Abstract
Background
The prevalence of Staphylococcus aureus varies depending on the healthcare facility, region and country. To understand its genetic diversity, transmission, dissemination, epidemiology and evolution in a particular geographical location, it is important to understand the similarities and variations in the population being studied. This can be achieved by using various molecular characterisation techniques. This study aimed to provide detailed molecular characterisation of South African mecA-positive S. aureus blood culture isolates by describing the SCCmec types, spa types and to lesser extent, the sequence types obtained from two consecutive national surveillance studies.
Methods
S. aureus blood culture isolates from a national laboratory-based and enhanced surveillance programme were identified and antimicrobial susceptibility testing was performed using automated systems. A real-time PCR assay confirmed the presence of the methicillin-resistance determinant, mecA. Conventional PCR assays were used to identify the SCCmec type and spa type, which was subsequently analysed using the Ridom StaphType™ software. Multilocus sequence typing was performed on selected isolates using conventional methods. MRSA clones were defined by their sequence type (ST), SCCmec type and spa type.
Results
A detailed description of findings is reported in this manuscript. SCCmec type III predominated overall followed by type IV. A total of 71 different spa types and 24 novel spa types were observed. Spa type t037 was the most common and predominated throughout followed by t1257. Isolates were multidrug resistant; isolates belonging to all SCCmec types were resistant to most of the antibiotics with the exception of type I; isolates with spa type t045 showed resistance to all antibiotics except vancomycin. The most diverse SCCmec-spa type complex was composed of the SCCmec type IV element and 53 different spa types.
Conclusion
Although ST data was limited, thereby limiting the number of clones that could be identified, the circulating clones were relatively diverse.
Keywords: Methicillin-resistant Staphylococcus aureus, SCCmec typing, Spa typing
Introduction
Staphylococcus aureus bacteraemia is an important cause of morbidity and mortality in both healthcare-associated (HA) and community-associated (CA) infections worldwide [1, 2]. S. aureus is responsible for an extensive range of human diseases, including bloodstream infections, pneumonia, endocarditis, food poisoning, toxic shock syndrome, skin and soft tissue infections, and bone and joint infections [3, 4]. The prevalence of S. aureus varies depending on the healthcare facility, region and country. Furthermore, the prevalence of methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) may also differ. In order to understand the genetic diversity, transmission, dissemination, epidemiology and evolution of MSSA and MRSA clones in a particular geographical location, it is important to acquire knowledge on the similarities and variations in the population being studied. This is not only important for epidemiological surveys but also for infection prevention and control policies [5]. This can be achieved by employing the use of various molecular characterisation techniques [2]. Reliable molecular techniques that have been used for typing S. aureus include Pulsed-field Gel Electrophoresis (PFGE), Multilocus Sequence Typing (MLST), Stapylococcal protein A (spa) typing and Staphylococcal Cassette Chromosome mec (SCCmec) typing [2, 6].
PFGE is based on the DNA banding pattern obtained after digesting the bacterial genome with a restriction enzyme [7]. MLST and its clustering algorithm, Based Upon Related Sequence Type (BURST) classifies isolates according to nucleotide variations in seven housekeeping/reference genes (loci) [5]. These genes are sequenced and a unique allele number is assigned using an online programme specific to the MLST scheme. A combination of the allele numbers (i.e. allelic profile) produces a particular sequence type (ST) for a bacterial strain. Those with similar STs are grouped together in a single clonal complex (CC) [6, 8]. Spa typing sequences the S. aureus-specific staphylococcal protein A (spa) gene which is one of the virulence factors on the surface of the organism preventing phagocytosis by the immune system [9]. Spa typing and its clustering algorithm, Based Upon Repeat Pattern (BURP) is based on the sequencing of a polymorphic 24 bp region of the spa gene. This is a variable-number tandem repeat (VNTR) sequence within the 3′ coding region [4]. The repeat regions are assigned a numerical code and the spa type is determined by the order of specific repeats [3]. Studies have shown that spa typing produced results that are notably comparable with that of MLST [6, 10]. Due to lower implementation costs and that only a single locus needs to be sequenced, spa typing has shown to be more efficient and results are consistent across different settings, specimen type and patient age [6]. Therefore spa typing has been shown to be appropriate for use in evolutionary and macro-epidemiology studies [4, 6, 11, 12]. However, as recombination events in a single locus can distort clonal relationships, there is the question of how a method that sequences only a single locus can be used for macro-epidemiology studies [13]. SCCmec typing classifies SCCmec elements according to their structural differences [5]. It involves the typing of the staphylococcal cassette chromosome mec, which is a mobile genetic element and harbours the methicillin-resistance determinant gene. This element is genetically diverse with many types, subtypes and variants being reported [14]. The molecular organisation of the cassette is complex, but it can be broken down into three structural components, which include: i) the cassette chromosome recombinase (ccr) gene complex, ii) the mec gene complex and iii) the joining (J) regions [15, 16]. The ccr gene complex encodes site-specific recombinases for the excision and insertion of the element into the chromosome [14, 16, 17]. This complex therefore affords the SCCmec element mobility and thus facilitates its transfer to other staphylococcal species [16]. The mec complex confers methicillin resistance as it consists of the mec gene, its regulatory genes, the mecI and the mecR1 genes and various insertion sequences [14, 18]. A combination of both the ccr gene complex and the mec gene class is used to assign the specific SCCmec type. Thirteen SCCmec types (I-XIII) have been defined in MRSA based on complete sequence data [17, 19–21]; International Working Group on the Staphylococcal Cassette Chromosome elements (IWG-SCC) (2015) Available online: http://www.sccmec.org).
Although we have previously described the MRSA population in South Africa [22–25], a detailed description of the SCCmec types and spa types is lacking. This study therefore reports on the various clones present in our MRSA study population by SCCmec and spa type combinations (SCCmec-spa type complexes). Moreover, although MLST data was lacking for the majority of our sample population, the predominating circulating clones (ST-SCCmec-spa type) based on the most common spa types were described.
Materials and methods
Bacterial strains and phenotypic methods
A case of S. aureus bacteraemia was defined as the isolation of S. aureus from a blood culture. Blood culture isolates, which formed part of the GERMS-SA laboratory-based and enhanced antimicrobial resistance surveillance studies from sentinel centres in South Africa were submitted and participation was voluntary. The first was a two-year laboratory-based surveillance study (June 2010 to July 2012); sites represented 13 sentinel centres from the Gauteng, KwaZulu-Natal, Free State and Western Cape provinces. The second was an enhanced surveillance study (August 2012 to December 2017); sites represented five sentinel centres from six large academic hospitals from the Gauteng and the Western Cape provinces. A 21-day exclusion period was applied to avoid duplicate isolates of the organism from the same patient.
In total, 5820 viable isolates [MSSA (n = 3801) and MRSA (n = 2019)] were submitted on Dorset transport media (Diagnostic Media Products (DMP), National Health Laboratory Service (NHLS), Johannesburg, South Africa). Each isolate was plated onto a 5% blood agar plate (DMP, NHLS, Johannesburg, South Africa) followed by organism identification and antimicrobial susceptibility testing using automated systems. Organism identification was done using VITEK® II (bioMèrieux, France) or MALDI-TOF MS (Microflex, Bruker Daltonics, MA, USA) and antimicrobial susceptibility testing (AST) was done using the MicroScan Walkaway system (Gram-positive panel PM33) (Siemens, Sacramento, CA, USA). Interpretation of susceptibility was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [26]. Bacterial cells were lysed at 95 °C for 25 min and the DNA was extracted and used in the genotypic assays.
