Abstract
Heat is a major abiotic stress that seriously affects watermelon (Citrullus lanatus) production. However, its effects may be mitigated through grafting watermelon to heat tolerant bottle gourd (Lagenaria siceraria) rootstocks. Understanding the genetic basis of heat tolerance and development of reliable DNA markers to indirectly select for the trait are necessary in breeding for new varieties with heat tolerance. The objectives of this study were to investigate the inheritance of heat tolerance and identify molecular markers associated with heat tolerance in bottle gourd. A segregating F2 population was developed from a cross between two heat tolerant and sensitive inbred lines. The population was phenotyped for relative electrical conductivity (REC) upon high temperature treatment which was used as an indicator for heat tolerance. QTL-seq was performed to identify regions associated with heat tolerance. We found that REC-based heat tolerance in this population exhibited recessive inheritance. Seven heat-tolerant quantitative trait loci (qHT1.1, qHT2.1, qHT2.2, qHT5.1, qHT6.1, qHT7.1, and qHT8.1) were identified with qHT2.1 being a promising major-effect QTL. In the qHT2.1 region, we identified three non-synonymous SNPs that were potentially associated with heat tolerance. These SNPs were located in the genes that may play roles in pollen sterility, intracellular transport, and signal recognition. Association of the three SNPs with heat tolerance was verified in segregating F2 populations, which could be candidate markers for marker assisted selection for heat tolerance in bottle gourd. The qHT2.1 region is an important finding that may be used for fine mapping and discovery of novel genes associated with heat tolerance in bottle gourd.
Introduction
Heat stress negatively affects physiological processes, reproduction, and adaptation in crop plants, which are exacerbated by global climate change [1,2]. Watermelon, Citrullus lanatus var. lanatus, is an important vegetable crop worldwide [3]. Despite of its tropical origin, watermelon production in many parts of the world is adversely affected by high temperatures (> 35°C) during the summer months [4]. Heat tolerance is a complex trait controlled by quantitative trait loci (QTL), which makes it difficult to introgress multiple favorable alleles into recipient susceptible varieties [5]. There have been attempts to deconstruct stress tolerance into measurable components for accurate phenotyping, with the aim that QTL associated with heat tolerance may then be identified and suitable alleles may be introgressed into elite genetic backgrounds [6]. For example, cell membrane stability as an indicator of heat stress may be quantified by relative electrical conductivity (REC). REC is highly sensitive to abiotic stress [7,8] and has been used in studies of abiotic stress tolerance in a range of crops, including salinity-alkalinity tolerance in muskmelon [9], drought tolerance in Perennial ryegrass [10], and cold tolerance in alfalfa [11].
One method to mitigate abiotic and biotic stresses in vegetable production is grafting [12]. For example, bottle gourd, Lagenaria siceraria (Mol.) Standl., has been used as the rootstock for watermelon to reduce heat stress and improve performance of plant growth [13,14]. Bottle gourd is a relative of watermelon in the Cucurbitaceae family, which originated from Africa but it is now widely distributed across the tropics [15,16]. In its long-term adaptation, bottle gourd has gained excellent tolerance to high temperatures and consequently, to heat stress [17,18]. In practice, due to the close genetic relatedness between bottle gourd and watermelon, there is a high degree of grafting compatibility between the two species [19]. However, little is known about the genetic basis of heat tolerance in bottle gourd.
Understanding the genetics of heat tolerance in bottle gourd and identification of DNA markers may facilitate development of novel bottle gourd rootstocks for heat adaptation of scion through marker-assisted selection. Recent progress in genetic and genomics resources in bottle gourd is also making this possible. For example, Xu et al. [20] reported partial sequencing of the bottle gourd genome using the 454 GS-FLX Titanium sequencing platform, from which 400 SSR markers were developed. The RAD-Seq [21] technology has also been applied to an F2 bottle gourd population for SNP and insertion-deletions marker development [22,23]. More recently, Wu et al. [16] reported a high-quality bottle gourd genome sequence which allowed reconstruction of the most recent common Cucurbitaceae ancestor genome through comparison with available extant modern cucurbit genome resources [24–26].
In addition, recent development of high throughput sequencing and genotyping technologies is also accelerating molecular marker development and genetic mapping studies for horticulturally important traits in vegetable crops. One such technique is QTL-Seq which combines bulked-segregant analysis (BSA) and high throughput genome sequencing for quick identification of QTLs [27]. QTL-seq has been widely used in a range of crops such as sunflower [28], rice [29], sorghum [30], potato [31], and cucumber [32,33] for the efficient detection of QTLs for complex quantitative traits. QTL detection for heat tolerance has also been studied in rice [34], chickpea [35], wheat [36], and tomato [37]. In this study, we used QTL-seq to identify major loci regulating heat tolerance in bottle gourd based on REC. Putative candidate genes controlling heat tolerance and SNPs markers that are highly associated with candidate genes were identified using the available genomic sequence for bottle gourd [21].
Materials and methods
Plant material and phenotyping
An F2 population was developed from the cross between two bottle gourd inbred lines, the heat tolerant L1 (P17) and the heat stress sensitive L6 (P23) [38]. Evaluation of heat tolerance was conducted in three experiments with 147, 56, and 60 F2 individuals, respectively. L1, L6 and F1 plants (10 each) were included in each experiment. The seedlings were planted in 32-hole plastic plugs (~ 230cm3/hole) filled with nursery substrate (2:1 mix of turf: vermiculite) and grown in a phytotron set to 16h light/8h dark cycle (30,000lux) at 25/18°C and 80% relative humidity. The first true leaf of the seedlings was collected for DNA extraction. When the third true leaf of the seedlings began to expand, heat stress was applied by moving the seedlings into a phytotron set at 40°C temperature under 80% relative humidity and 30,000lux for 6h of continuous heat exposure.
The electrical conductivity (EC) of leaves was measured before and after heat treatment to assess cell membrane damage, as described by Zhou and Leul [39] and He et al. [40] with modifications. In brief, sampled leaves were washed using deionized water, cut into 0.5-cm pieces, and immersed in deionized water for 30 min. Then, EC of the solution was measured using a conductivity meter (PHSJ-3F, Jingmi Instruments Co., Ltd., Shanghai, China) and recorded as S1. After boiling the leaf samples for 15 min, EC of the solution at room temperature was measured again and recorded as S2. A relative EC (REC) was calculated as . Leaf relative injury (LRI) was used as a metric of cell membrane damage, which was calculated as , where Lt and Lck are the REC values before and after heat exposure, respectively. A larger LRI value indicates less heat tolerance [41–43].
Analysis of variance (ANOVA) and t-tests were used to determine differences in heat tolerance, as indicated by mean (±SE) LRI, among P1, P2, and F1 plants at P < 0.05 using SPSS 13.0 software [16]. For the F2 population, the frequency distribution of LRI was plotted as a histogram and the normal distribution was fitted using the GaussAmp function in ORIGIN 9.1 software [44].
