Abstract
Bacterial cell division is mediated by a protein complex known as the divisome. Many protein–protein interactions in the divisome have been characterized. In this report, we analyse the role of the PASTA (Penicillin-binding protein And Serine Threonine kinase Associated) domains of Bacillus subtilis PBP2B. PBP2B itself is essential and cannot be deleted, but removing the PBP2B PASTA domains results in impaired cell division and a heat-sensitive phenotype. This resembles the deletion of divIB, a known interaction partner of PBP2B. Bacterial two-hybrid and co-immunoprecipitation analyses show that the interaction between PBP2B and DivIB is weakened when the PBP2B PASTA domains are removed. Combined, our results show that the PBP2B PASTA domains are required to strengthen the interaction between PBP2B and DivIB.
Keywords: peptidoglycan, cell division, divisome, penicillin-binding protein, bacterial two hybrid
Introduction
The synthesis of peptidoglycan during cell division is essential for the completion of division and in fact considered one of the drivers for constriction itself [1, 2]. Cell division is mediated by a complex of proteins collectively known as the divisome (Fig. 1). In most bacteria, the divisome contains two division-specific peptidoglycan synthesis proteins, FtsW, a protein from the SEDS-family with glycosyl transferase activity, and a division-specific class B penicillin-binding protein (bPBP) with transpeptidase activity [3, 4]. These proteins interact through their transmembrane segments, and the presence of the bPBP is required for the activation of the glycosyl transferase activity of FtsW [3]. A recent co-crystal structure from the homologous RodA–PBP2 elongasome complex from Thermus thermophilus revealed the activation of RodA glycosyl transferase activity by the extracytoplasmic ‘pedestal’ domain of PBP2, and large movements of the PBP2 cytoplasmic domain that allow this activation as well as transpeptidase activity [5]. In Bacillus subtilis , the divisome SEDS/bPBP proteins are FtsW and PBP2B, which are both essential [6, 7]. Recent work from our lab and work from Daniel and colleagues has shown that it is the presence of PBP2B that is essential, rather than its transpeptidase activity, which fits with the observed activation of FtsW transglycosylase activity [3, 5, 8, 9]. This was similar to a previous report on the Streptococcus pneumoniae homologue PBP2x, of which the transpeptidase activity is also not essential [10]. Both PBPs contain two PASTA – for Penicillin-binding protein And Serine Threonine kinase Associated – domains at their C-terminus. PASTA domains are exclusively found in Gram-positive bacteria, in some high molecular mass PBPs and in eukaryotic-like serine/threonine kinases (eSTKs) [11]. These domains contain 60–70 amino acids and have a characteristic secondary structure, which consists of three β strands and an α helix; the first and the second β strands are connected by a loop, but the sequence of the domain is not well conserved. Proteins can contain single or multiple PASTA domains. A truncated version of PBP2b (at residue 632), in which part of PASTA domain 1 and the entire PASTA domain 2 are deleted, has been reported to grow as filaments [12]. In PBP2x, loss of the PASTA domains abolishes the binding of Bocillin-FL, a fluorescent penicillin derivative [13], and localization of PBP2x to the division site [10], suggesting that PASTA domains mediate the interaction with peptidoglycan. This was nicely illustrated recently by a series of crystal structures that revealed that the PBP2x PASTA domains form an allosteric binding site for a pentapeptide stem in a nascent peptidoglycan strand, which positions another stempeptide on the same strand in the active site so that it can be cross-linked [14]. The allosteric binding site is formed at the interface of the two PASTA domains and the transpeptidase domain, and comprises the entire first and part of the second PASTA domain. Binding of the terminal d-Ala-d-Ala of the stempeptide at this side displaces a ‘gatekeeper’ arginine residue on the transpeptidase domain, which subsequently forms salt bridges with an aspartate and a glutamate residue on the first PASTA domain that opens up the active site, so that the donor stempeptide for transpeptidation on the same glycan strand can bind [14]. The PASTA domains of B. subtilis PBP2B lack all the residues required for this allosteric activation.
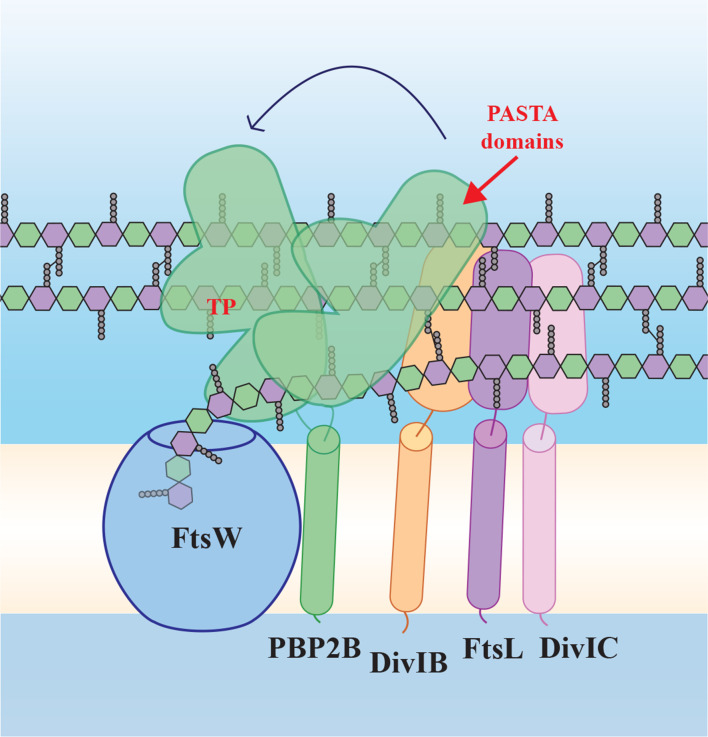
Fig. 1.
Schematic model of the ‘late division’ proteins based on the SEDS/bPBP structure [5] and our findings. FtsW and PBP2B form a SEDS/bPBP pair and interact via the transmembrane (TM) domain of PBP2B, but also via the extracytoplasmic part of PBP2B (containing the transpeptidase, TP, and PASTA domains), which has conformational flexibility. PBP2B interacts with DivIB and FtsL, which interacts with DivIC. According to our results, PBP2B PASTA domains play a role in strengthening the interaction between PBP2B and DivIB. FtsW polymerizes peptidoglycan building blocks, while PBP2B forms the cross-links between the strands. Yellow/white central band, membrane; peptidoglycan schematically drawn as hexagons (green, GlcNAc; purple, MurNAc) with attached pentapeptides.