Polymerase chain reaction (PCR) screening for mecA in MRSA isolates
The LightCycler 480 II (Roche Applied Science) instrument was used for the real-time PCR of mecA and nuc, which were amplified in a multiplex assay using the LightCycler 480 Probes Master kit (Roche Diagnostics, IN, USA) with previously published primers and probes [27].
SCCmec typing
All 2019 mecA-positive MRSA isolates were typed by a multiplex PCR assay using the Qiagen Multiplex PCR kit (Qiagen, Germany) and previously published primers [28].
Spa-typing
Spa-typing was performed on 1467 MRSA isolates. The spa gene was amplified using previously published primers [12] and the Amplitaq Gold DNA Polymerase kit (Applied Biosystems, CA, USA). Purified PCR products (Qiagen Purification kit; Qiagen, Germany) were sequenced (Inqaba Biotech, South Africa). Sequences were assembled using CLC Bio main workbench (Qiagen, Germany) and analysed using the Ridom StaphType™ software, (Ridom GmbH, Würzburg, Germany).
Multilocus sequence typing (MLST)
Multilocus Sequence Typing was performed on 48 isolates, which were selected randomly based on the most common spa-types. Primers [29] amplifying seven reference genes were used. Amplification was done using the Amplitaq Gold DNA Polymerase kit (Applied Biosystems, CA, USA). Purified PCR products were sequenced (Inqaba Biotech, South Africa). Sequences were assembled using the CLC Bio main workbench (Qiagen, Germany) and analysed using the online database (https://pubmlst.org/saureus/).
Results
SCCmec typing
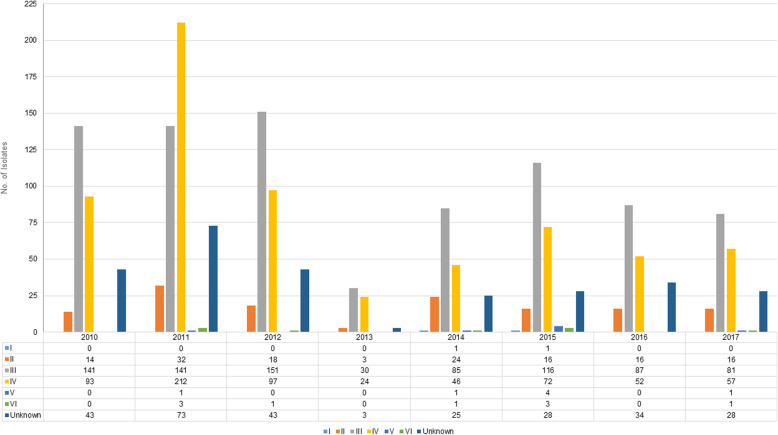
The distribution of SCCmec types per year in 2019 mecA-positive isolates is seen in Fig. 1. SCCmec type III predominated every year followed by type IV with the exception of 2011 where the opposite was seen. Type II was seen in multiple isolates throughout the study period and sporadic cases of types V and VI were noted from 2011 onwards. Only two cases of type I were seen in 2014 and 2015. A number of unknown types was noted from 2010 to 2017. We subsequently further investigated a proportion (n = 52) of the unknown types from 2013 to 2016 and found that the majority of the isolates were interpreted as type I-like, type II-like and type III-like [30].
Fig. 1.
Distribution of SCCmec types per year in mecA-positive isolates (n = 2019)
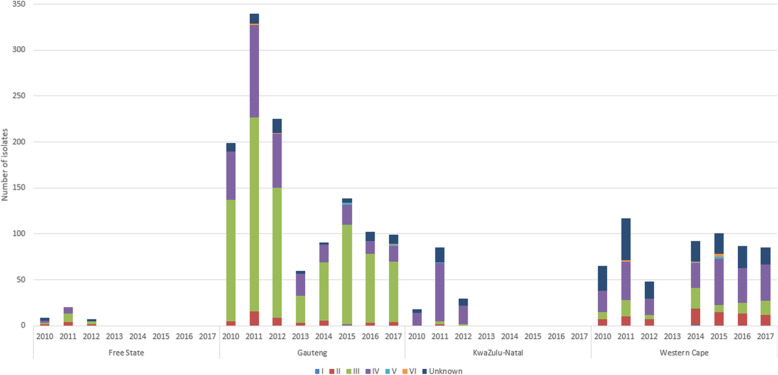
The distribution of SCCmec types per province per year is seen in Fig. 2. Type IV predominated in KwaZulu-Natal whereas type III predominated in the remaining three provinces. All six SCCmec types including unknown types were observed in Gauteng and the Western Cape provinces.
Fig. 2.
Distribution of SCCmec types per province per year in mecA-positive isolates (n = 2019)
Antibiotic non-susceptible phenotypes were examined and the distribution of SCCmec types per non-susceptible phenotype is seen in Table 1. Isolates belonging to all SCCmec types were resistant to most of the antibiotics with the exception of type I. All isolates were susceptible to vancomycin. Type III predominated in azithromycin-, erythromycin-, oxacillin-, cefoxitin-, penicillin-, trimethoprim/sulfamethoxazole-, daptomycin-, tetracycline-, ciprofloxacin-, levofloxacin-, moxyfloxacin- and gentamicin-non-susceptible isolates. Type II predominated in clindamycin-non-susceptible isolates and type IV predominated in rifampicin-non-susceptible isolates.
Table 1.
