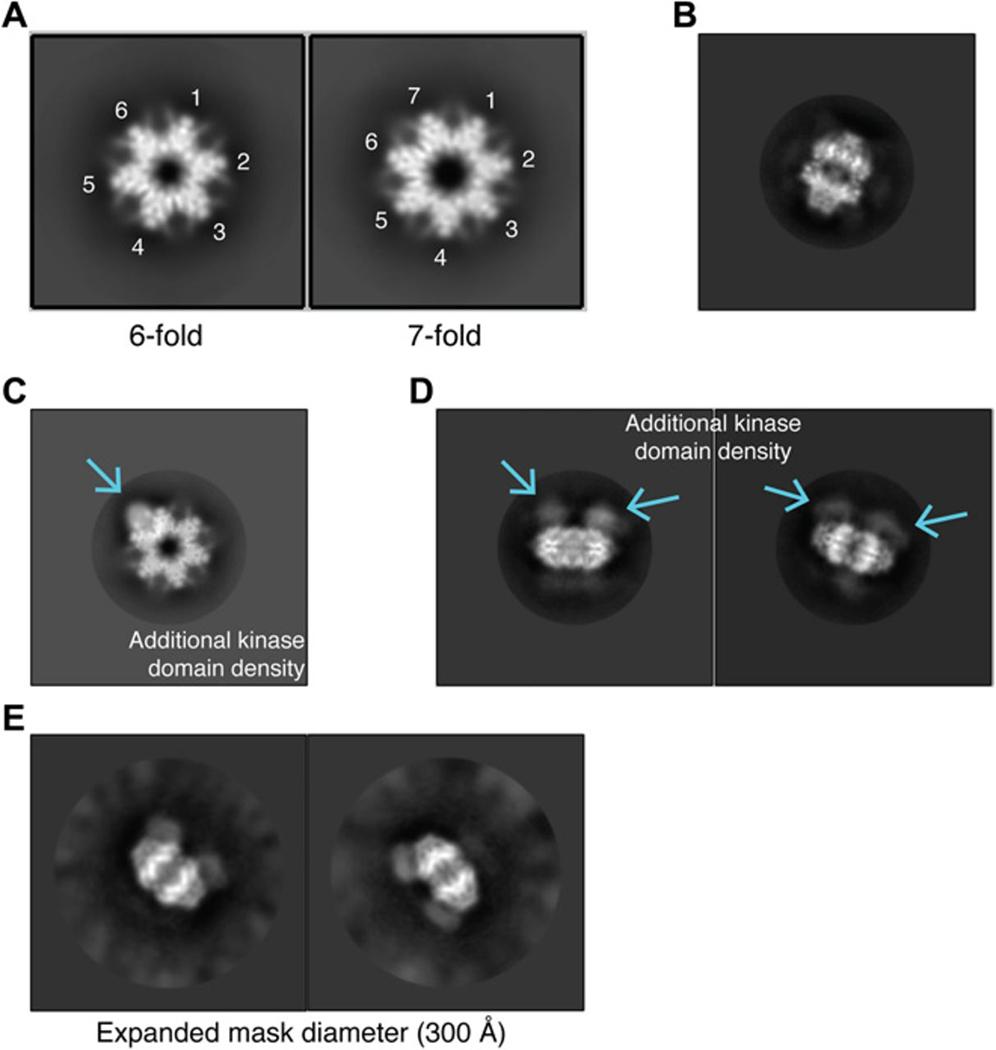
(A) Six- and seven-fold symmetric top views for CaMKII holoenzymes were observed in cryo-EM 2D classes. (B) Example of a clearly visible side view in the 2D classes. (C) Density for a single kinase domain (blue arrow) docked onto a dodecameric central hub seen in a top view. (D) Density for multiple kinase domains (blue arrows) above the hub midplace is visible in side views. (E) Density for kinase domains remained when the mask diameter was increased to 300 Å. No additional ordered density was visible at extended positions.
We generated initial models using Relion, and classified the full particle set, all without any assumptions of symmetry (see Materials and Methods for details). Both six-fold and seven-fold hub domains (without any ordered kinase domains) emerged during 3D classification. Ordered density of the kinase domains appeared in both the six-fold and seven-fold 3D classes. We focused on two representative six-fold final classes. The first class, which we refer to as Class A, was refined to 4.8 Å resolution [as evaluated by gold standard FSC (Fourier Shell Correlation) criterion]. In Class A, we observed one ordered kinase domain, which was docked against a six-fold hub in a position with the beta-clip engaged with the hub, as seen in the crystal structure of CaMKIIα-0 (PDB: 3SOA) (Fig. 5A). A second class (Class B) was refined to 6.6 Å resolution. Class B showed one kinase domain interacting with a six-fold hub in a previously unobserved orientation. In Class B, the kinase C-lobe (helices αEF and αG) engaged a loop in the hub domain (W373-P379), and the N-lobe was in a position closer to the hub mid-plane (Fig. 5B). The final reconstruction Class A (six-fold) included 160,000 particles. Final reconstruction Class B (six-fold) included 40,000 particles. In both reconstructions, the EM density (with recognizable secondary structure elements) was unambiguous for positioning the kinase domain. Both Class A and B exhibited preference for top views; however, these data showed angular distributions that enabled 3D reconstruction (fig. S4). Similar docked states as Class A and B were seen with seven-fold hubs. Enforcing D6 symmetry during refinement of the Class A or Class B particles resulted in reconstructions with a recognizable central hub, but loss of recognizable kinase density.
Fig. 5. Two kinase domain arrangements observed by cryo-EM.