Not all PASTA-domain containing proteins bind peptidoglycan, and various proteins contain multiple PASTA domains out of which only one can bind peptidoglycan [15]. Bioinformatics analyses revealed a key difference between PASTA domains that bind peptidoglycan and PASTA domains that do not – in a residue that determines the flexibility of the ‘putative binding pocket’, a conserved region localized at the end of the β strand β′1. Binder PASTA domains have an arginine or a glutamate residue at this position, while non-binders have a proline [15]. An example of this is B. subtilis PrkC, which induces spore germination upon peptidoglycan binding [16]. Only PrkC PASTA domain 3 contains such an arginine, and mutation of this residue completely abolishes peptidoglycan binding, indicating that PASTA domains 1 and 2 do not bind peptidoglycan [17]. Crystal structures of the three PrkC PASTA domains of Staphylococcus aureus [18] and of Mycobacterium tuberculosis PknB [19, 20], which has four PASTA domains, showed linear arrangements of the domains, in comparison with the more folded arrangement of the PBP2x two PASTA domains [18]. Other examples of proteins with PASTA domains are Staphylococcus aureus Stk1 [21] and Streptococcus pneumoniae StkP [22, 23]. PBP2B has prolines at both sites in its PASTA domains – as does PBP2x, but as explained above, PBP2x forms an allosteric binding site.
Thus, PBP2B does not have residues associated with peptidoglycan binding by its individual PASTA domains nor residues associated with an allosteric site formed between the PASTA domain and the transpeptidase domain. Combined with our previous observation that deletion of the PBP2B PASTA domains does not affect localization or binding of Bocillin-FL [8], this suggests that the PASTA domains of PBP2B have a different function than peptidoglycan binding.
Other reported functions for PASTA domains include protein localization and kinase activation [24]. In Streptococcus pneumoniae StkP, which contains four PASTA domains, the fourth domain is critical for localization through interaction with the peptidoglycan hydrolase LytB, whereas the first three PASTA domains function as a ruler that positions the fourth domain to control cell-wall thickness [25].
In this study, we have further investigated the role of the PASTA domains of PBP2B, and show that these domains strengthen the interaction between PBP2B and the divisome protein DivIB. This interaction becomes critical when cells are grown at higher temperatures.
Methods
Strains and media
Strains used in this study are listed in Table 1. All B. subtilis strains were grown in casein hydrolysate (CH) medium at 30 °C with shaking, unless other conditions are specified. When necessary, kanamycin (5 µg ml−1) and spectinomycin (50 µg ml−1) were added. To induce the expression of genes under control of the Pspac and Pxyl promoters, either IPTG (0.5 mM) or xylose (0.2 %, w/v) was added to the medium.
Table 1.
Strains
|
Strain |
Genetic features |
Source |
|---|---|---|
|
Bacillus strains | ||
|
168 |
trpC2 |
Laboratory collection |
|
3295 |
trpC2 chr::P spac -pbpB neo |
[41] |
|
4132 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA001(spc Pxyl-gfpmut-pbpB) |
[8] |
|
4133 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA002(spc Pxyl-gfpmut-pbpB1-1991) |
[8] |
|
4137 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA006(spc Pxyl-pbpB) |
[8] |
|
4138 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA007(spc Pxyl- pbpB1-1991) |
[8] |
|
4146 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA011(spc Pxyl-pbpB1-1991 prkC1068-1677) |
This work |
|
4147 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA012(spc Pxyl-gfpmut-pbpB1-1991 prkC1068-1677) |
This work |
|
4148 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA013(spc Pxyl-pbpb1-1991 spoVD1740-1890) |
This work |
|
4149 |
trpC2 chr::Pspac-pbpB neo amyE::pDMA014(spc Pxyl-gfpmut-pbpb1-1991 spoVD1740-1890) |
This work |
|
4174 |
divIB-3xFLAG ermC amyE::pDMA001(spc Pxyl‐gfpmut‐pbpB) |
This work |
|
4175 |
divIB-3xFLAG ermC amyE::pDMA002(spc Pxyl‐gfpmut‐pbpB1‐1991) |
This work |
|
GP2005 |
divIB-3xFLAG ermC |
Gift from Jörg Stülke |
|
ΔdivIB |
ΔdivIB tet |
Gift from Leendert Hamoen |
|
E. coli strains | ||
|
DH5α |
F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA–argF)U169 hsdR17 (rK − mK +) λ– |
Laboratory collection |
|
BTH101 |
F− cya-99 araD139 galE15 galK16 rpsL1 (StrR) hsdR2 mcrA1 mcrB1 |
[42] |
Construction of PBP2B chimeras
Chimeras (Fig. S1, available with the online version of this article) were constructed using restriction-free cloning [26]. Hybrid primers were used to amplify prkC and spoVD regions encoding PASTA domains from chromosomal DNA of B. subtilis . The hybrid primers were designed using the Restriction Free Cloning website (http://www.rf-cloning.org/), primers (Table S1) contained complementary sequences to prkC or spoVD and plasmids pDMA002 or pDMA007. A first PCR was performed using the hybrid primers to create a mega-primer that contains prkC or spoVD PASTA domains flanked by complementary sequences of pDMA002 or pDMA007. The mega-primers were used in a second PCR to replace the pbpB PASTA domains from pDMA002 or pDMA007 with prkC or spoVD PASTA domains. DpnI was added to the products obtained in the second PCR in order to degrade the original plasmid. After digestion, the PCR products were used to transform Escherichia coli DH5α cells. The resulting plasmids (Table 2) were sequenced and cloned into the amyE locus of B. subtilis 3295. Integration into the amyE locus was verified by growing the transformants on starch plates to check for amylase activity by iodine staining. A lack of clear zones around the colonies indicates that the construction was integrated into the amyE locus. Integration was confirmed by PCR.
Table 2.
Plasmids
|
Plasmid |
Genetic features |
Source |
|---|---|---|
|
pDMA002 |
bla amyE3′ spc Pxyl-gfpmut1-pbpB1-1991 amyE5′ |
[8] |
|
pDMA007 |
bla amyE3′ spc Pxyl-pbpB1-1991 amyE5′ |
[8] |
|
pDMA011 |
bla amyE3′ spc Pxyl-pbpB1-1991 prkC1068-1677 amyE5′ |
This work |
|
pDMA012 |
bla amyE3′ spc Pxyl-gfpmut1 pbpB1-1991 prkC1068-1677 amyE5′ |
This work |
|
pDMA013 |
bla amyE3′ spc Pxyl-pbpB1-1991 spoVD1735-1916 amyE5′ |
This work |
|
pDMA014 |
bla amyE3′ spc Pxyl-gfpmut1- pbpB1-1991 spoVD1735-1916 amyE5′ |
This work |
|
pKT25 |
Plasmid encoding T25 fragment of Bordetella pertussis cyaA, KmR |
[42] |
|
pUT18C |
Modified version of pUT18 with the polylinker located on the C-terminal end of T18, AmpR |
[42] |
|
pKT25-zip |
Derivative of pKT25 with a leucine zipper of GCN4 fused to the T25 fragment, KmR |
[42] |
|
pUT18C-zip |
Derivative of pUT18C with leucine zipper of GCN4 fused to the T18 fragment, AmpR |
[42] |
Growth curves
Strains were grown overnight in the presence of kanamycin (5 µg ml−1) and spectinomycin (50 µg ml−1) when necessary. IPTG (0.5 mM) was added to the medium to express wild-type pbpB and to ensure the proper growth of all strains before performing the growth curves. The following day, the strains were diluted to an OD600 0.05 and grown until early exponential phase. Next, cells were washed with CH medium to remove the IPTG. Cells were diluted to an OD600 0.001 in CH medium containing 0.2 % (w/v) xylose to express PBP2B, PBP2B-∆PASTA or PBP2B chimeras. A 200 µl aliquot of culture (in triplicate), of each condition under test, was loaded in a 96-well plate. The cultures were grown at 30 or 48 °C with shaking, and OD600 was measured every 10 min and recorded using a microplate spectrophotometer (Powerwave 340; Biotek).