Distribution of SCCmec types according to antibiotic non-susceptibility phenotypes for MRSA isolates (n = 2019)
| Antibiotic non-susceptibility phenotype | SCCmec type | |||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | Untypeable | Negative | |
| Erythromycin | 0 | 133 (6.58%) | 920 (45.56%) | 391 (19.36%) | 4 (0.19%) | 2 (0.09%) | 249 (12.33%) | 2 (0.09%) |
| Clindamycin | 1 (0.04%) | 129 (6.38%) | 106 (5.25%) | 31 (1.53%) | 0 | 4 (0.19%) | 30 (1.48%) | 0 |
| Oxacillin | 2 (0.09%) | 135 (6.68%) | 916 (45.36%) | 648 (32.09%) | 7 (0.34%) | 9 (0.44%) | 271 (13.42%) | 2 (0.09%) |
| Penicillin | 2 (0.09%) | 138 (6.83%) | 926 (45.86%) | 651 (32.24%) | 7 (0.34%) | 9 (0.44%) | 274 (13.57%) | 2 (0.09%) |
| Trimethoprim/Sulfamethoxazole | 0 | 12 (0.59%) | 883 (43.73%) | 542 (26.84%) | 2 (0.09%) | 3 (0.14%) | 42 (2.08%) | 2 (0.09%) |
| Daptomycin | 0 | 0 | 7 (0.34%) | 2 (0.09%) | 0 | 0 | 2 (0.09%) | 0 |
| Linezolid | 0 | 3 (0.14%) | 3 (0.14%) | 3 (0.14%) | 0 | 0 | 3 (0.14%) | 0 |
| Tetracycline | 0 | 14 (0.69%) | 913 (45.22%) | 573 (28.38%) | 3 (0.14%) | 2 (0.09%) | 79 (3.91%) | 2 (0.09%) |
| Rifampin | 0 | 14 (0.69%) | 62 (3.07%) | 571 (28.28%) | 1 (0.04%) | 2 (0.09%) | 19 (0.94%) | 2 (0.09%) |
| Ciprofloxacin | 0 | 134 (6.63%) | 917 (45.41%) | 589 (29.17%) | 4 (0.19%) | 3 (0.14%) | 76 (3.76%) | 2 (0.09%) |
| Levofloxacin | 0 | 134 (6.63%) | 916 (45.36%) | 512 (25.35%) | 3 (0.14%) | 3 (0.14%) | 75 (3.71%) | 2 (0.09%) |
| Moxyfloxacin | 0 | 133 (6.58%) | 918 (45.46%) | 510 (25.26%) | 3 (0.14%) | 2 (0.09%) | 75 (3.71%) | 2 (0.09%) |
| Gentamicin | 0 | 19 (0.94%) | 912 (45.17%) | 556 (27.53%) | 6 (0.29%) | 3 (0.14%) | 242 (11.98%) | 2 (0.09%) |
Susceptibility was classified according to CLSI guidelines [26]
Suggested antibiotics approved by the US Food and Drug Administration (FDA) for clinical use were included in the table. Antibiotics excluded were azithromycin as erythromycin is a surrogate for macrolides, cefoxtin as oxacillin is included for MRSA, those that are recommended for urine only as well as those that were not tested for using the MicroScan Gram-positive PM-33 panel. In addition, vancomycin was excluded as all isolates were susceptible
Majority of the isolates cultured were from adult patients (959/2019, 47.5%). Isolates from paediatric patients were represented by 44.8% (904/2019); the data for the remaining isolates (156/2019, 7.7%) was unknown. The predominating SCCmec type in isolates from adults was type IV (478/2019, 23.7%) followed by type III (265/2019, 13.1%), II (123/2019, 6.1%), unknown type (81/2019, 4.0%), V (4/2019, 0.2%) and VI (8/2019, 0.4%). Type I was not seen in isolates cultured from adult patients. The predominating SCCmec type in isolates from paediatric patients was type III (569/2019, 28.2%) followed by unknown types (188/2019, 9.3%), type IV (129/2019, 6.4%), II (13/2019, 0.6%), V (3/2019, 0.1%) and I (2/2019, 0.1%). Type VI was not seen in isolates cultured from paediatric patients. The predominating SCCmec types in isolates obtained from male and female patients were very similar. The predominating SCCmec type could not be correctly established from isolates obtained from patients that died versus those that recovered or were discharged due to the majority of cases having unknown data. The same is applicable for diagnosis.
Spa typing
Spa typing was performed on 1467 isolates; the remaining 552 isolates from the period 2010 to 2012 do not have spa types assigned. A total of 71 different spa types and 24 novel spa types were observed. Five isolates were untypable even upon repeat processing. Table 2 shows the distribution of predominating spa types over the seven and a half-year period. Spa type t037 was the most common and predominated throughout followed by t1257. Spa types t012, t045 and t064 were also constantly present over this time period. Spa type t4864 was seen only in 2014, t1467 was seen only in 2015, t718 was seen only in 2016 and t5691 emerged in 2017. The remaining spa types were seen in small numbers and not consistently throughout the seven and a half-year period.
Table 2.
Distribution of predominating spa types per year
| Spa type | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 |
|---|---|---|---|---|---|---|---|---|
| t012 | 11 | 11 | 1 | 3 | 18 | 12 | 14 | 14 |
| t037 | 136 | 116 | 53 | 28 | 82 | 114 | 85 | 83 |
| t045 | 31 | 10 | 4 | 1 | 17 | 27 | 22 | 19 |
| t064 | 18 | 15 | 2 | 4 | 3 | 3 | 2 | 2 |
| t1257 | 52 | 57 | 12 | 15 | 29 | 43 | 28 | 33 |
| t022 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 1 |
| t118 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 |
| t018 | 1 | 1 | 1 | 0 | 5 | 0 | 1 | 0 |
| t032 | 5 | 0 | 0 | 0 | 2 | 6 | 7 | 4 |
| t1971 | 0 | 0 | 1 | 1 | 1 | 4 | 7 | 7 |
| t1443 | 5 | 4 | 0 | 0 | 1 | 2 | 0 | 0 |
| t1467 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| t1476 | 0 | 1 | 0 | 0 | 1 | 3 | 6 | 1 |
| t5691 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| t021 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 2 |
| t148 | 3 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| t238 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| t294 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 1 |
| t451 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| t718 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| t891 | 1 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| t4833 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| t4864 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| t2029 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
Spa types not represented in the table are those that represent only a single isolate per year for all years
Table 3 shows the variation of spa types over the seven and a half-year period. The most number of spa types were seen in 2011 and the most number of novel spa types occurred in 2014, which also showed a high variation in the number of different spa types observed. No novel spa types were found in 2013.
Table 3.
Variation of spa types per year
| Year | No. of different spa types | No. of novel spa types |
|---|---|---|
| 2010 | 18 | 2 |
| 2011 | 29 | 7 |
| 2012 | 15 | 4 |
| 2013 | 11 | 0 |
| 2014 | 21 | 10 |
| 2015 | 23 | 6 |
| 2016 | 19 | 5 |
| 2017 | 22 | 2 |
The Gauteng province showed the most variation with 44 different spa types and 14 novel spa types followed by the Western Cape (n = 40 and n = 14), respectively. In KwaZulu-Natal 12 different spa types were seen and in the Free State eight different spa types were observed. One novel spa type was found in both KwaZulu-Natal and the Free State provinces but these spa types differed from each other. Only t012, t045, t064 and t1257 were observed in all four provinces; t037 was seen in all provinces except in KwaZulu-Natal and t1971 was seen in all provinces except in the Free State; t9061 was seen only in the Free State and t13165, t1555, t4268 and t951 were seen only in KwaZulu-Natal. Two spa types (t209 and t2293) were found in the Gauteng and Free State provinces, which also had one novel spa type. Three spa types (t148, t451 and t891) were found in the Gauteng and KwaZulu-Natal, which also had one novel spa type. Nine spa types (t008, t018, t021, t022, t032, t1443, t1476, t304, t718) and two novel spa types were observed in Gauteng and the Western Cape provinces. Twenty-four different spa types (t10304, t105, t1096, t1107, t118, t127, t174, t186, t1943, t272, t2724, t355, t421, t4410, t463, t4833, t4864, t5961, t701, t729, t7962, t840, t913 and t932) and 10 novel spa types were seen in Gauteng alone. Twenty-two different spa types (t015, t0121, t0379, t059, t11775, t1467, t1774, t1813, t223, t230, t238, t2409, t2526, t294, t324, t432, t498, t5483, t578, t6330, t6931 and t8636) and 10 novel spa types were seen in the Western Cape alone.
Antibiotic non-susceptible phenotypes were examined and the distribution of spa types representing majority of the isolates is seen in Table 4. One isolate belonging to spa type t10304 was non-susceptible to penicillin only (data not shown in table). All three isolates typed as t0379 displayed the same phenotypic profile and were non-susceptible to the fluoroquinolones and beta-lactam antibiotics only (data not shown in table). All four isolates typed as t2029 showed resistance to all antibiotics listed except for daptomycin, linezolid and rifampin (data not shown in table). All four isolates belonging to type t238 and t294 showed the same phenotypic profile and two isolates belonging to t304 and t421 displayed the same phenotypic profile (data not shown in table).
Table 4.
