Abstract
Containment of the COVID-19 pandemic requires reducing viral transmission. SARS-CoV-2 infection is initiated by membrane fusion between the viral and host cell membranes, mediated by the viral spike protein. We have designed a dimeric lipopeptide fusion inhibitor that blocks this critical first step of infection for emerging coronaviruses and document that it completely prevents SARS-CoV-2 infection in ferrets. Daily intranasal administration to ferrets completely prevented SARS-CoV-2 direct-contact transmission during 24-hour co-housing with infected animals, under stringent conditions that resulted in infection of 100% of untreated animals. These lipopeptides are highly stable and non-toxic and thus readily translate into a safe and effective intranasal prophylactic approach to reduce transmission of SARS-CoV-2.
One-sentence summary:
A dimeric form of a SARS-CoV-2-derived lipopeptide is a potent inhibitor of fusion and infection in vitro and transmission in vivo.
Infection by SARS-CoV-2 requires membrane fusion between the viral envelope and the host cell membrane, at either the cell surface or the endosomal membrane. The fusion process is mediated by the viral envelope spike glycoprotein, S. Upon viral attachment or uptake, host factors trigger large-scale conformational rearrangements in S, including a refolding step that leads directly to membrane fusion and viral entry (1–6). Peptides corresponding to the highly conserved heptad repeat (HR) domain at the C-terminus of the S protein (HRC peptides) may prevent this refolding and inhibit fusion, thereby preventing infection (7–12) (Fig. 1a–c). The HRC peptides form six-helix bundle (6HB)-like assemblies with the extended intermediate form of the S protein trimer, thereby disrupting the structural rearrangement of S that drives membrane fusion (7).
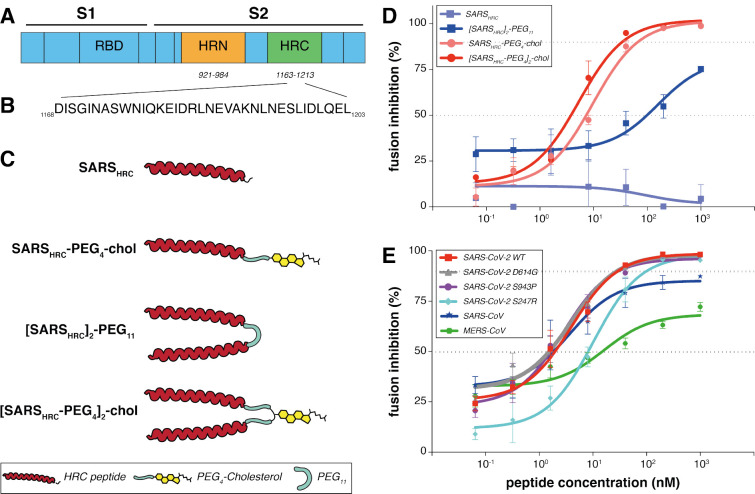
Figure 1: Peptide-lipid conjugates that inhibit SARS-CoV-2 spike (S)-mediated fusion.
(A) The functional domains of SARS-CoV-2 S protein: receptor-binding domain (RBD) and heptad repeats (HRN and HRC) are indicated. (B) Sequence of the peptides that derive from the HRC domain of SARS-CoV-2 S. (C) Monomeric and dimeric forms of lipid tagged SARS-CoV-2 inhibitory peptides that were assessed in cell-cell fusion assays. (D) Cell-cell fusion assays with different inhibitory peptides. The percentage inhibition is shown for four different peptides used at increasing concentrations. Inhibitory concentrations 50% and 90% were calculated (dotted lines). Percent inhibition was calculated as the ratio of relative luminescence units in the presence of a specific concentration of inhibitor and the relative luminescence units in the absence of inhibitor and corrected for background luminescence. Data are means ± standard deviation (SD) from three separate experiments. The difference between the results for [SARSHRC-PEG4]2-chol and SARSHRC-PEG4-chol lipopeptides are significant (Two-way ANOVA, **** p<0.0001). (E) Fusion inhibitory activity of [SARSHRC-PEG4]2-chol peptide against SARS-CoV-2 S variants, MERS-CoV-2 S, and SARS-CoV S. Data are means ± standard deviation (SD) from three separate experiments.