Microscopy
Cells were grown until exponential phase. Nile red (Sigma-Aldrich) (5 µg ml−1) and DAPI (Sigma-Aldrich) (1 µg ml−1) were used to stain membranes and DNA, respectively. Cells were spotted on agarose pads (1 %, w/v, in 1x PBS) and imaged using a Nikon Ti-E microscope (Nikon Instruments) equipped with a Hamamatsu Orca Flash4.0 camera. Image analysis was performed using the software packages ImageJ (http://rsb.info.nih.gov/ij/), ObjectJ (https://sils.fnwi.uva.nl/bcb/objectj/index.html), ChainTracer [27] and Adobe Photoshop (Adobe Systems). Automated image analysis was performed using ChainTracer. Box plots were generated using BoxPlotR (http://shiny.chemgrid.org/boxplotr/). All quantitative results were derived from at least two biological replicate experiments. In order to determine whether the cell-length distributions between the strain were statistically different, first a Kolmogorov–Smirnov test was performed, which indicated that the data did not follow a normal distribution. Therefore, the non-parametric Mann–Whitney U‐test (P<0.05) was used to compare the different groups. IBM SPSS Statistic V26 was used to perform the statistical analysis.
Protein stability
Membranes from strains 4132 and 4133 grown at 30 °C on CH medium with 0.2 % (w/v) xylose were isolated. Cells were grown until exponential phase and spun down (2095 g, 7 min, 4 °C). Pellets were washed in 1x PBS and then cells were lysed by sonication. Membranes were collected by centrifugation (110 000 g, 4 °C, 50 min) and resuspended in 1x PBS. The protein concentration was measured using a DC (detergent compatible) protein assay kit (Bio-Rad Laboratories) equalized for the two strain samples and aliquots of membrane material of the same volume were prepared. Aliquots were incubated at 30 or 48 °C for 5 min, 20 min, 1 h, 2 h and 14 h. Then, Bocillin 650/665 (Thermo Fisher Scientific) (5 µg ml−1), a near-infrared version of Bocillin that is compatible with GFP, was added to each sample. Samples were further incubated at room temperature for 10 min. After incubation, sample buffer was added to each sample to stop further protein degradation, and samples were analysed by SDS-PAGE gel (10%). GFP and Bocillin 650/665 were detected using a Typhoon FLA950 fluorescent image scanner (GE Healthcare Life Science). For GFP, the 473 nm laser and the LPB (Long Pass Blue) filter were used, and for Bocillin 650/665, the 635 nm laser and the LPR (Long Pass Red) filter were used.
After imaging, the same gels were used for immunoblotting. Proteins were transferred to a PVDF membrane. Primary antibodies were anti-GFP (Thermo Fisher Scientific). Anti-Rabbit IgG alkaline phosphatase conjugated secondary antibodies (Sigma-Aldrich) were used. Blots were developed using CDP-Star (Roche) and chemiluminescence was detected using a Fujifilm LAS 4000 imager (GE Healthcare Life Science).
Bacterial two-hybrid (BACTH) assays
BACTH assays were performed using the BACTH system components (kindly provided by Fabian Commichau, Göttingen University, Germany). Sequences from divIB, divIC, ftsL, pbpb and pbpb1-1991 were amplified from chromosomal DNA of B. subtilis 168. Primers contained XbaI and KpnI restriction sites (Table S1). Fragments were cloned into pKT25 and pUT18C using XbaI and KpnI. The resulting plasmids were sequence verified and were used to co-transform E. coli BTH101. To test for protein interactions, the transformants were plated on LB agar plates containing X-Gal (40 µg ml−1), IPTG (0.5 mM), kanamycin (50 µg ml−1) and ampicillin (100 µg ml−1). Plates were incubated at 30 °C for 36 h and scored for blue colour development. The β-galactosidase assay was performed as described elsewhere [28], with some modification. E. coli BTH101 containing the plasmids to be tested were grown as overnight cultures in LB containing IPTG (0.5 mM), kanamycin (50 µg ml−1) and ampicillin (100 µg ml−1) at 30 °C. The next day, 200 µl cells were transferred to a tube containing buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol, pH 7). To permeabilize the cells, 20 µl (0.01 %, w/v) SDS and 40 µl chloroform were added to each tube. After mixing, the chloroform was allowed to settle down and 50 µl permeabilized cells were transferred to a 96-well plate containing 150 µl buffer Z. Then, 40 µl (4 %, w/v) ONPG was added to start the enzymatic reaction. When the samples were yellow, the reaction time was recorded and reactions were stopped by adding 96 µl 1M Na2CO3. The absorbance at 420 nm and 550 nm was measured in a Powerwave 340 (Biotek) microplate reader and β-galactosidase activity was calculated as:
T=time in minutes; V=volume in millilitres
Co-immunoprecipitation
Overnight (O/N) cultures of strains GP2005 (mock), 4174 (expressing GFP-PP2B) and 4175 (expressing GFP-PBP2B-∆PASTA) were diluted 1 : 100 in LB, induced with 0.2 % (w/v) xylose and grown to an OD600 0.4. Co-immunoprecipitations were performed essentially as described previously [29]. Cells were harvested, resuspended in buffer I [10 mM Tris-HCl, 150 mM NaCl, pH 7.4, with cOmplete ULTRA tablets (mini EDTA-free, EASYpack) protease inhibitors (Roche)] and disrupted via sonication. Cell debris was removed by low-speed centrifugation and membranes were isolated through ultracentrifugation (100 000 g, 1 h, 4 °C) and solubilized with 1 % (w/v) n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) in buffer I by gentle shaking (4 °C, 30 min). Solubilized material was recovered as the supernatant from a second ultracentrifugation step (100 000 g, 30 min, 4 °C). The protein concentration was determined with the DC (detergent compatible) protein assay kit (Bio-Rad Laboratories) and 200 ng total membrane proteins were incubated for 1 h at 4 °C, with gentle shaking on a roller mix with 25 µl GFP-Trap agarose beads (Chromotek) in a final volume of 100 µl 1 % (w/v) DDM in buffer I, according to the manufacturer's recommendations. Beads had been previously blocked by 1 h incubation with 1 % (w/v) BSA in the corresponding buffer. After incubation, the flow-through fraction was collected (100 µl) using centrifugation (2500 g for 2 min) at 4 °C, beads were washed twice with buffer I with 1% (w/v) DDM, and resuspended in 40 µl 1× SDS-PAGE sample buffer. Low-binding tubes (Thermo Fisher Scientific) were used during the whole process. The input, flow-through and eluate fractions were analysed by SDS-PAGE and Western blotting. Blots were developed using anti-FLAG M2 mouse mAb (Sigma-Aldrich; 1 : 1000) or anti-GFP rabbit polyclonal antibody (Chromotek; 1 : 1000) and appropriate alkaline-phosphatase conjugated secondary antibodies (1:30000). Blots were developed with CDP-Star (Roche), chemiluminescence was detected using a Fujifilm LAS4000 luminescence imager (GE Healthcare Life Science) and analysed using ImageJ (http://rsb.info.nih.gov/ij/).
