Abstract
Biomaterial scaffold designs are needed for self-organizing features related to tissue formation while also simplifying the fabrication processes involved. Towards this goal, we report silk protein-based self-folding scaffolds to support 3D cell culture, while providing directional guidance and promotion of cell growth and differentiation. A simple and robust one-step self-folding approach was developed using bilayers consisting of a hydrogel and silk film in aqueous solution. The 3D silk rolls, with patterns transferred from the initially prepared 2D films, guided the directional outgrowth of neurites, and also promoted the osteogenic differentiation of human mesenchymal stem cells (hMSCs). The osteogenic outcomes were further supported by enhanced biomechanical performance. By utilizing this self-folding method, co-cultures of neurons and hMSCs were achieved by patterning cells on silk films and then converting these materials into a 3D format with rolling, mimicking aspects of the structure of osteons and providing physiologically relevant structures to promote bone regeneration. These results demonstrate the utility of self-folded silk rolls as efficient scaffold systems for tissue regeneration, while exploiting relatively simple 2D designs programmed to form more complex 3D structures.
Keywords: Self-folding, 3D cell culture, heterogeneous cell culture, neuron regeneration, osteogenesis
Graphical Abstract
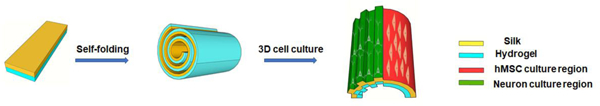
Biomaterial scaffold designs are needed to offer self-organizing features related to tissue formation while also simplifying the fabrication processes involved. Towards this goal, functional silk protein-based self-folding scaffolds are reported to support cell culture in 3D, while providing directional guidance and promotion of cell growth and differentiation.
Introduction
Tissues are complex systems that house cells which communicate in a heterogeneous environment. For successful tissue regeneration, bioengineered 3D scaffolds should provide features to promote cell growth and facilitate functional recovery, including: (1) adaption of the intrinsic 3D geometry, extracellular matrix (ECM) structure and chemistry of the target tissue, (2) biocompatibility, (3) adequate biomechanics to support tissue growth and function with a biodegradation rate commensurate with new tissue formation to transfer mechanical load, and (4) support for heterogenous cell growth. To address these needs, hydrogels have been widely used to support 3D cell growth [1]. With embedded cells inside hydrogels, these building blocks can be patterned to enable 3D tissues. Mixtures of culture medium and growth factors can be added to each building block, enabling heterogeneous cell cultures towards tissue-related outcomes. However, due to their relatively rapid degradation and poor mechanical properties with respect to orthopedic needs, hydrogel-based scaffolds often cannot support sustained cell growth and functional tissue formation. As an alternative, 3D-printed scaffolds have also emerged, including integrated polymers and inorganic systems, to provide structural control in 3D formats [2]. To achieve 3D cultures, these scaffolds are traditionally printed to mimic tissue geometry, followed by culturing a mixture of cells inside the printed scaffolds. In this approach, controlling the spatial distribution of each cell type is challenging. By mixing the cells with the printing precursors prior to deposition and cross-linking, the distribution of cells can be controlled within the scaffolds [3]. However, the cross-linking conditions to achieve such an outcome often limit the cell types and growth factors that can be used in the process[4]. More importantly, this technology continues to suffer from limits in spatial resolution, and thus nanoscale features in ECM systems cannot be duplicated in 3D printed scaffolds.
Self-folding is an effective approach to generate scaffolds enabling cell patterning in 3D formats [5]. Self-folding techniques rely on the difference in strains generated due to differences in swelling rates or extent of materials in aqueous solution, with and without external thermal stimulation [6]. For example, self-folded tubes were developed by using a modified gelatin gel as the hydrophilic layer and polycaprolactone as a hydrophobic portion, at room temperature. Multi-layered systems of p(N-isopropyl acrylamide- acrylic acid)/benzophenone acrylate/polyethylene glycol (PEG) self-folded at 33℃, close to physiological conditions [7]. Chemical surface functionalization with PEG was used to define the cell patterning. However, cells do not adhere to PEG-coated surfaces, which can limit cellular interactions and communication between the cells that are separated by the PEG-coated areas. Despite advances made to improve crosslinking with visible light and physical methods[8], cross-linking of synthetic polymers with UV light limits the cell types that can be used in the process. In addition, multiple steps of gelation, unstable matrices in culture media, and the release of crosslinker during degradation can be a concern when applying sensitive cell types and labile growth factors [9]. Thus, new systems and methods are sought to simplify or eliminate the crosslinking process to avoid the above challenges to develop bioengineered scaffolds for tissue engineering.
Multi-layered folding was previously achieved mainly using polydimethylsiloxane (PDMS). Semi-cured PDMS membranes with stretched PDMS membrane, through curing steps with curing agents or thermal stabilization, were used to fabricate rolling membranes [10]. Surface patterns were transferred onto the membranes using microstamping methods. Cells were cultured on the membranes to enable layered tubular structures to mimic blood vessels [11]. However, for these materials, it was still challenging to adjust the biomechanics and biocompatibility. Silk protein has been widely used in tissue engineering and regeneration [12] due to its biocompatibility, controllable biodegradability,[13] tunable biomechanical properties, [14], and low inflammatory effects [15]. Recently, spider silk and chitosan were coupled together to enable a self-folding tube, which showed potential as nerve guidance conduits, allowing the adherence and differentiation of neuronal cells inside the tubes, and the growth of neurites [16]. To further mimic the layered cylindrical feature of tissue structures, 3D multi-layered silk structures are needed. In addition, the design of new biomaterials with capabilities to adopt topological structures of the targeted tissue environment should significantly promote and guide cell growth [17].
The objective of the present work was to exploit self-folded silk protein rolls (SRs) as scaffolds for functional tissue regeneration. To enable the self-folding process, we used silk fibroin as the hydrophobic layer and coupled this with hydrogels as the hydrophilic layer. These rolled structures are unique in mimicking the layered cylindrical structures of tissues, in particular bone osteons. This self-folding approach converts 2D films into 3D structures, enabling additional features in the 3D structures critical for successful tissue regeneration. Topological patterns can be introduced to the 3D scaffolds through initial patterning of the 2D silk films with spatial features at the micron or nanoscales, which facilitates directional outgrowth of neurons [18]. In this work, micron-scale stripe patterns, previously found to be effective topographical cues to promote cell growth and provide directional guidance [18], were fabricated onto the silk films to obtain patterned silk rolls (pSRs). Heterogeneous cell co-cultures with different cell types defined at specific locations in 3D structures can also be achieved through initial patterning the 2D silk film with cells using microfluidics, followed by self-folding. SRs were evaluated for utility in neuronal and bone regeneration. Embryonic cortical neurons were cultured with the SRs and pSRs and exhibited directional growth by the pSRs. Organotypic spinal cord tissue slices, used as an ex vivo model for spinal cord injuries, were further cultured on both SRs and pSRs. With the directional guidance provided by pSRs, connections were formed between pairs of spinal cord tissue slices, placed 2–3 mm away, indicating a potential for spinal cord-related biomaterial needs. Osteogenic differentiation of human mesenchymal stem cells (hMSCs) was used as the model system for bone, where differentiation and enhanced calcium deposition were supported on the SRs and pSRs. Importantly, by integrating microfluidic-based cell patterning and self-folding, 3D heterogeneous cell cultures with neurons and hMSCs were demonstrated, with each cell type at the desired location in 3D, to mimic aspects of the osteon structure in the bone unit. Collectively, the self-folding 3D silk rolls provided linear guidance and promotion for cell growth through the surface patterning, offered new triggers for differentiation, and enabled 3D patterning of heterogeneous cell cultures through a self-folding approach.
Results and Discussion
One-step self-folding of agarose/silk bilayer films.
To achieve controlled self-folding, we designed silk/agarose bilayer systems. The swelling (the linear expansion) of the agarose layer in aqueous solution was significantly larger than the silk layer, which drives the self-folding process (Figure 1a) [19]. To illustrate the significance of the bilayer design, we compared the transformation of the agarose film only, silk film only and bilayer films (Supporting Figure S1). The agarose gel did not undergo folding when immersed in aqueous solution, while the silk layer folded randomly or crumbed, attributed to hydrophobic interactions to reduce the surface tension [19]. In the bilayer system, the hydrophobic silk layer bent and folded inside, while the hydrophilic agarose layer expanded and regulated the folding. Therefore, the silk concentration, the agarose concentration, and the agarose-to-silk film thickness ratios were parameters that can be exploited to control the self-folding process.
Figure 1. Self-folding strategy to control folding direction and number of layers.