We have previously demonstrated that lipid conjugation of HRC-derived inhibitory peptides markedly increases antiviral potency and in vivo half-life (13–16), and used this strategy to create entry inhibitors for prophylaxis and/or treatment of human parainfluenza virus type 3, measles virus, and Nipah virus infection (14, 15, 17–20). Both dimerization and peptide integration into cell membranes proved key to ensure respiratory tract protection and prevent systemic lipopeptide dissemination (16, 18). The lipidconjugated peptides administered intranasally to animals reached high concentrations both in the upper and lower respiratory tract, and the specific nature of the lipid can be designed to modulate the extent of transit from the lung to the systemic circulation and organs (18–22). Lipid conjugation also enabled activity against viruses that do not fuse until they have been taken up via endocytosis (23). Here, we show that a dimeric form of a SARS-CoV-2 S-specific lipopeptide is a potent inhibitor of fusion mediated by the SARS-CoV-2 S protein, prevents viral entry, and, when administered intranasally, completely prevents direct-contact transmission of SARS-CoV-2 in ferrets. We propose this compound as a candidate antiviral, to be administered by inhalation or intranasal spray, for pre-exposure or early post-exposure prophylaxis for SARS-CoV-2 transmission in humans.
To improve the antiviral potency of the previously assessed SARS-CoV-2 HRC-lipopeptide fusion inhibitor (7), we compared monomeric and dimeric derivatives of the SARS-CoV-2 S-derived HRC-peptide (Fig. 1d). Initial functional evaluation of the SARS-CoV-2 HRC lipopeptides was conducted with a cell-cell fusion assay based on β-galactosidase (β-gal) complementation that we adapted for assessment of SARS-CoV-2 S-mediated fusion. For this assay, cells expressing human angiotensin-converting enzyme 2 (hACE2) and the N-terminal fragment of β-gal were mixed with cells expressing the SARS-CoV-2 S protein and the C-terminal fragment of β-gal. When fusion mediated by S occurs, the two fragments of β-gal combine to generate a catalytically active species, and fusion is detected via the luminescence that results from substrate processing by β-gal. This assay format allows the assessment of potential SARS-CoV-2 S-mediated membrane fusion inhibitors without the use of infectious virus. The assay measures an inhibitor’s ability to block fusion of S-bearing cells with receptor-bearing target cells and is predictive of in vivo antiviral activity (14).
Fig. 1d shows the antiviral potency of two monomeric and two dimeric SARS-CoV-2 S-derived 36-amino acid (Fig. 1b) HRC-peptides, without (SARSHRC and [SARSHRC-PEG4]2) or with (SARSHRCPEG4-chol and [SARSHRC-PEG4]2-chol) appended cholesterol, in cell-cell fusion assays. The percentage inhibition corresponds to the extent of luminescence signal suppression observed in the absence of any inhibitor (i.e., 0% inhibition corresponds to maximum luminescence signal). Dimerization increased the peptide potency (see SARSHRC vs. [SARSHRC-PEG4]2 in Fig. 1d). The dimeric form of HRC lipopeptide was also more potent than its monomeric lipopeptide counterpart (SARSHRC-PEG4-chol IC50 ~10 nM and [SARSHRC-PEG4]2-chol IC50~3nM, (Two-way ANOVA, p<0.0001)). This dimeric cholesterolconjugated peptide ([SARSHRC-PEG4]2-chol; red line in Fig. 1d) is the most potent lipopeptide against SARS-CoV-2 that has been identified thus far. A lipopeptide based on the human parainfluenza virus type 3 (HPIV3) F protein HRC domain, used as a negative control, did not inhibit fusion at any concentration tested (Fig. S1). A cellular toxicity (MTT) assay was performed in parallel with this experiment to evaluate the potential toxicity of each lipopeptide (Fig. S2). Toxicity for each of the lipopeptides in this assay was minimal, even at the highest concentrations tested (<20% at 100 μM). No toxicity was observed for the dimeric SARS-CoV-2 lipopeptide at its IC90 entry inhibitory concentration (~350 nM).