Distribution of Spa types according to antibiotic non-susceptibility phenotypes for MRSA isolates (n = 1467)
| Spa type | Antibiotic non-susceptibility phenotype | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azithromycin | Erythromycin | Clindamycin | Oxacillin | Cefoxitin | Penicillin | Trimethoprim/ Sulfamethoxazole | Daptomycin | Linezolid | Tetracycline | Rifampin | Ciprofloxacin | Levofloxacin | Moxyfloxacin | Gentamicin | |
| t012 (n = 84) | 28 (1.90%) | 81 (5.52%) | 71 (4.83%) | 84 (5.72%) | 84 (5.72%) | 84 (5.72%) | 11 (0.74%) | 0 | 4 (0.27%) | 14 (0.95%) | 7 (0.47%) | 84 (5.72%) | 84 (5.72%) | 83 (5.65%) | 17 (1.15%) |
| t018 (n = 9) | 4 (0.27%) | 8 (0.54%) | 7 (0.47%) | 8 (0.54%) | 9 (0.61%) | 9 (0.61%) | 0 | 0 | 0 | 1 (0.06%) | 1 (0.06%) | 9 (0.61%) | 9 (0.61%) | 9 (0.61%) | 2 (0.13%) |
| t032 (n = 24) | 4 (0.27%) | 12 (0.81%) | 3 (0.20) | 24 (1.63%) | 24 (1.63%) | 24 (1.63%) | 1 (0.06%) | 0 | 0 | 2 (0.13%) | 43 (2.93%) | 23 (1.56%) | 23 (1.56%) | 23 (1.56%) | 3 (0.20) |
| t037 (n = 697) | 327 (22.29%) | 690 (47.03%) | 81 (5.52%) | 693 (47.23%) | 695 (47.37%) | 698 (47.58%) | 659 (44.92%) | 5 (0.34%) | 1 (0.06%) | 679 (46.28%) | 0 | 691 (47.10) | 688 (46.89%) | 690 (47.03%) | 681 (46.42%) |
| t045 (n = 131) | 53 (3.6%) | 130 (8.86%) | 13 (0.88%) | 130 (8.86%) | 130 (8.86%) | 130 (8.86%) | 11 (0.74%) | 2 (0.13%) | 4 (0.27%) | 18 (1.22%) | 6 (0.40%) | 25 (1.70%) | 24 (1.63%) | 26 (1.77%) | 123 (8.38%) |
| t064 (n = 49) | 21 (1.43%) | 29 (1.97%) | 0 | 49 (3.34%) | 49 (3.34%) | 49 (3.34%) | 44 (2.99%) | 0 | 0 | 47 (3.20%) | 47 (3.20%) | 30 (2.04%) | 25 (1.70_ | 26 (1.77%) | 47 (3.20%) |
| t1257 (n = 269) | 79 (5.3%) | 164 (11.17) | 14 (0.95%) | 268 (18.26%) | 268 (18.26%) | 268 (18.26%) | 249 (16.97%) | 0 | 2 (0.13%) | 263 (17.92) | 255 (17.38%) | 264 (17.99%) | 231 (15.74%) | 231 (15.74%) | 252 (17.17%) |
| t1443 (n = 14) | 1 (0.06%) | 1 (0.06%) | 0 | 14 (0.95%) | 13 (0.88%) | 14 (0.95%) | 13 (0.88%) | 0 | 0 | 13 (0.88%) | 14 (0.95%) | 13 (0.88%) | 13 (0.88%) | 13 (0.88%) | 13 (0.88%) |
| t1476 (n = 12) | 0 | 5 | 0 | 12 (0.81%) | 12 (0.81%) | 12 (0.81%) | 2 (0.13%) | 0 | 0 | 9 (0.61%) | 0 | 10 (0.68%) | 7 (0.47%) | 7 (0.47%) | 11 (0.74%) |
| t1971 (n = 23) | 2 (0.13%) | 19 (1.29%) | 1 (0.06%) | 23 (1.56%) | 23 (1.56%) | 23 (1.56%) | 23 (1.56%) | 0 | 0 | 23 (1.56%) | 23 (1.56%) | 23 (1.56%) | 21 (1.42%) | 21 (1.42%) | 23 (1.56%) |
| Novel spa types (n = 38) | 9 (0.61%) | 24 (1.63%) | 7 (0.47%) | 37 (2.52%) | 37 (2.52%) | 38 (2.59%) | 23 (1.56%) | 0 | 0 | 25 (1.70%) | 16 (1.09%) | 31 (2.11%) | 29 (1.97%) | 27 (1.84%) | 27 (1.84%) |
| Untypeable (n = 5) | 3 (0.20%) | 4 (0.27%) | 1 (0.06%) | 5 (0.34%) | 5 (0.34%) | 5 (0.34%) | 3 (0.20%) | 0 | 0 | 3 (0.20%) | 1 (0.06%) | 4 (0.27%) | 4 (0.27%) | 3 (0.20%) | 3 (0.20%) |
The following spa types were excluded from the table as they accounted for a small number of isolates: t008, t0121, t015, t021, t022, t0379, t059, t10304, t105, t1096, t1107, t11775, t118, t127, t13165, t1467, t148, t1555, t174, t1774, t1813, t186, t1994, t2029, t223, t2293, t230, t238, t2409, t2526, t272, t2724, t294, t304, t324, t355, t379, t421, t4268, t432, t4410, t451, t463, t4833, t4864, t498, t5483, t5691, t578, t6330, t6931, t701, t718, t729, t7962, t840, t8636, t891, t9061, t913, t932, t951
Susceptibility was classified according to CLSI guidelines [26]
Suggested antibiotics approved by the US Food and Drug Administration (FDA) for clinical use were included in the table. Antibiotics excluded were those that are recommended for urine only as well as those that were not tested for using the MicroScan Gram-positive PM-33 panel. In addition, vancomycin was excluded as all isolates were susceptible
A total of 552 isolates were not typed
Of the known adult vs paediatric information, the predominating spa type in isolates from adults was t1257 (195/1467, 13.3%) followed by t037 (189/1467, 12.7%), t012 (70/1467, 4.8%), t064 (32/1467, 2.2%), t1971 (20/1467, 1.4%), t032 (19/1467, 1.3%) and t045 (15/1467, 1%). The remaining spa types within this group individually represented less than 1%. This group consisted of 55 different spa types and 18 novel spa types. Two isolates were untypeable. The predominating spa type in isolates from paediatric patients was t037 (446/1467, 30.4%) followed by t045 (115/1467, 7.8%) and t1257 (53/1467, 3.6%). The remaining spa types within this group individually represented less than 1%. This group consisted of 32 different spa types and 10 novel spa types. Three isolates were untypeable. The following spa types were seen in isolates from adult patients only: t008, t0121, t018, t021, t0379, t059, t1175, t118, t1467, t174, t1774, t1813, t2029, t223, t2293, t230, t2409, t2526, t294, t304, t324, t379, t432, t4410, t463, t4864, t578, t6931, t701, t729, t7962, t840, t8636, t9061 and t913. There were 14 novel spa types in this group. The following spa types were seen in isolates from paediatric patients only: t10304, t1096, t127, t13165, t1555, t186, t1943, t272, t355, t4286, t498, t5483, t6330 and t932; six novel spa types were observed in this group. The predominating spa types in isolates obtained from male and female patients were very similar. Furthermore, the predominating spa type could not be correctly established from isolates obtained from patients that died versus those that recovered or were discharged due to the majority of cases having unknown data. The same is applicable for diagnosis.