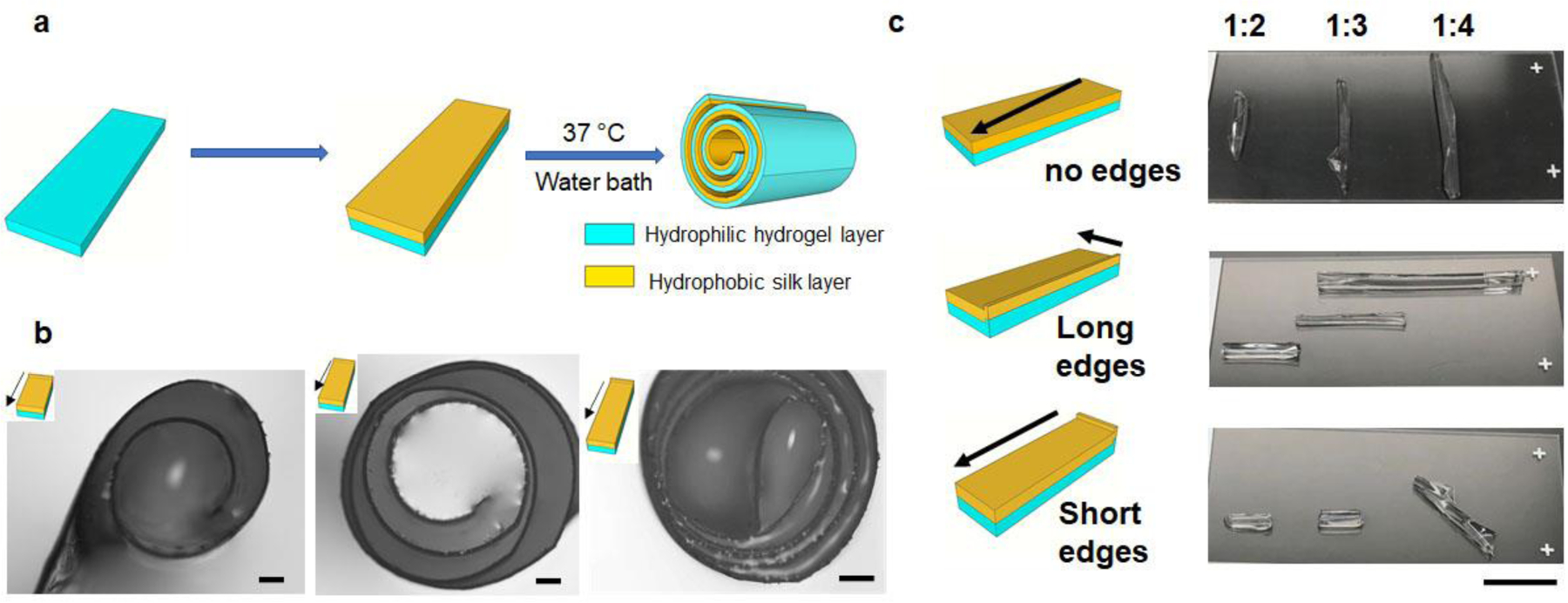
(a) Schematic of the self-folding process. (b) Layer number control by tuning the aspect ratio of the bilayer film with 1:1, 1:2 and 1:3, respectively. Scale bar: 200 μm. (c) Folding direction control by edge effects. Scale bar: 1 cm.
We initially fixed the thickness of the agarose film by applying 100 μL 1% w/v agarose solution onto 18 ×18 mm coverslips. We then varied the thickness of the silk layer by adjusting the volume of a 1% w/v silk solution from 50 to 400 μL (Supporting Figure S2a). No self-folding was achieved under these conditions (Supporting Figure S2b). We then increased the concentration of the silk solution to 2% w/v and 4% w/v, respectively. The bilayer films with a silk concentration of 2% started to fold into a roll with agarose-to-silk volume ratios of 1:3 and 1:4 (Supporting Figure S2c and d). As shown in the representative cross-section images, the roll formed by the ratio of 1:3 reached a 5-layer fold with an inner layer diameter of 0.59 ± 0.05 mm and outer layer diameter of 1.02 ± 0.01 mm (Supporting Figure S2e, N=3). For the 1:4 ratio group, a 4-layer folding was formed with an inner layer diameter of 0.77 ± 0.03 mm and outer layer diameter of 0.88 ± 0.01 mm (Supporting Figure S2f, N=3). At room temperature, the rolls remained folded over 6 months while in the PBS solution (Supporting Figure S3). Since the silk roll was designed as a bioengineered scaffold for interfacing with cellular systems, the more folding layers with the smallest inner layer diameter was preferred as this offered larger contacting area for cells. Therefore, we identified optimal conditions for folding ia 100 μL of 1% agarose solution and 300 μL of 2% silk solution on coverslips of 18 × 18 mm. These conditions resulted in a thickness of the dry bilayer film of 3.15 ± 0.03 μm, measured on cross-sections by scanning electron microscopy (Supporting Figure S4a), and corresponding to a wet film thickness of 30.36 ± 1.93 μm, measured by optical microscopy. In the drop casting process, the agarose gel was dried prior to casting the silk solution on top. The diffusion of silk solution into the agarose film formed a physical interlayer of mixed agarose/silk. This interlayer binds the two layers together without the need of crosslinkers. Cross-section SEM images (Supporting Figure S4b) demonstrated the binding between the bilayer films. The concentration of agarose solution was also tuned to evaluate the optimal folding conditions. The concentration of agarose solution was increased to 2%, 3% and 4% and fixed the concentration of silk at 2%. The volume used was 100 µL for the agarose solution and 300 µL for the silk solution, respectively. As shown in Supporting Figure S5, only single layer folding was achieved for 2% agarose, while for 3% agarose solution, the films formed half folds with a single layer. For the 4% solution, the roll failed to fold. This data suggests that the stiffness of the agarose film layer was a key parameter. When increasing the concentration of agarose solution, the film thickness increases but the stiffness also increases. Therefore, the self-folding process can only be achieved within a specific ratio of the two components in the bilayer films.Notably, the choice of the hydrophilic layer in the bilayer design was not limited to agarose, as gelatin gels (10% w/v) also produced similar self-folding silk rolls (Supporting Figure S6).
Second, the ratio between the width and length, i.e. the aspect ratio of the films, was also important for self-folding. The width of the bilayer films was maintained at 6 mm, and the length of the films was changed to tune the aspect ratios to 1:1, 1:2 and 1:3, respectively. The number of folding layers is defined as the number of bilayer films found along the radius of the silk rolls. As shown in Figure 1b, 4-layer folding was achieved on the film with a 1:1 aspect ratio, resulting in a roll with a total diameter of 0.6–0.8 mm. More folding layers were obtained by increasing the aspect ratio: 6 layers from a 1:2 aspect ratio and 8 layers from a 1:3 aspect ratio, with diameters that increased to 1.0–1.5 mm. Long-side rolling dominated for films with high aspect ratios, while diagonal and all-side rolling occurred for films with low aspect ratios. These findings were consistent with previous reports [6h]. Collectively, the diameter and number of layers formed in the silk rolls can be designed and controlled by tuning the aspect ratio of the bilayer films.
Additionally, the edges had a slightly higher thickness compared to the film center, formed during drying, due to surface tension. We hypothesized that this thick edge, in addition to the aspect ratio, could also define self-folding. To test the effect of these edges on self-folding, three groups of films were prepared for 3 given aspect ratios: no edge, long edge (edges along the long sides) and short edge (edges along the short sides). A representative image of each condition is displayed in Figure 1c. Each condition was repeated to confirm the reproducibility of the self-folding process (Supporting Figure S7). In the group with no edges, folding along the diagonal direction was preferred for all aspect ratios. In the long edge group, the presence of the edge regulated the initial folding direction, which was along the short side, as indicated by the arrow in the schematic, and regardless of the aspect ratio of the bilayer films. For the short edge, for aspect ratios greater than 1:3, the folding direction was dominated by the edge effect. However, by decreasing the aspect ratio further to 1:4, only the portion of the film that was close to the edge folded along the direction of the arrow, while the rest of the film folded along the diagonal direction [6h]. The folding direction was therefore determined by competitive effects attributed to the aspect ratios of the film and the edge effects. Specifically, the edge determines the initial folding direction, and a diagonal folding is more favored in the no edge area. Therefore, the self-folding can be controlled by designing film thickness, bilayer film aspect ratio, and thick edges.
To understand the self-folding process, we have further measured the expansion coefficient of the films. For 1% agarose films, the linear expansion coefficient was 0.43 ± 0.03 (N=5). For silk films, the expansion coefficients were: 0.14 ± 0.07 for 1% silk, 0.13 ± 0.03 for 2% silk and 0.09 ± 0.03 for 4% silk, respectively (N=5). The expansion coefficient of the bilayer film depends on the silk concentration and the ratio between agarose and silk in the bilayer films. As shown in Supporting Figure S8, the expansion coefficients of the bilayer films with high volume ratio between agarose and silk and low silk solution concentration were in the range of 0.33–0.41, closer to the agarose films. No folding was achieved in these films. When decreasing volume ratio between agarose and silk and increasing the silk concentration, the expansion coefficient dropped to 0.13–0.30, and the films self-folded.
To quantify the self-folding process, the measured radius of curvature of the rolled bilayer was compared to the theoretical values calculated from Timoshenko’s model [20].
Here ta and ts are the thickness of agarose and silk layers, respectively. Ea and Es are elastic modulus of agarose and silk layers. and are the linear expansion coefficients of agarose and silk layers where . l and represent the lengths of each layer at the wet state and dried state. The ratios m and n are defined as , . The thickness t was measured from the dried films by SEM imaging. Elastic modulus (E) was taken from the literature [21]. The expansion coefficient () was determined by measuring the length of individual layer from dried state to swollen state using optical microscopy. Values of these parameters used for the calculation are summarized in Supporting Table S1. Supporting Figure S9 shows the plot of the radius of the rolled bilayer as a function of the bilayer thickness (ta+ts). The plot indicated that radius measured from our fabricated silk rolls using optical microscopy fit well with the Timoshenko’s model.