Despite the overall stability of the SARS-CoV-2 genome, variants with mutations in S have spread globally (24–32). These mutations in S altered infectivity of cells (e.g., D614G (24)) or were located in the putative target domain of the HRC peptide (e.g., S943P). To determine the potency of the [SARSHRC PEG4]2-chol peptide for a range of variant SARS-CoV-2 viruses, we examined fusion inhibition mediated by each of these emerging S protein mutants. In addition, to assess the potential for broad-spectrum activity we assessed potency against the S of SARS-CoV and MERS-CoV (using dipeptidyl peptidase 4 (DPP4) receptor-bearing cells as the target for the latter). Fig. 1e shows the IC50 and IC90 of [SARSHRC-PEG4]2-chol for inhibition of fusion by the S mutants and in addition, for SARS-CoV S and MERS-CoV S. The [SARSHRC-PEG4]2-chol lipopeptide inhibited all SARS-CoV-2 strains with S mutations at comparable potency and showed considerable potency against both SARS-CoV and MERSCoV.
The lead peptide, [SARSHRC-PEG4]2-chol, was subsequently assessed for its ability to block entry of live SARS-CoV-2 in VeroE6 and VeroE6 cells overexpressing the protease TMPRSS2 (33), one of the host factors thought to facilitate viral entry at the cell membrane (2). The TMPRSS2-expressing cells accurately represent the entry route in airway cells, an important feature highlighted by the failure of chloroquine to inhibit infection in TMPRSS2-expressing Vero cells and in human lung cells (34). The [SARSHRC-PEG4]2-chol peptide was dissolved in an aqueous buffer containing 2% dimethylsulfoxide (DMSO), was incubated with cells for 1 hr at 37 °C, after which a fixed concentration of SARS-CoV-2 was added. After 8 hrs at 37 °C, fusion events were quantified by SARS-CoV-2 nucleoprotein (NP) staining. The [SARSHRC-PEG4]2-chol peptide inhibited live virus entry with an IC50 ~300 nM in cells not overexpressing TMPRSS and ~5 nM in VeroE6-TMPRSS2, with an