SCCmec and spa types complexes
The SCCmec-spa type combinations are referred to as complexes. A total of 1467 SCCmec-spa type complexs were obtained. The five isolates that were not typeable for spa type were excluded from the analysis; SCCmec types for each of these varied (SCCmec II, III, IV, V and unknown type). The most diverse complex was composed of the SCCmec type IV element and 53 different spa types. Next were the isolates with unknown SCCmec type; these were associated with 28 different spa types. SCCmec type III was associated with 24 different spa types and SCCmec type II was associated with 20 different spa types. There were smaller numbers of SCCmec type I, V and VI isolates and predominance was therefore inconsequential; the isolates varied with regard to spa type. The SCCmec-spa type combinations constituting the complexes are shown in Table 5.
Table 5.
SCCmec- spa type combinations
| SCCmec type, n | Spa type, n (%) |
|---|---|
| SCCmec type I isolates (n = 2) | t015, t186 (n = 1, 50%, each). |
| SCCmec type II (n = 104) | t012 (n = 71, 67.6%); t037 (n = 7, 6.7%); t021 (n = 4, 3.8%); t238 (n = 4, 3.8%); t1257 (n = 3, 2.9%); t018 (n = 2, 1.9%); t0121 (n = 1, 0.9%); t045 (n = 1, 0.9%); t064 (n = 1, 0.9%); t2526 (n = 1, 0.9%); t4864 (n = 1, 0.9%); t6330 (n = 1, 0.9%); t729 (n = 1, 0.9%); t840 (n = 1, 0.9%); t8636 (n = 1, 0.9%); t913 (n = 1, 0.9%); novel spa types: txAF, txAK, txAO, txAQ (n = 1, 0.9%, each). |
| SCCmec type III (n = 709) | t037 (n = 656, 92.5%); t045 (n = 12, 1.7%); t1257 (n = 8, 1.1%); t012 (n = 8, 1.1%); t2029 (n = 4, 0.6%); t0421 (n = 2, 0.3%); t1476 (n = 2, 0.3%); t032 (n = 1, 0.1%); t127 (n = 1, 0.1%); t2293 (n = 1, 0.1%); t355 (n = 1, 0.1%); t932 (n = 1, 0.1%); t7962 (n = 1, 0.1%); t701 (n = 1, 0.1%); t1943 (n = 1, 0.1%); t4410 (n = 1, 0.1%); t5691 (n = 1, 0.1%); novel spa types: txAB, txAF, txAM, txAN, txAP, txAZ, txBD (n = 1, 0.1%, each). |
| SCCmec type IV (n = 451) | IV-t1257 (n = 255, 56.5%); t064 (n = 47, 10.4%); t1973 (n = 23, 5.1%), t032 (n = 21, 4,7%); t1443 (n = 14, 3.1%); t037 (n = 12, 2.7%); t022 (n = 5, 1.1%); t1467 (n = 5, 1.1%), t118 (n = 4, 0.9%,); t294 (n = 4, 0.9%,); t4833 (n = 4, 0.9%); t451 (n = 3, 0.7%); t891 (n = 3, 0.7%); t012 (n = 2, 0.4%); t105 (n = 2, 0.4%); t2293 (n = 2, 0.4%); t304 (n = 2, 0.4%); t718 (n = 2, 0.4%); t008 (n = 1, 0.2%); t015 (n = 1, 0.2%); t018 (n = 1, 0.2%); t0379 (n = 1, 0.2%); t045 (n = 1, 0.2%); t059 (n = 1, 0.2%); t1555 (n = 1, 0.2%); t1774 (n = 1, 0.2%); t230 (n = 1, 0.2%); t272 (n = 1, 0.2%); t2724 (n = 1, 0.2%); t324 (n = 1, 0.2%); t379 (n = 1, 0.2%); t4268 (n = 1, 0.2%); t432 (n = 1, 0.2%); t4864 (n = 1, 0.2%); t5691 (n = 1, 0.2%); t578 (n = 1, 0.2%); t951 (n = 1, 0.2%); novel spa types: txAA (n = 5, 1.1%), txAC and txAI (n = 2, 0.4%), each), txAD, txAE, txAG, txAJ, txAL, txAM, txAN, txAS, txAO, txBA, txBB, txBC txBE (n = 1, 0.2%, each). |
| SCCmec type V (n = 3) | t1476, t045, t037 (n = 1, 33.3%, each) |
| SCCmec type VI (n = 5) | t1813 (n = 2, 80%); t174 (n = 1, 20%); t223 (n = 1, 20%); novel spa type txAD (n = 1, 20%). |
| Unknown SCCmec types (n = 190) | t045 (n = 114, 60%); t037 (n = 22, 11.6%); t1476 (n = 9, 4.7%); t018 (n = 6, 3.2%); t148 (n = 5, 2.6%); t012 (n = 3, 1.6%); t1257 (n = 3, 1.6%); t13165 (n = 3, 1.6%);, t008 (n = 2, 1.1%,); t032 (n = 2, 1.1%,); t5691 (n = 2, 1.1%,); t891 (n = 2, 1.1%,); t021 (n = 1, 0.5%); t064 (n = 1, 0.5%); t10304 (n = 1, 0.5%); t1096 (n = 1, 0.5%); t1107 (n = 1, 0.5%); t11775 (n = 1, 0.5%); t2409 (n = 1, 0.5%); t463 (n = 1, 0.5%); t498 (n = 1, 0.5%); t5483 (n = 1, 0.5%); t6931 (n = 1, 0.5%); t9061 (n = 1, 0.5%); novel spa types: txAH (n = 2, 1.1%), txAD (n = 1, 0.5%), txAF (n = 1, 0.5%), txAL (n = 1, 0.5%). |
Predominating circulating clones
MRSA clones were defined by their sequence type (ST), SCCmec type and spa type. Multilocus Sequence Typing was performed on 48 isolates only. Isolates were selected randomly based on the most common spa-types (t037, n = 9; t1257, n = 10; t012, n = 9; t064, n = 9; t045, n = 8; t032, n = 3). The predominating circulating clones based on common spa types are seen in Table 6. Although only data for 48 isolates are present, the circulating clones are relatively diverse. As MLST was only done on a few selected isolates we could not confidently establish the circulating clones that are representative of entire surveillance population. We can therefore not comment on the evolution of MRSA clones in our setting.
Table 6.
Predominating circulating clones
| ST (CC) | SCCmec type | Spa type | No. of isolates |
|---|---|---|---|
| 5 (5) | III | t045 | 1 |
| 5 (5) | V | t045 | 2 |
| 5 (5) | Unknown | t045 | 4 |
| 5 (5) | Unknown | t1257 | 1 |
| 22 (22) | IV | t012 | 1 |
| 22 (22) | IV | t032 | 2 |
| 4121 (22) | IV | t032 | 1 |
| 36 (30) | II | t012 | 5 |
| 36 (30) | II | t037 | 1 |
| 36 (30) | II | t064 | 1 |
| 36 (30) | III | t045 | 1 |
| 239 (8) | II | t1257 | 1 |
| 239 (8) | III | t1257 | 1 |
| 239 (8) | III | t012 | 1 |
| 239 (8) | III | t037 | 6 |
| 239 (8) | IV | t037 | 1 |
| 239 (8) | Unknown | t012 | 1 |
| 239 (8) | Unknown | t037 | 1 |
| 612 (8) | III | t012 | 1 |
| 612 (8) | IV | t064 | 8 |
| 612 (8) | IV | t1257 | 6 |
| Unknowna | IV | t1257 | 1 |
a ST is unknown due to new allele for the pta gene; at position 277 the nucleotide adenine (A) is present and not the expected nucleotide, guanine (G)
Discussion
This study is a detailed description of the molecular characterisation of MRSA isolates with specific focus on SCCmec types and spa types and, to a lesser extent, sequence types. It is important to have a genetic understanding of the circulating strains in a geographical region to establish genetic diversity, transmission, dissemination, epidemiology and evolution. Antimicrobial susceptibility profiles were also reported; apart from using antimicrobial susceptibility results for treatment regimens, antimicrobial susceptibility profiles are also important in identifying a link to specific genotypes, which could potentially identify virulence patterns. Antimicrobial selection may potentially also be a key factor in the dissemination of predominating MRSA clones within a hospital environment [31].