(A) In reconstruction Class A, a single kinase domain was resolved between two hub domain subunits. A kinase domain from the CaMKIIα-0 holoenzyme crystal structure (PDB: 3SOA) was fitted into the additional density. (B) In a second reconstruction, Class B, a kinase domain was visible interacting with a hub subunit with the kinase C-lobe making contact with a loop at the top of the hub domain. The CaMKIIα kinase domain (PDB: 3SOA) was docked into the additional density, and the CaMKII hub domain (PDB: 5IG3) was docked into the hub density. (C) Unmasked maps of Class A are shown below in gray, where adjusted contour levels reveal EM density for additional kinase domains. (D) Sub-classification of Class B particles results in the 3D model shown in pink where EM density for two docked kinases is evident.
Discussion
We unambiguously identified at least 70 distinctly spliced CaMKII transcripts present in three individual human hippocampal samples using Illumina sequencing. Note that excised hippocampal tissue samples are necessarily comprised of multiple cell types from the hippocampus, including neurons, glia, and interneurons. Therefore, the transcripts that we report here are derived from this mixture of cell types, not from excitatory neurons alone. From our experiment, it is impossible to tell which transcripts are co-expressed specifically in excitatory neurons or any other cell type. Addressing this important question requires precise extraction of individual neurons from the hippocampus followed by single-cell transcript sequencing. The alternative, culturing neurons outside of brain tissue, disrupts numerous in vivo cell-cell interactions, including synapses, quite possibly leading to non-physiological changes in CaMKII transcription and splicing.
Using the sequences that we obtained, and those that others previously annotated from various tissues, we examined the differences in kinase activity regulation between CaMKII splice variants. We discovered that, whereas CaMKIIα regulation was dependent on the variable linker region, CaMKIIβ regulation was not. Furthermore, we showed a role for the hub domain in the regulation of CaMKII activation by Ca2+/CaM. Our data provide insights into the heterogeneity of CaMKII transcripts in a human tissue and uncover a previously uncharacterized role for the hub domain in regulating CaMKII activity through an allosteric mechanism. This finding opens a window into understanding allosteric control of CaMKII activity through modulation of the hub domain.
We sequenced a large number of transcripts from all four CaMKII genes from three separate samples of human hippocampal tissue. Note that CaMKIIα produced a no-linker transcript (CaMKIIα-0) that accounted for 0.5 to 25% of mapped reads. This sequence was previously crystallized to obtain the only full-length atomic resolution structure of CaMKII. Our data demonstrate the potential physiological relevance of the compact conformation observed in that structure (19). The sheer number of variants detected in the hippocampus (70 across the four CaMKII genes) suggests that (i) either splicing is not tightly regulated at every exon, or (ii) there are complex mechanisms of regulation responsible for every transcript that we detected. In either case, certain exons (CaMKIIβ exons 19-21 encoding proline-rich protein sequences and CaMKIIγ exon 13 encoding an F-actin interaction protein sequence) appear to be specifically suppressed in the hippocampus.
In terms of the abundance of detected transcripts, systematic PCR bias is unlikely to be solely responsible for the approximately 1000-fold differences between the high- and trace-level detection categories of detected variants of CaMKIIβ, CaMKIIγ, and CaMKIIδ (tables S1, S5, S9, and S13). Physiological differences in transcript copy number between variants likely contributed to these differences in detection level. Furthermore, given the overall consistency in variant detection between donors, the few instances of extensive differences in detection level may reflect physiological differences between individuals.
We found that agarose gel separation of transcripts followed by Sanger sequencing of individual bands could not resolve CaMKII variants from the hippocampus, because sequencing traces from most bands indicated a mixture of templates present in the sequenced sample. In many cases, careful analysis of the Sanger sequencing trace enabled us to identify at least two distinct splice variants (fig. S1). Therefore, it is difficult to interpret the presence or absence of an expected gel band as indicative of the presence or absence of a particular CaMKII transcript in a sample. Because each Illumina read obtained by our approach reflects a single template molecule, it produces a more robust survey of the CaMKII transcripts. We suggest that this approach should be applied to other tissues to begin to catalog the diversity in CaMKII transcripts. We expect that in other tissues, the identity and relative abundance of transcripts will vary from those we observed in the hippocampus. With this information in hand, we will certainly improve our understanding of the regulation of CaMKII alternative splicing, and ultimately be able to link this regulation to the Ca2+ signaling requirements within each tissue.
Our results comparing the EC50 values between CaMKIIα and CaMKIIβ splice variants showed that the regulation of CaMKII activation was homolog-dependent and, specifically, affected by the sequence of the hub domain. Our measurements characterized CaMKII activation at equilibrium Ca2+/CaM concentrations. CaMKII activity is also dependent on the frequency of Ca2+ spikes, which is how Ca2+ is delivered to endogenous CaMKII in postsynaptic neuron (19,30,37). Future experiments will be needed to carefully dissect this kinetic contribution in the activation of different splice variants. The variable linker in CaMKIIα does play a role in regulating CaMKIIα activity, which is consistent with previous work (19). We used cartoon holoenzymes to illustrate the compact and extended autoinhibited states of CaMKII (Fig. 3), which correspond to lower and higher sensitivities to Ca2+/CaM, respectively. For CaMKIIα, these cartoons are based on a crystal structure (PDB: 3SOA) and SAXS measurements, respectively (19). Unexpectedly, the variable linker in CaMKIIβ did not play a role in regulating CaMKIIβ activity. Our SAXS measurements directly comparing CaMKIIα-0 and CaMKIIβ-0 suggest that α-0 adopts a compact conformation, whereas β-0 adopts a more extended conformation as evidenced by its larger Rg and Dmax values (fig. S3). These data corroborate the trend that was previously observed comparing α-0 and α-30 where the more compact conformation requires more Ca2+/CaM for activation (19), which is what we also observed in our experiments.