IC90 ~1 uM in both cell types (Fig. 2a). In addition, we assessed the efficacy of the [SARSHRC-PEG4]2-chol peptide dissolved in a sucrose solution instead of DMSO, which would strengthen translational potential for human use. [SARSHRC-PEG4]2-chol peptide retained its potency in this formulation, with an IC50 ~300 nM in cells not overexpressing TMPRSS2 and ~5 nM in VeroE6-TMPRSS2 (Fig. 2b). The efficacy data are summarized in Fig. 2c.
Figure 2. Inhibition of live SARS-CoV-2 entry by [SARS-CoV-2-HRC-peg4]2-chol peptide.
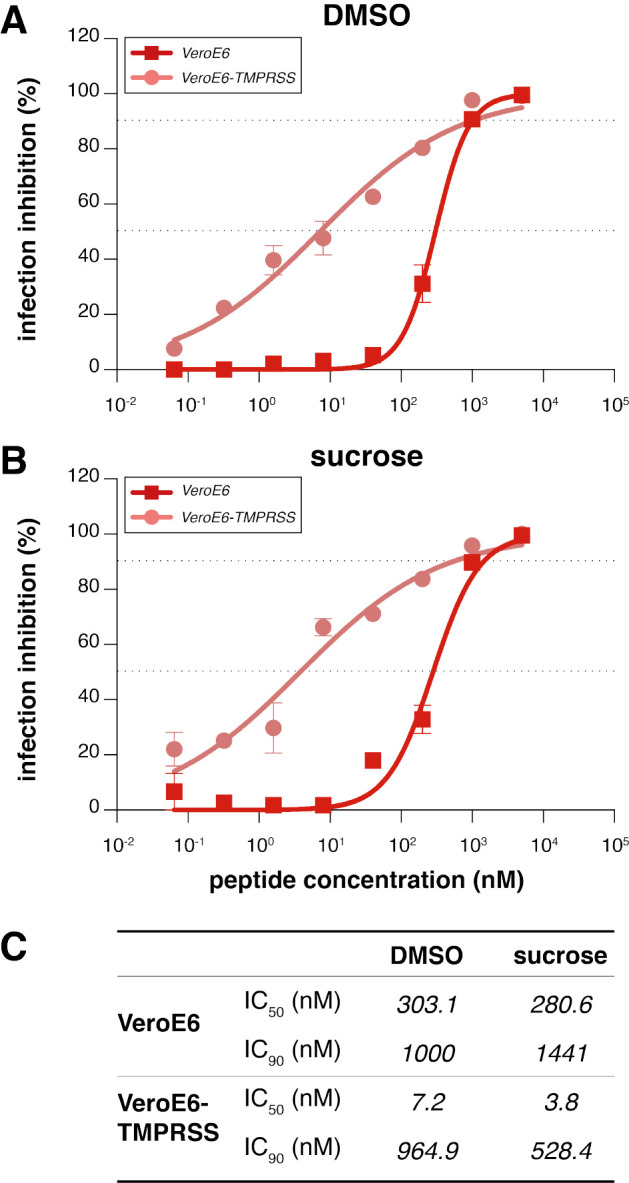
The percentage inhibition of infection is shown on VeroE6 and VeroE6-TMPRSS2 cells with increasing concentrations of [SARS-CoV-2-HRC-peg4]2-chol. A DMSO-diluted stock (A, as used in ferrets) and sucrose-diluted stock (B, potential formulation for human use) were tested side-by-side. Inhibitory concentrations 50% and 90% were calculated (dotted lines). (C) Inhibitory concentrations 50% and 90% of [SARSHRC-PEG4]2-chol in live SARS-CoV-2 viral infection assays in VeroE6 cells with or without TMPRSS2 protease overexpression.
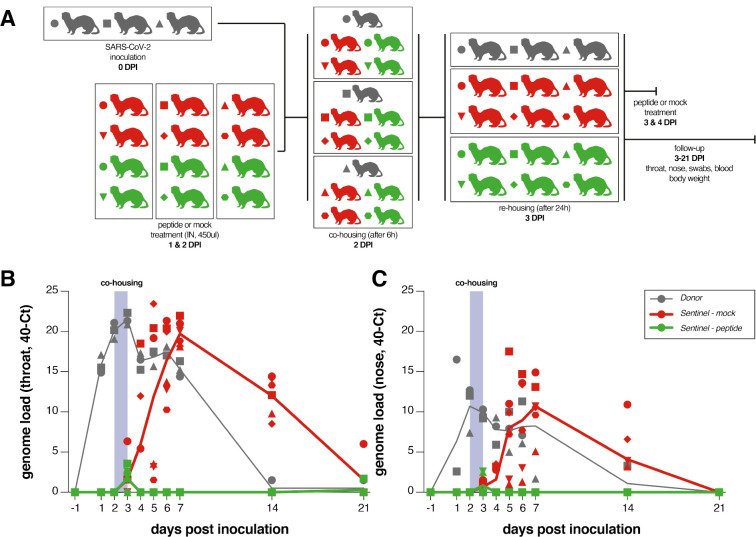
Ferrets are an ideal model for assessing respiratory virus transmission, either by direct contact or by aerosol transmission (35–40). Mustelids are highly susceptible to infection with SARS-CoV-2, as also illustrated by frequent COVID-19 outbreaks at mink farms (41). Direct contact transmission of SARS-CoV in ferrets was demonstrated in 2003 (42), and both direct contact and airborne transmission have recently been shown in ferrets for SARS-CoV-2 (36, 43, 44). Direct contact transmission in the ferret model is highly reproducible (100% transmission from donor to acceptor animals), but ferrets display limited clinical signs. After infection via direct inoculation or transmission, SARS-CoV-2 can readily be detected in and isolated from the throat and nose, and viral replication leads to seroconversion.