SCCmec type III was the most predominant SCCmec type followed by type IV. Type III was also the most frequent SCCmec type in studies in Iran [32, 33], Serbia [34], Brazil [35] and Europe [36]. The most prevalent spa type in our study was t037. This is in keeping with a review conducted in 2018 of European, Asian, American, Australian and African studies from 2007 onwards including 18 studies from Africa which showed that the most prevalent spa type was t037 [5]. The review also showed that t084 and t064 were common in Africa. In contrast to our study, t064 was present in a small number of isolates (n = 49) and t084 was not observed at all. Interestingly this review also showed that the most prevalent spa type in America was t008, which was reported only in America and Canada. Our current study has shown the occurrence of t008 in three isolates from Gauteng and the Western Cape provinces.
Isolates harbouring SCCmec type III and IV elements were the most resistant as evidenced by the large number of non-susceptible phenotypes to majority of the antimicrobial agents (Table 1). A 2014 study in Iran showed similar findings; they further molecularly characterised resistance genes and found that their type III isolates contained different resistance genes [37]. In contrast, an Indian study in 2016 showed more phenotypic resistance to non-beta-lactam antibiotics in their type I isolates [38].
A 2017 Chinese study on 120 MRSA isolates showed differences to the current study; 100 % of their spa type t037 isolates were resistant to clindamycin, erythromycin, ciprofloxacin, gentamicin, tetracycline and trimethoprim/sulfamethoxazole whereas only 6% of our isolates were resistant to clindamycin and 45 to 47% were resistant to the remaining antibiotics. However, in keeping with the study from China, none of our t037 isolates were resistant to rifampin and vancomycin (Table 4) [39]. Another Chinese study with 106 t037 isolates showed predominant resistance to clindamycin, erythromycin, ciprofloxacin, gentamicin, tetracycline, trimethoprim/sulfamethoxazole and chloramphenicol [40]. Of six Nigerian t037 isolates, all were resistant to clindamycin, erythromycin, ciprofloxacin, gentamicin, tetracycline and trimethoprim/sulfamethoxazole in addition to penicillin, oxacillin and moxifloxacin [41]; in the current study, almost 50% (47–48%) of the t037 isolates were resistant to penicillin, oxacillin and moxifloxacin.
The study of circulating clones and clonal evolution is important because it is used to assess the relationship between clonal types, disease symptoms, antibiotic choice and clinical outcomes [42]. Clones are bacterial strains that have descended from a common ancestor and through point mutations, recombination, acquisition and deletion of mobile genetic elements they diversify resulting in wide-ranging genotypes and phenotypes [43]. In order to establish circulating clones and clonal evolution, multiple molecular tools should be employed; the combination of ST, SCCmec type and spa type would ideally be preferred. However, as MLST is more costly, we were not able to perform this technique on all isolates. Studies have shown that SCCmec typing is not a very discriminatory method and that spa typing alone was not able to clearly predict ST or PFGE type but when combined with BURP analysis producing spa CCs, it is sufficient for describing the clonal structure of S. aureus [6, 10]. Although useful, it should be noted that spa typing takes only one gene into consideration in relation to the entire genome and therefore does not reflect mutational events occurring throughout the genome [5]. Nevertheless, spa typing is extremely useful and we have coupled it with SCCmec typing and sequence typing to a lesser extent, to provide information on the circulating S. aureus strains in our population.
A review manuscript by Asadollahi et. al., in 2018 [5] showed that from five African studies, t037 was most associated with SCCmec type III (106 isolates) and least associated with type V (one isolate). Our study showed similar findings; t037 was mostly associated with SCCmec type III (656 isolates) and least associated with type V (one isolate). In another study of German, French, Japanese and Finnish isolates in 2007, majority of the t037 isolates (n = 8) were also associated with SCCmec type III [44]. This was also seen in seven isolates from a 2014 Iranian study but two t037 isolates were also associated with SCCmec type IV and one was associated with SCCmec type I [37].
The Asadollahi et. al., review manuscript further showed that t037 was associated with ST239 and t064 was associated with ST8 [5]. In our study, t037 was mainly associated with ST239 but one isolate was associated with ST36. The isolates belonging to t064 were mainly associated with ST612 and one isolate was associated with ST36. The review further showed that t032 was always associated with ST22 irrespective of the continent in which it was observed; one of the t032 isolates in our study also showed this finding whereas the second t032 isolate was associated with ST4122. Both ST22 and ST4121 belong to MLST CC22. As MLST was only performed on a few selected isolates, the results could have potentially differed if ST data was available for more isolates.
Other publications have used ST and the SCCmec element to define clonal types [45, 46]. In the current study, the Brazilian/Hungarian clone (ST239-MRSA-III) accounted for eight out of the 48 (17%) isolates typed. This is also a common MRSA strain in New Zealand, where the most common associated spa type is t037. Alternative clone names include EMRSA-1, EMRSA-4, EMRSA-11, Por/Bra, Vienna, AUS-2 EMRSA and AUS-3 EMRSA) (http://esr.cri.nz/assets/HEALTH-CONTENT/Images-and-PDFs/MRSAdescriptions.pdf), [45]. Of the eight isolates in the current study, six were spa type t037. This clone has also been observed in Finland, Germany, Greece, Ireland, Netherlands, Poland, Portugal, Slovenia, Sweden, United Kingdom and the United States of America [45]. Another common MRSA strain in New Zealand is ST22-MRSA-IV (EMRSA-15, Barnim) (http://esr.cri.nz/assets/HEALTH-CONTENT/Images-and-PDFs/MRSAdescriptions.pdf), most associated with spa types t032, t1401 and t5501. In our study, two of the three isolates were t032 and the remaining one being t012. This clone has also been seen in Germany Ireland, Sweden and the United Kingdom [45]. Strain ST-36-MRSA-II (EMRSA-16) also common in New Zealand and most associated with t018 (http://esr.cri.nz/assets/HEALTH-CONTENT/Images-and-PDFs/MRSAdescriptions.pdf) was also seen in seven isolates in the current study; however none were associated with spa type t018, five were t012 and one each for t037 and t064. This clone was also seen in Finland and the United Kingdom [45]. Another clonal type observed in our study included ST5-MRSA-III (n = 1) which is a Belgian clone [45].
As spa typing was not done on all MRSA isolates and as MLST was only performed on a few selected isolates we could not confidently establish the circulating clones that are representative of entire surveillance population. We therefore cannot comment on the evolution of MRSA clones in our setting. However, although ST data was available for 48 isolates only (which also had spa and SCCmec type data), the circulating clones are relatively diverse and if the ST was omitted and only SCCmec and spa types considered, the diversity of the circulating strains increases. Nevertheless, of the 48 clones we have observed taking ST, SCCmec type and spa type into consideration, the most common were ST612-IV-t064 (n = 8), ST612-IV-t1257 (n = 6), ST239-III-t037 (n = 6) and ST36-II-t012. Multiple introductions of ST612 was observed in Western Australia in both human and equine reservoirs [47]. ST612 was also recently observed in the clone ST612-CC8-t1257-SCCmec_IVd(2B) obtained from the poultry food chain in South Africa [48]. In addition to the studies mentioned above, the Brazilian/Hungarian clone ST239-III-t037 was commonly found over a 15 year period in a study in China beginning in 1994 [40]. The presence of this clone was also observed in various continents [49]. Therefore, this clone is very well established globally.
