Abstract
As a versatile intracellular second messenger, calcium ion (Ca2+) regulates a plethora of physiological processes. To achieve precise control over Ca2+ signals in living cells and organisms, a set of optogenetic tools have recently been crafted by engineering photosensitive domains into intracellular signaling proteins, G-protein coupled receptors (GPCRs), receptor tyrosine kinases (RTKs), and Ca2+ channels. We highlight herein the optogenetic engineering strategies, kinetic properties, advantages and limitations of these genetically-encoded Ca2+ channel actuators (GECAs) and modulators. In parallel, we present exemplary applications in both excitable and non-excitable cells and tissues. Furthermore, we briefly discuss potential solutions for wireless optogenetics to accelerate the in vivo applications of GECAs under physiological conditions, with an emphasis on integrating near-infrared (NIR) light-excitable upconversion nanoparticles (UCNPs) and bioluminescence with optogenetics.
Keywords: optogenetics, Calcium signaling, protein design and engineering, LOV2, CRY2, Near-infrared light, upconversion nanoparticles, genetically-encoded calcium channel actuators, immune response, neuromodulation, CRISPRa, NFAT, synthetic biology, Ion channel
Graphical Abstract
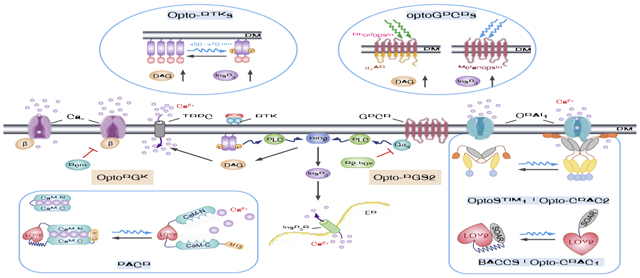
1. Introduction
Ca2+, as a signal for life and death and secondary messenger, controls a myriad of cellular events, including muscle contraction, neurotransmitter release, lymphocyte activation, gene expression and metabolism1–3. In mammalian cells, the intracellular Ca2+ concentration is tightly regulated through coordinated actions of a repertoire of Ca2+-signaling components, including G-protein coupled receptors (GPCRs), the receptor tyrosine kinases (RTKs), various Ca2+ channels and transporters/exchangers, and intracellular Ca2+ sensing and buffering proteins1–3. Upon engagement of membrane receptors by ligands or antigens, phospholipase C gamma (PLCγ) is phosphorylated and activated to catalyze the hydrolysis of phosphatidylinositol 4,5-biphosphate (PI(4,5)P2), yielding two important secondary messengers, diacylglycerol (DAG) and inositol 1,4,5-triphosphate (InsP3). InsP3 further binds to ER-resident InsP3 receptors to induce Ca2+ release from the ER Ca2+ store, followed by the activation of store-operated Ca2+ entry (SOCE)2. SOCE is best exemplified by the Ca2+ release-activated Ca2+ (CRAC) channel composed of ORAI1 and the stromal interaction molecule 1 (STIM1), and constitutes the primary route of Ca2+ entry in non-excitable tissues4–6. By taking advantage of naturally-existing photoreceptors derived from microbes, plants or mammals, these signalling events can be recapitulated through optogenetic engineering to achieve tailored function. Compared to pharmacological, chemogenetic and genetic approaches, these genetically-encoded Ca2+ channel actuators (GECAs) have the advantages of rapid reversibility, superior spatiotemporal resolution, and non-invasiveness4. In this review, we present the latest progress in this endeavor, with a focus on the design principles, kinetic features and caveats to be taken into consideration during the application of these tools. When used in combination with genetically-encoded Ca2+ indicators, GECAs permit the simultaneous monitoring and interrogation of Ca2+-modulated physiological processes both in vitro and in vivo.
2. Engineered photoresponsive GPCRs
As the founding member of the optogenetic field, channelrhodopsin-2 (ChR2) derived from the green alga Chlamydomonas reinhardtii has transformed the neuroscience field by enabling precise control of neural circuits with light both in vitro and in vivo7–9. As a cation permeant channel, ChR2 permits the non-selective entry of Na+, H+, and Ca2+ into the cytosol upon blue-green light stimulation (450–510 nm), thereby depolarizing the plasma membrane (PM) to trigger neuronal firing10 (Fig. 1a). However, given the over 75-fold higher concentrations of extracellular Na+ over Ca2+ (136–151 mM versus 1.4–1.8 mM), the Ca2+ permeability of ChR2 under physiological conditions is very low (PCa/PNa: 0.15)11, thus greatly hampering its application in non-excitable cells and tissues. One ChR2 variant, designated CatCh for Calcium translocating Channelrhodopsin, harbors a point mutation L132C with the Ca2+ permeability enhanced by 1.6-fold (PCa/PNA: 0.24)11, 12, but still falls far behind voltage- or store-operated Ca2+ channels (PCa/PNa: > 1000)6. More Ca2+-specific ChR2 variants are yet to be developed.
Figure 1. Optogenetic tools for remote activation of intracellular Ca2+ signaling.