Fabrication of patterned silk rolls.
Patterned topographical cues can modulate cell growth[22]. A micron-stripe pattern guides the directional growth of axons and can promote axonal growth, as we have previously reported [18]. This modulated growth facilitated the functional connection of injured neurons, using embryonic spinal cord tissue slices used as an injury model. To make the pSRs, we first fabricated micron-stripe patterns on silicon wafers as a mold. The patterns were a set of 20 μm stripes with 20 μm spacing, as previously shown to guide and promote axonal growth [18]. Deep reactive ion etching (DRIE) was used to control the depth of the patterns on the silicon substrate, to ensure that the patterns could be transferred onto the bilayer films [23]. As shown in Supporting Figure S10, the patterns on the silicon wafers were successfully fabricated with a depth of 31.63 ± 1.56 μm. By casting silk fibroin on the patterned silicon surface, as confirmed by SEM, pSRs with micron-stripe patterns were transferred onto both sides of the film (Figure 2a, b). As shown in Figure 2c and 2d, self-folding was achieved for the patterned silk films.
Figure 2. Pattern design on self-folding silk rolls.
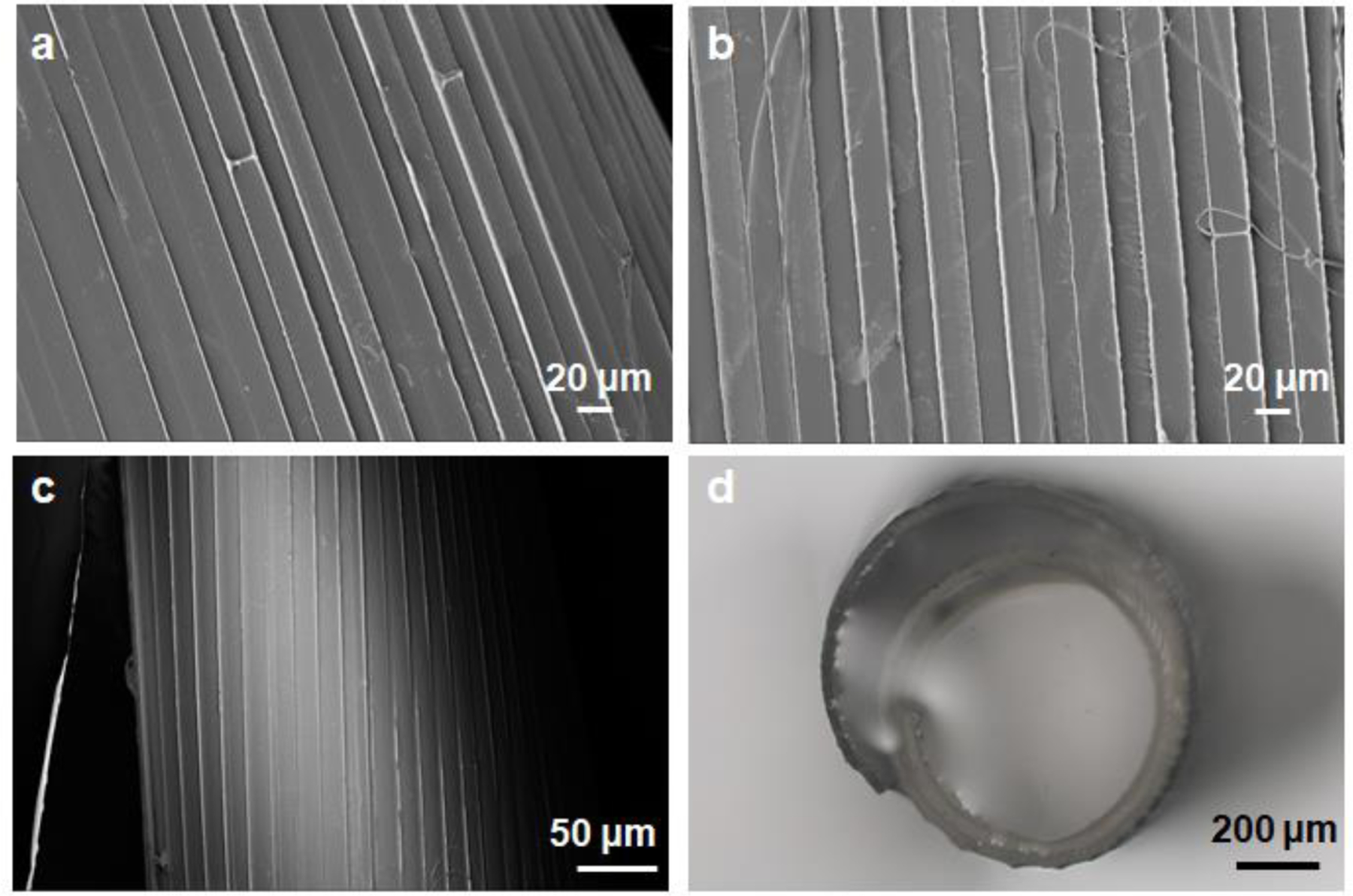
(a,b) SEM images of the patterned transferred onto agarose layer (a) and silk layer (b). (c) SEM images of the patterned silk rolls. (d) Optical images of patterned silk rolls.
The degradation of methanol-annealed silk films like these used in this study has been previously reported [24]. No weight change was observed in methanol-annealed silk films with 1% w/v after 2 weeks in PBS [25]. With higher concentration of silk, such as 2% and 4% in this study, the silk films are expected to be more stable under these conditions [26]. Further, the biodegradation rates of silk films can be further tuned if needed depending on the desired tissue engineering application [13b, 27]. To characterize the silk rolls, we also studied the biomechanics of the rolls (Supporting Figure S11). The compressive modulus was 5.86 ± 0.36 MPa for SRs and 4.65 ± 1.63 MPa for pSRs, respectively, considerably higher than that for the hydrogel systems. This data indicates that SRs and pSRs can serve as scaffolds to support cell growth.
Patterned silk rolls facilitated directional growth of axons.
First, we showed that SRs as 3D scaffolds were compatible with neuron cultures. Rat E14–16 embryonic cortical neurons were cultured on the SRs. To increase neuron adhesion, the SRs were precoated with poly-d-lysine (PDL). Cell seeding was achieved through the capillary effect of the SRs (Supporting Figure S12). The neurons were cultured on the SRs and pSRs for 7 days and then fixed for imaging. To visualize the neurons and axons, anti-Tau immunofluorescent (IF) staining was used and imaged using confocal microscopy. As shown in the 3D-reconstructed fluorescence images (Figure 3a), neurons attached and grew within the SRs, with stained cells detected on all layers with a uniform distribution.
Figure 3. Silk rolls for neuron directional growth.