To assess the efficacy of [SARSHRC-PEG4]2-chol in preventing SARS-CoV-2 transmission, naive ferrets were treated prophylactically with the lipopeptide before being co-housed with SARS-CoV-2 infected ferrets. In this setup, transmission via multiple routes can theoretically occur (aerosol, orofecal, and scratching or biting during play or fight), and ferrets are continuously exposed to infectious virus during the period of co-housing, providing a stringent test for antiviral efficacy. The study design is shown in Fig. 3a. Three donor ferrets (grey in diagram) were inoculated intranasally with 5.4 × 105 TCID50 SARS-CoV-2 on day 0. Twelve recipient ferrets housed separately were treated by nose drops with a mock preparation (red) or [SARSHRC-PEG4]2-chol peptide (green) on 1- and 2-days post-inoculation (DPI) of the donor animals. The [SARSHRC-PEG4]2-chol peptides for intranasal administration were dissolved to a concentration of 6 mg/mL in an aqueous buffer containing 2% DMSO, administering a final dose of 2.7 mg/kg to ferrets (450 uL, equally divided over both nostrils). Six hours after the second treatment on 2 DPI, one infected donor ferret was co-housed with four naive recipient ferrets (two mock-treated, two peptide-treated). The experiment was performed in three separate, negatively pressurized HEPA-filtered ABSL3-isolator cages. After a 24-hour transmission period, co-housing was stopped and donor, mock-treated and peptide-treated ferrets were housed as separate groups. Additional [SARSHRC-PEG4]2-chol peptide treatments were given to recipient animals on 3 and 4 DPI. Peptide stocks and working dilutions had similar IC50’s, confirming that peptide-treated ferrets were always dosed with comparable amounts (Fig. S3a and 3b).
Figure 3. SARSHRC-PEG4]2-chol prevents SARS-CoV-2 transmission in vivo.
(a) Experimental design. (b) Viral loads detected in throat and nose swabs via RT-PCR. Viral loads are displayed as 40-Ct. Donor animals shown in grey, mock-treated animals in red, peptide-treated animals in green. 3/3 donor animals, 6/6 mock-treated animals and 0/6 lipopeptide-treated animals were productively infected. Symbols correspond to individual animals and are consistent throughout figures.
Throat and nose swabs were collected from ferrets daily for the first week, and additionally on 14 and 21 DPI, for assessment of viral replication. Small volume blood samples were collected on 0, 7, 14, and 21 DPI for assessing the presence of neutralizing antibodies in serum. Fig. 3b and 3c shows the viral loads (detection of viral genomes via RT-qPCR) for directly inoculated donor animals (grey), mock-treated recipient animals (red) and lipopeptide-treated recipient animals (green). All directly inoculated donor ferrets were productively infected, as shown by SARS-CoV-2 genome detection in throat and nose swabs, and efficiently and reproducibly transmitted the virus to all mock-treated acceptor ferrets (Fig. 3b and 3c, red curves). Notably, productive SARS-CoV-2 infection was not detected in the throat or nose of any of the peptide-treated recipient animals (Fig. 3b and 3c, green curves). A slight rise in viral loads in samples collected at 3DPI was detected, at the end of the co-housing, confirming that peptide-treated animals were exposed to SARS-CoV-2. Strikingly, from 4 DPI onwards, there was a clear treatment effect in which the [SARSHRC-PEG4]2-chol peptide protected 6/6 ferrets from transmission and productive infection. Donor ferrets and 6/6 mock-treated recipient animals seroconverted on 21 DPI. None of the peptide-treated animals seroconverted, demonstrating that in-host virus replication was completely blocked by [SARSHRC-PEG4]2-chol) (Fig. 4). None of the animals showed clinical signs as a result of infection or treatment over the course of the experiment, and body weights remained stable (Fig. S4).
Figure 4. [SARSHRC-PEG4]2-chol-treated animals do not seroconvert.