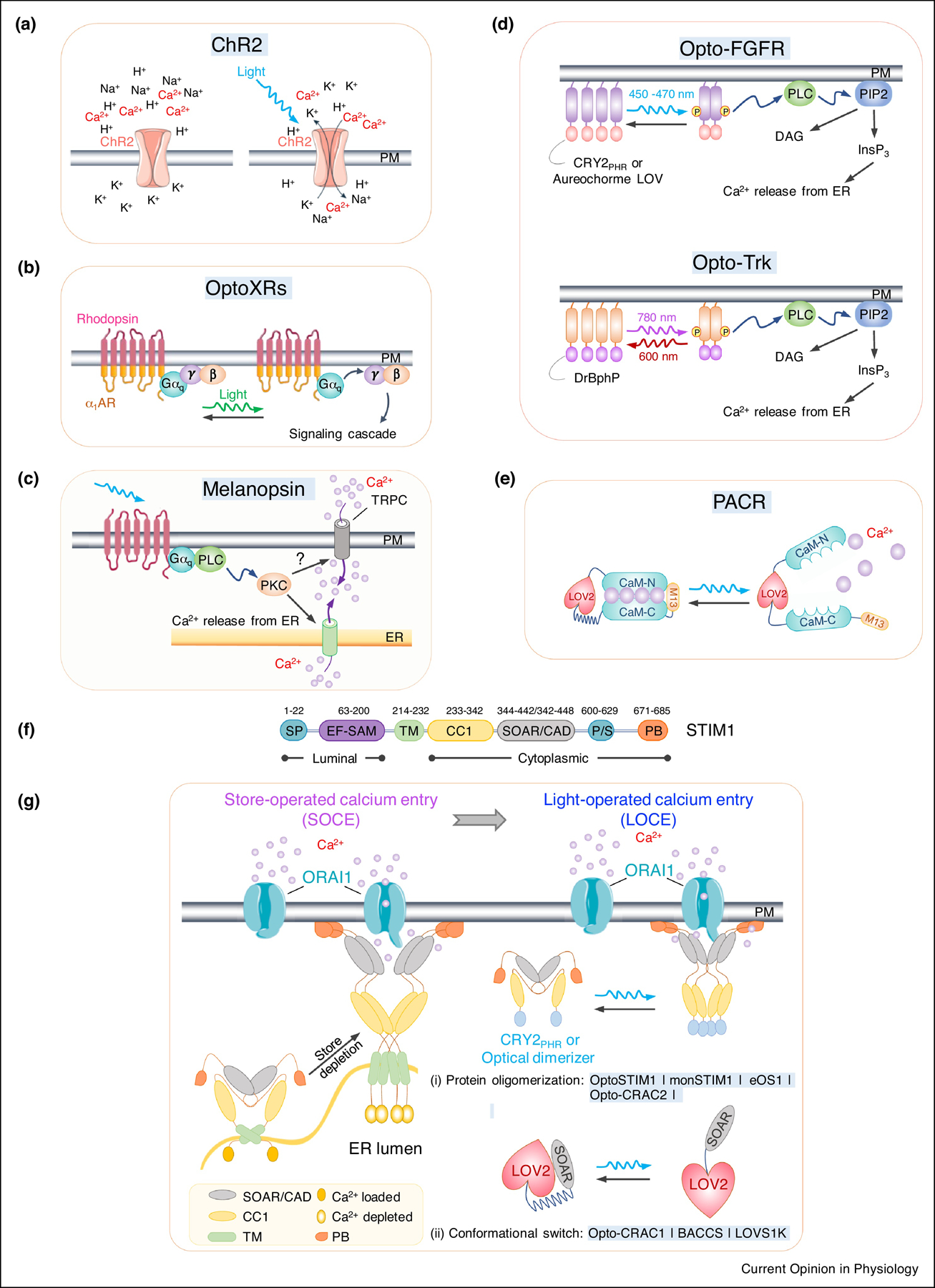
a. Channelrhodopsin-2 (ChR2) as a cation-permeant channel is non-selective toward H+, K+, Na+, and Ca2+.
b. OptoXRs as light-sensing rhodopsin-GPCR chimeras. The extracellular and TM regions are derived from a light-sensitive rhodopsin, while intracellular regions from different adrenergic receptors (β2AR or α1AR).
c. Blue light-mediated isomerization of the light-absorbing retinal cofactor triggers the conformation change of melanopsin (OPN4) and subsequent activation of Gαq-phospholipase C (PLC) to induce Ca2+ influx via TRPC channel.
d. Opto-RTKs as photoactivatable membrane-anchored receptor tyrosine kinases. The intracellular domain of RTKs (FGFR or TrK), is fused to CRY2 or other optical dimerizers, thereby enabling light-inducible clustering to activate downstream signaling, including the PLC pathway and subsequent Ca2+ mobilization.
e. LOV2-CaM-M13 hybrid protein as a photoactivatable Ca2+ releaser (PACR). Photostimulation results in the release of bound Ca2+ into the surrounding environment.
f. Domain architecture of STIM1. SP, signal peptide; EF, EF-hand Ca2+-binding motif; SAM, sterile alpha motif; TM, transmembrane domain; CC1, coiled-coil domain 1; SOAR, STIM-Orai activating region; PS, proline-serine rich domain; PB, polybasic domain.
g. Genetically-encoded Ca2+ channel actuators (GECAs) derived from STIM1.
(Left) schematic illustrating functional STIM1-ORAI1 coupling in response to store depletion. Ca2+ depletion within the ER lumen induces oligomerization of the luminal EF-SAM domain to initiate STIM1 activation and triggers conformational switch to expose SOAR/CAD and the C terminal PB domain. Next, STIM1 undergoes further oligomerization and migration toward the PM, a process facilitated by the association between SOAR/CAD and ORAI1, as well as the interaction between positively-charged PB and negatively-charged phosphoinositides embedded in PM. SOAR/CAD is responsible for direct engagement and activation of ORAI channels to mediate Ca2+ influx.
(right) Optogenetic engineering to convert store-operated Ca2+ entry (SOCE) into light-operated Ca2+ entry (LOCE). (i) A protein oligomerization-based engineering strategy using various optical dimerizers (Opto-CRAC2) and CRY2-STIM1 fusion proteins (OptoSTIM1, monSTIM1, and eOS1). Upon light illumination, CRY2 or optical dimerizers undergoes oligomerization, mimicking Ca2+ -depletion induced EF-SAM multimerization in ER lumen, to trigger STIM1 activation and the subsequent opening of ORAI Ca2+ channels in the PM. (ii) The design of GECAs based on LOV2 conformational switch (Opto-CRAC1, LOVS1K or BACCS), in which the CC1-SOAR interaction-mediated intramolecular autoinhibition is mimicked by LOV2-SOAR fusion in the dark. Upon photostimulation, LOV2 undergoes conformational changes to expose SOAR/CAD, which engages PM-resident ORAI Ca2+ channel to induce Ca2+ influx.
Instead of site-directed mutagenesis, Airan et al. explored an alternative approach by generating a series of rhodopsin-GPCR chimeras, designated “OptoXRs”13, to photo-manipulate intracellular second messengers, including cAMP and Ca2+. In a typical OptoXR, the intracellular loops of rhodopsin were replaced with the intracellular regions of Gq-coupled adrenergic receptors (Fig. 1b). Upon photostimulation at 504 nm, OptoXR evoked cytosolic Ca2+ elevation via the activation of the PLC pathway in HEK cells, and was further applied to photo-control behaviors in awake mice13. A limitation of ChR2 variants and OptoXRs is that the broad rhodopsin absorption peak (440–590 nm) may potentially conflict with biosensors used to report the functional consequences of optogenetic tools14, 15. In addition, owing to the rapid trans-to-cis isomerization of the retinal co-factor (tens of ms), sustained Ca2+ entry requires constant photostimulation and tends to cause undesired photo-toxicity. The latter concern is partially mitigated by the use of naturally-existing melanopsin (encoded by OPN4), a photopigment protein found in human retinal ganglion cells and the iris of mice and primates16 (Fig. 1c). Melanopsin can generate both sustained and oscillatory Ca2+ signals within seconds (t1/2, ON: < 10 s) to activate a downstream Ca2+-responsive master transcription factor, the nuclear factor of activated-T cells (NFAT)17. Melanopsin has been introduced into mouse embryonic stem cell-derived embryoid bodies (EBs) to generate pacemaker-like activity in cardiomyocytes within EBs following photostimulation18. When expressed in non-excitable primary T cells, melanopsin could photo-boost tumor killing in a mouse model of hepatocarcinoma19. Similar to the design of OptoXRs, a chimeric receptor, Opto-mGluR6, composed of melanopsin and metabotropic glutamate receptor 6 (mGluR6), has been devised and expressed in retina to restore the vision of blind mice with retinal degeneration20. One potential caveat associated with these GPCR-based optogenetic actuators is the paralleled stimulation of the DAG/PKC pathway following PLC activation, which could cause non-Ca2+-related functional crosstalk to complicate data interpretation.