Results and Discussion
Absence of PBP2B PASTA domains results in a temperature-sensitive phenotype
In a previous study [8], we created a series of strains expressing PBP2B variants from which the PASTA domains were removed (Fig. S1). As pbpB is an essential gene, we generated strains in which the expression of wild-type pbpB is under control of IPTG with an extra copy of the pbpB variant (with/without PASTA, with/without gfp) inserted in the amyE locus under control of the Pxyl promoter. This strategy allows cultivation of the strains while expressing wild-type pbpB, followed by depletion of PBP2B (Fig. 4a – Pspac pbpB strain without IPTG) and a switch to PBP2B variant production by the removal of IPTG and the addition of xylose; thus, ensuring that the observed phenotype is not a product of a suppressor mutation. Previously, we showed that PBP2B-∆PASTA was able to complement PBP2B depletion under standard conditions (CH medium, 30 °C), indicating that PASTA domains are not essential under standard conditions [8]. However, we noted that the cells were slightly elongated, which we have now quantified. The strain producing PBP2B has a mean length of 3.34 µm (n=200 cells), while the strain producing PBP2B-∆PASTA has a mean length of 4.85 µm (n=200 cells), which is ~1.5 times longer (Mann–Whitney U‐test, P=0) (Fig. 2, Tables S2 and S3). In addition, there is more variation in the length distribution of the PBP2B-∆PASTA-producing strain, as can be observed in the boxplot. When the temperature was increased to 37 °C, the mean length of the strain producing PBP2B-∆PASTA increased to 5.20 µm (n=200 cells), whereas the strain expressing PBP2B (n=200 cells) was slightly shorter than at 30 °C (Fig. 2, Table S2). In other work, we observed that some phenotypes associated with peptidoglycan synthesis defects at the septum are not observed when cells are grown on minimal medium [30]. Therefore, we also analysed the effect of the deletion of PASTA domains in cells grown in a defined minimal medium (SM medium). This revealed that the elongated phenotype for the strain expressing PBP2B-∆PASTA is still observed (Fig. S2a–c, Tables S2 and S3). At 37 °C, cells grown on SM medium were shorter overall but, again, the strain expressing PBP2B-∆PASTA displayed an elongated phenotype (Mann–Whitney U‐test, P=0).
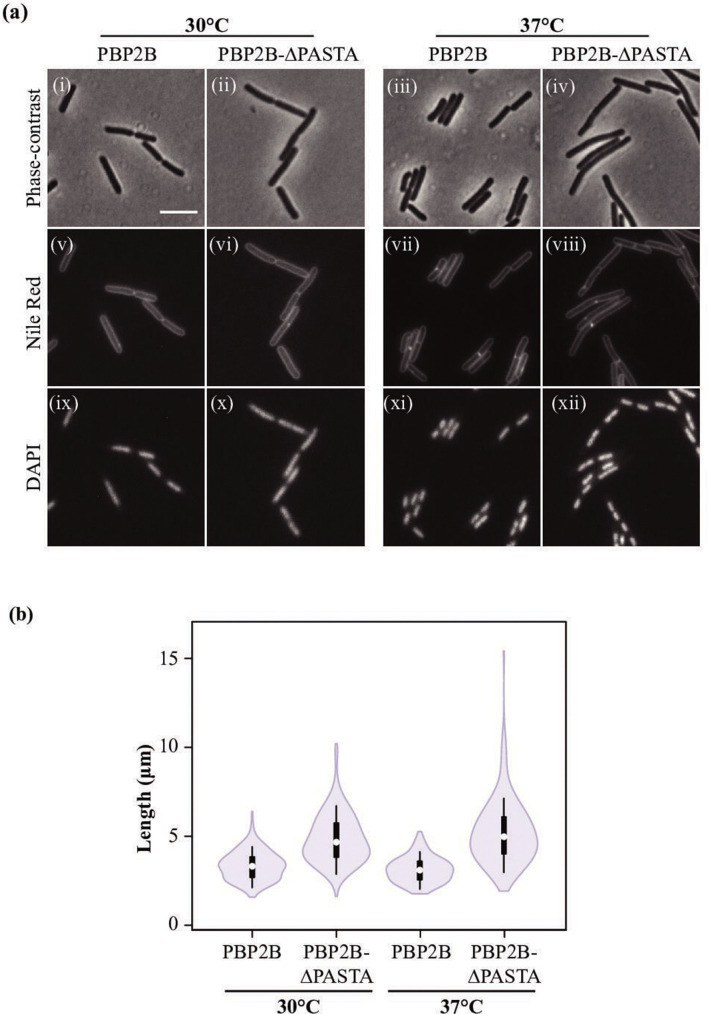
Fig. 2.
Phenotype of strains producing PBP2B and PBP2B-∆PASTA. (a) Phase-contrast microscopy of the strains producing PBP2B (strain 4137) and PBP2B-ΔPASTA (strain 4138). Cultures were grown in CH medium at 30 and 37 °C until exponential phase. Membranes and DNA were labelled with Nile red (v-viii) and DAPI (ix-xii), respectively. (i, v, ix) PBP2B (strain 4137) at 30 °C; (ii, vi, x) PBP2B-∆PASTA (strain 4138) at 30 °C; (iii, vii, xi) PBP2B at 37 °C; (iv, viii, xii) PBP2B-∆PASTA at 37 °C. Bar, 5 µm; the same for all panels. (b) Length distribution of cells. Cells were grown in CH medium at 30 or 37 °C until exponential phase. As B. subtilis forms chains, cells were labelled with Nile red in order to determine the boundaries of single cells. The lengths of the cells were obtained using ChainTracer [27]. The values obtained (n=200 per strain) are shown as box plots. White circles show the medians (PBP2B 30°C, 3.32 µm; PBP2B-ΔPASTA 30 °C, 4.67 µm; PBP2B 37 °C, 3.10 µm; PBP2B-PASTA 37 °C, 4.97 µm); box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values.
The increase in cell length is a characteristic phenotype indicative of a problem in cell division. To discard the possibility that the delay in cell division was a consequence of problems with chromosome segregation, DAPI was used to stain DNA. The PBP2B and PBP2B-∆PASTA strains grown at 30 and 37 °C presented condensed nucleoids in all cells (Fig. 2), indicating that chromosome segregation was not affected. Finally, strains expressing GFP-fusions to PBP2B and PBP2B-∆PASTA, grown at 30 °C in LB with 0.5% xylose, were scored for the presence of the GFP-PBP2B variant at the division site (Fig. S2d). GFP-PBP2B was present at the division site in 58.8 % (±4.9 %, n=609) of the cells, whereas GFP-PBP2B-∆PASTA was present at the division site in 37.8 %(±1.0 %, n=606) of the cells (Chi-square test, P<0.01). This again indicates that cell division is delayed/impaired when the PASTA domains are absent.