QTL-Seq and data analysis
Genomic DNA was isolated from young leaves of parental lines L1 and L6, and 147 F2 plants from the first experiment using the CTAB method [45]. For QTL-seq, two DNA pools (heat tolerance: T-pool; heat sensitive: S-pool) were constructed by mixing an equal amount of DNA from nine heat tolerant (LRI = 1 − 5%) and nine heat sensitive (LRI = 55 − 77%) individuals from the F2 population. Sequencing libraries of ~ 500 bp insert size of the two pools were constructed and pair-end sequenced (150 bp) at ~ 15× coverage for the two parents and ~ 20×coverage for each pool on an Illumina Hi-Seq 4000 at Shanghai Biozeron Co., Ltd. The raw sequence data were generated by Illumina base calling software CASAVA v1.8.2 [46] and raw paired end reads were trimmed and quality controlled by Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic) with default setting. The high-quality clean reads from L1, L6, T-pool and S-pool were aligned against the reference genome of bottle gourd cv. Hangzhou Gourd (http://cucurbitgenomics.org/organism/13, [21]) using BWA software (http://bio-bwa.sourceforge.net/, [47]). After removing the duplicate reads using Picard Tools (http://picard.sourceforge.net/), SNPs were detected from the valid BAM file using the GATK “UnifiedGenotyper” function (http://www.broadinstitute.org/gatk/). Low-quality SNPs with quality value < 20 and read depth coverage < 4× or > 200× were excluded [48]. SNPs with the consistent differential base type with two parents were remained.
The SNP-index for each SNP position was calculated for the T-pool and S-pool using the formula:
[49]. To identify candidate regions for heat tolerance QTLs, the Δ(SNP-index) for all the SNP positions with given read depths under the null hypothesis of no QTLs was obtained by subtracting the SNP-index of T-pool from the S-pool [32]. The statistical confidence intervals of Δ(SNP-index) were plotted [27]. For each read depth, 95% and 99% confidence intervals of Δ(SNP-index) were obtained following Takagi et al. [27]. The regions (P < 0.01) were then designated as QTLs.
To identify the parent that contributed to a putative QTL, the profile of L1 allele frequency difference (L1AFD) between the T-pool and S-pool was plotted using a 1 Mbp sliding window moving across the genome with a fixed step length of smoothing window size of 10 kb. The L1 allele frequency within each window in each pool was estimated using the formula developed by Yang et al. [50]. For a putative QTL, a positive value for L1AFD indicated that L1 increased heat tolerance and a negative value indicated that L1 decreased heat tolerance [33,50].
For the polymorphic SNPs located in the genomic regions that harbored the major-effect heat tolerance QTL, further functional annotation was completed using the available bottle gourd genome data (http://cucurbitgenomics.org/organism/13) and Uniprot database (www.uniprot.org), using ANNOVAR analysis (http://www.openbioinformatics.org/annovar/) to detect putative candidate genes [21].
SNP marker development and selection
Based on gene annotations and putative functions, five nonsynonymous candidate SNPs located in the qHT2.1 interval that are related to heat stress tolerance were selected for marker development. Flanking sequences were used for PCR primer development using Primer 6.0 soft (http://www.PromerBiosoft.com). The five SNPs were genotyped for four heat tolerant and four sensitive F2 individuals which were used to generate the two DNA pools. If the SNPs showed polymorphism between the heat tolerant and sensitive lines, they were used to genotype additional plants. Eighteen new individuals including six heat tolerant individuals (LRI < 10%) and twelve sensitive individuals (LRI > 10%) were selected from the same F2 population used for DNA pool construction. The LRI threshold was set to 10% because the mean LRI value of the tolerance parent L1 was 9.88±5.00%. PCR and Sanger sequencing were used and sequence results were visualized and checked using the SnapGene software. We used trait-marker association to select the most promising SNP markers.
Results
LRI as an indicator of heat tolerance
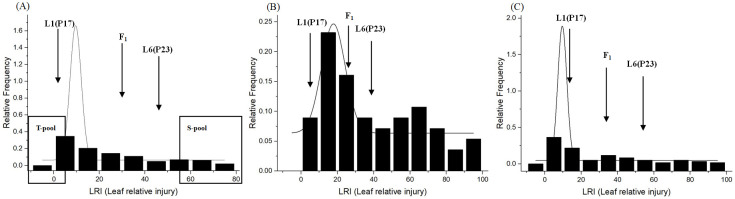
There were significant differences in the mean LRI values between L1 (P17: 9.88±5.00%) and L6 (P23: 47.26±10.78%) under heat stress conditions (Table 1). The mean LRI value of F1 (31.98±6.64%) was significantly higher than that of L1 but not significantly different with that of L6, suggesting the recessive nature of heat tolerance. The LRI of the F2 progeny ranged from 0.34% to 97.49%, with mean LRI value of 29.68±8.52% (Table 1). Transgressive segregation was observed on both sides of the distribution. Overall, the distributions of the three F2 populations were slightly skewed towards L1 (P17) and showed Gaussian segregation (Fig 1A–1C).
Table 1. Mean leaf relative injury (LRI) values of parental lines, F1 and three F2 populations under heat stress conditions.
| Type | Code | LRI | Reaction | |
|---|---|---|---|---|
| Mean±SE | Range | |||
| Tolerance parent | L1 (P17) | 9.88±5.00a* | 4.28 − 20.25 | Tolerant |
| Sensitive parent | L6 (P23) | 47.26±10.78b | 33.52 − 59.32 | Sensitive |
| Mid-parent value | - | 28.32±7.89 | - | - |
| F1 population | L1 × L6 | 31.98±6.64b | 19.95 − 43.95 | Sensitive |
| F2 population | - | 29.68±8.52 | 0.34 − 97.49 | - |
*P < 0.05.
Fig 1. Frequency distribution of leaf relative injury (LRI) among three F2 populations with 147 individuals (A), 56 individuals (B), and 60 individuals (C).
L1: heat tolerant parent P17; L6: heat sensitive parent P23. Distribution of L1 near the origin of the x-axis indicates negative transgressive segregants, while distribution of L6 indicates positive transgressive segregants under heat-stress conditions. DNA of eighteen seedlings was selected from an F2 population with 147 individuals (A) with extreme phenotypes (low and high LRI values) to develop tolerant and sensitive pools.
Sequence data
High throughput Illumina sequencing yielded 31.9, 32.7, 40.9, and 40.9 M 150-bp paired-end raw reads from L1, L6, T-pool, and S-pool, respectively. After trimming and filtering, more than 80% of reads were mapped to the reference genome of bottle gourd cv. Hangzhou Gourd (313.4 Mbp, Table 2). Specifically, 23.0, 24.4, 29.0, and 29.5 million short reads were mapped for L1 (10.10×depth coverage or 92.75% coverage), L6 (10.85×depth coverage or 92.85% coverage), T-pool (12.71×depth coverage or 93.25% coverage), and S-pool (12.92×depth coverage or 93.25% coverage), respectively. Lower sequencing depth (about 10×) and higher coverage (about 92%) of L1 and L6 showed the close genetic relationship between the parental lines and cv. Hangzhou Gourd. In our study, the high-quality bottle gourd Hangzhou Gourd genome sequence [21] was used to map each DNA pool. The sequence data of parental lines were used to verify SNPs detected from two pools. Thus, SNPs between the two pools with alleles not inherited from either parent were filtered out.
Table 2. Main statistics of resequencing and SNP calling in two parental lines and two pools.
| Sample | Number of raw reads | Trimmed and filtered reads | Uniquely mapped reads | Average Depth | Coverage (%)a | Number of SNPs | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Total | Homozygous | Heterozygous | ||||
| L1 | 31,863,484 | 28,368,754 | 89.03 | 22,974,995 | 80.99 | 10.10 | 92.75 | 476,940 | 345,242 | 131,698 |
| L6 | 32,686,118 | 29,026,164 | 88.80 | 24,419,983 | 84.13 | 10.85 | 92.85 | 513,552 | 418,749 | 94,803 |
| T-pool | 40,647,374 | 35,340,578 | 86.94 | 29,026,863 | 82.13 | 12.71 | 93.25 | 543,798 | 359,267 | 184,531 |
| S-pool | 40,930,720 | 35,936,306 | 87.80 | 29,457,284 | 81.97 | 12.92 | 93.25 | 549,415 | 361,111 | 188,304 |
a size of the bottle gourd reference genome is 313,397,697 bp (Xu et al. 2014 [22]).