3. Photoactivatable RTKs
Similar to GPCR, the engagement of RTKs by their cognate ligands often leads to receptor multimerization to activate the PLCγ pathway for Ca2+ mobilization. To faithfully recapitulate this process, optical multimerizers, including Arabidopsis cryptochrome 2 (CRY2), the light-oxygen-voltage domains (LOV) of aureochorme, and Deinococcus phytochrome BphP (DrBphP), have been fused to the catalytic intracellular kinase domain of RTKs to generate Opto-RTKs21–24 (Fig. 1d). Most engineering efforts have been devoted to two RTKs, the fibroblast growth factor receptor 1 (FGFR1)21, 22 and the tropomyosin-related kinase (Trk)24. Upon light stimulation, the photosensitive moieties undergo dimerization or oligomerization to drive the activation of the intracellular kinase domain of RTKs. Repetitive Ca2+ signals can be generated by switching on and off blue light for CRY2 or LOV-based tools21–24, or toggling between near-infrared (780 nm) and far-red light (660 nm) for the case of DrBphP25. Opto-RTKs can thus be applied to control intracellular Ca2+ signals using both visible and non-visible light. The use of NIR light further enables multiplexed experiments using fluorescent probes and biosensors in the blue-green range without activation of the engineered receptors. Opto-RTKs, however, are fraught with the lack of Ca2+ signaling fidelity because of simultaneous activation of multiple downstream pathways. For example, aside from the Ca2+-associated PLC pathway, photostimulation of Opto-FGFR1 could co-activate proliferation-and survival-related RAS-MAPK, PI3K-AKT and STAT-dependent signaling cascades21–24. Extra cautions, therefore, have to be taken when using Opto-RTKs to interrogate Ca2+-regulated signaling events.
4. Photoswitchable Ca2+ releaser
Optogenetic engineering has also been extended to intracellular Ca2+ sensor proteins, such as the four EF-hand-containing calmodulin. A photoactivatable Ca2+ releaser (PACR) has been generated via optimized insertion of LOV2 into an engineered calmodulin-M13 fusion protein (CaM-M13) (Fig. 1e). In the dark, PACR interacts with Ca2+ with a dissociation constant (Kd) of approximately 16 nM. Upon blue light stimulation, structural changes within LOV2 disrupt the CaM-M13 interaction, which in turn causes Ca2+ release from PACR because of a pronounced reduction in the affinity for Ca2+ (Kd = 3.75 μM)26, thereby leading to an elevation of [Ca2+]cyto. Even though highly specific for Ca2+, the application of PACR faces three drawbacks: (i) the limited Ca2+ releasing capability (<90 nM increase of [Ca2+]cyto) and (ii) cell-to-cell variations because of varying levels of PACR expression; and (iii) the potential perturbation to resting cytosolic Ca2+ level and complications from other CaM-regulated targets.
5. Optogenetic actuators engineered from the CRAC channel
During SOCE activation, STIM1 dynamically couples to ORAI1 via two major molecular steps (Fig. 1f–g): (i) the initiation of STIM1 activation via Ca2+-depletion induced oligomerization of the luminal domain containing the EF-SAM domain; (ii) inside-out signal propagation toward the STIM1 cytoplasmic domain (STIM1ct) to overcome an intramolecular inhibition primarily mediated by the interaction between the coiled-coil 1 region (CC1) and the STIM-Orai activating domain (SOAR)2, 5, 6, 27. Two major optogenetic engineering approaches, using either CRY2 or LOV2 photosensory domains derived from plants, have been applied to reconstruct these key molecular steps to enable optical control of Ca2+ influx through CRAC channels (Fig. 1g).
5.1. CRY2-based OptoSTIM1 and variants
To recapitulate EF-SAM oligomerization during initiation of SOCE, the luminal and transmembrane domains of STIM1 have been replaced by the photolyase-homology region (PHR) of CRY2 to yield OptoSTIM128, which has an activation kinetics of <1 min in response to blue light stimulation and a deactivation kinetics of around 4–6 min upon the withdrawal of light (Fig 1g and Table 1). Very recently, two further improved versions, monster-OptoSTIM1 (monSTIM1) and enhanced OptoSTIM1 (eOS1), have been generated to enhance the light sensitivity and dynamic range of Ca2+ response by 55-fold and 5~10-fold, respectively. The deactivation half-life of the OptoSTIM1 toolset generally ranges from 4 to 7 min28, 29. For eOS1, it takes about 10 min to return to the basal level30. In addition to CRY2-mediated homo-oligomerization, the fusion of optical dimerizers, made of iLID/SspB or CRY2/CIBN, with STIM1ct can likewise photo-trigger Ca2+ influx in mammalian cells, with the activation and deactivation half-lives falling in the range of ~30 sec and 40–190 sec, respectively31. Furthermore, ER-tethered versions were developed to manipulate membrane contact site (MCS) assembly at ER-PM junctions and induce localized Ca2+ influx (t1/2, on = 37.8 ± 5.3 s and t1/2, off = 97.5 ± 16.5 s) in response to blue light stimulation31. These tools set the stage for more physiologically-relevant mimicry of native STIM1 puncta formation and Ca2+ microdomain generation.
Table 1.