It has been shown that the transformation from a MRSA clone to a MSSA clone can occur through the excision of the SCCmec element and consequently the loss of methicillin resistance. Therefore, it is possible for a clone to evolve from MSSA into MRSA through the acquisition of the SCCmec element or from MRSA to MSSA through the excision of the SCCmec element [50]. Molecular typing is extremely useful in studying genetic diversity and a study on a collection of isolates from 19 countries in Europe, the United Kingdom, The United States and Latin America has shown that MRSA and MSSA differ with regards to the diversity of their genetic backgrounds as MSSA has shown to be more diverse [10]. A limitation of the current study is that molecular typing was performed on MRSA isolates only; results for MSSA is therefore lacking and we cannot make any remarks on this matter. To add to genetic diversity, clones responsible for causing HA infections and CA infections may differ and the recombination between HA and CA clones does occur [50]. A detailed investigation taking into consideration aspects like virulence factors such as surface proteins, invasins, biochemical properties, membrane-damaging toxins, exotoxins e.g. Panton-Valentine Leukocidin (PVL), biofilm production, antimicrobial resistance genes and clinical syndromes [42, 43, 50, 51] would be beneficial.
Conclusion
This study reports a large dataset of isolates collected from various provinces in South Africa from 2010 to 2017. A variety of spa types were observed in this study; this is in keeping with other reports showing the presence of multiple spa types in the MRSA population. Moreover, data from Africa is not abundant. It is evident that MRSA clones are diverse; they disseminate both rapidly and efficiently and it is important to understand why particular clones dominate in a specific geographical location in order to develop effective strategies to control the spread of S. aureus infections.
Acknowledgments
We thank Wilhelmina Strasheim, Rubeina Badat, Naseema Bulbulia, Rosah Mobokachaba, Gloria Molaba and Marshagne Smith for assistance with the laboratory work and Boniwe Makwakwa for assistance with the database.
Abbreviations
- HA
Healthcare-associated
- CA
Community-associated
- MSSA
Methicillin-susceptible S. aureus
- MRSA
Methicillin-resistant S. aureus
- PFGE
Pulsed-field Gel Electrophoresis
- MLST
Multilocus Sequence Typing
- spa
Stapylococcal protein A
- SCCmec
Staphylococcal Cassette Chromosome mec
- BURST
Based Upon Related Sequence Type
- ST
Sequence type
- CC
Clonal complex
- BURP
Based Upon Repeat Pattern
- VNTR
Variable-number tandem repeat
- ccr
Cassette chromosome recombinase
- DMP
Diagnostic Media Products
- NHLS
National Health Laboratory Service
- NICD
National Institute for Communicable Diseases
- AST
Antimicrobial susceptibility testing
- CLSI
Clinical and Laboratory Standards Institute
- PCR
Polymerase Chain reaction
Authors’ contributions
ASM conceived and designed the experiments, performed the experiments, analysed the data and wrote the manuscript. OP was responsible for the study, its overall design and coordination and in editing of the manuscript. RM performed laboratory experiments and assisted in the editing of the manuscript. ML assisted in laboratory experiments and in the editing of the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Institute for Communicable Diseases and received no specific grant from any funding agency.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. Access to the National Institute for Comminicable Diseases GERMS-SA database cannot be granted due to the presence of patient identifiers.
Ethics approval and consent to participate
Ethical clearance was obtained from the University of the Witwatersrand Human Research Committee (Protocol number M10464). Samples used in the study were bacterial isolates. No patient specimens were used. Therefore, informed consent was not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bassetti M, Trecarichi EM, Mesini A, Spanu T, Giacobbe DR, Rossi M, et al. Risk factors and mortality of healthcare-associated and community-acquired Staphylococcus aureus bacteraemia. Clin Microbiol Infect. 2012;18(9):862–869. doi: 10.1111/j.1469-0691.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 2.Singh G, Broor S, Agarwal P. Molecular characterisation of Staphylococcus aureus using spa typing as a diagnostic tool in Haryana, India. Indian J Med Microbiol. 2018;36(1):26–31. doi: 10.4103/ijmm.IJMM_17_330. [DOI] [PubMed] [Google Scholar]
- 3.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42(2):792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, van Belkum A, Asadollahi K, et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: a review. Front Microbiol. 2018;9:163. doi: 10.3389/fmicb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hara FP, Suaya JA, Ray T, Baxter R, Brown ML, Mera RB, et al. Spa typing and multilocus sequence typing show comparable performance in a macroepidemiologic study of Staphylococcus aureus in the United States. Microb Drug Resist. 2016;22(1):88–96. doi: 10.1089/mdr.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma-Kuinkel BK, Rude TH, Fowler VG., Jr Pulse field gel electrophoresis. Methods Mol Biol. 2016;1373:117–130. doi: 10.1007/7651_2014_191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urwin R, Maiden MC. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;11(10):479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Votintseva AA, Fung R, Miller RR, Knox K, Godwin H, Wyllie DH, et al. Prevalence of Staphylococcus aureus protein A (spa) mutants in the community and hospitals in Oxfordshire. BMC Microbiol. 2014;14:63. doi: 10.1186/1471-2180-14-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faria NA, Carrico JA, Oliveira DC, Ramirez M, de Lencastre H. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46(1):136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallin M, Deplano A, Denis O, De Mendonca R, De Ryck R, Struelens MJ. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J Clin Microbiol. 2007;45(1):127–133. doi: 10.1128/JCM.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, et al. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46(2):574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nubel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105(37):14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T, Hiramatsu K, Oliveira DC, De Lencastre H, Zhang K, Westh H, et al. Classification of Staphylococcal Cassette Chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker K, Ballhausen B, Köck R, Kriegeskorte A. Methicillin-resistance in Staphylococcus isolates: the ‘mec’ alphabet with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int J Med Microbiol. 2014;304:794–804. doi: 10.1016/j.ijmm.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Turlej A, Hryniewicz W, Empel J. Staphylococcal cassette chromosome mec (Sccmec) classification and typing methods: an overview. Pol J Microbiol. 2011;60(2):95–103. doi: 10.33073/pjm-2011-013. [DOI] [PubMed] [Google Scholar]