Based on the uniquely mapped reads, 543,798 and 549,415 SNP were identified from the T-pool and S-pool, respectively. Among them, 359,267 and 361,111 were homozygous in T-pool and S-pool; 184,531 and 188,304 SNPs were heterozygous in the both pools, respectively (Table 2). After filtering our low quality SNPs, 153808 SNPs between the T-pool and S-pool were kept for further analysis.
Identification of QTL for heat tolerance from QTL-Seq
The SNP-index graphs were generated for the T-pool (Fig 2A) and S-pool (Fig 2B). The Δ(SNP-index) was determined by subtracting the SNP-index of T-pool from the S-pool and plotted against the genome positions (Fig 2C). By examining the Δ(SNP-index) plot, we identified seven genomic regions that exhibited high Δ(SNP-index) values (Table 3) on chromosomes 1, 2, 5, 6, 7 and 8. These were candidate regions harboring heat tolerance QTLs (Fig 2C). Among them, the peak on Chr 5 was the highest, followed by the peaks on Chr 2 (qHT2.1), Chr 1, Chr 7, Chr 6, Chr 8 and Chr 2 (qHT2.2). The two adjacent peaks on Chr 2 contained most of the SNPs, 9052 and 1355, respectively. The peak region on Chr 5 only contained two SNPs (Table 3). The QTL associated with these regions were designated as qHT1.1, qHT2.1, qHT2.2, qHT6.1, qHT7.1, and qHT8.1, respectively, hereinafter.
Fig 2.
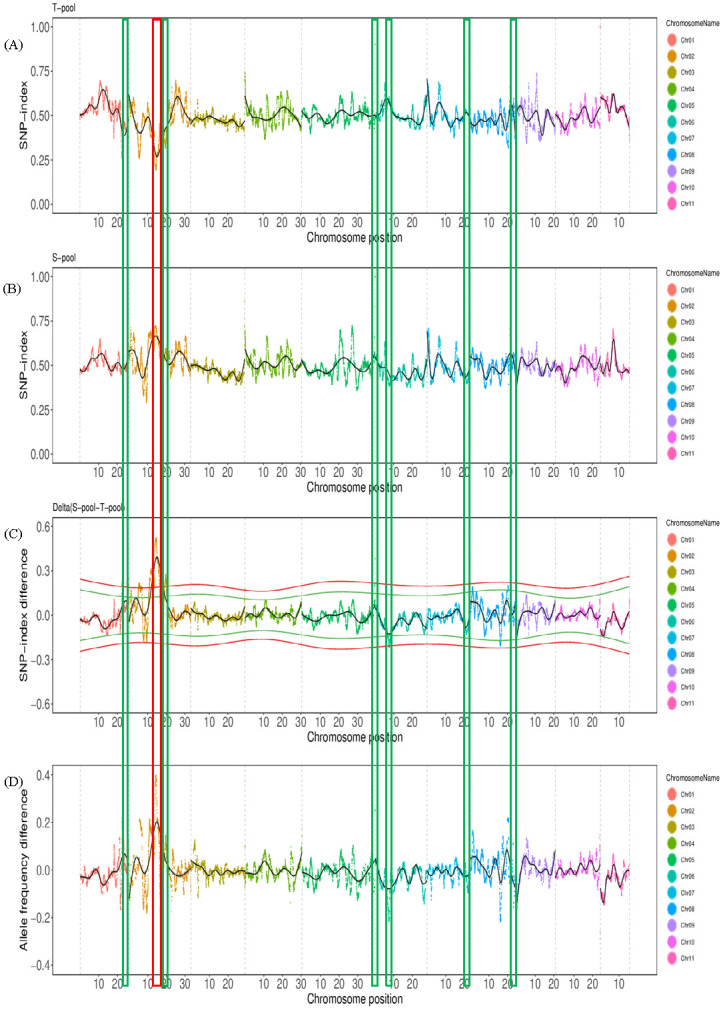
SNP-index plots of T-pool (A) and S-pool (B) and Δ(SNP-index) plot (C) from the QTL-Seq analysis. The x-axis represents the position of eleven chromosomes and the y-axis represents SNP-index or Δ(SNP-index) value. The Δ(SNP-index) plot (C) shows statistical confidence intervals under the null hypothesis of no QTL (P < 0.01). The red and green wavy line means 99% and 95% confidence intervals, respectively. The promising genomic region identified for LRI associated with heat tolerance is highlighted at 11.03–19.25 Mb on Chromosome 2 by red frame. The other six QTL region with high Δ(SNP-index) value are highlighted by green frame. (D) Profile of the tolerant parent (L1) allele frequency difference (L1AFD).
Table 3. QTLs identified from QTL-Seq that conferred heat tolerance in bottle gourd.
| Region | QTL | SNP Number | SNP-index | Δ(SNP-index) | P-value | Interval | L1AFD | |
|---|---|---|---|---|---|---|---|---|
| T-pool | S-pool | |||||||
| Chr01:26,260,000–27,279,999 | qHT1.1 | 12 | 0.3713 | 0.622 | 0.25 | 0.000413811 | 1,019,999 | 0.13 |
| Chr02:11,030,000–19,249,999 | qHT2.1 | 9052 | 0.3092 | 0.6285 | 0.32 | 9.79E-06 | 8,219,999 | 0.24 |
| Chr02:19,410,000–20,989,999 | qHT2.2 | 1355 | 0.3579 | 0.5463 | 0.19 | 0.006172464 | 1,579,999 | 0.11 |
| Chr05:39,390,000–40,419,999 | qHT5.1 | 2 | 0.45 | 0.8333 | 0.38 | 1.42E-07 | 1,029,999 | 0.25 |
| Chr06:7,460,000–8,459,999 | qHT6.1 | 934 | 0.5917 | 0.379 | -0.21 | 0.001783751 | 999,999 | -0.21 |
| Chr07:23,220,000–24,229,999 | qHT7.1 | 5 | 0.3648 | 0.5812 | 0.22 | 0.001986976 | 1,009,999 | 0.23 |
| Chr08:24,880,000–25,889,999 | qHT8.1 | 4 | 0.1853 | 0.3988 | 0.21 | 0.002248176 | 1,009,999 | 0.19 |
The L1AFD value of a QTL reflects the magnitude and direction of L1 allele effect of the associated QTL. Thus, qHT5.1 seemed to have the largest effect on REC, followed by qHT2.1, qHT7.1, qHT6.1, qHT8.1, qHT1.1 and qHT2.2. The tolerant alleles of qHT5.1, qHT2.1, qHT6.1, qHT8.1, qHT1.1 and qHT2.2 were derived from the tolerant parent L1 (L1AFD value > 0) while the qHT7.1 (L1AFD value < 0) was from the susceptible parent L6 (Fig 2D and Table 3).
Although qHT5.1 showed the highest values of Δ(SNP-index) and L1AFD, only two SNPs were associated with this QTL, both of which were located in intergenic regions. Thus, qHT2.1, with Δ(SNP-index) value of 0.32 (P < 0.01), L1AFD value of 0.24, and containing 9052 SNPs, was selected as a promising major QTL controlling heat tolerance.