As we noticed that the elongation phenotype in CH medium was more severe at 37 °C than at 30 °C, the temperature was increased to 48 °C. Surprisingly, the PBP2B-∆PASTA strain did not grow at 48 °C (Fig. 3a). This result suggests that the PBP2B-∆PASTA strain is temperature sensitive. We also noted that after prolonged incubation, the control depletion strain with pbpB controlled by Pspac started growing (Fig. 3a). This also happened at lower temperatures and analysis of several depletion strains revealed that this is due to the appearance of suppressor mutations in Pspac, the promoter used to control pbpB (not shown). To get more insight into the effects of high temperature on the phenotype, the strains were grown under normal conditions (30 °C, CH medium) to make sure that the cells were growing healthy. Then, cultures were shifted to 48 °C and pictures were taken every 20 min. The strain expressing PBP2B showed no drastic changes in the phenotype during the course of the experiment (Fig. 3b). However, after 40 min, the strain expressing PBP2B-∆PASTA started to display cells with decreased contrast, a characteristic of dying cells (Fig. 3b). After 1 h at 48 °C, we observed that the amount of dying cells in the culture of the strain expressing PBP2B-∆PASTA increased. These observations confirm that the deletion of the PASTA domains from PBP2B confers a temperature-sensitive phenotype.
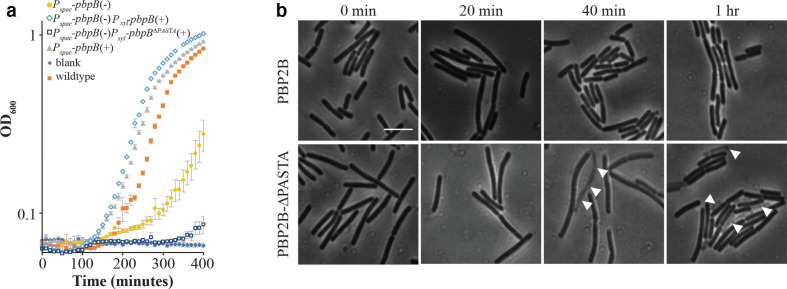
Fig. 3.
The PBP2B-∆PASTA strain is thermosensitive. (a) Growth curves in CH medium at 48 °C, OD600 was measured every 10 min. Genotypes are shown, and presence (+) or absence (–) of inducer is indicated. ♦ CH medium, blank; ■ wild-type (strain 168); ▲ Pspac pbpB(+) (strain 3295); ● Pspac pbpB(–) (strain 3295); ◊ Pspac pbpB(–) Pxyl pbpB(+) (strain 4137); □ Pspac pbpB(–) Pxyl pbpBΔPASTA(+) (strain 4138). Representative results from three independent experiments are shown. All experiments were performed in triplicate. The resulting mean and se are shown for each time point. (b) Lysis of cells at 48 °C. Cells from strains 4137 (PBP2B) and 4138 (PBP2B-ΔPASTA) were grown at 30 °C with xylose and without IPTG until early exponential phase, then cells were shifted to 48 °C and followed by microscopy every 20 min. White arrowheads indicate dead cells. Bar, 5 µm. Representative results from three independent experiments are shown.
PASTA domains of PBP2B can be partially replaced by PASTA domains from other Bacillus proteins
B. subtilis has two other proteins that contain PASTA domains, SpoVD and PrkC. SpoVD is a PBP paralogous to PBP2B. It is crucial for spore cortex synthesis and contains a single PASTA domain [31]. PrkC, which contains three PASTA domains, is a eukaryotic-like serine/threonine kinase that is involved in processes like germination and biofilm formation, WalR activation and that localizes to the septum [16, 32, 33]. In order to test whether the PASTA domains of SpoVD and PrkC were able to replace the function of the PASTA domains of PBP2B, the PBP2B PASTA domains were exchanged for PASTA domains from SpoVD or PrkC (Fig. S1). Growth of the strains expressing the chimera proteins was followed at 30 °C in CH medium (Fig. 4a), and was found to be similar to the background deletion strain. This indicates that the exchange of the PASTA domains did not interfere with the essential function of PBP2B. The cells expressing the chimera proteins were examined by microscopy, which showed that the cells expressing the chimeras were longer (Table S3) than cells expressing wild-type PBP2B [3.34 µm (±0.06)]. The PBP2B-PASTASpoVD chimera [3.57 µm (±0.07)] resulted in mild elongation, whereas the PBP2B-PASTAPrkC chimera [4.55 µm (±0.12)] gave a more striking elongation, although not to the same extent as PBP2B-∆PASTA cells [4.85 µm (±0.10)] (Fig. 4c, e, Tables S2 and S3).
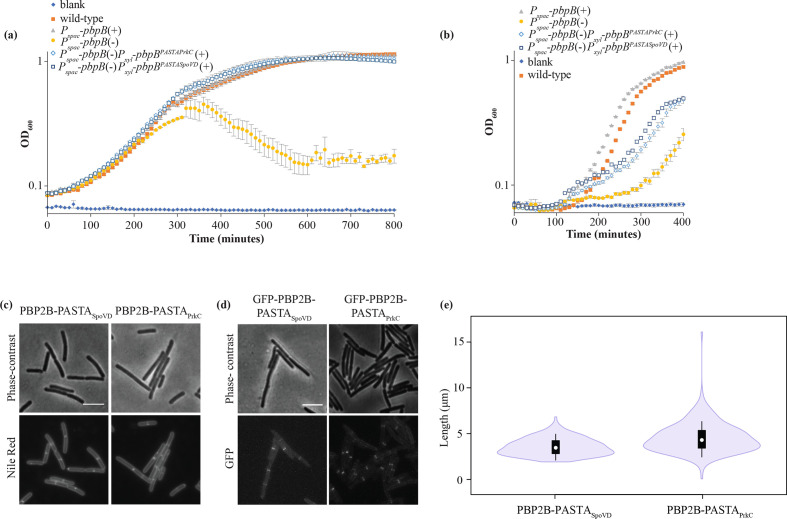
Fig. 4.