- 17.Shore AC, Coleman DC. Staphylococcal cassette chromosome mec: recent advances and new insights. Int J Med Microbiol. 2013;303(6–7):350–359. doi: 10.1016/j.ijmm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Hiramatsu K, Tomasz A, De Lencastre H, Oerreten V, Holden MTG, et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 2012;56:4997–4999. doi: 10.1128/AAC.01199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2018;61:74–76. doi: 10.1016/j.meegid.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z, Li F, Liu D, Xue H, Zhao X. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob Agents Chemother. 2015;59(12):7597–7601. doi: 10.1128/AAC.01692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortuin-de Smidt MC, Singh-Moodley A, Badat R, Quan V, Kularatne R, Nana T, et al. Staphylococcus aureus bacteraemia in Gauteng academic hospitals, South Africa. Int J Infect Dis. 2015;30:41–48. doi: 10.1016/j.ijid.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Perovic O, Iyaloo S, Kularatne R, Lowman W, Bosman N, Wadula J, et al. Prevalence and trends of Staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012: laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One. 2015;10(12):e0145429. doi: 10.1371/journal.pone.0145429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perovic O, Singh-Moodley A, Govender NP, Kularatne R, Whitelaw A, Chibabhai V, et al. A small proportion of community-associated methicillin-resistant Staphylococcus aureus bacteraemia, compared to healthcare-associated cases, in two South African provinces. Eur J Clin Microbiol Infect Dis. 2017;36(12):2519–2532. doi: 10.1007/s10096-017-3096-3. [DOI] [PubMed] [Google Scholar]
- 25.Singh-Moodley A, Marais E, Perovic O. Discrepancies in the identification of methicillinresistant Staphylococcus aureus and the absence of mecC in surveillance isolates in South Africa. S Afr J Infect Dis. 2015;1(1):1–3. [Google Scholar]
- 26.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing: twenty ninth informational supplement. 2019. [Google Scholar]
- 27.Thomas LC, Gidding HF, Ginn AN, Olma T, Iredell J. Development of a real-time Staphylococcus aureus and MRSA (SAM-) PCR for routine blood culture. J Microbiol Methods. 2007;68(2):296–302. doi: 10.1016/j.mimet.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh-Moodley A, Strasheim W, Mogokotleng R, Ismail H, Perovic O. Unconventional SCCmec types and low prevalence of the Panton-Valentine Leukocidin exotoxin in South African blood culture Staphylococcus aureus surveillance isolates, 2013-2016. PLoS One. 2019;14(11):e0225726. doi: 10.1371/journal.pone.0225726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shopsin B, Mathema B, Zhao X, Martinez J, Kornblum J, Krieswirth BN. Resistance rather than virulence selects for the clonal spread of methicillin-resistant Staphylococcus aureus: implications for MRSA transmission. Microb Drug Resist. 2000;6(3):239–244. doi: 10.1089/mdr.2000.6.239. [DOI] [PubMed] [Google Scholar]
- 32.Abbasian S, Farahani NN, Mir Z, Alinejad F, Haeili M, Dahmardehei M, et al. Genotypic characterization of Staphylococcus aureus isolated from a burn centre by using agr, spa and SCCmec typing methods. New Microbes New Infect. 2018;26:15–19. doi: 10.1016/j.nmni.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taherirad A, Jahanbakhsh R, Shakeri F, Anvary S, Ghaemi EA. Staphylococcal cassette chromosome mec types among methicillin-resistant Staphylococcus aureus in Northern Iran. Jundishapur J Microbiol. 2016;8(8):e33933. [DOI] [PMC free article] [PubMed]
- 34.Asanin J, Misic D, Aksentijevic K, Tambur Z, Rakonjac B, Kovacevic I, et al. Genetic profiling and comparison of human and animal methicillin-resistant Staphylococcus aureus (MRSA) isolates from Serbia. Antibiotics. 2019;8:26. doi: 10.3390/antibiotics8010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beatriz A, Machado MP, Reiter KC, Paiva RM, Barth AL. Distribution of staphylococcal cassette chromosome mec (SCCmec) types I, II, III and IV in coagulase-negative staphylococci from patients attending a tertiary hospital in southern Brazil. J Med Microbiol. 2007;56:1328–1333. doi: 10.1099/jmm.0.47294-0. [DOI] [PubMed] [Google Scholar]
- 36.Davies TA, Shang W, Amsler KM, Bajaksouzian S, Jacobs MR, Bush K. Molecular characterisation of meticillin-resistant Staphylococcus aureus isolates from two ceftobiprole phase 3 complicated skin and skin-structure infection clinical trials. Int J Antimicrob Agents. 2009;34(2):166–168. doi: 10.1016/j.ijantimicag.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi S, Sekawi Z, Monjezi A, Maleki MH, Soroush S, Sadeghifard N, et al. Emergence of SCCmec type III with variable antimicrobial resistance profiles and spa types among methicillin-resistant Staphylococcus aureus isolated from healthcare- and community-acquired infections in the west of Iran. Int J Infect Dis. 2014;25:152–158. doi: 10.1016/j.ijid.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh A, Singh Y, Kapil A, Dhawan B. Staphylococcal cassette chromosome mec (SCCmec) typing of clinical isolates of coagulase-negative staphylocci (CoNS) from a tertiary care hospital in New Delhi, India. Indian J Med Res. 2016;143(3):365–370. doi: 10.4103/0971-5916.182629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong H, Yu F, Zhang W, Li X, Wang H. Molecular epidemiology and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus strains in a tertiary hospital in China. Front Microbiol. 2017;8:838. doi: 10.3389/fmicb.2017.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Liu Y, Jiang X, Chen M, Wang H. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob Agents Chemother. 2010;54(5):1842–1847. doi: 10.1128/AAC.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shittu AO, Okon K, Adesida S, Oyedara O, Witte W, Strommenger B, et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011;11:92. doi: 10.1186/1471-2180-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Noriega E, Seas C. The changing pattern of methicillin-resistant staphylococcus aureus clones in Latin America: implications for clinical practice in the region. Braz J Infect Dis. 2010;14(Suppl 2):S87–S96. doi: 10.1590/S1413-86702010000800004. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Noriega E, Seas C, Guzman-Blanco M, Mejia C, Alvarez C, Bavestrello L, et al. Evolution of methicillin-resistant Staphylococcus aureus clones in Latin America. Int J Infect Dis. 2010;14(7):e560–e566. doi: 10.1016/j.ijid.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Werbick C, Becker K, Mellmann A, Juuti KM, von Eiff C, Peters G, et al. Staphylococcal chromosomal cassette mec type I, spa type, and expression of Pls are determinants of reduced cellular invasiveness of methicillin-resistant Staphylococcus aureus isolates. J Infect Dis. 2007;195(11):1678–1685. doi: 10.1086/517517. [DOI] [PubMed] [Google Scholar]
- 45.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99(11):7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vivas MC, Gutierrez A. In: Open I, editor. Typification methods and molecular epidemiology of Staphylococcus aureus with methicillin resistance: We of Science. London: Web of Science™ Core Collection (BKCI); 2018.
- 47.Murphy RJT, Ramsay JP, Lee YT, Pang S, O'DeabJulie MA, Pearson C, et al. Multiple introductions of methicillin-resistant Staphylococcus aureus ST612 into Western Australia associated both with human and equine reservoirs. Int J Antimicrob Agents. 2019;54(6):681–685. doi: 10.1016/j.ijantimicag.2019.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Amoako DG, Somboro AM, Abia ALK, Allam M, Ismail A, Bester LA, et al. Genome mining and comparative pathogenomic analysis of an endemic methicillin-resistant Staphylococcus aureus (MRSA) clone, ST612-CC8-t1257-SCCmec_IVd(2B), isolated in South Africa. Pathogens. 2019;8(4):166. doi: 10.3390/pathogens8040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18. [DOI] [PMC free article] [PubMed]
- 50.Donnio PY, Fevrier F, Bifani P, Dehem M, Kervegant C, Wilhelm N, et al. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob Agents Chemother. 2007;51(12):4342–4350. doi: 10.1128/AAC.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baldan R, Testa F, Lore NI, Bragonzi A, Cichero P, Ossi C, et al. Factors contributing to epidemic MRSA clones replacement in a hospital setting. PLoS One. 2012;7(8):e43153. doi: 10.1371/journal.pone.0043153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. Access to the National Institute for Comminicable Diseases GERMS-SA database cannot be granted due to the presence of patient identifiers.