Summary of optogenetic tools for noninvasive and precise control Ca2+ signals in mammals.
| Names | Engineering templates | Photosensitive components | Stimulation | Activation (t1/2, sec) | Deactivation (t1/2, sec) | Cell / Tissue Types | Refs |
|---|---|---|---|---|---|---|---|
| Photoswitchable GPCRs and G proteins | |||||||
| ChR2 | Channelrhodopsin-2 | opsin | Blue light | ~0.002 | ~0.01 | Multiple mammalian cells, model organism, rodents and primates | 7 |
| OptoGPCR / OptoXRs | Gq-coupled α1AR | opsin | Blue/green light | N/A | N/A | HEK293 and mice | 13 |
| Melanopsin | opsin | Blue light | < 10 | N/A | Multiple mammalian cells and mice | 47, 48 | |
| Photoactivatable RTKs | |||||||
| OptoRTKs | FGFR1398–822 | CRY2phr | Blue light | <30 | N/A | HeLa/HUVECs | 21, 22 |
| TrkA/B/C | HeLa | 24 | |||||
| TrkA | DrBphP-PCM | nir | ~7 | ~10 | HeLa | 25 | |
| STIM1-based GECAs | |||||||
| BACCS variants | STIM1347–448 | LOV2 | Blue light | < 30 | ~30–60 | HEK293T | 33 |
| STIM1347–448 | 2*LOV2 | ||||||
| ORAI1 + STIM1347–448 | 2*LOV2 | ||||||
| dOrai + dmStim413–514 | 2*LOV2 | S2 | |||||
| LOVS1K | STIM1233–450 | LOV2 | Blue light | N/A | N/A | HeLa | 34 |
| Opto-CRAC1 variants | Mouse STIM1336–486 | LOV2 | Blue light | 23.4 ± 4.2 | 24.9 ± 4.8 | HeLa and multiple cell lines | 32 |
| NIR | <30 | <30 | BMDCs and mice | ||||
| Human STIM1336–685 | LOV2 | Blue light | 36.3 ± 6.4 | n/a | HeLa | ||
| chicken STIM1262–412 | LOV2 | Blue light | ~10 | <30 | Skin mesenchymal cells | 35 | |
| zebrafish STIM1341–442 | LOV2 | Blue light | 18.7 | 24.5 | HeLa | 36 | |
| Opto-CRAC2 variants | STIM1233–685 | iLID/SspB | Blue light | 28.5 ± 3.2 | 48.6 ± 5.4 | HeLa | 31 |
| STIM1233–685 | CRY2PHR/CIBN | 23.4 ± 2.6 | 153.0 ± 26.2 | ||||
| ER-tethered-STIM1233–685 | CRY2PHR | 37.8 ± 5.3 | 97.5 ± 16.5 | ||||
| OptoSTIM1 variants | STIM1238–685 | CRY2PHR | Blue light | 64.5 ± 4.8 | 274 ± 23.7 | HeLa and multiple cell lines | 4, 28 |
| STIM1238–463 | 43.2 ± 3.8 | 383 ± 52.3 | |||||
| STIM1342–685 | 48.2 ± 5.4 | 344 ± 34.6 | |||||
| monSTIM1 | STIM1238–685 | CRY2PHR-E281A-9A | Blue light | 24.8 ± 1.0 | 513.5 ± 25.8 | HEK and neurons and mice | 29 |
| eOS1 | STIM1238–685 | CRY2PHR-E490G | Blue light | <60 | <600 | B3Z T cell | 30 |
| Two-photon | In vivo CD8+ T dells | ||||||
| Light-switchable inhibition of Ca2+ signaling | |||||||
| OptoRGK | Rem1–266 | iLID/SspB | Blue light | 3.2 ± 1.0* | 23.0 ± 2.4* | HEK293; HL-1 | 37 |
cytosol-plasma membrane translocation
Abbreviations: BACCS, blue light-activated Ca2+ channel switch; BMDCs, bone marrow-derived DCs; CaM-M13, calmodulin fused with the M13 peptide; ChR2, Channelrhodopsin-2; CIBN, the N-terminal domain of CIB1; CRY2, cryptochrome2; eOS1, enhanced OptoSTIM1; FGFR1, fibroblast growth factor receptor 1; GECA, genetically encoded Ca2+ actuator; HUVECs, human umbilical vein endothelial cells; iLID, improved light-induced dimer (LOV2-SsrA); LOV2, light-oxygen-voltage sensing) domain; LOVS1K, LOV2 fused STIM1 fragments (233–450); monSTIM1, monster-OptoSTIM1; PACR, photoactivatable Ca2+ releaser; Opto-α1AR, Optogenetic Gq-coupled α1-adrenergic receptor; Opto-CRAC, Optogenetic Ca2+ release-activated Ca2+ channel; OptoRGK, Optogenetic Ras-like GTPase Rad/Rem/Gem/Kir; Opto-RGS2, Optogenetic regulators of G-protein signaling; Opto-RTKs, Optogenetic receptor tyrosine kinases; OptoSTIM1, Optogenetic stromal interaction molecule 1; OptoXRs, optogenetic control of intracellular signal transduction; S1, somatosensory cortex; S2, Drosophila S2 cells; SP, Signal peptide of STIM1; NIR, near-infrared; TM, transmembrane domain; Trk, tropomyosin-related kinase;
5.2. Opto-CRAC series and other LOV2-based GECAs
To mimic the intramolecular autoinhibition of STIM1 mediated by the juxtamembrane CC1 and SOAR, CC1 was replaced by LOV2 and various STIM1ct fragments were optimized to generate LOV2-STIM1ct chimeras (Opto-CRAC31, 32, BACCS33, and LOVS1K34). These LOV2-based GECAs enable more rapid and readily reversible control of Ca2+ signals, with the activation kinetics ranging from 10 sec to 30 sec and the deactivation half-lives in the range of 30–50 sec (Table 1). In these chimeric constructs, LOV2 cages the fused STIM1ct fragment in a largely inactive conformation presumably through steric hindrance. Upon photostimulation, the photochemical reaction within LOV2 leads to the unfolding of the C-terminal Jα helix to unleash the caged STIM1ct fragments, thereby exposing the SOAR/CAD region to engage and gate ORAI Ca2+ channels in the PM (Fig. 1g). Opto-CRAC32 or BACCS variants33, either anchored to PM or using STIM1ct fragments derived from species other than human and rodent (fruit fly33, chicken35, and zebrafish36) further diversify the kinetic properties of the LOV2-based GECAs. In addition, these LOV2-based GECAs can be directly fused to ORAI1 channels, in the form of a single or two copies32, 35, to enable optogenetic control of Ca2+ signaling in cells or tissues with no or little expression of endogenous ORAI proteins.
Collectively, compared to Opto-GPCRs and Opto-RTKs, STIM1-based GECAs directly gate one of the most Ca2+-selective channels, the ORAI protein family, and therefore, retain high Ca2+ selectivity over other cations32. Nonetheless, one has to bear in mind the caveat that STIM1, and presumably STIM1-derived GECAs too, might cross-talk with other types of Ca2+ channels (e.g., certain types of transient receptor potential (TRP) channels and voltage gated Ca2+ channels) in selected cell types.