Phenotype of strains expressing PBP2B PASTA chimeras. (a, b) Growth curves in CH medium at 30 °C (a) and at 48 °C (b), OD600 was measured every 10 min. Genotypes are shown and presence (+) or absence (−) of inducer is indicated. ♦, CH medium, blank; ■, wild-type (strain 168); ▲, Pspac-pbpB(+) (strain 3295); ●, Pspac pbpB(−) (strain 3295); ◊, Pspac-pbpB (−) Pxyl-pbpBPASTAPrkC(+) (strain 4146); □, Pspac-pbpB (−) Pxyl-pbpBPASTASpoVD(+) (strain 4148). Representative results from three independent experiments are shown. All experiments were performed in triplicate. The resulting mean and se are shown for each time point. (c–e) Microscopy and length distributions of cells grown at 30 °C in CH medium with xylose (no IPTG) until exponential phase. (c) Cells expressing PBPB2-PASTASpoVD (strain 4148) and PBP2B-PASTAPrkC (strain 4146), labelled with Nile red and imaged by phase-contrast and fluorescence microscopy. Representative results from three independent experiments are shown. Bar, 5 µm. (d) Cells producing GPF-PBP2B-PASTASpoVD (strain 4149) and GFP-PBP2B-PASTAPrkC (strain 4147), imaged with phase-contrast and fluorescence microscopy. Bar, 5 µm. (e) Length distribution of cells producing PBP2B-PASTASpoVD (strain 4148) and PBP2B-PASTAPrkC (strain 4146), the chimeras imaged in (c). The length of cells was obtained using ChainTracer [27]. The measured lengths (n = 200 per strain) are shown as box plots. White circles show the medians (PBP2B-SpoVD, 3.46 µm; PBP2B-PrkC, 4.30 µm); box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values.
GFP-fusions to the chimera proteins showed that both chimeras localize to division sites, as expected from the observation that the chimeras do not interfere with the essential function of PBP2B (Fig. 4d). Subsequently, these strains were grown at 48 °C to see whether the chimeric proteins complemented the temperature-sensitive phenotype. Although both chimeric proteins did allow some growth at 48 °C, the lag phase of the cells was longer compared to the PBP2B strain and cells did not reach similar OD600 values (Fig. 4b). Again, the strain expressing the PBP2B-PASTAPrkC chimera was most affected. This is not wholly surprising, as the structural organization of PrkC PASTA domains seemed different from that of PBP PASTA domains [18], while SpoVD is a sporulation-specific PBP that, like PBP2B, is partnered with a cognate SEDS protein, SpoVE [31, 34]. These results indicate that the PASTA domains from other B. subtilis proteins can partially, but not fully, complement the absence of the PBP2B PASTA domains.
PBP2B PASTA domains strengthen the interaction with DivIB
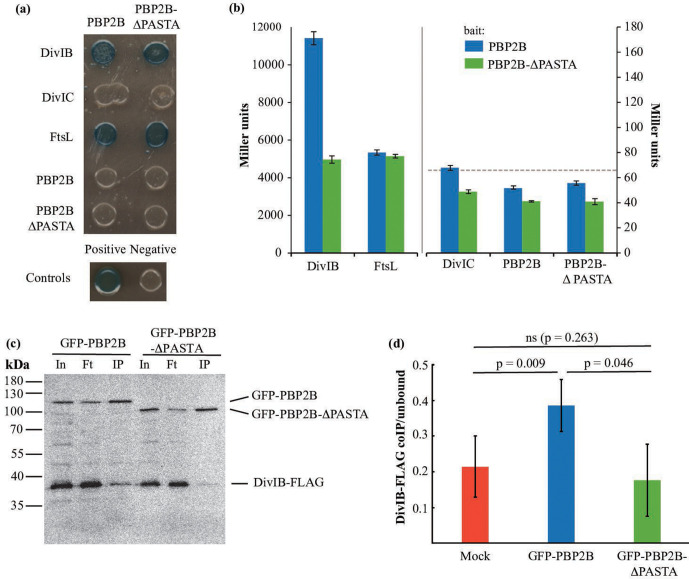
A possible explanation for the temperature-sensitive phenotype of the strain expressing PBP2B-∆PASTA is that the PBP2B-∆PASTA protein becomes more labile at increased temperatures. An analysis of PBP2B and PBP2B-∆PASTA stability in isolated membranes at 30 and 48 °C revealed that although PBP2B is less stable at 48 °C, both PBP2B and PBP2B-∆PASTA are degraded at similar rates (Fig. S3). This indicates that the in vitro intrinsic stability of these proteins is the same. It could be that in vivo there is a difference, but such a difference would be indistinguishable from increased degradation caused by a secondary effect such as divisome instability resulting in degradation [35]. We noted that the temperature sensitivity of the PBP2B-∆PASTA strain was similar to the phenotype of a divIB deletion strain [36]. DivIB (in other organisms FtsQ) is a divisome protein that interacts with DivIC (in other organisms FtsB) and FtsL, and that regulates the turnover of FtsL and DivIC [35, 37]. This turnover is regulated by PBP2B and the transpeptidase domain of PBP2B has been shown to interact with the C-terminus of DivIB [37–39]. We hypothesized that the absence of the PASTA domains from PBP2B might influence the interaction with DivIB and/or other proteins. To test this, we performed a BACTH assay, in which we tested the ability of PBP2B and PBP2B-∆PASTA to interact with DivIB, DivIC, FtsL and itself. On plates, we confirmed the previous result from Daniel and colleagues [37] that PBP2B interacts with DivIB and FtsL, but not with DivIC, and found no apparent difference between PBP2B and PBP2B-∆PASTA (Fig. 5a). Notably, we did not detect a PBP2B self-interaction. Also, we only found positive results when the PBP2B variants were expressed from the pKT25 plasmid (Fig. S4) – this is probably due to the difference in copy numbers between the two plasmids used in the assay and not uncommon in BACTH screens of interactions between PBPs and other proteins [40]. We also analysed the interactions using a β-galactosidase assay (Fig. 5b), which has the added benefit of providing a quantitative result, which can give a hint about the strength of the interaction. It has to be noted that the 'strength' of an interaction does not scale 1 : 1 with β-galactosidase activity and, thus, changes in activity are only indicative of a change in interaction. The β-galactosidase assay confirmed the interactions of PBP2B with DivIB and FtsL, and the observation that the interaction with DivIB ‘appears’ the strongest fit with the observation of Robichon and colleagues who used an artificial targeting assay in E. coli to show that the interaction with DivIB is the strongest interaction between PBP2B and other division proteins [39]. In the absence of the PASTA domains, the activity resulting from the interaction with DivIB was roughly halved, whereas the activity resulting from the interaction with FtsL was unchanged (Fig. 5b). This result suggests that the PASTA domains of PBP2B are not required for the interaction with DivIB, but that they do increase the strength of the interaction.
Fig. 5.