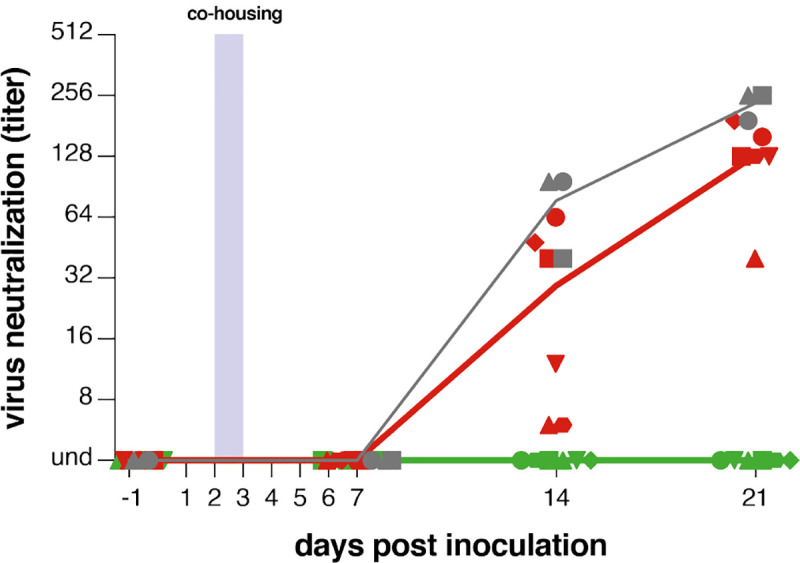
Presence of neutralizing antibodies was determined in a live virus neutralization assay. Virus neutralizing antibodies are displayed as endpoint serum dilution factor to block SARS-CoV-2 replication. Donor animals shown in grey, mock-treated animals in red, peptide-treated animals in green. 3/3 donor animals, 6/6 mock-treated animals and 0/6 lipopeptide-treated animals seroconverted. Symbols correspond to individual animals and are consistent throughout figures.
Based on the in vitro and in vivo results shown here, we expect that prophylactic intranasal administration of the [SARSHRC-PEG4]2-chol peptide prevents transmission from infected to uninfected individuals, even during a 24-hour period of intense direct contact. In vitro data suggest that this lipopeptide will be effective against emerging variants with mutations in S and possibly against other coronaviruses. This efficacy can be readily assessed in real time and adjustments made if needed.
Parallel approaches to prevent transmission that target ACE2 or the interaction between S and ACE2 have also shown promise in vitro (e.g. the “miniprotein” approach recently reported by Cao et al (45)). In distinction to various antibody or nanobody products (46) the [SARSHRC-PEG4]2-chol peptide is inexpensive to produce, has a long shelf life, and does not require refrigeration. Moreover, this is the first compound to convincingly prevent SARS-CoV-2 transmission in a relevant animal model. We envision the use of fusion inhibitory lipopeptides as complementary to other pandemic mitigation strategies. In addition to the nasal drop administration for the [SARSHRC-PEG4]2-chol peptide, other routes that would be equally or more acceptable to humans, for example inhalation devices, are being explored. This HRC lipopeptide fusion inhibitor is feasible for advancement to human use and should readily translate into a safe and effective nasal spray or inhalation administered fusion inhibitor for anti-SARS-CoV-2 prophylaxis, thus supporting containment of the current COVID-19 pandemic.
Supplementary Material
Acknowledgements
We thank Mart Lamers, Sander Herfst, Elwin Verveer, Anna Mykytyn and Marion Koopmans for their contributions to this study.
Funding: This work was supported by funding from the National Institutes of Health (AI146980, AI121349, and NS091263 to MP, AI114736 to AM), the Sharon Golub Fund at Columbia University Medical Center, and a Harrington Discovery Institute COVID-19 Award to AM.
Footnotes
Data and materials availability: All data is available in the manuscript or the supplementary materials.
Competing interest: RDdV, FTB, RLdS, AM and MP are listed as inventors on a provisional patent application covering findings reported in this manuscript.