6. Genetically-encoded photoswitchable inhibitors of Ca2+ signaling
Complementary to the GECA toolkit, two optogenetic tools capable of suppressing Ca2+ entry and/or ER Ca2+ release have been generated. To develop a light-inducible OFF-switch for voltage-gated Ca2+ (CaV) channel, Ma et al. developed an optogenetic construct, termed OptoRGK37 (Fig. 2a), by using a light-sensitive heterodimerization system (iLID/SspB) to control the subcellular localization of a small G protein, Rem1–266. Rem belongs to the RGK family of G proteins and functions as negative regulator of Ca2+ entry mediated by CaV channels in excitable cells. OptoRGK has been successfully used to perturb Ca2+ oscillations in HL-1 cardiomyocytes, and shows promise to rescue cardiac arrhythmia37. In a second case, Hannanta-anan et al. utilized a similar strategy using the CRY2/CIBN optical dimerizer to recruit regulators of G-protein signaling (RGS) toward PM, thereby reducing the lifespan of GTP-bound α-subunits to terminate agonist-evoked calcium oscillations via inhibition of Gαq-mediated downstream signaling (including Ca2+ mobilization from internal stores) in response to blue light stimulation38 (Fig. 2b). Complementary to the genetic approach, photopharmacological tools building upon azopyrazoles have also been invented to selectively suppress CRAC channel activity in a light-dependent manner39.
Figure 2. Genetically-encoded photoswitchable inhibitors of intracellular Ca2+ signaling.
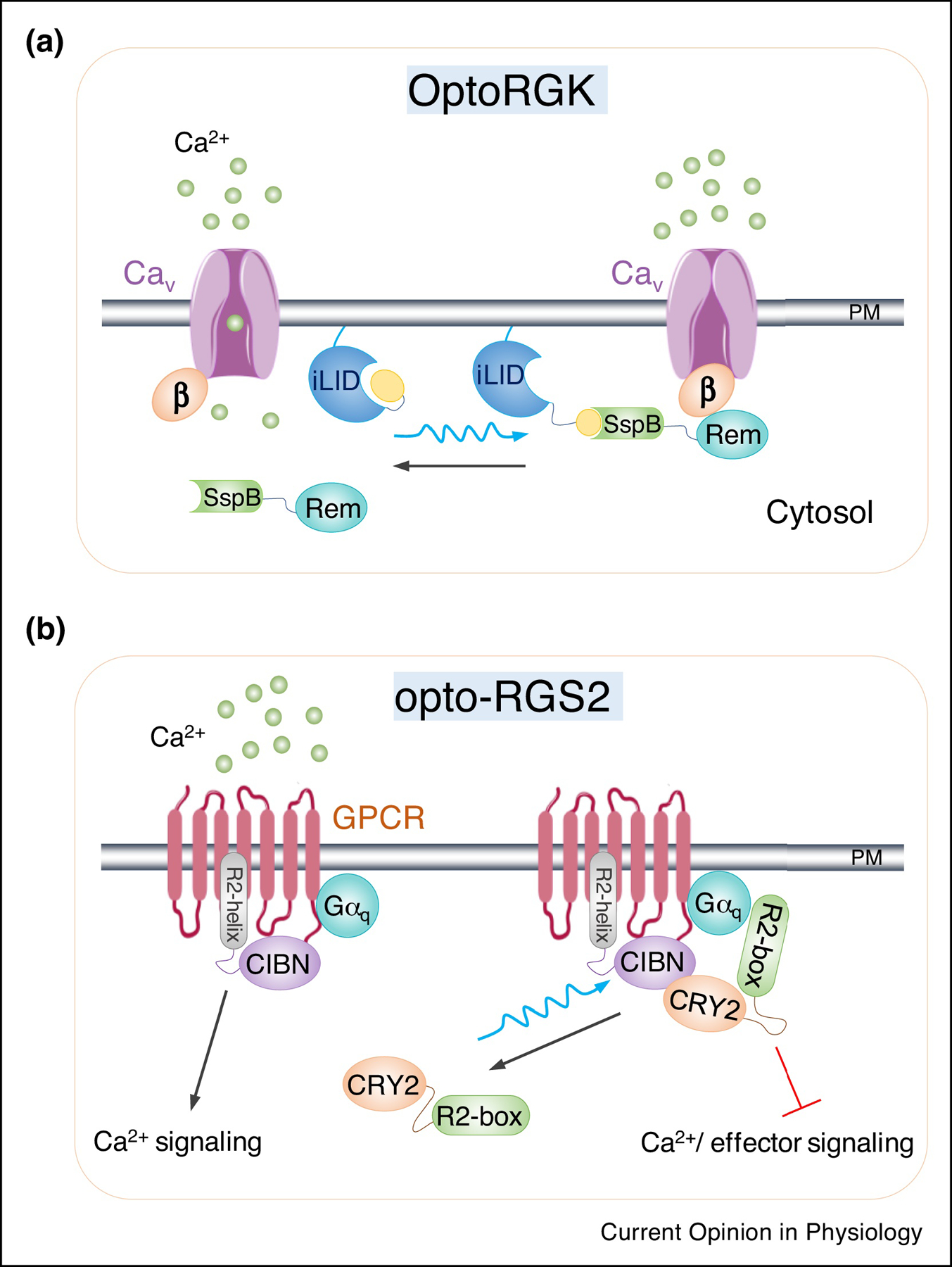
a. Cartoon illustration of OptoRGK to photo-control CaV channel activity in cardiomyocytes. An optical dimerizer pair made of LOV2-SsrA (iLID) and SspB is utilized to induce the translocation of Rem1 core domain (a member of the RGK family of GTP-binding proteins) from the cytosol to PM, where it engages the auxiliary beta subunits of CaV channel to suppress its activity upon photostimulation.
b. Design of optogenetic regulators of G-protein signaling (Opto-RGS2). In this design, RGS2 is split into two components, including the catalytic box (R2-box) which is critical for inhibition of Gαq signaling and the N-terminal amphipathic helix (R2-helix). The two split parts are fused with either CRY2PHR or its binding partner CIBN. In the dark, R2-box is distributed in the cytoplasm. Upon blue light illumination, the forced dimerization between CIBN-R2-helix and CRY2-R2-box triggers reconstitution of the active protein and subsequently inhibit Gαq signaling and terminates Ca2+ release originated from GPCR signaling.
7. Applications of GECAs and wireless optogenetics
Given the strict Ca2+ selectivity and high signalling fidelity, STIM1-derived GECAs have been used ex vivo to control Ca2+/NFAT-dependent gene expression (Fig. 3a) and cytokine production in cells of the immune system (Fig. 3b). When coupled with the CRISPR activation (CRISPRa) technique40, Opto-CRAC can further be repurposed to control the expression of endogenous genes. The calcium-responsive transcriptional reprogramming tool (CaRROT)36, made of NFAT1–460-dCas9-VP64, has been devised to serve this purpose, which allows precise control of transcriptional outputs at user-defined genomic loci, thus expanding the Ca2+-responsive effectors to almost any endogenous genes (Fig. 3c). Moving beyond in vitro applications, recent studies have reported encouraging progress in the use of non-opsin-based optogenetics in living animals.