The interaction between PBP2B and DivIB is diminished in the absence of the PASTA domains. (a, b) BACTH assay. (a) Interaction assay on plates containing X-Gal. PBP2B, PBP2B∆-PASTA, DivIB, DivIC and FtsL were cloned into plasmids pKT25 and pUT18C and used to co-transform E. coli BTH101. Co-transformants were grown on LB plates containing X-Gal and incubated at 30 °C for 36 h. Blue colonies are considered indicative of protein–protein interaction. PBP2B and PBP2B ∆PASTA were used as bait in pKT25, while prey proteins were expressed in pUT18C. Positive control, transformants containing pKT25-zip and pUT18C-zip; negative control, transformants containing empty pKT25 and pUT18. Representative results from three independent experiments are shown. (b) β-Galactosidase assay. Interaction between PBP2B and PBP2B-∆PASTA cloned into pKT25 in combination with the late division protein cloned into pUT18C. The positive control showed an activity of 63 278 Miller units and the negative control 66 (shown as a dotted line). Note the different scales on the left and right y-axes, and the discontinued x-axis, indicated by a break and a vertical grey line. Representative results from three independent experiments are shown. All experiments were performed in triplicate. The resulting mean and se are shown for each interaction. (c, d) Co-immunoprecipitation assay. (c) Western blot of a co-immunoprecipitation experiment. Solubilized membranes from strains producing DivIB-FLAG and GFP-PBP2B (4174) or GFP-PBP2B-∆PASTA (4175) were immunoprecipitated using GFP-Trap agarose beads. Input (In), flow-through (Ft) and eluted (IP) material was analysed by SDS-PAGE/Western blot, and the blot was simultaneously developed using anti-FLAG and anti-GFP antibodies. (d) Quantification of the fraction of DivIB-FLAG co-immunoprecipitated with either a mock control, GFP-PBP2B or GFP-PBP2B-∆PASTA, expressed as the ratio of the signal of DivIB-FLAG in the immunoprecipitate (IP) fraction to the signal of DivIB-FLAG in the flowthrough sample. Bars show the mean of three independent biological replicates with sd. The Western blots used for the quantification are shown in Fig. S4(c). ns is not significant.
To validate the results from the BACTH experiments, co-immunoprecipitation experiments were performed. GFP-PBP2B and GFP-PBP2B-∆PASTA were produced in a B. subtilis strain that produces a FLAG-tagged version of DivIB at the native locus under control of the wild-type promoter (GP2005, a kind gift from Jörg Stülke). DivIB-FLAG is functional as the GP2005 strain is not thermosensitive (not shown). Anti-GFP nanobodies coupled to agarose (GFP-Trap) were used to immunoprecipitate GFP-PBP2B and GFP-PBPB2B-∆PASTA, and the immunoprecipitates were analysed by Western blot, with detection using anti-FLAG and anti-GFP antibodies. The amount of DivIB-FLAG immunoprecipitated from cells producing GFP-PBP2B appeared significantly higher than that from cells producing GFP-PBP2B-∆PASTA, although the overall recovery in both cases was low (Fig. 5c). This was confirmed by quantification of the amount of immunoprecipitated DivIB-FLAG as a fraction of the unbound material in the flowthrough of the sample, in an experiment where strain GP2005, which produces only DivIB-FLAG, was included as a mock control and the blots were developed with anti-FLAG only, to avoid potential cross-reactivity (Fig. 5d and S4). Recovery of DivIB-FLAG by GFP-PBP2B was significantly higher than from the mock and the GFP-PBP2B-∆PASTA producing strain, where levels were not higher than the background. Combined, the BACTH and co-immunoprecipitation results indicate that the PASTA domains of PBP2B strengthen the DivIB-PBP2B interaction.
Conclusions
In this paper, we show that although the PASTA domains are not absolutely essential for the scaffolding role of PBP2B, they do become essential at elevated temperatures. This phenotype is similar to the phenotype described for a divIB deletion [36], suggesting the PASTA domains are involved in the same pathway. We could show that the PASTA domains are involved in the interaction between PBP2B and DivIB using both a BACTH and a co-immunoprecipitation approach. Earlier, King and colleagues identified an interaction between the C-terminal part of DivIB and the transpeptidase domain of PBP2B [38]. In the modelled structure of PBP2B, the transpeptidase domain and the DivIB C-terminus are at similar distance from the membrane, but this is also the distance at which the PASTA domains can be found [38]. It is possible that the PASTA domains strengthen the interaction between DivIB and the transpeptidase domain, but alternatively the PASTA domains interact with another region of DivIB. Given the broad range of conformations recently reported for the elongasome bPBP bound to its SEDS partner, and the influence on these conformations by interaction partners such as MreC [5], it is tempting to speculate that the PASTA domains may be required to stabilize one of these conformations in PBP2B, to control peptidoglycan synthesis (Fig. 1). Our results show that PASTA domains can have distinct functions in similar proteins – whereas the PASTA domains in Streptococcus pneumoniae clearly function to allosterically activate the transpeptidase activity of the protein [14], their function in B. subtilis PBP2B seems to be the stabilization of an important protein–protein interaction in the divisome.
Supplementary Data
Funding information
This work was partially supported by a VIDI fellowship (864.09.010) from the Netherlands Organisation for Scientific Research (NWO).
Acknowledgements
We would like to thank Fabian Commichau and Jörg Stülke (Göttingen University, Germany) for the kind gifts of the BACTH plasmids and strain GP2005, respectively. We thank Leendert Hamoen (University of Amsterdam, The Netherlands) for the ΔdivIB tet strain. We also thank Tal Shamia (Chromotek GmbH) for advice on the use of GFP-Trap beads.
Author contributions
Conceptualization – D.M.A., D.-J.S. Methodology – D.M.A., A.M.-V., L.C.B. Validation – D.M.A., A.M.-V., L.C.B. Formal analysis – D.M.A., A.M.-V. Investigation – D.M.A., A.M.-V., L.C.B. Writing – original draft preparation – D.M.A., D.-J.S. Writing – review and editing – D.M.A., A.M.-V., L.C.B., D.-J.S. Visualization – D.M.A., A.M.-V., D.-J.S. Supervision – D.-J.S. Project administration – D.-J.S. Funding – D.-J.S.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: BACTH, bacterial two-hybrid; PASTA, penicillin-binding protein and serine threonine kinase associated; PBP, penicillin-binding protein.
Three supplementary tables and four supplementary figures are available with the online version of this article.