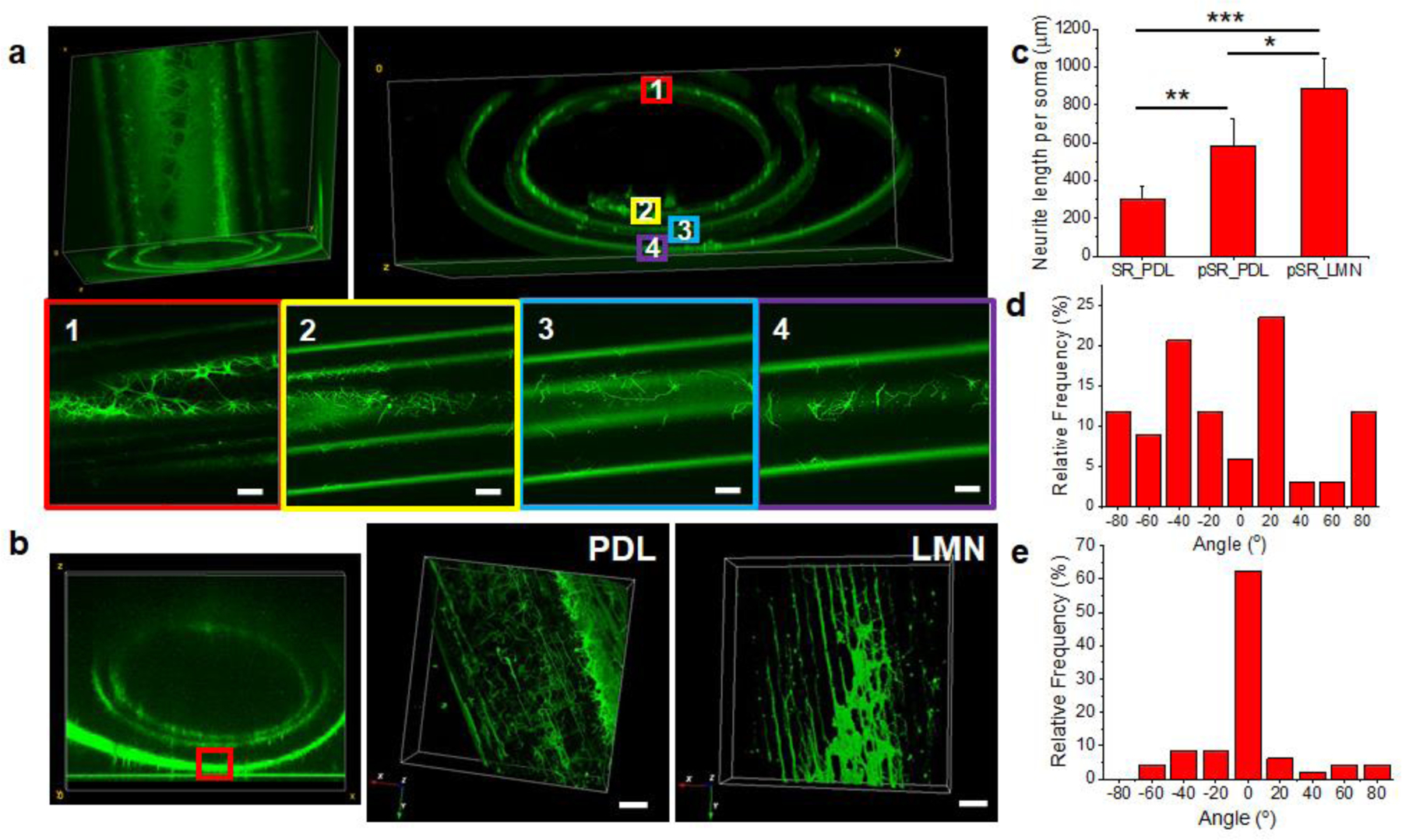
(a) Reconstructed confocal images of neurons cultured on silk rolls within each layer. (b) Reconstructed confocal images (left) of neurons cultured on patterned silk rolls within PDL (middle) and LMN coating (right). Scale bars: 20 μm. (c) Neurite length measurement (N=36 for SR PDL coated group, N=55 for pSR PDL coated group and N=60 for pSR LMN coated group, respectively ). (PDL: poly-d-lysine, LMN: laminin; SR: silk roll; pSR: patterned silk roll. ) (d,e) Neurite angular analysis on silk roll (d) and patterned silk roll (e). (n.s.: non-significant, p > 0.5; *: p < 0.5; **: p < 0.01; ***: p < 0.001)
To further validate whether the patterns on the pSRs promoted directional growth of neurites, embryonic neurons were seeded into the pSRs and IF staining was used to visualize cell growth patterns. In addition, the pSRs were coated with PDL and laminin (LMN), respectively, to test the effect of surface coatings on cell growth. Consistent with our previous findings on 2D patterned glass or silicon surfaces, a directional growth of axons was found on the 3D pSRs (Figure 3b). To quantify the growth, morphometric and statistical analysis of the confocal images was were used. As shown in Figure 3c, the SRs coated with PDL had an average neurite length of 301.13 ± 66.18 μm (N=36), while the pSRs showed an average neurite length of 580.86 ± 147.51 μm (N=55) for the PDL coating and 882.77 ± 158.04 μm (N=60) for the LMN coating. The patterns on the pSRs, along with the PDL/LMN surface coating, displayed a synergistic effect in facilitating neurite outgrowth. An analysis of the directionality of neurite growth was also performed (Figure 3d, e). The direction of the micron-stripe parallel patterns was defined as 0° and the angle of the neurites with respect to the pattern direction was measured. As shown in Figure 3d, the SRs without surface patterns did not provide directional guidance to the neurons, as the angular distribution was uniform in all directions from −90° to 90°. In comparison, in the pSRs coated with LMN, over 85% of neurites were distributed between −30° to 30° of the pattern direction, indicating a preferred growth direction defined by the micron-stripe patterns on pSRs. These results demonstrated that the patterned 3D silk rolls successfully facilitated the directional outgrowth of neurites.
To further highlight the significance of the patterned silk rolls in promoting neuronal growth, rat E14–16 embryonic spinal cord tissue slices were cultured with SRs and pSRs. The spinal cord tissue slices are widely used as an ex-vivo spinal cord injury model to study neuronal regeneration [28]. To stabilize the tissue slices in the silk rolls, on a silk roll two grooves were prepared with a width of 400 – 500 μm, depth of 500 – 700 μm and 2–3 mm apart on a silk roll. A 350 μm-thick spinal cord tissue slice was placed in each groove in the SRs and pSRs. The experiments were reaped for 3 times and the representative images were shown for SRs (Figure 4a) and pSRs (Figure 4e), respectively. After 14 days in culture, the slices were stained with anti-SMI31 antibody and imaged by confocal microscopy. In SRs, the growth of the tissue slices resulted in interconnected neurite meshes around the initially seeded tissues, but no connections were formed between the two slices (Figure 4 b–d). In pSRs, guided by the patterns, the regenerated neural filaments aligned in the direction of the patterns, and connections were achieved between the two tissue slices (Figure 4f and g). This connection is critical for enabling the propagation of electrical signals, as previously demonstrated [18]. More importantly, as shown in the cross-sectional images along the axial direction of the rolls (Figure 4h), the connections were established on multiple layers within the pSRs. These results indicated that the rolls promote tissue regeneration and reconnection in a 3D environment, with relevance for repairing injured nerves, like in spinal cord injuries.
Figure 4. Spinal cord tissue slices cultured in the silk rolls.
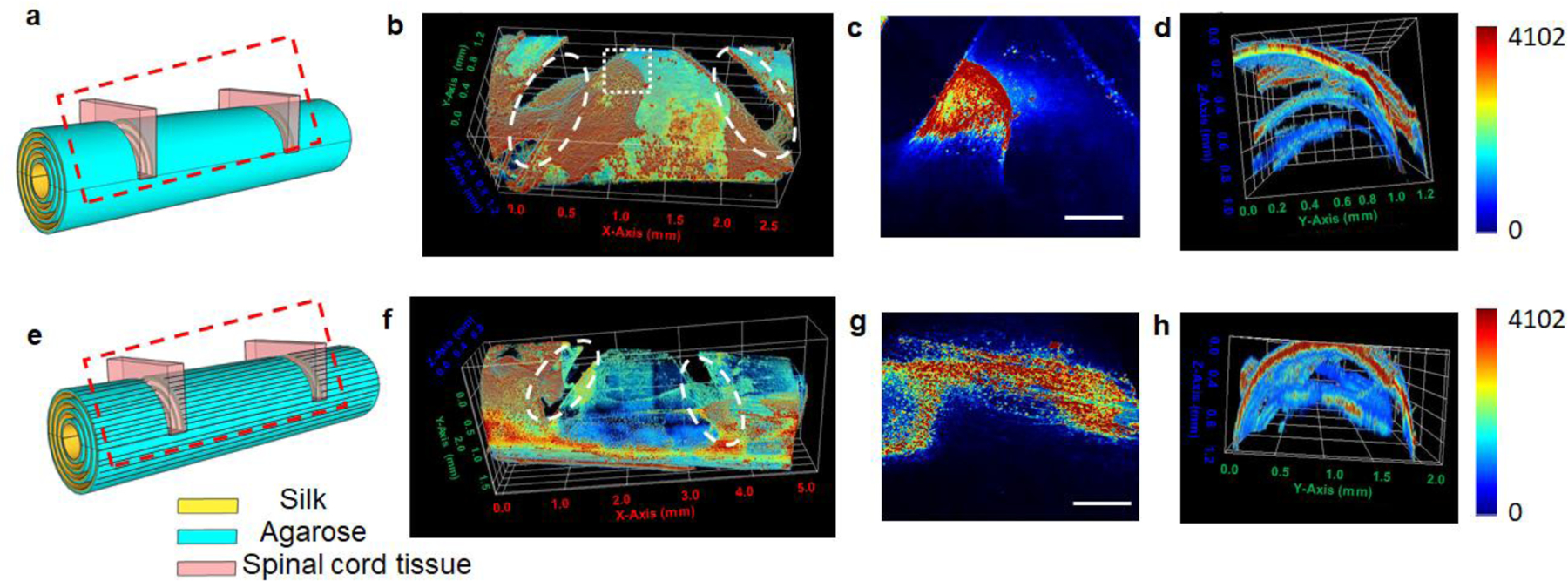
(a,e) Schematic of culturing spinal cord tissue slices in SRs (a) and pSRs (e) (N=3). Red dash box: region imaged by confocal microscopy. (b-d) Reconstructed confocal images of spinal cord tissues cultured on SRs with a top view (b), zoomed-in view at the top layer (c, indicated by the white dash box) and cross-section view (d). White dash circles indicated the location of the spinal cord tissue slices placed in SRs and pSRs. (f-h) Reconstructed confocal images of spinal cord tissues cultured on pSRs with a top view (f), zoomed-in view (g, indicated by the white dash box) and cross-section view (h). Scale bars: 500 μm.
Osteogenic differentiation of hMSCs on the silk rolls.
The growth and osteoblastic differentiation of human mesenchymal stem cells (hMSCs) were assessed on the 3D silk structures to validate their utility as scaffolds for bone regeneration. Mesenchymal stem cells for bone regeneration are well-described in the literature and crucial for neo-osteogenesis, the formation of new bone at the injured site[29]. From a materials perspective, scaffolds for bone repair should be biocompatible, biodegradable and osteoinductive/osteoconductive to promote cellular proliferation and osteogenic differentiation. hMSCs were cultured with the as-designed silk rolls to investigate whether the SRs promoted the osteogenic differentiation of hMSCs.
Seeding of hMSCs was achieved through the capillary effect of the SRs (Supporting Figure S12). SRs and pSRs were coated with 0.1% gelatin to promote cell attachment. After one week of culture, the hMSCs were fixed and stained with phalloidin for confocal imaging. The cells adhered, spread, and proliferated on all layers of the SRs (Figure 5a) and pSRs (Figure 5b). Unlike the neurons, no directional growth or morphological differences of hMSCs were observed on the SRs versus the pSRs scaffolds, indicating that the growth pattern of hMSCs was not sensitive to the surface topography provided by the pSRs.