The results from our experiments with CaMKIIα and CaMKIIβ chimeras provide evidence that the hub identity is a determinant of sensitivity to Ca2+/CaM. We observed a large difference in the activation of the CaMKIIβ kinase domain when it was fused to the CaMKIIα hub domain. Substitutions between CaMKIIα and CaMKIIβ hub domain sequences are distributed quite uniformly throughout the domain (Fig. 6A). This led us to hypothesize that there is a dynamic component to this regulation. We turned to single particle cryo-EM to gain a better understanding of the structural mechanism of activity regulation.
Fig. 6. Allosteric contributions from the hub domain regulate CaMKII activation.

(A) The 32 positions that differ between the CaMKIIα and CaMKIIβ hubs are represented by spheres at the Cα position on the structure of CaMKIIα hub (PDB: 3SOA). The hub is shown in cyan, the regulatory segment is in orange, and the kinase domain is in green. (B) Sequence alignment of the hub domains of human CaMKIIs, colored by degree of conservation (dark to light corresponds to from most to least conserved). For convenience, all hub sequences start at residue 1. Residues in the CaMKIIα hub that differ in the CaMKIIβ hub are highlighted by red boxes. The loop mediating kinase docking in the Class B cryo-EM reconstruction is shown in the black box.
Our cryo-EM data showed that the kinase domains were in close physical contact with the hub and suggest two specific sources for hub-dependent regulation observed in the chimera experiments. These cryo-EM data reveal two conformations sampled by the kinase domain under frozen hydrated conditions. First, we observed the same docked conformation in single-particle cryo-EM reconstructions using CaMKIIα-30 (Fig. 5A, Class A) that was previously observed in a crystal structure of CaMKIIα-0 (19). The crystal structure shows a compact state of the holoenzyme in which the kinase domain is docked onto the hub domain and the calmodulin binding segment is occluded. The conformation we observed by cryo-EM (Class A) suggests that with the holoenzyme in solution, kinase domains may sample a similar compact state, even in the presence of a 30-residue linker. Single amino acid substitutions from CaMKIIα to CaMKIIβ are not found at this docking interface (Fig. 6A), thereby confounding straightforward tests of this interface in hub-dependent regulation. Our cryo-EM analysis also revealed a second, distinct docked state, Class B (Fig. 5B). In this reconstruction, the kinase domain is flipped relative to Class A, with the C-lobe oriented above the N-lobe, relative to the hub. A loop in the hub domain mediating interaction between the kinase and the hub differs in sequence between CaMKIIα and CamKIIβ (Fig. 6B, black box). In Class B, the regulatory segment is accessible for calmodulin binding, but there remains extensive contact between the kinase C-lobe and the hub, which may influence activation. These two cryo-EM reconstructions advance our understanding of holoenzyme architecture because they show that in a holoenzyme with a 30-residue linker, the kinase domains make intimate contact with the hub, to the extent that two distinct cryo-EM reconstructions can be observed with specific docked conformations.
Note that in both Class A and Class B reconstructions of CaMKIIα −30, the final model shows only one well-resolved kinase domain per holoenzyme. The final high-resolution maps were generated with masks that only included a single kinase domain and excluded regions for other potential kinase domains. However, we do not interpret these data as suggesting other positions are unoccupied. Rather, the other positions showed lower occupancy, or may be less well ordered, as suggested by maps contoured to show additional noise. In these maps, we observed density, again below and above the hub mid-plane. As previously mentioned, the 2D class averages also showed density above and below the hub mid-plane (Fig. 4D). In these unmasked maps, when adjusting the contour level (Fig. 5C), we observed that although the density was more poorly resolved, the N and C-lobes of the kinase domains can be distinguished and correspond to an equivalent docked position. The position of the unmodeled density is consistent with kinase domain positions along the surface of the hub, and inconsistent with the extended positions proposed from negative stain reconstructions (35). Further sub-classification of Class B can generate classes with more than one site occupied; however, the overall resolution is poorer (Fig. 5D). For these reasons, we present Class A and Class B as representative of kinase-hub interactions, indicative of potential binding modes.
The position of the kinase domain in Class A is compatible with the packing of all 12 kinase domains in the same orientation. A docked state with Class A and Class B next to one another is also compatible when occurring at adjacent sites within a ring, suggesting that a kinase domain in a given holoenzyme may engage and interact with the hub in multiple orientations. In future studies, it will be interesting to investigate whether constructs such as the CaMKIIα kinase/β hub chimera show this kinase proximity to the hub. The 30-residue linker in CaMKIIα-30 is predicted to be a disordered segment. This disordered linker endows the kinase domains with many degrees of freedom. Locking the kinase into a docked conformation is likely to be entropically unfavorable and transient, making coincident docking statistically unlikely. The conformational diversity of kinase domain positions and their relative distribution may be a rich area for future exploration, especially as advances in computational methods facilitate better modeling of flexible protein regions.
Our cryo-EM analysis showed two 3D reconstructions of CaMKIIα-30 with different docked states in which the kinase domain makes direct contact with the hub. The kinase and hub domains have evolved together, therefore it is not surprising that they have also evolved ways to interact (38). We speculate that these interactions may mediate autoinhibition or calmodulin-dependent activation. Because the interactions that we observed may vary depending on the identity of the CaMKII hub domain, they do not exclude other interactions, which may be uncovered when kinase domains are less likely to interact with the hub (34). With the rich diversity of splice variants described here, a given holoenzyme may have an entire set of regulatory interactions that it can sample. Thoughtful future experiments will be needed to specifically distinguish the role of these dynamic interactions in holoenzyme activation, to more fully elucidate the intricate mechanistic details of a remarkably versatile enzyme.