Edited by: J. Stülke and E. L Denham
References
- 1.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature. 2018;554:528–532. doi: 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.den Blaauwen T, Hamoen LW, Levin PA. The divisome at 25: the road ahead. Curr Opin Microbiol. 2017;36:85–94. doi: 10.1016/j.mib.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol. 2019;4:587–594. doi: 10.1038/s41564-018-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, Patel V, Helmann JD, Dörr T. Don't let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Mol Microbiol. 2017;106:847–860. doi: 10.1111/mmi.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjodt M, Rohs PDA, Gilman MSA, Erlandson SC, Zheng S, et al. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat Microbiol. 2020;5:813–820. doi: 10.1038/s41564-020-0687-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel RA, Williams AM, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis . J Bacteriol. 1996;178:2343–2350. doi: 10.1128/JB.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales Angeles D, Liu Y, Hartman AM, Borisova M, de Sousa Borges A, et al. Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol Microbiol. 2017;104:319. doi: 10.1111/mmi.13629. [DOI] [PubMed] [Google Scholar]
- 9.Sassine J, Xu M, Sidiq KR, Emmins R, Errington J, et al. Functional redundancy of division specific penicillin-binding proteins in Bacillus subtilis . Mol Microbiol. 2017;106:304–318. doi: 10.1111/mmi.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters K, Schweizer I, Beilharz K, Stahlmann C, Veening J-W, et al. Streptococcus pneumoniae PBP2x mid-cell localization requires the C-terminal PASTA domains and is essential for cell shape maintenance. Mol Microbiol. 2014;92:733–755. doi: 10.1111/mmi.12588. [DOI] [PubMed] [Google Scholar]
- 11.Yeats C, Finn RD, Bateman A. The PASTA domain: a β-lactam-binding domain. Trends Biochem Sci. 2002;27:438. doi: 10.1016/S0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 12.Yanouri A, Daniel RA, Errington J, Buchanan CE. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis . J Bacteriol. 1993;175:7604–7616. doi: 10.1128/JB.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer P, Todorova K, Sauerbier J, Hakenbeck R. Mutations in Streptococcus pneumoniae penicillin-binding protein 2x: importance of the C-terminal penicillin-binding protein and serine/threonine kinase-associated domains for beta-lactam binding. Microb Drug Resist. 2012;18:314–321. doi: 10.1089/mdr.2012.0022. [DOI] [PubMed] [Google Scholar]
- 14.Bernardo-García N, Mahasenan KV, Batuecas MT, Lee M, Hesek D, et al. Allostery, recognition of nascent peptidoglycan, and cross-linking of the cell wall by the essential penicillin-binding protein 2x of Streptococcus pneumoniae . ACS Chem Biol. 2018;13:694–702. doi: 10.1021/acschembio.7b00817. [DOI] [PubMed] [Google Scholar]
- 15.Calvanese L, Falcigno L, Squeglia F, D'Auria G, Berisio R. Structural and dynamic features of PASTA domains with different functional roles. J Biomol Struct Dyn. 2017;35:2293–2300. doi: 10.1080/07391102.2016.1217274. [DOI] [PubMed] [Google Scholar]
- 16.Shah IM, Laaberki M-H, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Squeglia F, Marchetti R, Ruggiero A, Lanzetta R, Marasco D, et al. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J Am Chem Soc. 2011;133:20676–20679. doi: 10.1021/ja208080r. [DOI] [PubMed] [Google Scholar]
- 18.Ruggiero A, Squeglia F, Marasco D, Marchetti R, Molinaro A, et al. X-ray structural studies of the entire extracellular region of the serine/threonine kinase PrkC from Staphylococcus aureus . Biochem J. 2011;435:33–41. doi: 10.1042/BJ20101643. [DOI] [PubMed] [Google Scholar]
- 19.Mir M, Asong J, Li X, Cardot J, Boons G-J, et al. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barthe P, Mukamolova GV, Roumestand C, Cohen-Gonsaud M. The structure of PknB extracellular PASTA domain from Mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure. 2010;18:606–615. doi: 10.1016/j.str.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Paracuellos P, Ballandras A, Robert X, Kahn R, Hervé M, et al. The extended conformation of the 2.9-Å crystal structure of the three-PASTA domain of a Ser/Thr kinase from the human pathogen Staphylococcus aureus . J Mol Biol. 2010;404:847–858. doi: 10.1016/j.jmb.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Maestro B, Novaková L, Hesek D, Lee M, Leyva E, et al. Recognition of peptidoglycan and β-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae . FEBS Lett. 2011;585:357–363. doi: 10.1016/j.febslet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morlot C, Bayle L, Jacq M, Fleurie A, Tourcier G, et al. Interaction of penicillin-binding protein 2x and Ser/Thr protein kinase StkP, two key players in Streptococcus pneumoniae R6 morphogenesis. Mol Microbiol. 2013;90:88–102. doi: 10.1111/mmi.12348. [DOI] [PubMed] [Google Scholar]
- 24.Pensinger DA, Schaenzer AJ, Sauer J-D. Do shoot the messenger: PASTA kinases as virulence determinants and antibiotic targets. Trends Microbiol. 2018;26:56–69. doi: 10.1016/j.tim.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zucchini L, Mercy C, Garcia PS, Cluzel C, Gueguen-Chaignon V, et al. PASTA repeats of the protein kinase StkP interconnect cell constriction and separation of Streptococcus pneumoniae . Nat Microbiol. 2018;3:197–209. doi: 10.1038/s41564-017-0069-3. [DOI] [PubMed] [Google Scholar]
- 26.Bond SR, Naus CC. RF-Cloning.org: an online tool for the design of restriction-free cloning projects. Nucleic Acids Res. 2012;40:W209–W213. doi: 10.1093/nar/gks396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syvertsson S, Vischer NOE, Gao Y, Hamoen LW. When phase contrast fails: ChainTracer and NucTracer, two ImageJ methods for semi-automated single cell analysis using membrane or DNA staining. PLoS One. 2016;11:e0151267. doi: 10.1371/journal.pone.0151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battesti A, Bouveret E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli . Methods. 2012;58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Scheffers D-J, Robichon C, Haan GJ, den Blaauwen T, Koningstein G, et al. Contribution of the FtsQ transmembrane segment to localization to the cell division site. J Bacteriol. 2007;189:7273–7280. doi: 10.1128/JB.00723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielińska A, Savietto A, de Sousa Borges A, Roelofsen JR, Hartman AM, et al. Membrane fluidity controls peptidoglycan synthesis and MreB movement. bioRxiv. 2019:736819. doi: 10.1101/736819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniel RA, Drake S, Buchanan CE, Scholle R, Errington J. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol. 1994;235:209–220. doi: 10.1016/S0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- 32.Libby EA, Goss LA, Dworkin J. The eukaryotic-like Ser/Thr kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis . PLoS Genet. 2015;11:e1005275. doi: 10.1371/journal.pgen.1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pompeo F, Foulquier E, Serrano B, Grangeasse C, Galinier A. Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis . Mol Microbiol. 2015;97:139–150. doi: 10.1111/mmi.13015. [DOI] [PubMed] [Google Scholar]
- 34.Fay A, Meyer P, Dworkin J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol. 2010;399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel RA, Errington J. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol Microbiol. 2000;36:278–289. doi: 10.1046/j.1365-2958.2000.01857.x. [DOI] [PubMed] [Google Scholar]
- 36.Beall B, Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/JB.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel RA, Noirot-Gros M-F, Noirot P, Errington J. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J Bacteriol. 2006;188:7396–7404. doi: 10.1128/JB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowland SL, Wadsworth KD, Robson SA, Robichon C, Beckwith J, et al. Evidence from artificial septal targeting and site-directed mutagenesis that residues in the extracytoplasmic β domain of DivIB mediate its interaction with the divisomal transpeptidase PBP 2B. J Bacteriol. 2010;192:6116–6125. doi: 10.1128/JB.00783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robichon C, King GF, Goehring NW, Beckwith J. Artificial septal targeting of Bacillus subtilis cell division proteins in Escherichia coli: an interspecies approach to the study of protein-protein interactions in multiprotein complexes. J Bacteriol. 2008;190:6048–6059. doi: 10.1128/JB.00462-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fenton AK, Manuse S, Flores-Kim J, Garcia PS, Mercy C, et al. Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc Natl Acad Sci USA. 2018;115:2812–2817. doi: 10.1073/pnas.1715218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamoen LW, Errington J. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J Bacteriol. 2003;185:693–697. doi: 10.1128/JB.185.2.693-697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.