Identification of heat tolerance candidate genes in qHT2.1 interval
In general, the confidence intervals of the QTL regions based on Δ(SNP-index) peaks were large, with a maximum of 8.22 Mbp for qHT2.1 (Table 3). There were 9052 SNPs between T-pool and S-pool in this region, of which 527 were in genic regions. Regions harboring the 279 SNPs were annotated by BLASTx against the non-redundant protein database (S1 Table, [51]), which identified 62 non-synonymous SNPs with Δ(SNP-index) ≥ 0.5 (S2 Table). Gene ontology classification revealed these 62 genes were mainly associated with biological processes (S1 Fig), such as metabolism; cellular components, such as the cell membrane; and molecular function, such as catalytic activity. The biological processes of the 62 candidate genes were retrieved from the available bottle gourd genome data and Uniprot database. Thirty four of the 62 genes seemed to be related with heat stress and are involved in biological processes, pollen and flower sterility, oxidative stress response, autophagy, and abiotic stress response (S3 Table).
Development of SNP markers associated with heat tolerance
Among the 34 candidate genes potentially associated with tolerance, five genes harboring five nonsynonymous SNPs were selected for primer design (Table 4). Among them, SNP 2 (BG_GLEAN_10022642, annexin 5) was related to pollen and flower sterility. SNP 15 (BG_GLEAN_10022339, PQL1 and PQL2) and SNP 16 (BG_GLEAN_10022589, serine/threonine-protein kinase) were related to signal recognition. SNP 26 (BG_GLEAN_10022734, MuDR) and SNP 31 (BG_GLEAN_10022727, AP-3 adaptor complex) were related to intracellular transport. All these five SNPs altered the predicted protein sequences of the genes (Table 4).
Table 4. Primer design, position of SNPs located in their respective gene and the protein sequence alteration.
| SNP | Gene | Mutation | Chr | Pos | Protein | Primer-F | Tm-R | Primer-R | Tm-F |
|---|---|---|---|---|---|---|---|---|---|
| 15 | BG_GLEAN_10022339 | C->T | 2 | 12878537 | V → I | TACTCCTCGGCGTCTAACCA | 60.0 | TGAGAAGCAGCCTCTCGTTG | 60.0 |
| 26 | BG_GLEAN_10022734 | A->G | 2 | 17634545 | T → A | AGGTAGGCCCGAAGACTCAT | 60.0 | GGGAGGGCAGCTTGTTACTT | 60.0 |
| 2 | BG_GLEAN_10022642 | C->A | 2 | 16355258 | S → Y | CGCGCTATGAAGGTCCAGAA | 60.2 | CCCAGGATTCTCAGCACACA | 60.0 |
| 16 | BG_GLEAN_10022589 | A->G | 2 | 15709303 | K → E | GAGCCCAGTTGGTTCCTTGA | 60.0 | CATCTGCGATCTCCGTCCTG | 60.0 |
| 31 | BG_GLEAN_10022727 | T->A | 2 | 17457363 | C → S | CGTCTATGGGGGTTGGCAAT | 60.1 | CGAGAGTCCTGCAGCAGAAA | 60.0 |
Genotyping of 10 heat tolerant and 16 sensitive F2 individuals (S2 and S3 Figs, Table 5) showed two of the five SNPs (SNP 15 and SNP 26) had no polymorphism between the heat tolerant and sensitivity lines. SNP 2 (CC), SNP 16 (GG) and SNP 31 (TT) were homozygous in heat tolerant individuals, which were with homozygous (AA, AA, AA) or heterozygous genotype (AC, AG, AT) in heat sensitive plants, respectively. The alleles of the three SNPs (SNP 2, SNP 16 and SNP 31) showed good correlations with heat tolerance in the two parents, the two pools and tested F2 plants (Table 5) suggesting their potential value as makers for marker-assisted selection of heat tolerance.
Table 5. Sanger sequencing verification of five candidate SNP loci in 10 heat tolerant and 16 sensitive F2 individuals.
| SNP | Gene | L1 | L6 | Heat tolerant individuals | Heat sensitive individuals | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||||
| 15 | BG_GLEAN_10022339 | T | C | C | C | C | C | -* | - | - | - | - | - | C | C | C | C | - | - | - | - | - | - | - | - | - | - | - | - |
| 26 | BG_GLEAN_10022734 | G | A | AG | AG | AG | AG | - | - | - | - | - | - | AG | AG | AG | AG | - | - | - | - | - | - | - | - | - | - | - | - |
| 2 | BG_GLEAN_10022642 | C | A | C | C | C | C | C | C | C | C | C | C | A | AC | AC | A | A | A | AC | A | AC | AC | AC | AC | A | AC | AC | AC |
| 16 | BG_GLEAN_10022589 | G | A | G | G | G | G | G | G | G | G | G | G | A | A | A | A | A | A | AG | A | AG | AG | AG | AG | A | AG | AG | AG |
| 31 | BG_GLEAN_10022727 | T | A | T | T | T | T | T | T | T | T | T | T | A | AT | AT | A | A | A | AT | A | AT | AT | AT | AT | A | AT | AT | AT |
*SNP 15 and SNP 26 had no difference between the four heat tolerant lines and four heat sensitivity lines, so they were not genotyped for the 18 lines additionally.
Discussion
Parental lines were not used as reference genome
Based on QTL-seq protocol [27], the genome sequence of either of the two parents can be used as a reference to map the DNA pool [32,52]. This would make it easier to identify the source of the SNPs and to tell from which parent the allele responsible for the QTL region came from. But since there is a close genetic relationship between the parental lines used in this study and cv. Hangzhou Gourd, the cultivar used to develop the high-quality reference genome for bottle gourd [19], the available genome sequence for the latter was used as reference in our study. The sequence data of parental lines were mainly applied to ensure the differential SNPs detected from two pools were inherited from either parent.
Moreover, the L1 allele frequency difference (L1AFD) was plotted to identify the size of the effect and direction of action of the parental allele for each QTL [50]. The profile of L1AFD indicated that except for qHT7.1 (L1AFD < 0) derived from the susceptible parent L6, the other six tolerant alleles of qHT5.1, qHT2.1, qHT6.1, qHT8.1, qHT1.1 and qHT2.2 (L1AFD value > 0) were all from the tolerant parent L1.
The qHT2.1 had higher values of Δ(SNP-index) and L1AFD, and contained the largest number of detected SNPs (9052). Thus, qHT2.1 was identified as the most promising major-effect QTL for heat tolerance. For cucumber [33], credible QTLs associated with downy mildew resistance were detected via QTL-Seq method and double checked by conventional method. In their research, the reads of two DNA pools were mapped against the reference genome of cucumber cv. Chinese Long, not mapped against the parental line of TH118FLM or WME. The genetic relationship among cucumber was narrow. The polymorphism information content value of SSR and SCAR was only 0.65 [53]. For sunflower [54], the reference genome for Helianthus annuus was used and not the parental lines 902R or 906R, for the identification of candidate resistance gene to broomrape by BSA-seq.