Materials and Methods
Initial amplification of transcript variable regions
Two human total RNA samples and one human cDNA sample were purchased from BioChain. The samples were derived from complete hippocampi of three human donors (donor 1: 82 year old male; donor 2: 26 year old male; donor 3: 66 year old male) without neurological or psychiatric diagnoses. BioChain advised that the tissue from donor 2 did not pass all internal histological quality controls, although the RNA derived from that tissue did pass all RNA quality controls. We detected many fewer low-expression variants (less than 1% of mapped reads) of CaMKIIβ, γ, and δ in the sample from donor 2, compared to donors 1 and 3. However, robustly expressed variants were highly consistent between all three donors (tables S1 to S16). This is consistent with nonspecific partial degradation of RNA in the donor 2 sample. Whereas low copy number transcripts fell below the threshold of detection due to partial degradation across all mRNAs, robustly expressed transcripts remained detectable, despite losing a fraction of their initial copy number. cDNA was reverse-transcribed from donor 1 and donor 2 RNA samples using the ProtoScript II first strand cDNA synthesis kit (NEB #E6560) with oligo dT priming. Variable regions of transcripts were PCR-amplified for 35 cycles with high-specificity primer pairs designed to minimize cross-hybridization between CaMKII genes (file S2) with either Phusion polymerase (NEB #M0530) or KAPA HiFi polymerase (Roche). This yielded a mixed population of variable-length amplicons reflecting the splice variants of the targeted gene present in the starting sample.
Construction of Illumina sequencing libraries
Illumina sequencing libraries were prepared from the initial variable-length amplicon pools with an adapted PCR-based construction protocol (39). First, gene-specific primers with overhangs containing Illumina-specified sequences (Fig. 2B, file S2) were used to sub-amplify transcript variable regions. Second, generic primers annealing to the overhangs introduced in the first reaction were used to incorporate annealing sites for Illumina sequencing primers, NEBNext library indices (NEB #E7335 single-index set, #E7600 dual-index set), and P5/P7 flow cell annealing sequences. The resulting amplicons were agarose gel-purified with size selection based on the theoretical minimal (no linker exons spliced in) and maximal (all linker exons spliced in) lengths of the variable region inserts.
Analysis of Illumina sequencing
Illumina reads were grouped by sequence similarity, producing clusters with >90% sequence identity and two or fewer base pair insertions/deletions. The consensus sequence of each cluster was aligned to exon sequences from the corresponding CaMKII gene obtained from the reference human genome (GRCh38.p12, Ensembl release 95) to map exon order in the transcript represented by the cluster. More than 90% of clustered reads from each Illumina library were unambiguously mapped with reference exons of the corresponding gene, with all splice junctions matching those observed in previously annotated splice variants. We refer to these as mapped reads. Python2 code implementing read clustering is available at: github.com/romansloutsky/illumina_read_clustering. Two technical replicates of libraries were prepared from the donor 1 tissue sample, starting with the initial reverse transcription step, and sequenced. One replicate each of donors 2 and 3 was sequenced. For each gene, different collections of splice variants were detected in each of the four cDNA samples, so calculating the correlation between replicates across all detected variants was not possible. Instead, for each gene, we calculated pairwise Pearson correlation in variant detection levels (fraction of mapped reads which mapped to that variant) and Spearman rank-based correlation between replicates over only those variants that were detected in all four replicates (table S19). Whereas the Pearson correlation between replicates and donors was more than 90% for most pairwise comparisons, it was dominated by the magnitude of differences between the strongest- and weakest-detected variants. When the highest detected variants differ between samples, the Pearson correlation decreases. Technical replicates are no better correlated than biological replicates, indicating substantial variability between library preps and sequencing reactions. This is not unexpected, given the 93 total cycles of PCR amplification, as well as the known run-to-run variability in flow cell clustering efficiency when sequencing mixtures of inserts of different lengths. Spearman correlation generally tracks with Pearson correlation, but is less skewed by the magnitude of highest-detected variants. On the whole, Spearman correlation indicates a limited ability to accurately order variants according to detection level. We believe the highest resolution of detection level quantification possible from these sequencing experiments is the categorization of variants into several detection level ranges. We used ‘high’ (approximately 20% or more of mapped reads), ‘medium’ (between about 20% and 5%), ‘low’ (approximately between 5% and 1%), and ‘trace’ (below 1%) categories. We then classified transcripts as consistent (in the same detection level category) or inconsistent (in different detection level categories) between donors. Allowing for observed variation between technical replicates and considering ‘trace’ detection (typically fewer than 50, but always more than 5, reads) in some replicates consistent with no detection in others, the overwhelming majority of variants were detected at a consistent level between all three individuals (table S9). We believe that the remaining inconsistent variants reflect physiological differences between individuals.
Plasmid construction
cDNAs encoding full-length CaMKII variants were subcloned into a pET vector containing N-terminal 6xHis followed by a SUMO tag. CaMKII chimeras were constructed as follows. The cDNAs encoding the kinase/regulatory segment and hub domains of CaMKIIα and CaMKIIβ were amplified from full-length constructs. The kinase domain includes the regulatory segment of its corresponding kinase. Each domain was amplified with a 20-bp overhang that was the corresponding CaMKII gene. The chimeras were assembled using Gibson assembly.