References
- 1.Li F., Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol 3, 237–261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M. et al. , SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol 94, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P. et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch B. J., van der Zee R., de Haan C. A., Rottier P. J., The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77, 8801–8811 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls A. C. et al. , Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A 114, 11157–11162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Outlaw V. K. et al. , Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain. mBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia S., Wang Q., Liu S. W., Lu L., Jiang S. B., [Development of peptidic MERS-CoV entry inhibitors]. Yao Xue Xue Bao 50, 1513–1519 (2015). [PubMed] [Google Scholar]
- 9.Xia S. et al. , A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv 5, eaav4580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S. et al. , Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30, 343–355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y., Yu D., Yan H., Chong H., He Y., Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J Virol 94, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X. et al. , Broad-Spectrum Coronavirus Fusion Inhibitors to Combat COVID-19 and Other Emerging Coronavirus Diseases. Int J Mol Sci 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porotto M. et al. , Viral entry inhibitors targeted to the membrane site of action. J Virol 84, 6760–6768 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porotto M. et al. , Inhibition of Nipah virus infection in vivo: targeting an early stage of paramyxovirus fusion activation during viral entry. PLoS Pathog 6, e1001168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pessi A. et al. , A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. PLoS One 7, e36833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welsch J. C. et al. , Fatal measles virus infection prevented by brain-penetrant fusion inhibitors. J Virol 87, 13785–13794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Outlaw V. K. et al. , Dual Inhibition of Human Parainfluenza Type 3 and Respiratory Syncytial Virus Infectivity with a Single Agent. J Am Chem Soc 141, 12648–12656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueira T. N. et al. , Structure-Stability-Function Mechanistic Links in the Anti-Measles Virus Action of Tocopherol-Derivatized Peptide Nanoparticles. ACS Nano 12, 9855–9865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueira T. N. et al. , Effective in Vivo Targeting of Influenza Virus through a Cell-Penetrating/Fusion Inhibitor Tandem Peptide Anchored to the Plasma Membrane. Bioconjug Chem 29, 3362–3376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathieu C. et al. , Broad spectrum antiviral activity for paramyxoviruses is modulated by biophysical properties of fusion inhibitory peptides. Sci Rep 7, 43610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueira T. N. et al. , In Vivo Efficacy of Measles Virus Fusion Protein-Derived Peptides Is Modulated by the Properties of Self-Assembly and Membrane Residence. J Virol 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figueira T. N. et al. , Quantitative analysis of molecular partition towards lipid membranes using surface plasmon resonance. Sci Rep 7, 45647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K. K. et al. , Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol conjugated peptide, a new antiviral strategy for influenza virus. J Biol Chem 286, 42141–42149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L. et al. , The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv, (2020). [Google Scholar]
- 25.Daniloski Z., Guo X., Sanjana N. E., The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. bioRxiv, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Y. N. et al. , SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg Microbes Infect, 1–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maitra A. et al. , Mutations in SARS-CoV-2 viral RNA identified in Eastern India: Possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci 45, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karacan I. et al. , The origin of SARS-CoV-2 in Istanbul: Sequencing findings from the epicenter of the pandemic in Turkey. North Clin Istanb 7, 203–209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas N. K., Majumder P. P., Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J Med Res, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaaswarkhanth M., Al Madhoun A., Al-Mulla F., Could the D614G substitution in the SARS CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int J Infect Dis 96, 459–460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S. J., Nguyen V. G., Park Y. H., Park B. K., Chung H. C., A Novel Synonymous Mutation of SARS-CoV-2: Is This Possible to Affect Their Antigenicity and Immunogenicity? Vaccines (Basel) 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koyama T., Weeraratne D., Snowdon J. L., Parida L., Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mykytyn A. Z. et al. , The SARS-CoV-2 multibasic cleavage site facilitates early serine protease-mediated entry into organoid-derived human airway cells. bioRxiv, 2020.2009.2007.286120 (2020). [Google Scholar]
- 34.Hoffmann M. et al. , Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 585, 588–590 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Munoz-Fontela C. et al. , Animal models for COVID-19. Nature, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richard M. et al. , SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun 11, 3496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munster V. J. et al. , Practical considerations for high-throughput influenza A virus surveillance studies of wild birds by use of molecular diagnostic tests. J Clin Microbiol 47, 666–673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munster V. J., Fouchier R. A., Avian influenza virus: of virus and bird ecology. Vaccine 27, 6340–6344 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Herfst S. et al. , Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries R. D. et al. , Delineating morbillivirus entry, dissemination and airborne transmission by studying in vivo competition of multicolor canine distemper viruses in ferrets. PLoS Pathog 13, e1006371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molenaar R. J. et al. , Clinical and Pathological Findings in SARS-CoV-2 Disease Outbreaks in Farmed Mink (Neovison vison). Vet Pathol 57, 653–657 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Martina B. E. et al. , Virology: SARS virus infection of cats and ferrets. Nature 425, 915 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S. J. et al. , Antiviral Efficacies of FDA-Approved Drugs against SARS-CoV-2 Infection in Ferrets. MBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y. I. et al. , Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 27, 704–709 e702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao L. et al. , De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoof M. et al. , An ultra-high affinity synthetic nanobody blocks SARS-CoV-2 infection by locking Spike into an inactive conformation. bioRxiv, (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.