Figure 5. Silk rolls as osteoconductive substrates for hMSCs.
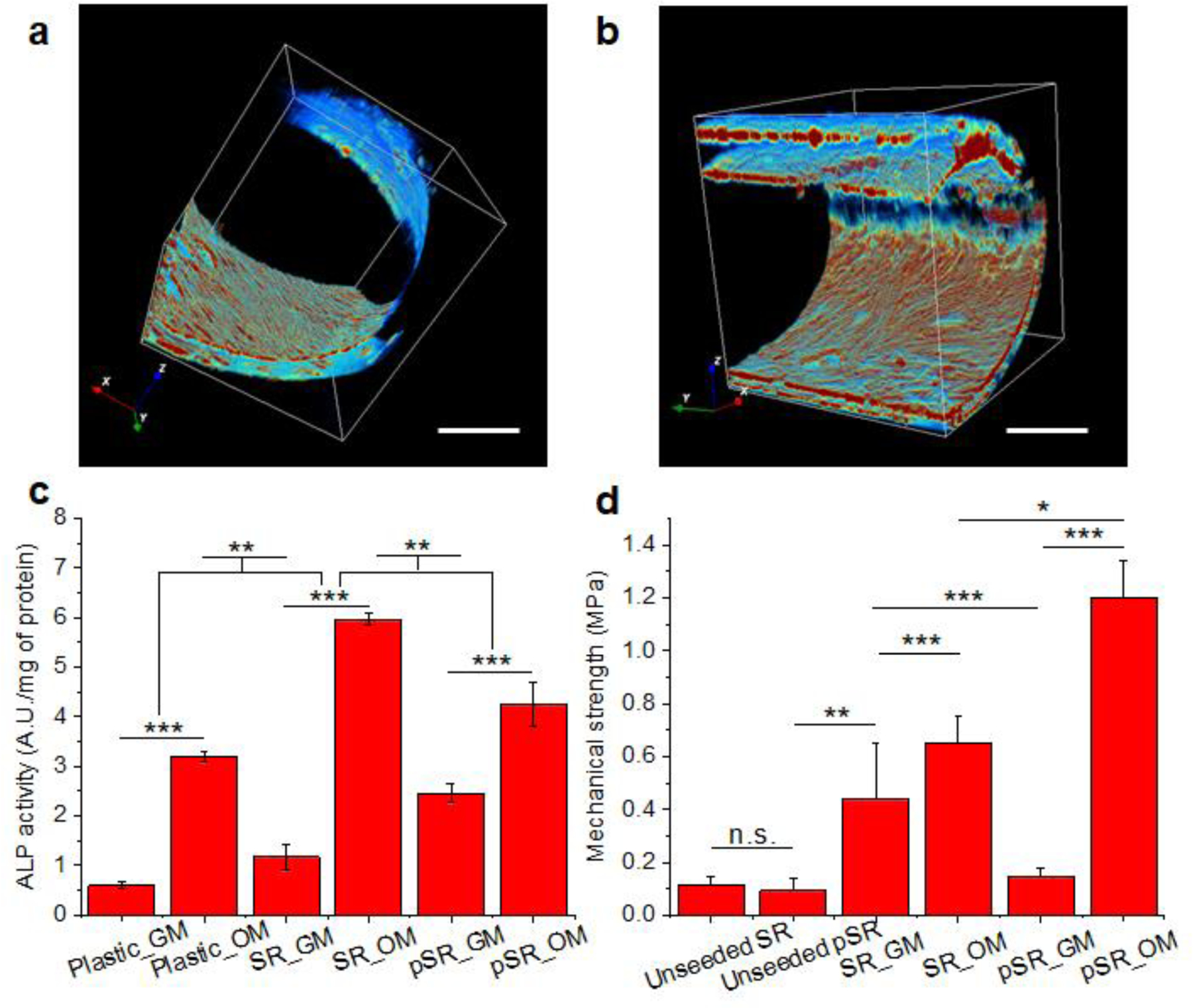
(a,b) 3D reconstructed confocal images of hMSCs cultured with SRs (a) and pSRs (b). Scale bars: 500 μm. (c) ALP activity. (d) Biomechanical measurements of silk rolls. (SR: silk rolls, pSR: patterned silk rolls; GM: growth medium; OM: osteogenic medium. N=5 samples for each group. n.s.: non-significant, p > 0.5; *: p < 0.5; **: p < 0.01; ***: p < 0.001)
To quantify the osteogenic differentiation of the hMSCs on the silk scaffolds, alkaline phosphatase (ALP) activity was measured in the hMSCs as an indicator of differentiation into osteoblasts[30]. Three groups were designed: the plastic culture dish (control group), SRs, and pSRs (N=5 for each group). The hMSCs were cultured in growth medium (GM) or osteogenic medium (OM), to investigate the osteoconductivity and osteoinductivity of the devices. ALP activity was normalized to total protein content, determined by the bicinchoninic acid assay. In Figure 5c, in the growth medium, the order of ALP enzymatic activity after 21 days of culture was: pSRs > SRs > plastic, demonstrating that the silk rolls had a positive impact on the differentiation process. In the osteogenic medium, all groups showed increased enzymatic activity compared to the growth medium, with SRs and pSRs showing higher activity than the plastic group. Interestingly, the positive effect of the patterning on ALP activity is less marked in osteogenic medium versus growth medium. Nevertheless, the results revealed a synergistic effect between the osteogenic medium and the silk rolls to promote osteogenesis. Osteogenic differentiation of the hMSCs was confirmed by assessing calcium deposition using Von Kossa staining. As shown in Supporting Figure S13, the SR and pSR groups showed strong positive staining in osteogenic medium, indicating calcium deposition, while little to no calcium deposition could be observed in the growth medium. Nuclear fast red counterstaining confirmed the presence of cells in all groups.
To assess the quality of the tissue formed within the scaffolds, the biomechanical properties of the silk rolls were assessed before and after cell culture (Figure 5d). All groups were tested in a hydrated state using phosphate-buffered saline (PBS). For the empty rolls, the compressive strengths measured for SRs and pSRs were 0.11 ± 0.04, 0.10 ± 0.05 MPa, respectively, and not significantly different. After 35 days of culture with hMSCs, the compressive strength of the SRs increased to 0.44 ± 0.21 MPa in growth medium and 0.65 ± 0.11 MPa in osteogenic medium (N=5 for each group). These data support that SRs promoted the osteoblastic differentiation of hMSCs, mineral deposition, and increases in biomechanical properties. For pSRs, the compressive strength was 0.15 ± 0.03 MPa after 35 days of culture in growth medium, similar to the empty roll. When the samples were switched to osteogenic medium, the compressive strength increased to 1.20 ± 1.14 MPa. These results are consistent with the Von Kossa results, indicating increased calcium deposition in the SR and pSR groups, and suggest that SRs and pSRs are promising scaffolds for bone regeneration.
Patterned heterogeneous co-culture of neurons/hMSCs on silk rolls.
A distinct benefit of these self-folding 3D silk structures is that a heterogenous 3D cell culture with different cell types at specific locations can be achieved by patterning cells on the surface of the 2D silk film, followed by 2D-to-3D folding. As a proof-of-concept, a 3D co-culture of neurons and hMSCs was demonstrated, in which the final locations of these two cell types in 3D were pre-designed based on their locations on the 2D silk films. A silk bilayer film was prepared with part of its surface patterned with micron-stripes and the other part smooth. Compartments were then defined using polydimethylsiloxane (PDMS) on the surface of the as-prepared silk film to introduce region-specific cell cultures. The micron-striped patterned region was coated with PDL for neuron culture, while the smooth area was coated with 0.1% gelatin for hMSCs adhesion. After seeding neurons and hMSCs into the two regions, respectively, the whole system was incubated overnight to ensure cell attachment. Then, the PDMS compartment was removed, the entire silk film was released from the silicon wafer and immersed in the culture medium. (Figure 6a). Film self-folding was achieved during cell incubation at 37℃, as shown in the schematic in Figure 6b. The cells were kept in culture for 7 days. Neurons were stained with anti-Tau using IF staining, and hMSCs were stained with anti-CD73, to show cell localization. Only a portion of the silk rolls was imaged (indicated as the black box in Figure 6b), due to the limitations in the focal length of the confocal microscope.
Figure 6. Silk rolls as scaffolds for functional bone segments with nerve and osteogenesis.