Figure 3. Applications of GECAs and potential solutions for wireless optogenetics.
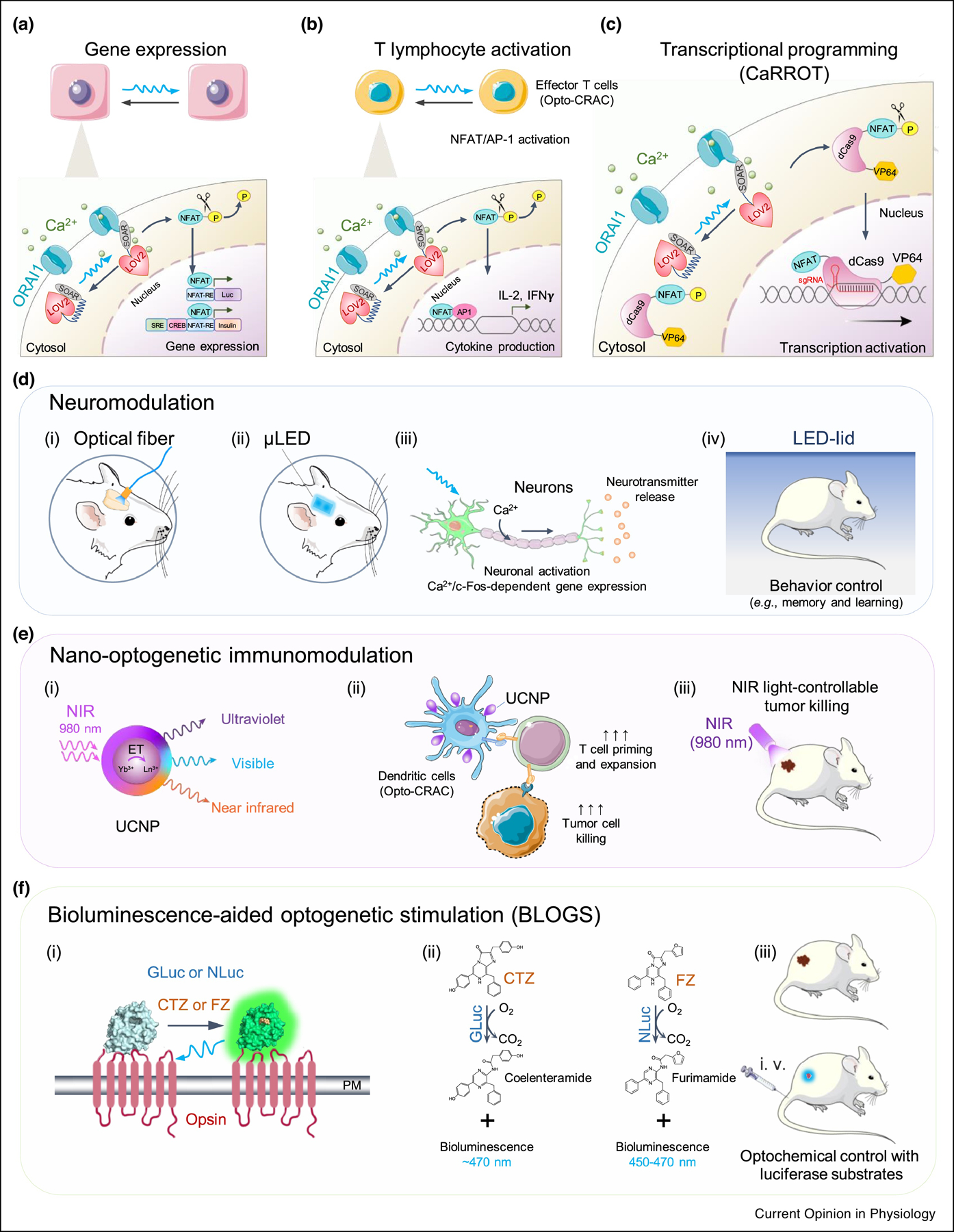
a. GECAs (e.g., Opto-CRAC) utilized to control Ca2+-dependent gene expression using synthetic response elements (RE) derived from various Ca2+-responsive transcription factors. NFAT, nuclear factor of activated-T cells; CREB: cAMP response element-binding domain; SRE, serum response element.
b. GECAs used to control Ca2+/NFAT-driven cytokine production in T cells in the presence of co-stimulatory signals to activate activator protein 1 (AP-1).
c. Coupling GECAs with CRISPRa to enable light-tunable transcriptional programming. Upon blue light illumination, Opto-CRAC1 induces cytosolic Ca2+ influx, which in turn activates CaM/calcineurin to dephosphorylate the N-terminal non-DNA binding fragment of NFAT (NFAT1–460) and eventually drive the nuclear entry of the fusion protein (NFAT1–460-dCas9-VP64) – a calcium-responsive transcriptional reprogramming tool (designated as CaRROT). In the presence of small guide RNAs (sgRNAs), CaRROT can be precisely targeted to the promoter regions of any desired genomic loci to photo-tune the expression of endogenous genes.
d. Optogenetic neuromodulation with GECAs (e.g., OptoSTIM1 or monSTIM1). Blue light can be delivered into a specific brain region via implantation of optical fiber (i) or μLED (ii). Photoactivatable Ca2+ influx can lead to induction of Ca2+-responsive gene expression (e.g., c-Fos) in both neurons and astrocytes in deep-brain regions such as hippocampal and thalamic regions (iii), thereby modulating the behavior (e.g., social fear learning) in awake mice (iv).
e. A nano-optogenetic platform for wireless optogenetic immunomodulation in a mouse model of melanoma. (i) schematic illustration of the core/shell structure of lanthanide doped upconversion nanoparticles (UCNPs), which act as nanoilluminator to emit visible light upon near-infrared irradiation (NIR); (ii-iii) NIR light-controllable activation of Ca2+ influx in dendritic cells (DCs) expressing Opto-CRAC1 could boost antigen presentation and DC maturation to more efficiently prime T cells in the draining lymph nodes, thereby facilitating dendritic cell-based immunomodulatory therapy against melanoma.
f. Bioluminescence-aided optogenetic stimulation (BLOGS). (i) Cartoon illustrating the photoactivation of a chimeric luminopsin (LMO) receptor comprising a microbial opsin and Gaussia luciferase (GLuc) or NanoLuc luciferase (NLuc) in the presence of their cognate substrates coelenterazine (CTZ) or furimazine (FZ). (ii) Photochemical reactions catalyzed by GLuc or NLuc. (iii) Schematic depiction of wireless optogenetics using BLOGS in mice administered with luciferase substrates.