Protein expression and purification
All CaMKII variants used for this study were recombinantly expressed and purified as previously described (40). Plasmids encoding CaMKII variants were co-transformed with lambda phosphatase into Rosetta 2(DE3)pLysS competent cells (Millipore). Expression was induced overnight with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, GoldBio) and cultures were grown overnight at 18 °C. Cell pellets were resuspended in Buffer A [25 mM Tris-HCl (pH 8.5), 150 mM KCl, 50 mM Imidazole, 10% glycerol, Sigma] with 25 mM magnesium chloride, containing a cocktail of protease inhibitors and DNase (0.2 mM AEBSF, 5.0 μM Leupeptin, 1 µg/mL Pepstatin, 1 µg/mL Aprotinin, 0.1 mg/mL Trypsin Inhibitor, 0.5 mM Benzamidine, 1 µg/mL DNAse) (Sigma) and lysed. All subsequent purification steps were performed with an ÄKTA pure chromatography system at 4°C. Clarified cell lysate was loaded onto a 5-mL HisTrap FF NiNTA Sepharose column (GE), and eluted with a combination of 50% Buffer A and 50% Buffer B ([25 mM Tris-HCl (pH 8.5), 150 mM KCl, 1M imidazole, 10% glycerol) for a final concentration of 0.5 M imidazole. The protein was desalted of residual imidazole with a HiPrep 26/10 Desalting column, and His SUMO tags were cleaved with Ulp1 protease overnight at 4°C in Buffer C [25 mM Tris-HCl (pH 8.5), 150 mM KCl, 2 mM TCEP (GoldBio), 50 mM Imidazole, 10% glycerol]. Cleaved tags were separated by a subtractive NiNTA step. Next, an anion exchange step was performed with a 5-mL HiTrap Q-FF and protein was eluted with a KCl gradient. Eluted proteins were concentrated and further purified in gel filtration buffer [25 mM Tris-HCl (pH 8.0), 150 mM KCl, 1 mM TCEP, 10% glycerol] using a Superose 6 Increase 10/300 size exclusion column. Pure fractions were then concentrated, aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C until further use. All columns were purchased from GE. Calmodulin (Gallus gallus) was recombinantly expressed from a pET-15b vector (a generous gift from Angus Nairn, Yale School of Medicine) in BL21(DE3) cells (Millipore) and purified as previously described (41). To quantify the calmodulin concentration for making stocks to use in the kinase assays, we used circular dichroism on a Jasco J-1500 Spectrophotometer to make a measurement in triplicate for our purified sample scanning a wavelength spectrum between 250 and 215 nm to measure the characteristic wavelength of 222 nm as previously described (42).
We calculated the calmodulin concentration as follows:
where the circular dichroism at 222 nm (CD222nm) is expressed in mdeg, Θ is the molar ellipticity, and l is the path length in cm.
Coupled kinase assays
Kinase activity was monitored under previously described conditions (29,43) with a Synergy H1 microplate reader (Biotek). Each well of the test plate contained master mix composed of the following (all concentrations listed as final, working concentrations): 5 mM Tris (pH 7.5, Fisher Scientific), 150 mM KCl (Sigma), 10X Tris/MgCl2 buffer 50 mM/10 mM respectively (Fisher Scientific), 0.2 mM CaCl2 (Sigma), 1 mM phosphoenolpyruvate (Alfa Aesar), nicotinamide adenine dinucleotide (0.2 mg/ml, Sigma), pyruvate kinase (55.7 U/ml, Sigma), lactate dehydrogenase (78 U/ml, Millipore Sigma), 0.5 mM ATP (Sigma), 0.3 mM syntide (LifeTein). The final pH in each well of the reaction was ~ 7.5 to 8, and the final enzyme concentration was 13.3 nM. The addition of calmodulin (concentrations ranging from 0.4 nM to 2 μM) to the reaction was used to initiate CaMKII activity, after which absorbance was measured at 15-s intervals for 10 min. The change in absorbance over time was fitted with a straight line (y = mx + c) to obtain a slope (m) proportional to the kinetic rate of the reaction. For each time series, slopes were fitted to a sliding window of 5 points (1 min 15 s) and the maximum observed slope was used to represent the kinetic rate of that reaction. Kinetic rates across a series of calmodulin concentrations were fitted with the following equation:
to obtain EC50 (defined as the calmodulin concentration needed to reach the half-maximal reaction velocity) and cooperativity values (Hill coefficients, nH). 95% confidence intervals for fit parameters (EC50 and nH) were determined using the following bootstrap procedure. 10,000 replicate calmodulin concentration series were generated by randomly selecting one observed kinetic rate at each measured calmodulin concentration from the set of replicates for that variant. Each generated concentration series was fit with the equation described earlier. Parameter values at the 2.5th and 97.5th quantiles of the 10,000 fits were taken as the boundaries of the 95% confidence interval.
Small-angle X-ray scattering
Small-angle X-ray scattering (SAXS) data were collected at the mail-in high-throughput SIBYLS beamline 12.3.1 at the Advanced Light Source (ALS) using a Pilatus3 2M pixel array detector (Dectris). Samples (20 μl) were loaded in a 1.5-mm thick chamber by an automated Tecan Freedom Evo liquid handling robot (44). Incident X-rays were tuned to a wavelength of 11keV/1.27Å, samples were exposed for 10 s, and data collected every 0.1 or 0.33 s. SAXS data were collected for CaMKIIα-0 at concentrations of 1, 3, and 7.2 mg/ml, whereas those for CaMKIIβ-0 were collected at concentrations of 3, 7, and 11.5 mg/ml. Frames were chosen using SAXS Frameslice (https://bl1231.als.lbl.gov/ran). Data were then scaled and merged using the low q region from the lowest concentration datasets. Rg and Dmax values were determined using both Scatter 3.0 and primus (ATSAS 3.0).