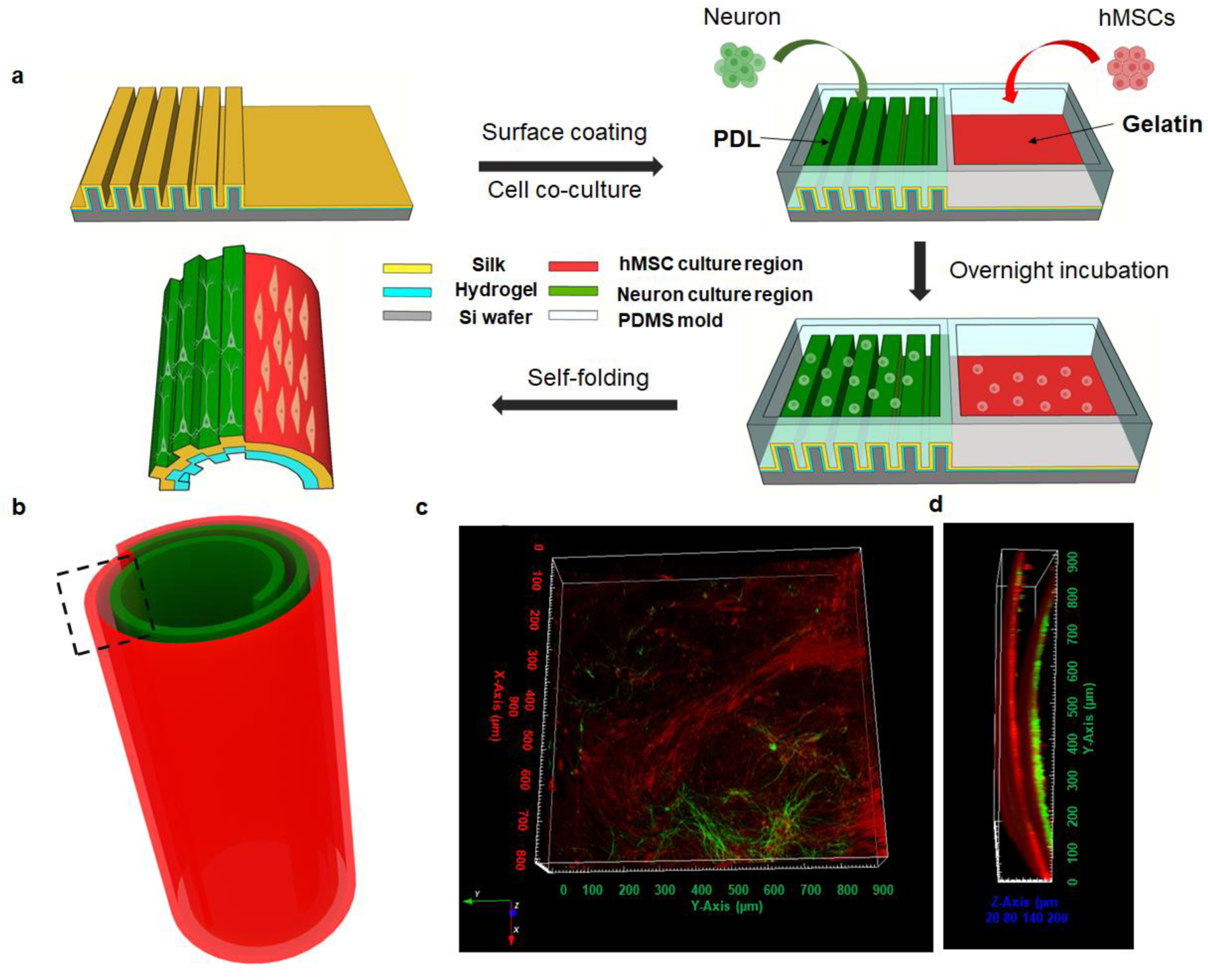
(a) Schematic of heterogeneous cell seeding and co-culture. (b) Schematic of heterogeneous cell culture in the self-folding silk roll. (c,d) Reconstructed confocal images of co-culture in the silk roll, indicated in the black dashed region. (c) top view and (d) cross-section view.
With spatial separation and location achieved by the PDMS compartments, heterogeneous cell cultures were achieved by simply coupling compartments onto the bilayer films before the self-folding process and seeding the cells. Additional topological structures on the surface of the silk films can be introduced independently. Neurons were cultured on the micron-stripe patterned area for directional growth, while hMSCs were grown on the smooth regions of the films. The experiments were repated for 5 times. As shown in the reprentative 3D reconstructed images from top and cross-sectional views along the axial direction of the rolls (Figure 6c,d), 3D cell patterning was achieved. Since the folding direction is controlled by the edge effects, the self-folding was initiated on the neuron side. Neurons were thus centered in the 3D rolls and surrounded by hMSCs, to mimic the structure of osteons. With the advantage of simple self-folding, this method is easy to manipulate with no need for further chemical modifications. Also, the process is compatible with cell growth conditions, which holds promise for expanding the approach to more cell types and more complicated designs, to mimic hierarchical tissue conditions.
Conclusions
Starting from agarose/silk bilayer films, 3D silk roll structures were achieved using a one-step folding method at 37℃. The self-folding can be designed and controlled by tuning film thickness, aspect ratio and edge effects, to produce multi-layer silk rolls with a controlled and predictable diameter and number of layers. Topological cues that modulate cell growth behavior, such as micron-stripe patterns, can be introduced into the silk roll to form patterned silk rolls. We demonstrated that these self-folded concentric multi-layer silk rolls can serve as 3D scaffolds for neuron cultures with directional axonal outgrowth on the pSRs. Spinal cord tissue slices cultured on the multi-layer pSRs exhibited connections through all layers between slices that were ~2 mm apart. Both of these results are critical for functional reconnection in injured neuronal systems. 3D cultured hMSCs on the SRs exhibited osteoblastic differentiation and mineral deposition. Heterogenous 3D cell cultures with neurons and hMSCs in the silk rolls were achieved with selective cell patterning, leveraging the features of SRs, including surface structure patterning of 2D films, 2D-to-3D conversion, and compatibility with microfluidics. Overall, these self-folded silk rolls are unique functional scaffolds able to promote directional growth, differentiation and regionalized heterogenous 3D culture outcomes, providing the potential for tissue regeneration and engineering.
Experimental Section
Materials.
The photoresist S1318, SU8, and MF319 developer were purchased from Electronic Materials, MA. Poly-D-lysine, penicillin/streptomycin, bovine serum albumin (BSA), Triton X-100, OsO4, strychnine, and bicuculline were purchased from Sigma-Aldrich Chemical Co, MO. Glutamine-Dulbecco’s Modified Eagle Medium (DMEM), neurobasal medium, B27, N2, glutamine, anti-MAP2, anti-Tau, anti-chicken secondary antibody, and goat anti-mouse antibody were purchased from Thermo Fisher Scientific Inc., MA. Trypsin/ethylenediaminetetraacetic acid (EDTA) and phosphate-buffered saline (PBS) were purchased from VWR, PA. Fetal bovine serum (FBS) and horse serum (HS) were purchased from Atlanta Biologicals, GA. All other chemicals were purchased from Sigma-Aldrich without further purification.
Silk fibroin preparation.
The silk fibroin solution used for the silk roll fabrication was prepared using our published protocols[31]. Briefly, pieces of Bombyx mori cocoons were boiled in 0.02 M aqueous Na2CO3 for 30 minutes. The degummed silk was extensively rinsed in distilled water, dried overnight, and dissolved in 9.3 M LiBr at 60°C for 4 hours. The silk/LiBr solution was dialyzed against distilled water for 2 days with 10 changes of water. The solution was centrifuged at 9,000 rpm for 2 × 20 min. For the subsequent silk roll fabrication, the silk concentration was determined using an analytical balance by evaporating water from a solution of known weight and weighing the remaining solid.
Silk roll fabrication.
The bilayer silk films were prepared by coupling agarose layers and silk layers. A bilayer film was first fabricated on a cover glass by drop-casting agarose (1%) and silk solution (1 – 4%) sequentially and then immersed in an 80% methanol solution to ensure insolubility [32]. The bilayer film was cut to control the size and aspect ratio on the cover glass in the dry state. The self-folding was achieved by peeling the film off from the cover glass and immersing in water at 37°C.
Fabrication and characterization of patterned silk rolls.
All roll fabrication was carried out in the Optoelectronic Processing Facility at Boston University. Silicon wafers (P-type, <100>, Addison Engineering, Inc.) were cut into substrates in the size of 20 mm × 20 mm. Silicon substrates were cleaned through multiple steps of solvent rinse, in the sequence of toluene, acetone, isopropanol, and deionized water before the photolithography process. For photolithography, photoresist S1813 was spin-coated onto the cover glasses at a speed of 4,000 rpm/s for 45s to achieve a thickness around 1 μm. After the silicon substrates were soft baked at 98°C for 180 s, they were exposed to UV light using a mask aligner (MA6, Suss) with an exposure power of 10.0 mW for 8 s, followed with post-baking for 2 minutes at 118°C. The developing process was carried out by applying the MF319 developer. Then, micron-stripes were fabricated onto the silicon substrates via deep reactive ion etching (Instrument model number: STS-ASE DRIE). For the etching process, the etching step used SF6 as the reagent gas with flow rate at 450 sccm for 2s and a passivation step used C4F8 with a flow rate of 150 sccm for 1.5s. Chamber pressure was set to be 18 mTorr for etching step and 9 mT for the passivation step. The radio frequency power was set for 1.1 kW for etching step and 0.75 kW for the passivation step. A high-frequency mode of 13.56 MHz was used. A platen power was fixed for 40 W for etching step and 10 W for the passivation step and 300 cycles were used to reach an etching depth of ~ 35 μm. For SEM imaging (Zeiss Supra 55VP), the dried bilayer films and silk rolls were dried and sputtered with Pd/Au before SEM imaging.
Timoshenko’s model fitting.