7.1. Neuromodulation in awake mice
A bottleneck with the existing optogenetic approach is a situation where the depth of tissue penetration of visible light is shallow (<2–3 mm)41, requiring optogenetic protocols to use invasive fiber optic probes or microLED to deliver visible light (in the range of 400–500 nm) into tissues (Fig. 3d). These invasive procedures are technically demanding, tend to elicit inflammatory responses, and remain incompatible with chronic experiments. To overcome these issues, monSTIM1 was generated by using a hyper-sensitive CRY2 variant with one mutation and nine additional amino acids in the C-terminus of CRY2PHR (CRY2E281A-A9) to boost its clustering capability. The new construct displayed 55-fold increase in light-sensitivity compared to the parental OptoSTIM129 (Fig 1g). Hence, monSTIM1 was utilized to manipulate Ca2+ signaling and activate Ca2+-dependent gene expression in deep-brain regions (hippocampal and thalamic regions), and to boost the social learning capability of awake mice without invasive surgical procedures29 (Fig. 3d). However, upon non-invasive light stimulation, how deep the tissues can be reached to effectively photoactivate monSTIM1 remains elusive. Furthermore, blue light emitting at 450–470 nm may generate local heat and oxidative stress while STIM1 is also sensitive to both heat and ROS27. It remains to be rigorously validated if the observed behavior changes are indeed caused by Ca2+ signals per se rather than undesired side effects of photostimulation. Another attractive strategy to overcome this bottleneck is to exploit optogenetic modules that can be switched on and off by NIR that has a greater depth of tissue penetration (up to centimeters)42.
7.2. Immunomodulation in living animals
STIM1-based GECAs have been successfully applied in vivo to control the motility and function of cells of the immune system. eOS1 has been employed to manipulate Ca2+ signals in single T cells and photo-tune T cell migration, adhesion and chemokine release in vivo via two-photon-based photoactivation30. Although two-photon optogenetics provide a better spatial specificity and deep penetration in scattering tissue43, potential heat generation during photostimulation and special equipment requirements make it non-ideal for daily use in the laboratory setting, let alone further clinical applications. A wireless nano-optogenetic approach based on NIR-excitable upconversion nanoparticles (UCNPs; Fig. 3e) has been developed to accelerate in vivo application using immune cells. The most significant advantage of UCNPs is their unusual inverse excitation and emission profiles: UCNPs are excited using low power (mW) and deep tissue-penetrating (1–2 cm) NIR light. This low energy input is efficiently converted to a higher energy output emission at diverse shorter wavelengths, including blue light emission at 470 nm that can activate most optogenetic constructs (Fig. 3e). Since UCNPs can be illuminated from NIR sources located outside the body, they can act as “nano-illuminator relays” to activate optogenetic constructs in a wireless manner. The nano-optogenetic concept has been validated in a dendritic cell-based immunomodulatory therapy against tumor in a mouse mode of melanoma32 (Fig. 3e).
8. Conclusions
In summary, a variety of optogenetic tools, based on both opsin or non-opsin photoreceptors, have been crafted to enable remote control of intracellular Ca2+ signaling both in cellulo and in living organisms (summarized in Table 1). Most tools are engineered from membrane receptors (GPCRS and RTKs) and signaling proteins (CaM, G proteins and STIM1) that are directly or indirectly involved in Ca2+ signaling. Among these tools, GECAs inspired by STIM1 are regarded as the most Ca2+ selective and have been most widely applied to modulate the central nervous system and the immune system. The in vivo application of these tools will likely be accelerated by developing NIR-responsive optogenetic modules and combining existing tools with nanotechnologies or bioluminescence to enable in vivo optogenetics. The feasibility of bioluminescence-aided optogenetic stimulation (Fig. 3f), at the cost of partially sacrificing the spatial resolution, has been validated in several recent studies44–46. These innovative platforms might represent alternative solutions for future wireless optogenetics.
Highlights.
Overview of optogenetic tools to control Ca2+-modulated physiological processes
Engineering strategies for genetically-encoded Ca2+ channel actuators (GECAs)
Guidelines for selecting appropriate GECAs to achieve tailored function
Insights into potential solutions for wireless optogenetics in vivo
Acknowledgements
We thank the financial supports from the Welch Foundation (BE-1913-20190330 to YZ), the John S. Dunn Foundation (to YZ), the National Institutes of Health (R01GM112003, R21GM126532, and R01CA232017 to Y.Z.), and the American Cancer Society (RSG-16-215-01-TBE to YZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors have no competing interests or conflict of interest to declare.
References
- 1.Clapham DE Calcium signaling. Cell 131, 1047–1058 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Hogan PG, Lewis RS & Rao A Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol 28, 491–533 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berridge MJ, Bootman MD & Roderick HL Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4, 517–529 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Ma G et al. Optogenetic toolkit for precise control of calcium signaling. Cell Calcium 64, 36–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen NT et al. CRAC channel-based optogenetics. Cell Calcium 75, 79–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakriya M & Lewis RS Store-Operated Calcium Channels. Physiol Rev 95, 1383–1436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagel G et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyden ES, Zhang F, Bamberg E, Nagel G & Deisseroth K Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]; *A pioneering study describing ChR2 as the founding member of the optogenetic field, which revolutionizes the way neuroscientists to interrogate neuronal activities and dissect neural circuits.
- 9.Zhang F et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Fenno L, Yizhar O & Deisseroth K The development and application of optogenetics. Annu Rev Neurosci 34, 389–412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck S et al. Synthetic Light-Activated Ion Channels for Optogenetic Activation and Inhibition. Front Neurosci 12, 643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinlogel S et al. Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nat Neurosci 14, 513–518 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Airan RD, Thompson KR, Fenno LE, Bernstein H & Deisseroth K Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 (2009). [DOI] [PubMed] [Google Scholar]; *The first study describes the design of rhodopsin-GPCR chimeras, termed optoXRs, to remotely control GPCR signaling with high spatiotemporal precision.