Cryo-electron microscopy data collection and analysis
C-flat Holey Carbon grids with gold support (CF-1.2/1.3-3Au-50, EMS) were made hydrophilic by glow-discharging at 30 mA for 30 s using PELCO easiGlow (Ted Pella). Purified CaMKII (3 μl) at 0.5 to 5 mg/ml was applied to these grids at ~96% relative humidity, blotted for 5 s, and plunge-frozen into liquid ethane using Cryoplunge 3 (Gatan). Frozen hydrated samples were imaged on an FEI Talos Arctica at 200 kV with a K2 Summit direct detection camera in super-resolution mode with a total exposure dose of approximately 52 electrons. Thirty-five frames per movie were collected at a magnification corresponding to 0.455 Å per pixel using SerialEM (45). Micrographs (6510) were collected at defocus values ranging from −0.5 to −2.5 μm. Movie frames were motion-corrected and dose-weighted by MotionCor2 (46), sampled to 0.91 Å per pixel, and contrast transfer function (CTF) parameters were estimated by CTFFIND4 (47). Particle picking was performed with crYOLO giving 1,500,734 initial particles. Initial ab-initio models were generated in Relion (48). After successive rounds of 2D and 3D classification, 290,980 particles were selected. Additional rounds of 3D classification led to the reconstructions of 4.8 Å from 160,211 particles (reconstruction Class A), of 6.6 Å from 40,329 particles (reconstruction Class B, fig. S4). Maps used for figures were filtered according to local resolution with b-factor sharpening within Relion. Models were fit into the map using Chimera (49) and Coot (50) and rigid body refined with PHENIX (51).
Supplementary Material
Fig. S1. PCR amplicons from multiple CaMKIIβ variants do not separate on an agarose gel.
Fig. S2. Data and EC50 fits for all tested CaMKII variants.
Fig. S3. SAXS data and analyses of CaMKIIα-0 and CaMKIIβ-0.
Fig. S4. Cryo-EM data.
Table S1. Detected CaMKIIα splice variants.
Table S2. Read counts of CaMKIIα splice variants from donor 1.
Table S3. Read counts of CaMKIIα splice variants from donor 2.
Table S4. Read counts of CaMKIIα splice variants from donor 3.
Table S5. Detected CaMKIIβ splice variants.
Table S6. Read counts of CaMKIIβ splice variants from donor 1.
Table S7. Read counts of CaMKIIβ splice variants from donor 2.
Table S8. Read counts of CaMKIIβ splice variants from donor 3.
Table S9. Detected CaMKIIγ splice variants.
Table S10. Read counts of CaMKIIγ splice variants from donor 1.
Table S11. Read counts of CaMKIIγ splice variants from donor 2.
Table S12. Read counts of CaMKIIγ splice variants from donor 3.
Table S13. Detected CaMKIIδ splice variants.
Table S14. Read counts of CaMKIIδ splice variants from donor 1.
Table S15. Read counts of CaMKIIδ splice variants from donor 2.
Table S16. Read counts of CaMKIIδ splice variants from donor 3.
Table S17. Pearson and Spearman correlation matrices for CaMKIIβ variants.
Table S18. Pearson and Spearman correlation matrices for CaMKIIγ variants.
Table S19. Pearson and Spearman correlation matrices for CaMKIIδ variants.
File S1. Exon sequences for all CaMKII genes.
File S2. Primers for initial amplification of variably spliced transcript regions.
Acknowledgments:
We thank F. Heyd and A. Franz for helpful discussions on data and the manuscript. We also thank P. Chien, E. Strieter, and S. Garman for helpful discussions, and J. Kuriyan for helpful comments on the manuscript. We thank G. Hura and K. Burnett for SAXS data collection and help with analysis. SAXS data were collected at the Advanced Light Source (ALS), SIBYLS beamline on behalf of US DOE-BER, through the Integrated Diffraction Analysis Technologies (IDAT) program. Additional support comes from the NIGMS project ALS-ENABLE (P30 GM124169) and a High-End Instrumentation Grant S10OD018483. Cryo-EM data were collected at the Harvard Cryo-EM Center for Structural Biology. We are grateful for support from S. Sterling, R. Walsh, S. Rawson, and Z. Li for cryo-EM data collection. We thank A. Nairn for providing the calmodulin expression plasmid.