To fit the Timoshenko’s model, we measured the following parameters: (1) film thickness from agarose and silk layers using SEM. Agarose films were dried on cover glass slips (18 mm X 18 mm) achieved using 1% agrose solution with a volume of 100 µL. Silk films were obtained via drying silk solution with a concentration of 2% and a volume of 200 – 400 µL on cover glass slips (18 mm X 18 mm). (2) Linear expansion was measured from agarose and silk films in the wet state post 80% methanol annealing. (3) The radius of the self-folded roll was measured from the cross-section images captured using optical microscopy (Nikon, NX90). The model was simulated using MATLAB.
Animals.
Embryonic day (E) 14−16 rats, obtained from female, pregnant Sprague−Dawley rats bred were used. All animal care was carried out in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80−23; revised 1996). The procedures used involved with animals were operated under the protocol #201800681 approved by Boston University Animal Care and Use Committee.
Embryonic neurons and spinal cord tissue slices culture.
All substrates used in the embryonic neuron cell cultures were immersed in 0.01% poly-D-lysine and laminin (Sigma-Aldrich, MO) overnight at 4°C and washed in PBS before culture. Primary spinal cord neurons were obtained from the Sprague−Dawley rat E15 embryos. The spinal cords were isolated and placed in the L15 medium. Meninges were removed and spinal cords were cut into small pieces, dissociated by incubation in 0.05% trypsin/ethylenediaminetetraacetic acid (EDTA, VWR, PA) 15 min at 37°C and triturated every 5 min. Dissociated cells were washed with and triturated in 10% heat-inactivated fetal bovine serum (FBS, Atlanta Biologicals, GA), 5% heat-inactivated horse serum (HS, Atlanta Biologicals, GA), 2 mM Glutamine-Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific Inc., MA), and cultured in cell culture dishes (100 mm diameter) for 30 min at 37°C to eliminate glial cells and fibroblasts [17]. The supernatant containing neurons was collected and seeded on poly-D-lysine coated cover glasses and incubated in a humidified atmosphere containing 5% CO2 at 37°C with 10% FBS + 5% HS + 2 mM glutamine DMEM. After 16 h, the medium was replaced with Neurobasal medium (Thermo Fisher Scientific Inc., MA) containing 2% B27 (Thermo Fisher Scientific Inc., MA), 1% N2 (Thermo Fisher Scientific Inc., MA), and 2 mM glutamine (Thermo Fisher Scientific Inc., MA).
For organotypic tissue culture, spinal cords or cortex from Rat E15 embryos were isolated and sliced into slices in the thickness of 250 μm with a tissue slicer. Slices were then placed on coverslips. Organotypic slices were cultured with 1 mL of medium containing 67% DMEM, 8% sterile water, 25% fetal bovine serum, and 25 ng/mL nerve growth factor; adjusted to 300 mOsm and pH 7.35; and incubated in a humidified atmosphere containing 5% CO2 at 37°C for 7 to 14 days before fixation, staining, and imaging.
hMSCs culture.
Human mesenchymal stem cells (hMSCs) were isolated from total bone marrow aspirate from a healthy, non-smoking male under the age of 25 (Lonza, USA). After isolation, the hMSCs were allowed to reach 80% confluence, after which they were trypsinized, suspended in FBS containing 10% DMSO and stored in liquid nitrogen. Prior to cell seeding, silk rolls were sterilized by being immersed in 70% ethanol and treated with UV under the fume hood for 1 hour. They were then coated with 0.1% gelatin overnight at 37℃ in an incubator and washed three times with PBS. hMSCs (passage number < 5) were seeded into the silk rolls using a cell suspension with a density of 100 × 103 cells/mL. The rolls were then transferred into a fresh well containing 2,500 cells/mL and left in growth medium (DMEM high glucose GLUTAMAX (Gibco), 10% Fetal Bovine Serum (Gibco), 1% antibiotic-antimycotic (Gibco), 1% Non-Essential Amino Acids (Gibco) and 5ng/mL basic fibroblast growth factor (Gibco)). After 2 days the growth medium was either replaced with fresh growth medium or with osteogenic medium and changed every 2–3 days for the duration of the study. Osteogenic medium consisted of DMEM high glucose GLUTAMAX (Gibco), 10% Fetal Bovine Serum (Gibco), 1% antibiotic-antimycotic (Gibco), 100 nM dexamethasone (Sigma-Aldrich), 10 mM sodium β-glycerophosphate (Sigma-Aldrich), 0.05 mM L-ascorbic acid and 5 ng/mL basic fibroblast growth factor (Gibco).
Neuron/hMSC coculture.
For the co-cultures, the half-patterned silk films were prepared by drop-casting silk fibroin onto a half-patterned silicon substrate. A PDMS pattern with two compartments was placed on the silk films on the silicon substrate to form a device for the co-cultures. The whole device was immersed in 70% ethanol under UV light in a fume hood for 1hr for sterilization. The silk surface was coated with 0.01% PDL/LMN for neuron culture and 0.1% gelatin for hMSC culture overnight at 37℃ in an incubator and washed three times with PBS. Neurons and hMSCs were seeded into each compartment with a cell density of 3,000 cells/cm2 and 5,000 cells/cm2, respectively, in coculture medium (Neuron basal + 1% N2 + 2% B27 + 1% Glutamax + 1% p/s + 1% non-essential amino acid + 1 ng/mL nerve growth factor) [33], in a humidified atmosphere containing 5% CO2 at 37 ℃ for 7 days before fixation, staining and imaging .
Immunofluorescence staining and confocal imaging.
To track neurite outgrowth, cells were fixed with 4% paraformaldehyde for 20 min. After 3 washes, cells were blocked in 5% bovine serum albumin (Sigma-Aldrich, MO) for 30 min and permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, MO). Then cells were incubated with mouse monoclonal anti-Tau (1:1000) antibody for 2 h, and then with goat anti-mouse antibody (1:1000, Thermo Fisher Scientific Inc., MA) for 1 h at room temperature. The IF images were taken using a confocal microscope (Olympus, FV1000).
Alkaline phosphatase (ALP) activity assay.
Early osteogenic differentiation of the hMSCs was evaluated by measuring the Alkaline Phosphatase (ALP) activity after 21 days of culture, using Alkaline Phosphatase Colorimetric Assay Kit (Abcam, ab83369). Briefly the silk rolls were washed in cold PBS, and the cells were treated with 0.2% Triton X 100 (X110, Sigma) for 10 minutes, followed by sonication. Samples were centrifuged at 4℃ for 15 minutes to remove insoluble materials, and the supernatant was collected and kept on ice. Cell lysates and the assay buffer solution (5 mM p-nitrophenylphosphate) were added to a 96-well plate, incubated for 1 hour, and the absorbance was read at 405 nm using a microplate reader. A standard curve was prepared from standards (0–20 μM) prepared with a pNPP solution. To detect ALP expression, nitro-blue tetrazolium/indolylphosphate (NBT/BCIP) (Thermo Scientific) staining was also performed. Before staining, the cells were washed, NBT/BCIP was added and the samples were incubated at 37°C in a humidified chamber containing 5% CO2. After 30 mins, the samples were washed with PBS and imaged using a BZ-X700 series microscope (Keyence, Itasca, IL).
Von-Kossa staining.
Late osteogenic differentiation of the hMSCs was assessed using Von Kossa staining, using a commercially available kit (Kit KTVKO, American MasterTech). After 5 weeks of culture in growth or osteogenic cell culture medium, the silk rolls were rinsed in PBS, fixed with 4% paraformaldehyde for 30 minutes, rinsed extensively with distilled water, and placed in a 5% silver nitrate solution and exposed to UV light for approximately 1 minute until a dark staining could be observed. The rolls were then rinsed extensively in distilled water, placed for 2–3 minutes in 5% sodium thiosulfate, and counterstained with a nuclear fast red stain for 5 minutes. The silk rolls were once again rinsed extensively with distilled water before being imaged using a BZ-X700 series microscope (Keyence, Itasca, IL).
Statistical analysis.
For neurite length and angular distribution analysis, at least N=36 neurons were used. Data analysis from ALP activities and mechanical analysis was carried out for at least N=5 samples. Data shown are mean ± SD. P values were determined as: n.s.: non-significant, p > 0.5; *: p < 0.5; **: p < 0.01; ***: p < 0.001. Fluorescence images were analyzed using Image J. The reconstructed confocal images were processed using Icy. Neurite length, neurite angular distribution, ALP activity and biomechanical measurements were analyzed and poltted using Origin. The Timoshenko’s model fitting was analyzed using MATLAB.
Supplementary Material
Acknowledgements
Y.H and V.F contributed equally to this work. This work was supported by grants from Boston University Nanotechnology Innovation Center Cross-Disciplinary fellowship for Yimin Huang, Boston University Clinical & Translational Science Institute Pilot Award (funded by NIH 1UL1TR001430) for Nan Zheng, and the NIH (P41EB027062 - DLK).