- 14.Deisseroth K & Hegemann P The form and function of channelrhodopsin. Science 357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shichi H Spectrum and purity of bovine rhodopsin. Biochemistry 9, 1973–1977 (1970). [DOI] [PubMed] [Google Scholar]
- 16.Hankins MW, Peirson SN & Foster RG Melanopsin: an exciting photopigment. Trends Neurosci 31, 27–36 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Hannanta-Anan P & Chow BY Optogenetic Control of Calcium Oscillation Waveform Defines NFAT as an Integrator of Calcium Load. Cell Syst 2, 283–288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beiert T, Bruegmann T & Sasse P Optogenetic activation of Gq signalling modulates pacemaker activity of cardiomyocytes. Cardiovasc Res 102, 507–516 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Zhao B et al. An Optogenetic Controllable T Cell System for Hepatocellular Carcinoma Immunotherapy. Theranostics 9, 1837–1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Wyk M, Pielecka-Fortuna J, Lowel S & Kleinlogel S Restoring the ON Switch in Blind Retinas: Opto-mGluR6, a Next-Generation, Cell-Tailored Optogenetic Tool. PLoS Biol 13, e1002143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JM, Lee M, Kim N & Heo WD Optogenetic toolkit reveals the role of Ca2+ sparklets in coordinated cell migration. Proc Natl Acad Sci U S A 113, 5952–5957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim N et al. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem Biol 21, 903–912 (2014). [DOI] [PubMed] [Google Scholar]; * One of the first studies to describe the design of optoFGFR1 to control intracellular Ca2+ signals and other downstream effectors associated with RTK signaling.
- 23.Grusch M et al. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J 33, 1713–1726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; * One of the first studies to describe the use of LOV domains from aureochrome to photo-manipulate RTK signaling.
- 24.Chang KY et al. Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat Commun 5, 4057 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Leopold AV, Chernov KG, Shemetov AA & Verkhusha VV Neurotrophin receptor tyrosine kinases regulated with near-infrared light. Nat Commun 10, 1129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors describe the use of NIR light-controllable optogenetic modules derived from a bacterial phytochrome to modulate RTK signaling.
- 26.Fukuda N, Matsuda T & Nagai T Optical control of the Ca2+ concentration in a live specimen with a genetically encoded Ca2+-releasing molecular tool. ACS Chem Biol 9, 1197–1203 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Soboloff J, Rothberg BS, Madesh M & Gill DL STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol 13, 549–565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyung T et al. Optogenetic control of endogenous Ca(2+) channels in vivo. Nat Biotechnol 33, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]; * This study describes the first development of CRY2-STIM1 chimeras to enable optical control of Ca2+ influx by taking advantage of endogenous ORAI channels into mammalian cells. The authors further apply the tool to remotely modulate neuronal activities both in vitro and in vivo.
- 29.Kim S et al. Non-invasive optical control of endogenous Ca(2+) channels in awake mice. Nat Commun 11, 210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohineust A, Garcia Z, Corre B, Lemaitre F & Bousso P Optogenetic manipulation of calcium signals in single T cells in vivo. Nat Commun 11, 1143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study describes an improved version of optoSTIM1 termed eOS1 to control the migration, adhesion and chemokine production in single T cells by utilizing two-photon photoactivation.
- 31.Ma G et al. Optogenetic engineering to probe the molecular choreography of STIM1-mediated cell signaling. Nat Commun 11, 1039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; * A comprehensive study illustrating the use of optogenetic engineering approaches to recapitulate key signaling events during store-operated calcium entry, which validates oligomerization and conformational switch as two essential steps to drive CRAC channel activation.
- 32.He L et al. Near-infrared photoactivatable control of Ca(2+) signaling and optogenetic immunomodulation. Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; *One of the first studies demonstrating a LOV2-fusion strategy to mimic STIM1 intramolecular trapping. The first proof-of-concept study demonstrates the feasibility of wireless nano-optogenetics to modulate the immune system in living animals.
- 33.Ishii T et al. Light generation of intracellular Ca(2+) signals by a genetically encoded protein BACCS. Nat Commun 6, 8021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; *One of the first studies illustrating the design of LOV2-STIM1 chimeras to reversibly control Ca2+ signaling.
- 34.Pham E, Mills E & Truong K A synthetic photoactivated protein to generate local or global Ca(2+) signals. Chem Biol 18, 880–890 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Li A et al. Calcium oscillations coordinate feather mesenchymal cell movement by SHH dependent modulation of gap junction networks. Nat Commun 9, 5377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen NT, He L, Martinez-Moczygemba M, Huang Y & Zhou Y Rewiring Calcium Signaling for Precise Transcriptional Reprogramming. ACS Synth Biol 7, 814–821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study integrates genetically-encoded calcium channel actuators with the CRISPR-based transcriptional activation technique to generate CaRROT, which extends Ca2+-controllable gene transcription toward any genomic loci in a light-dependent manner.
- 37.Ma G et al. Optogenetic Control of Voltage-Gated Calcium Channels. Angew Chem Int Ed Engl 57, 7019–7022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; *One of the few studies describing an OFF-switch to photo-suppress calcium influx mediated by voltage-gated calcium channel in excitable cells.
- 38.Hannanta-Anan P & Chow BY Optogenetic Inhibition of Galphaq Protein Signaling Reduces Calcium Oscillation Stochasticity. ACS Synth Biol 7, 1488–1495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X et al. Optical Control of CRAC Channels Using Photoswitchable Azopyrazoles. J Am Chem Soc 142, 9460–9470 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.BASHKATOV AN, GENINA EA & TUCHIN VV OPTICAL PROPERTIES OF SKIN, SUBCUTANEOUS, AND MUSCLE TISSUES: A REVIEW. Journal of Innovative Optical Health Sciences 04, 9–38 (2011). [Google Scholar]
- 42.Redchuk TA, Omelina ES, Chernov KG & Verkhusha VV Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat Chem Biol 13, 633–639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oron D, Papagiakoumou E, Anselmi F & Emiliani V Two-photon optogenetics. Prog Brain Res 196, 119–143 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Tan P, He L, Han G & Zhou Y Optogenetic Immunomodulation: Shedding Light on Antitumor Immunity. Trends Biotechnol 35, 215–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.England CG, Ehlerding EB & Cai W NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug Chem 27, 1175–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim CK, Cho KF, Kim MW & Ting AY Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study demonstrates the feasibility of combining NanoLuc-catalyzed bioluminescence with LOV2-based optogenetic devices to photo-stimulate cell signaling.
- 47.Ye H, Daoud-El Baba M, Peng RW & Fussenegger M A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1568 (2011). [DOI] [PubMed] [Google Scholar]; *A pioneering study describing the use of melanopsin as an optogenetic tool to control Ca2+ signaling and blood glucose metabolism in awake mice.
- 48.Kumbalasiri T, Rollag MD, Isoldi MC, Castrucci AM & Provencio I Melanopsin triggers the release of internal calcium stores in response to light. Photochem Photobiol 83, 273–279 (2007). [DOI] [PubMed] [Google Scholar]