Funding: Funding was provided by the University of Massachusetts, Amherst and a grant to M.M.S. from the National Institute of General Medical Sciences (R01GM123157). N.D. and A.T. were supported by National Research Service Award (T32GM008515) from the National Institutes of Health, as part of the UMass Chemistry-Biology Interface Training Program.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Cryo-EM maps and models were deposited, respectively, into EMDB under identifiers EMD-21535 (Class A) and EMD-21536 (Class B), and the PDB under identifiers 6W4O (Class A) and 6W4P (Class B). Illumina sequencing data for all hippocampal CaMKII transcripts are available on figshare (xxx). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Lin YC, and Redmond L (2009) Neuronal CaMKII acts as a structural kinase. Communicative & integrative biology 2, 40–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escoffier J, Lee HC, Yassine S, Zouari R, Martinez G, Karaouzene T, Coutton C, Kherraf ZE, Halouani L, Triki C, Nef S, Thierry-Mieg N, Savinov SN, Fissore R, Ray PF, and Arnoult C (2016) Homozygous mutation of PLCZ1 leads to defective human oocyte activation and infertility that is not rescued by the WW-binding protein PAWP. Hum Mol Genet 25, 878–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon SY, Jellerette T, Salicioni AM, Lee HC, Yoo MS, Coward K, Parrington J, Grow D, Cibelli JB, Visconti PE, Mager J, and Fissore RA (2008) Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. The Journal of clinical investigation 118, 3671–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backs J, Backs T, Neef S, Kreusser MM, Lehmann LH, Patrick DM, Grueter CE, Qi X, Richardson JA, Hill JA, Katus HA, Bassel-Duby R, Maier LS, and Olson EN (2009) The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A 106, 2342–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook SG, Bourke AM, O’Leary H, Zaegel V, Lasda E, Mize-Berge J, Quillinan N, Tucker CL, Coultrap SJ, Herson PS, and Bayer KU (2018) Analysis of the CaMKII alpha and beta splice-variant distribution among brain regions reveals isoform-specific differences in holoenzyme formation. Sci Rep-Uk 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier LS, and Bers DM (2002) Calcium, calmodulin, and calcium-calmodulin kinase II: heartbeat to heartbeat and beyond. Journal of molecular and cellular cardiology 34, 919–939 [DOI] [PubMed] [Google Scholar]
- 7.Tombes RM, Faison MO, and Turbeville JM (2003) Organization and evolution of multifunctional Ca(2+)/CaM-dependent protein kinase genes. Gene 322, 17–31 [DOI] [PubMed] [Google Scholar]
- 8.Lisman J, Yasuda R, and Raghavachari S (2012) Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci 13, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva AJ, Paylor R, Wehner JM, and Tonegawa S (1992) Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science 257, 206–211 [DOI] [PubMed] [Google Scholar]
- 10.Borgesius NZ, van Woerden GM, Buitendijk GH, Keijzer N, Jaarsma D, Hoogenraad CC, and Elgersma Y (2011) betaCaMKII plays a nonenzymatic role in hippocampal synaptic plasticity and learning by targeting alphaCaMKII to synapses. J Neurosci 31, 10141–10148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giese KP, Fedorov NB, Filipkowski RK, and Silva AJ (1998) Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science 279, 870–873 [DOI] [PubMed] [Google Scholar]
- 12.Hanson PI, and Schulman H (1992) Inhibitory autophosphorylation of multifunctional Ca2+/calmodulin-dependent protein kinase analyzed by site-directed mutagenesis. J Biol Chem 267, 17216–17224 [PubMed] [Google Scholar]
- 13.Rellos P, Pike AC, Niesen FH, Salah E, Lee WH, von Delft F, and Knapp S (2010) Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol 8, e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patton BL, Miller SG, and Kennedy MB (1990) Activation of type II calcium/calmodulin-dependent protein kinase by Ca2+/calmodulin is inhibited by autophosphorylation of threonine within the calmodulin-binding domain. J Biol Chem 265, 11204–11212 [PubMed] [Google Scholar]
- 15.Meyer T, Hanson PI, Stryer L, and Schulman H (1992) Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science 256, 1199–1202 [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharyya M, Stratton MM, Going CC, McSpadden ED, Huang Y, Susa AC, Elleman A, Cao YM, Pappireddi N, Burkhardt P, Gee CL, Barros T, Schulman H, Williams ER, and Kuriyan J (2016) Molecular mechanism of activation-triggered subunit exchange in Ca/calmodulin-dependent protein kinase II. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg OS, Deindl S, Comolli LR, Hoelz A, Downing KH, Nairn AC, and Kuriyan J (2006) Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J 273, 682–694 [DOI] [PubMed] [Google Scholar]
- 18.Myers JB, Zaegel V, Coultrap SJ, Miller AP, Bayer KU, and Reichow SL (2017) The CaMKII holoenzyme structure in activation-competent conformations. Nat Commun 8, 15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao LH, Stratton MM, Lee IH, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H, and Kuriyan J (2011) A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin-dependent kinase II holoenzyme. Cell 146, 732–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, Zhang G, Neubert TA, and Tsien RW (2014) gammaCaMKII shuttles Ca(2)(+)/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell 159, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brocke L, Srinivasan M, and Schulman H (1995) Developmental and Regional Expression of Multifunctional Ca2+/Calmodulin-Dependent Protein-Kinase Isoforms in Rat-Brain. Journal of Neuroscience 15, 6797–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heist EK, Srinivasan M, and Schulman H (1998) Phosphorylation at the nuclear localization signal of Ca2+/calmodulin-dependent protein kinase II blocks its nuclear targeting. Journal of Biological Chemistry 273, 19763–19771 [DOI] [PubMed] [Google Scholar]