Genetics basis of heat tolerance in bottle gourd
Plants have evolved a range of metabolic responses, such as antioxidant activity, membrane lipid unsaturation, protein stability, gene expression and translation, and accumulation of compatible solutes [55], to cope with heat stress through the activation of stress-response genes [56]. Heat tolerance, which could be characterized by different phenotypic and physiological parameters, has been shown to be a quantitative trait [57]. The quantitative nature of heat tolerance has been revealed in a number of crop plants, like wheat [58,59], rice [34], and broccoli [60,61]. However, there are also reports of monogenic and oligogenic responses to heat tolerance [62,63]. Little is known about the inheritance of heat tolerance in bottle gourd. In this study, the F1 of the L1×L6 was heat sensitive suggesting that heat tolerance is a recessive trait in bottle gourd. We also identified major-effect QTL for this trait. Thus, our work represented the first to report REC related to heat tolerance in this important vegetable crop.
In the present study, qHT2.1 was identified as the most promising major QTL for heat tolerance. Similar results have been reported for African rice (Oryza glaberrima Steud.) [64]. It was reported that a major QTL that controls the survival rate of seedlings exposed to high temperatures was governed by the Thermo tolerance 1 gene. The gene encodes a 23S proteasome subunit that degrades ubiquitinated cytotoxic denatured proteins formed due to high temperature stress. Major-effect QTL control of complex traits, such as heat tolerance, may be explained by inheritance patterns of the trait [13] and action of major regulator genes that may switch off subordinate genes, if a key gene is mutated [65].
Annotation of heat tolerance candidate genes
Functional genomics studies in plants facilitate the elucidation of candidate genes and their relationship with traits [35]. For example, heat stress affects pollen and flower sterility [66]. Of the 34 candidate genes, four were found to have key roles in gametogenesis [67], pollen development [68,69], or Ogura cytoplasmic male sterility [70]. Heat-stress also causes cellular damage by oxidative stress and toxicity due to reactive oxygen species (ROS) formation [71,72]. Antioxidants scavenge ROS to mitigate oxidative stress. We found four putative candidate genes that had a role in defying oxidative stress and recovering plants from heat-stress damage [73–76]. Many reports have mentioned transcription factors [77], binding factors [78,79], intracellular transporters [80,81], and enzymes [82,83] that are responsible for abiotic tolerance in different crops. The signaling molecules [84] and autophagy [85] also play important roles in plant responses to stress.
Markers and candidate genes associated with heat tolerance in bottle gourd
In this study, three SNPs with contrasting functional annotation were found to be associated with heat tolerance in bottle gourd. The three SNPs were located in three genes including those encoding for homologs of annexin 5 (BG_GLEAN_10022642, SNP 2), AP-3 adaptor complex protein (BG_GLEAN_10022727, SNP 31), and serine/threonine-protein kinase (BG_GLEAN_10022589, SNP 16). A common response of crop plants to temperature stress is significant yield loss, as a result of effects on spikelet fertility in rice [86], pod set in lentil [65], and pollen sterility in canola [66]. Annexins are members of the ubiquitous family of proteins present in eukaryotic organisms [87] and localized in various subcellular compartments [88]. They are known to be involved in a variety of cellular processes, such as maintenance of vesicular trafficking, cellular redox homeostasis, actin binding, and ion transport [89], due to their calcium- and membrane-binding capacity. Annexin 5 is involved in pollen grain development and germination, and pollen tube growth through the promotion of calcium-modulated endo-membrane trafficking [90]. For example, down-regulation of Arabidopsis annexin 5 in transgenic Ann5-RNAi lines has been shown to cause sterility in pollen grains [68].
Adaptor proteins that are involved in protein trafficking and sorting [91–93] may recognize cargo and coat proteins during vesicle formation [94]. AP-3 was first identified in mammalian cells and probably functions as a clathrin adaptor [95]. Losses in AP-3 function reduce seed germination potential [96,97], due to mistargeted protein S-ACYL transferase10 that is critical for pollen tube growth during dynamic vacuolar organization in Arabidopsis [98].
Serine/threonine-protein kinases (STKs) are involved in signal transduction networks to coordinate growth and differentiation of cell responses to extracellular stimuli [99]. In a range of smut pathogenesis-related biological processes, Huang et al. [84] reported that STKs may act as receptors or signaling factors, such as in Ca2+ signaling, that then activate defense responses. Umezawa et al. [100] reported that SRK2C is annotated as a putative and osmotic-stress-activated STK in Arabidopsis. It might mediate signal transduction and regulate a series of drought stress-response genes that enhance expression of 35S:SRK2C-GFP to improve drought tolerance in plants.
Conclusions
Understanding the genetic basis of heat tolerance and development of reliable DNA markers to indirectly select for the trait are important in breeding for new varieties with heat tolerance. In this study, we show that REC could be used as an indicator for heat tolerance, which exhibited recessive inheritance. We also identified three SNPs that are associated with heat tolerance. The three SNPs were identified in genes regulating pollen sterility, intracellular transport, and signal recognition. These SNPs can be used in marker-assisted selection for heat tolerance in bottle gourd. Intensity of global warming will increase over the next two to three decades and will exacerbate challenges for the cultivation of Cucurbitaceae plants. Identification of heat tolerant genotypes of bottle gourds that can be used to improve heat tolerance of other Cucurbitaceae plants, such as watermelon, through grafting techniques may mitigate these challenges. These results revealed the novel region of 11.03 − 19.25 Mb on Chr 2 harboring qHT2.1 that may provide the basis for further exploration and fine mapping of novel genes associated with heat tolerance in bottle gourd.
Supporting information
(TIF)
Eight individuals were the member of tow DNA pools including four heat tolerant F2 (A, C, E, G, I) and four heat sensitive F2 (B, D, F, H, J). (A, B) SNP15: BG_GLEAN_10022339. (C, D) SNP26: BG_GLEAN_10022734. (E, F) SNP 2: BG_GLEAN_10022642. (G, H) SNP 16:BG_GLEAN_10022589. (I, J) SNP 31: BG_GLEAN_10022727. The SNP loci are shaded and indicated with a red arrow.