Reference
- [1].a) Griffin DR, Weaver WM, Scumpia PO, Di Carlo D, Segura T, Nature Materials 2015, 14, 737–744; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kato-Negishi M, Morimoto Y, Onoe H, Takeuchi S, Advanced Healthcare Materials 2013, 2, 1564–1570. [DOI] [PubMed] [Google Scholar]
- [2].a) Inzana JA, Olvera D, Fuller SM, Kelly JP, Graeve OA, Schwarz EM, Kates SL, Awad HA, Biomaterials 2014, 35, 4026–4034; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Murphy SV, Atala A, Nature Biotechnology 2014, 32, 773–785; [DOI] [PubMed] [Google Scholar]; c) Do AV, Khorsand B, Geary SM, Salem AK, Advanced Healthcare Materials 2015, 4, 1742–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Advanced Materials 2014, 26, 3124–3130; [DOI] [PubMed] [Google Scholar]; b) Colosi C, Costantini M, Barbetta A, Dentini M, Methods in Molecular Biology 2017, 1612, 369–380. [DOI] [PubMed] [Google Scholar]
- [4].Hanson Shepherd JN, Parker ST, Shepherd RF, Gillette MU, Lewis JA, Nuzzo RG, Advanced Functional Materials 2011, 21, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ionov L, Soft Matterials 2011, 7, 6786–6791. [Google Scholar]
- [6].a) Guan J, He H, Hansford DJ, Lee LJ, The Journal of Physical Chemistry B 2005, 109, 23134–23137; [DOI] [PubMed] [Google Scholar]; b) Deng T, Yoon C, Jin Q, Li M, Liu Z, Gracias DH, Applied Physics Letters 2015, 106;; c) Jeong K-U, Jang J-H, Kim D-Y, Nah C, Lee JH, Lee M-H, Sun H-J, Wang C-L, Cheng SZD, Thomas EL, Journal of Materials Chemistry 2011, 21, 6824–6830; [Google Scholar]; d) Jamal M, Bassik N, Cho JH, Randall CL, Gracias DH, Biomaterials 2010, 31, 1683–1690; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Malachowski K, Jamal M, Jin Q, Polat B, Morris CJ, Gracias DH, Nano Letters 2014, 14, 4164–4170; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Liu Y, Boyles JK, Genzer J, Dickey MD, Soft Matterials 2012, 8, 1764–1769; [Google Scholar]; g) Ionov L, Polymer Reviews 2013, 53, 92–107; [Google Scholar]; h) Stoychev G, Zakharchenko S, Turcaud S, Dunlop JW, Ionov L, Acs Nano 2012, 6, 3925–3934. [DOI] [PubMed] [Google Scholar]
- [7].Stroganov V, Pant J, Stoychev G, Janke A, Jehnichen D, Fery A, Handa H, Ionov L, Advanced Functional Materials 2018, 28. [Google Scholar]
- [8].a) Noshadi I, Hong S, Sullivan KE, Shirzaei Sani E, Portillo-Lara R, Tamayol A, Shin SR, Gao AE, Stoppel WL, Black LD III, Khademhosseini A, Annabi N, Biomater Sci 2017, 5, 2093–2105; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, Camci-Unal G, Dokmeci MR, Peppas NA, Khademhosseini A, Advanced Materials 2014, 26, 85–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Annabi N, Mithieux SM, Zorlutuna P, Camci-Unal G, Weiss AS, Khademhosseini A, Biomaterials 2013, 34, 5496–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cheng S, Jin Y, Wang N, Cao F, Zhang W, Bai W, Zheng W, Jiang X, Advanced Materials 2017, 29, 1700171. [DOI] [PubMed] [Google Scholar]
- [11].Gong P, Wenfu Z, Huang Z, Zhang W, Xiao D, Jiang X, Advanced Functional Materials 2013, 23.
- [12].a) Meinel L, Karageorgiou V, Hofmann S, Fajardo R, Snyder B, Li C, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL, Journal of Biomedical Materials Research Part A 2004, 71, 25–34; [DOI] [PubMed] [Google Scholar]; b) Wang Y, Kim HJ, Vunjak-Novakovic G, Kaplan DL, Biomaterials 2006, 27, 6064–6082; [DOI] [PubMed] [Google Scholar]; c) Hofmann S, Knecht S, Langer R, Kaplan DL, Vunjak-Novakovic G, Merkle HP, Meinel L, Tissue Engineering Part A 2006, 12, 2729–2738. [DOI] [PubMed] [Google Scholar]
- [13].a) Zhang L, Liu X, Li G, Wang P, Yang Y, Journal of Biomedical Materials Research Part A 2019, 107, 104–113; [DOI] [PubMed] [Google Scholar]; b) Lu Q, Zhang B, Li M, Zuo B, Kaplan DL, Huang Y, Zhu H, Biomacromolecules 2011, 12, 1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Bai S, Han H, Huang X, Xu W, Kaplan DL, Zhu H, Lu Q, Acta Biomaterialia 2015, 20, 22–31; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Maziz A, Leprette O, Boyer L, Blatché C, Bergaud C, Biomedical Physics & Engineering Express 2018, 4, 065012. [Google Scholar]
- [15].Farokhi M, Mottaghitalab F, Fatahi Y, Khademhosseini A, Kaplan DL, Trends in Biotechnology 2018, 36, 907–922. [DOI] [PubMed] [Google Scholar]
- [16].a) Aigner T, Scheibel T, Acs Applied Materials & Interfaces 2019, 11, 15290–15297; [DOI] [PubMed] [Google Scholar]; b) Aigner T, Haynl C, Salehi S, O’Connor A, Scheibel T, Mater Today Bio 2020, 5, 100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang Y, Jiang Y, Wu Q, Wu X, An X, Chubykin AA, Cheng J-X, Xu X-M, Yang C, Acs Biomaterials Science & Engineering 2018, 4, 1037–1045. [DOI] [PubMed] [Google Scholar]
- [18].Huang YM, Jiang Y, Wu QY, Wu XB, An XD, Chubykin AA, Cheng JX, Xu XM, Yang C, Acs Biomaterials Science & Engineering 2018, 4, 1037–1045. [DOI] [PubMed] [Google Scholar]
- [19].Zhang F, Fan J, Zhang P, Liu M, Meng J, Jiang L, Wang S, Npg Asia Materials 2017, 9, e380–e380. [Google Scholar]
- [20].a) Gomez LP, Bollgruen P, Egunov AI, Mager D, Malloggi F, Korvink JG, Luchnikov VA, Lab on a Chip 2013, 13, 3827–3831; [DOI] [PubMed] [Google Scholar]; b) Timoshenko S, Journal of the Optical Society of America 1925, 11, 233–255; [Google Scholar]; c) Kang D, Yu H, Kim J, International Journal of Polymer Science 2015, 2015, 758474. [Google Scholar]
- [21].a) Normand V, Lootens DL, Amici E, Plucknett KP, Aymard P, Biomacromolecules 2000, 1, 730–738; [DOI] [PubMed] [Google Scholar]; b) Lawrence BD, Wharram S, Kluge JA, Leisk GG, Omenetto FG, Rosenblatt MI, Kaplan DL, Macromol Biosci 2010, 10, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a) Loesberg WA, te Riet J, van Delft FC, Schon P, Figdor CG, Speller S, van Loon JJ, Walboomers XF, Jansen JA, Biomaterials 2007, 28, 3944–3951; [DOI] [PubMed] [Google Scholar]; b) Kang K, Park YS, Park M, Jang MJ, Kim SM, Lee J, Choi JY, Jung DH, Chang YT, Yoon MH, Lee JS, Nam Y, Choi IS, Nano Letters 2016, 16, 675–680; [DOI] [PubMed] [Google Scholar]; c) Pan F, Zhang M, Wu G, Lai Y, Greber B, Scholer HR, Chi L, Biomaterials 2013, 34, 8131–8139. [DOI] [PubMed] [Google Scholar]
- [23].Laermer F, Urban A, Microelectronic Engineering 2003, 67–68, 349–355.
- [24].Jin HJ, Park J, Karageorgiou V, Kim UJ, Valluzzi R, Cebe P, Kaplan DL, Advanced Functional Materials 2005, 15, 1241–1247. [Google Scholar]
- [25].a) Minoura N, Tsukada M, Nagura M, Biomaterials 1990, 11, 430–434; [DOI] [PubMed] [Google Scholar]; b) Shang K, Rnjak-Kovacina J, Lin Y, Hayden RS, Tao H, Kaplan DL, Translational Vision Science & Technology 2013, 2, 2–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li AB, Kluge JA, Guziewicz NA, Omenetto FG, Kaplan DL, Journal of controlled release : official journal of the Controlled Release Society 2015, 219, 416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lu Q, Hu X, Wang X, Kluge JA, Lu S, Cebe P, Kaplan DL, Acta Biomaterialia 2010, 6, 1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Usmani S, Aurand ER, Medelin M, Fabbro A, Scaini D, Laishram J, Rosselli FB, Ansuini A, Zoccolan D, Scarselli M, De Crescenzi M, Bosi S, Prato M, Ballerini L, Science Advance 2016, 2, e1600087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].a) Zheng C, Chen J, Liu S, Jin Y, International Journal of Oral Science 2019, 11, 23; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Iaquinta MR, Mazzoni E, Bononi I, Rotondo JC, Mazziotta C, Montesi M, Sprio S, Tampieri A, Tognon M, Martini F, Frontiers in Cell and Developmental Biology 2019, 7, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Birmingham E, Niebur GL, McHugh PE, et al. , European Cells & Materials Ltd, 2012. [DOI] [PubMed]
- [31].Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, Kaplan DL, Nature Protocols 2011, 6, 1612–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tsukada M, Gotoh Y, Nagura M, Minoura N, Kasai N, Freddi G, Journal of Polymer Science Part B: Polymer Physics 1994, 32, 961–968. [Google Scholar]
- [33].Sun H, Benardais K, Stanslowsky N, Thau-Habermann N, Hensel N, Huang D, Claus P, Dengler R, Stangel M, Petri S, Plos One 2013, 8, e72926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