- 23.Kalderon D, Roberts BL, Richardson WD, and Smith AE (1984) A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 24.O’Leary H, Lasda E, and Bayer KU (2006) CaMKIIbeta association with the actin cytoskeleton is regulated by alternative splicing. Mol Biol Cell 17, 4656–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan S, Downing KH, and Molloy JE (2019) Architectural Dynamics of CaMKII-Actin Networks. Biophysical Journal 116, 104–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto K, Narayanan R, Lee SH, Murata K, and Hayashi Y (2007) The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A 104, 6418–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayer KU, Lohler J, and Harbers K (1996) An alternative, nonkinase product of the brain-specifically expressed Ca2+/calmodulin-dependent kinase II alpha isoform gene in skeletal muscle. Mol Cell Biol 16, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayer KU, Harbers K, and Schulman H (1998) alpha KAP is an anchoring protein for a novel CaM kinase II isoform in skeletal muscle. Embo Journal 17, 5598–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao LH, Pellicena P, Deindl S, Barclay LA, Schulman H, and Kuriyan J (2010) Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation. Nat Struct Mol Biol 17, 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Koninck P, and Schulman H (1998) Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science 279, 227–230 [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya M, Karandur D, and Kuriyan J (2019) Structural Insights into the Regulation of Ca(2+)/Calmodulin-Dependent Protein Kinase II (CaMKII). Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolodziej SJ, Hudmon A, Waxham MN, and Stoops JK (2000) Three-dimensional reconstructions of calcium/calmodulin-dependent (CaM) kinase IIalpha and truncated CaM kinase IIalpha reveal a unique organization for its structural core and functional domains. J Biol Chem 275, 14354–14359 [DOI] [PubMed] [Google Scholar]
- 33.Morris EP, and Torok K (2001) Oligomeric structure of alpha-calmodulin-dependent protein kinase II. J Mol Biol 308, 1–8 [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, and Kuriyan J (2005) Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell 123, 849–860 [DOI] [PubMed] [Google Scholar]
- 35.Myers JB, Zaegel V, Coultrap SJ, Miller AP, Bayer KU, and Reichow SL (2018) The CaMKII holoenzyme structure in activation-competent conformations (vol 8, 15742, 2017). Nat Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharyya M, Stratton MM, Going CC, McSpadden ED, Huang Y, Susa AC, Elleman A, Cao YM, Pappireddi N, Burkhardt P, Gee CL, Barros T, Schulman H, Williams ER, and Kurivan J (2016) Molecular mechanism of activation-triggered subunit exchange in Ca2+/calmodulin-dependent protein kinase II. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayer KU, De Koninck P, and Schulman H (2002) Alternative splicing modulates the frequency-dependent response of CaMKII to Ca(2+) oscillations. EMBO J 21, 3590–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuriyan J, and Eisenberg D (2007) The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 [DOI] [PubMed] [Google Scholar]
- 39.Kanizay LB, Jacobs TB, Gillespie K, Newsome JA, Spaid BN, and Parrott WA (2015) HtStuf: High-Throughput Sequencing to Locate Unknown DNA Junction Fragments. Plant Genome-Us 8. [DOI] [PubMed] [Google Scholar]
- 40.Stratton M, Lee IH, Bhattacharyya M, Christensen SM, Chao LH, Schulman H, Groves JT, and Kuriyan J (2013) Activation-triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. eLife 3, e01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putkey JA, and Waxham MN (1996) A peptide model for calmodulin trapping by calcium/calmodulin-dependent protein kinase II. J Biol Chem 271, 29619–29623 [DOI] [PubMed] [Google Scholar]
- 42.Harmat V, Bocskei Z, Naray-Szabo G, Bata I, Csutor AS, Hermecz I, Aranyi P, Szabo B, Liliom K, Vertessy BG, and Ovadi J (2000) A new potent calmodulin antagonist with arylalkylamine structure: crystallographic, spectroscopic and functional studies. J Mol Biol 297, 747–755 [DOI] [PubMed] [Google Scholar]
- 43.Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, and Knight WB (1995) Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry 34, 14843–14851 [DOI] [PubMed] [Google Scholar]
- 44.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL 2nd, Tsutakawa SE, Jenney FE Jr., Classen S, Frankel KA., Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, and Tainer JA (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nature methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology 152, 36–51 [DOI] [PubMed] [Google Scholar]
- 46.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng YF, and Agard DA (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A, and Grigorieff N (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology 192, 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, and Scheres SHW (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta Crystallogr D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Structural Biology 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gascuel O (1997). BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution 14, 685–95. [DOI] [PubMed] [Google Scholar]
- 53.Katoh K, Rozewicki J, and Yamada K (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20, 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sloutsky R, and Stratton MM (2020). European Journal of Neuroscience, doi: 10.1111/ejn.14761 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. PCR amplicons from multiple CaMKIIβ variants do not separate on an agarose gel.
Fig. S2. Data and EC50 fits for all tested CaMKII variants.
Fig. S3. SAXS data and analyses of CaMKIIα-0 and CaMKIIβ-0.
Fig. S4. Cryo-EM data.
Table S1. Detected CaMKIIα splice variants.
Table S2. Read counts of CaMKIIα splice variants from donor 1.
Table S3. Read counts of CaMKIIα splice variants from donor 2.
Table S4. Read counts of CaMKIIα splice variants from donor 3.
Table S5. Detected CaMKIIβ splice variants.
Table S6. Read counts of CaMKIIβ splice variants from donor 1.
Table S7. Read counts of CaMKIIβ splice variants from donor 2.
Table S8. Read counts of CaMKIIβ splice variants from donor 3.
Table S9. Detected CaMKIIγ splice variants.
Table S10. Read counts of CaMKIIγ splice variants from donor 1.
Table S11. Read counts of CaMKIIγ splice variants from donor 2.
Table S12. Read counts of CaMKIIγ splice variants from donor 3.
Table S13. Detected CaMKIIδ splice variants.
Table S14. Read counts of CaMKIIδ splice variants from donor 1.
Table S15. Read counts of CaMKIIδ splice variants from donor 2.
Table S16. Read counts of CaMKIIδ splice variants from donor 3.
Table S17. Pearson and Spearman correlation matrices for CaMKIIβ variants.
Table S18. Pearson and Spearman correlation matrices for CaMKIIγ variants.
Table S19. Pearson and Spearman correlation matrices for CaMKIIδ variants.
File S1. Exon sequences for all CaMKII genes.
File S2. Primers for initial amplification of variably spliced transcript regions.