(TIF)
Eighteen individuals were the additional new lines, including six heat tolerant F2 (A, C, E) and twelve sensitive F2 (B, D, F). (A, B) SNP 2: BG_GLEAN_10022642. (C, D) SNP 16:BG_GLEAN_10022589. (E, F) SNP 31: BG_GLEAN_10022727. The SNP loci are shaded and indicated with a red arrow.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are grateful to Q.F. Lou and Y. Q. Weng for critical comments on this work.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
Funding for this research comes from the National Natural Science Foundation of China and Special Fund for Agro-Scientific Research in the Public Interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hall JL. Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot. 2002; 53: 1–11. [PubMed] [Google Scholar]
- 2.Fedoroff NV, Battisti DS, Beachy RN, Cooper PJ, Fischhoff DA, Hodges CN, et al. Radically rethinking agriculture for the 21st century. Science. 2010; 327: 833–834. 10.1126/science.1186834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian SW, Jiang LJ, Cui XX, Zhang J, Guo SG, Li MY, et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018; 37: 1353–1356. 10.1007/s00299-018-2299-0 [DOI] [PubMed] [Google Scholar]
- 4.Brinen GH. Plant and row spacing, mulch and fertilizer rate effects on watermelon production. J Am Soc Hort Sci. 1979; 104: 724–726. [Google Scholar]
- 5.Asthir B. Protective mechanisms of heat tolerance in crop plants. J Plant Interact. 2015; 10: 202–210. [Google Scholar]
- 6.Gilliham M, Able JA, Roy SJ. Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 2017; 90: 898–917. 10.1111/tpj.13456 [DOI] [PubMed] [Google Scholar]
- 7.Ibrahim E, Saadalla M, Baenziger S, Bockelman H, Morsy S. Cell membrane stability and association mapping for drought and heat tolerance in a worldwide wheat collection. Sustainability. 2017; 9: 1606–1022. 10.3390/su9091606 [DOI] [Google Scholar]
- 8.Arvin MJ, Donnelly DJ. Screening potato cultivars and wild species to abiotic stresses using an electrolyte leakage bioassay. J. Agr Sci Tech. 2008; 10: 33–42. [Google Scholar]
- 9.Jin XQ, Liu T, Xu JJ, Gao ZX, Hu XH. Exogenous GABA enhances muskmelon tolerance to salinity-alkalinity stress by regulating redox balance and chlorophyll biosynthesis. BMC Plant Bio. 2019; l19: 48–65. 10.1186/s12870-019-1660-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su AY, Niu SQ, Liu YZ, He AL, Zhao Q, Paré PW, et al. Synergistic effects of Bacillus amyloliquefaciens (GB03) and water retaining agent on drought tolerance of Perennial ryegrass. Int J Mol Sci. 2017; 18: 2651–2664. 10.3390/ijms18122651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulc RM, Albrecht KA, Duke SH. Leakage of intracellular substances as an indicator of freezing injury in alfalfa. Crop Sci. 1991; 31: 430–435. [Google Scholar]
- 12.Davis AR, Perkins-Veazie P, Sakata Y, López-Galarza S, Maroto JV, Lee S, et al. Cucurbit grafting. Crit Rev in Plant Sci. 2008; 27: 50–74. 10.1080/07352680802053940 [DOI] [Google Scholar]
- 13.Lee JM, Oda M. Grafting of herbaceous vegetable and ornamental crops. Hortic Rev Am Soc Hortic Sci. 2003; 28: 61–124. [Google Scholar]
- 14.Yang Y, Lu X, Yan B, Li B, Sun J, Guo S, et al. Bottle gourd rootstock-grafting affects nitrogen metabolism in NaCl-stressed watermelon leaves and enhances short-term salt tolerance. J Plant Physiol. 2013; 170: 653–661. 10.1016/j.jplph.2012.12.013 [DOI] [PubMed] [Google Scholar]
- 15.Schaefer H, Heibl C, Renner SS. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc R Soc London B Biol Sci. 2009; 276: 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Shamimuzzaman M, Sun HH, Salse J, Sui XL, Wilder A, et al. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 2017; 92: 963–975. 10.1111/tpj.13722 [DOI] [PubMed] [Google Scholar]
- 17.Samadia DK. Performance of bottle gourd genotypes under hot arid environment. Indian J Hort. 2002; 59: 167–170. [Google Scholar]
- 18.Mashilo J, Shimelis H, Odindo A. Yield-based selection indices for drought tolerance evaluation in selected bottle gourd (Lagenaria siceraria (Molina) Standl.) landraces. Acta Agr Scand S-B P. 2017; 67: 43–50. 10.1080/09064710.2016.1215518 [DOI] [Google Scholar]
- 19.Liu N, Yang J, Fu X, Zhang L, Tang K, Guy KM, et al. Genome‑wide identification and comparative analysis of grafting‑responsive mRNA in watermelon grafted onto bottle gourd and squash rootstocks by high‑throughput sequencing. Mol Genet Genomics. 2016; 291: 621–633. 10.1007/s00438-015-1132-5 [DOI] [PubMed] [Google Scholar]
- 20.Xu P, Wu XH, Luo J, Wang BG, Liu YH, Ehlers JD, et al. Partial sequencing of the bottle gourd genome reveals markers useful for phylogenetic analysis and breeding. BMC Genomics. 2011; 12: 467–480. 10.1186/1471-2164-12-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008; 3: e3376 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu P, Xu SZ, Wu XH, Tao Y, Wang BG, Wang S, et al. Population genomic analyses from low-coverage RAD-Seq data: A case study on the non-model cucurbit bottle gourd. Plant J. 2014; 77: 430–442. 10.1111/tpj.12370 [DOI] [PubMed] [Google Scholar]
- 23.Wu XY, Xu P, Wu XH, Wang BG, Lu ZF, Li GJ. Development of insertion and deletion markers for bottle gourd based on restriction site-associated DNA sequencing data. Hort Plant J. 2017; 3: 13–16. [Google Scholar]
- 24.Huang SW, Li RQ, Zhang ZH, Li L, Gu XF, Fan W, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009; 41: 1275–1281. 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, et al. The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA. 2012; 109: 11872–11877. 10.1073/pnas.1205415109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo SG, Zhang JG, Sun HH, Salse J, Lucas WJ, Zhang HY, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013; 45: 51–58. 10.1038/ng.2470 [DOI] [PubMed] [Google Scholar]
- 27.Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013; 74: 174–183. 10.1111/tpj.12105 [DOI] [PubMed] [Google Scholar]
- 28.Livaja M, Wang Y, Wieckhorst S, Haseneyer G, Seidel M, Hahn V, et al. BSTA: a targeted approach combines bulked segregant analysis with next generation sequencing and de novo transcriptome assembly for SNP discovery in sunflower. BMC Genomics. 2013; 14: 628–638. 10.1186/1471-2164-14-628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Huang D, Tang W, Zheng Y, Liang K, Cutler AJ, et al. Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One. 2013; 8: e68433 10.1371/journal.pone.0068433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Y, Lv P, Hou S, Li S, Ji G, Ma X, et al. Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in sorghum (Sorghum bicolor L. Moench). PLoS One. 2015; 10: e0127065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminski KP, Korup K, Andersen MN, Sonderkaer M, Andersen MS, Kirk HG, et al. Next generation sequencing bulk segregant analysis of potato support that differential flux into the cholesterol and stigmasterol metabolite pools is important for steroidal glycoalkaloid content. Potato Res. 2016; 59: 81–97. [Google Scholar]
- 32.Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, et al. QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor Appl Genet. 2014; 127: 1491–1499. 10.1007/s00122-014-2313-z [DOI] [PubMed] [Google Scholar]
- 33.Win KT, Vegas J, Zhang CY, Song K, Lee S. QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor Appl Genet. 2017; 130: 199–211. 10.1007/s00122-016-2806-z [DOI] [PubMed] [Google Scholar]
- 34.Shanmugavadivel P S, Amitha MSV, Chandra P, Ramkumar MK, Tiwari R, Mohapatra T, et al. High resolution mapping of QTLs for heat tolerance in rice using a 5K SNP array. Rice. 2017; 10: 28–39. 10.1186/s12284-017-0167-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul PJ, Samineni S, Thudi M, Sajja SB, Rathore A, Das RR, et al. Molecular mapping of QTLs for heat tolerance in chickpea. Mol Sci. 2018; 19: 2166–2186. 10.3390/ijms19082166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mondal S, Mason RE, Huggins T, Hays DB. QTL on wheat (Triticum aestivum L.) chromosomes 1B, 3D and 5A are associated with constitutive production of leaf cuticular wax and may contribute to lower leaf temperatures under heat stress. Euphytica. 2015; 201: 123–130. 10.1007/s10681-014-1193-2 [DOI] [Google Scholar]
- 37.Jha UC, Bohra A, Singh NP. Heat stress in crop plants: Its nature, impacts and integrated breeding strategies to improve heat tolerance. Plant Breed. 2014; 133: 679–701. [Google Scholar]
- 38.Wang YH, Song H, Quan QS, Wang YE, Zhang XQ, Yan LY, et al. A method of heat tolerance identification and selection of germplasm in bottle gourd type rootstocks. Chinese Patent No 2012100939951. 2014; Beijing: National Intellectual Property Administration, PRC.
- 39.Zhou WJ, Leul M. Uniconazole-induced alleviation of freezing injury in relation to changes in hormonal balance, enzyme activities and lipid peroxidation in winter rape. Plant Growth Regul. 1998; 26: 41–47. [Google Scholar]
- 40.He AL, Niu SQ, Zhao Q, Li YS, Gou JY, Gao HJ, et al. Induced salt tolerance of Perennial ryegrass by a novel bacterium strain from the rhizosphere of a desert shrub Haloxylon ammodendron. Int J Mol Sci. 2018; 19: 469–472. 10.3390/ijms19020469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun CX, Cao HX, Chen ST, Feng ML, Li RS, Ma ZL. Study on cold resistance of snake fruit by application of electrical conductivity and logistic equation. Acta Agr Jiangxi. 2009; 21: 33–35 (in Chinese). [Google Scholar]
- 42.Ling F, Li C, Hui Z, Jiao J, Lyu P. Measurement of cold tolerance by electrical conductivity method in associated with the logistic equation on different varieties of Olea europaea L. Acta Agr Guangdong. 2015; 1: 13–17 (in Chinese). [Google Scholar]
- 43.Saadalla MM, Shanahan JF, Quick JS. Heat tolerance in winter wheat: I. hardening and genetic effects on membrane thermostability. Crop Sci. 1990; 30: 1243–1247. 10.2135/cropsci1990.0011183X003000060017x [DOI] [Google Scholar]
- 44.Sui Y, Miao Y, Hu N, Zhao Y, Zhou Y. Genetic analyzing yield traits of chilli pepper. J Nanjing Agri Univ. 2014; 37: 46–52. [Google Scholar]
- 45.Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980; 8: 4321–4326. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soldi S, Vasileiadis S, Uggeri F, Campanale M, Morelli L, Fogli MV, et al. Modulation of the gut microbiota composition by rifaximin in non-constipated irritable bowel syndrome patients: a molecular approach. Clin Exp Gastroenterol. 2015; 8: 309–325. 10.2147/CEG.S89999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuryn S, Gras LS, Jamet K, Jarriault S. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics. 2010; 186: 427–430. 10.1534/genetics.110.119230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh VK, Khan AW, Jaganathan D, Thudi M, Roorkiwal M, Takagi H, et al. QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnology Journal. 2016; 14: 2110–2119. 10.1111/pbi.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z, Huang D, Tang W, Zheng Y, Liang K, Cutler AJ, et al. Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS ONE. 2013; 8(7): e68433 10.1371/journal.pone.0068433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007; 35: 61–65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandey MK, Khan AW, Singh VK, Vishwakarma MK, Shasidhar Y, Kumar V, et al. QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnology Journal. 2017; 15: 927–941. 10.1111/pbi.12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staub JE, Chung SM, Fazio G. Conformity and genetic relatedness estimation in crop species having a narrow genetic base: the case of cucumber (Cucumis sativus L.). Plant Breeding. 2005; 124(1): 44–53. 10.1111/j.1439-0523.2004.01061.x [DOI] [Google Scholar]
- 54.Liu S, Wang P, Liu Y, Wang P. Identification of candidate gene for resistance to broomrape (Orobanche cumana) in sunflower by BSA-seq. Oil Crop Science. 10.1016/j.ocsci.2020.05.003 [DOI] [Google Scholar]
- 55.Kaya H, Ichi-Shibahara K, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assemblyfactor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001; 104: 131–142. 10.1016/s0092-8674(01)00197-0 [DOI] [PubMed] [Google Scholar]
- 56.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010; 61: 1041–1052. 10.1111/j.1365-313X.2010.04124.x [DOI] [PubMed] [Google Scholar]
- 57.Paliwal R, Reoder MS, Kumar U, Srivastava JP, Joshi AK. QTL mapping of terminal heat tolerance in hexaploid wheat (T. aestivum L.). Theor Appl Genet. 2012; 125: 561–575. 10.1007/s00122-012-1853-3 [DOI] [PubMed] [Google Scholar]
- 58.Barakat MN, Al-Doss AA, Elshafei AA, Moustafa KA. Identification of new microsatellite marker linked to the grain filling rate as indicator for heat tolerance genes in F2 wheat population. Aust J Crop Sci. 2011; 5: 104–110. [Google Scholar]
- 59.Talukder SK, Md A, Babar K, Vijayalakshmi J, Poland PVV, Bowden R, et al. Mapping QTL for the traits associated with heat tolerance in wheat (Triticum aestivum L.). BMC Genet. 2014; 15: 97–110. 10.1186/s12863-014-0097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farnham M, Björkman T. Evaluation of experimental broccolihybrids developed for summer production in the Eastern United States. Hort Science. 2011; 46: 858–863. [Google Scholar]
- 61.Branham SE, Stansell ZJ, Couillard DM, Farnham MW. Quantitative trait loci mapping of heat tolerance in broccoli (Brassica oleracea var. italica) using genotyping‑by‑sequencing. Theor Appl Genet. 2017; 130: 529–538. 10.1007/s00122-016-2832-x [DOI] [PubMed] [Google Scholar]
- 62.Bouwkamp JC, Summers WL. Inheritance of resistance to temperature-drought stress in the snap bean. J Hered. 1982; 73: 385–386. [Google Scholar]
- 63.Marfo KO, Hal AE. Inheritance of heat tolerance during pod set in cowpea. Crop Sci. 1992; 32: 912–918. [Google Scholar]
- 64.Li XM, Chao DY, Wu Y, Huang X, Chen K, Cui LG, et al. Natural alleles of a proteasome a2 subunit gene contribute to thermo tolerance and adaptation of African rice. Nat Genet. 2015; 47: 827–833. 10.1038/ng.3305 [DOI] [PubMed] [Google Scholar]
- 65.Singh D, Singh CK, Tomar RSS, Pal M. Genetics and molecular mapping of heat tolerance for seedling survival and pod set in lentil. Crop Sci. 2017; 57: 3059–3067. 10.2135/cropsci2017.05.0284 [DOI] [Google Scholar]
- 66.Rahamana M, Mamidib S, Rahmana M. Genome-wide association study of heat stress tolerance traits in spring-type Brassica napus L. under controlled conditions. Crop J. 2018; 6: 115–125. [Google Scholar]
- 67.Gull IS, Hulpiau P, Saeys Y, Roy F. Metazoan evolution of the armadillo repeat super family. Cell Mol Life Sci. 2017; 74: 525–541. 10.1007/s00018-016-2319-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lichocka M, Rymaszewski W, Morgiewicz K, Barymow-Filoniuk I, Chlebowski A, Sobczak M, et al. Nucleus- and plastid-targeted annexin 5 promotes reproductive development in Arabidopsis and is essential for pollen and embryo formation. BMC Plant Biol. 2018; 18: 183–198. 10.1186/s12870-018-1405-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hruba P, Honys D, Twell D, Capkova V, Tupy J. Expression of β-galactosidase and β-xylosidase genes during microspore and pollen development. Planta. 2005; 220: 931–940. 10.1007/s00425-004-1409-0 [DOI] [PubMed] [Google Scholar]
- 70.Wang CG, Chen XQ, Lan TY, Li H, Song WQ. Cloning and transcript analyses of the chimeric gene associated with cytoplasmic male sterility in cauliflower (Brassica oleracea var. botrytis). Euphytica. 2006; 151: 111–119. [Google Scholar]
- 71.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: An overview. Environ Exp Bot. 2007; 61: 199–223. [Google Scholar]
- 72.Hamada A, Zintal G, Hegab MM, Pandey R, Asard H, Abuelsoud W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front Plant Sci. 2016; 26: 1–11. 10.3389/fpls.2016.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Idänheimo N, Gauthier A, Salojärvi J, Siligato R, Brosché M, Kollist H, et al. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against a poplastic oxidative stress. Biochem Bioph Res Co. 2014; 445: 457–462. 10.1016/j.bbrc.2014.02.013 [DOI] [PubMed] [Google Scholar]
- 74.Umate P, Tuteja R, Tuteja N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol. 2010; 73: 449–465. 10.1007/s11103-010-9632-5 [DOI] [PubMed] [Google Scholar]
- 75.Xiao WF, Chen P, Xiao JS, Wang L, Liu TH, Wu YF, et al. Comparative transcriptome profiling of a thermal resistant vs. sensitive silkworm strain in response to high temperature under stressful humidity condition. PLoS One. 2017; 12: e0177641 10.1371/journal.pone.0177641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yabuta S, Ifuku K, Takabayashi A, Ishihara S, Ido K, Ishikawa N, et al. Three PsbQ-Like proteins are required for the function of the chloroplast NAD (P) H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 2010; 51: 866–876. 10.1093/pcp/pcq060 [DOI] [PubMed] [Google Scholar]
- 77.Rashkova S, Karam SE, Pardue ML. Element-specific localization of drosophila retrotransposon gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci USA. 2002; 99: 3621–3626. 10.1073/pnas.032071999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donlin MJ, Lisch D, Freeling M. Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the mutator transposable element family. Plant Cell. 1995; 7: 1989–2000. 10.1105/tpc.7.12.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abeliovich H, Tzfati Y, Shlomai J. A trypanosomal CCHC-Type zinc finger protein which binds the conserved universal sequence of kinetoplast DNA minicircles: isolation and analysis of the complete cDNA from Crithidia fasciculata. Mol Cell Biol. 1993; 13: 7766–7773. 10.1128/mcb.13.12.7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kraus B, Boller K, Reuter A, Schnierle BS. Characterization of the human endogenous retrovirus K Gag protein: identification of protease cleavage sites. Retrovirology. 2011; 8: 21–29. 10.1186/1742-4690-8-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997; 91: 109–118. 10.1016/s0092-8674(01)80013-1 [DOI] [PubMed] [Google Scholar]
- 82.Parsiegla G, Noguere C, Santell L, Lazarus RA, Bourne Y. The structure of human DNase I bound to magnesium and phosphate ions points to a catalytic mechanism common to members of the DNase I-like super family. Biochemistry. 2012; 51: 10250–10258. 10.1021/bi300873f [DOI] [PubMed] [Google Scholar]
- 83.Du YL, Singh R, Alkhalaf LM, Kuatsjah E, He HY, Eltis LD, et al. A pyridoxal phosphate-dependent enzyme that oxidizes an unactivated carbon-carbon bond. Nat Chem Biol. 2016; 12: 194–199. 10.1038/nchembio.2009 [DOI] [PubMed] [Google Scholar]
- 84.Huang N, Ling H, Su YC, Liu F, Xu LP, Su WH, et al. Transcriptional analysis identifies major pathways as response components to Sporisorium scitamineum stress in sugarcane. Gene. 2018; 678: 207–218. 10.1016/j.gene.2018.08.043 [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Cai SY, Yin LL, Shi K, Xia XJ, Zhou YH, et al. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015; 11: 2033–2047. 10.1080/15548627.2015.1098798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ye CR, Tenoriol FA, Argayoso MA, Laza MA, Koh HJ, Redoña ED, et al. Identifying and confirming quantitative trait loci associated with heat tolerance at flowering stage in different rice populations. BMC Genet. 2015; 16: 41–51. 10.1186/s12863-015-0199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davies JM. Annexin-mediated calcium signalling in plants. Plants (Basel). 2014; 3: 128–140. 10.3390/plants3010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laohavisit A, Davies JM. Annexins. New Phytol. 2011; 189: 40–53. 10.1111/j.1469-8137.2010.03533.x [DOI] [PubMed] [Google Scholar]
- 89.Laohavisit A, Brown AT, Cicuta P, Davies JM. Annexins: components of the calcium and reactive oxygen signaling network. Plant Physiol. 2010; 152: 1824–1289. 10.1104/pp.109.145458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu J, Wu X, Yuan S, Qian D, Nan Q, An L, et al. Annexin5 plays a vital role in Arabidopsis pollen development via Ca2+-dependent membrane trafficking. PLoS One. 2014; 9: e102407 10.1371/journal.pone.0102407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marks MS, Ohno H, Kirchhausen T, Bonifacino JS. Protein sorting by tyrosine-based signals. Trends Cell Biol. 1997; 7: 124–128. 10.1016/S0962-8924(96)10057-X [DOI] [PubMed] [Google Scholar]
- 92.Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, et al. Dynamic analysis of Arabidopsis AP2 s subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development. 2013; 140: 3826–3837. 10.1242/dev.095711 [DOI] [PubMed] [Google Scholar]
- 93.Park M, Song K, Reichardt I, Kim H, Mayer U, Stierhof YD, et al. Arabidopsis u-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc Natl Acad Sci USA. 2013; 110: 10318–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bassham DC, Brandizzi F, Otegui MS, Sanderfoot AA. The secretory system of Arabidopsis. Arabidopsis Book. 2008; 6: e0116 10.1199/tab.0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996; 133: 749–760. 10.1083/jcb.133.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feraru E, Paciorek T, Feraru MI, Zwiewka M, De Groodt R, De Rycke R, et al. The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell. 2010; 22: 2812–2824. 10.1105/tpc.110.075424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zwiewka M, Feraru E, Möller B, Hwang I, Feraru MI, Kleine-Vehn J, et al. The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 2011; 21: 1711–1722. 10.1038/cr.2011.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feng QN, Liang X, Li S, Zhang Y. The adaptor protein-3 complex mediates pollen tube growth by coordinating vacuolar targeting and organization. Plant Physiol. 2018; 177: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holzman LB, Merritt SE, Fan G. Identification, molecular cloning, and characterization of dual leucine zipper bearing kinase. J Biol Chem. 1994; 269: 30808–30817. [PubMed] [Google Scholar]
- 100.Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004; 101: 17306–17311. 10.1073/pnas.0407758101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Eight individuals were the member of tow DNA pools including four heat tolerant F2 (A, C, E, G, I) and four heat sensitive F2 (B, D, F, H, J). (A, B) SNP15: BG_GLEAN_10022339. (C, D) SNP26: BG_GLEAN_10022734. (E, F) SNP 2: BG_GLEAN_10022642. (G, H) SNP 16:BG_GLEAN_10022589. (I, J) SNP 31: BG_GLEAN_10022727. The SNP loci are shaded and indicated with a red arrow.
(TIF)
Eighteen individuals were the additional new lines, including six heat tolerant F2 (A, C, E) and twelve sensitive F2 (B, D, F). (A, B) SNP 2: BG_GLEAN_10022642. (C, D) SNP 16:BG_GLEAN_10022589. (E, F) SNP 31: BG_GLEAN_10022727. The SNP loci are shaded and indicated with a red arrow.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.