Abstract
Animal and cellular models are essential tools for all areas of biological research including neuroscience. Model systems can also be used to investigate the pathophysiology of psychiatric disorders such as attention-deficit/hyperactivity disorder (ADHD) and autism spectrum disorder (ASD). In this review, we provide a summary of animal and cellular models for three genes linked to ADHD and ASD in human patients – CNTNAP2, ADGRL3, and PARK2. We also highlight the strengths and weaknesses of each model system. By bringing together behavioral and neurobiological data, we demonstrate how a cross-species approach can provide integrated insights into gene function and the pathogenesis of ADHD and ASD. The knowledge gained from transgenic models will be essential to discover and validate new treatment targets for these disorders.
Keywords: ADGRL3, attention-deficit/hyperactivity disorder, autism spectrum disorder, CNTNAP2, cross-species, Drosophila, human induced pluripotent stem cells, mouse, PARK2, zebrafish
Introduction
Psychiatric disorders
Psychiatric disorders can affect any aspect of mental health, including cognition, emotionality, and sociability. They are a leading cause of disability worldwide and represent a significant economic burden to the society (Bloom et al., 2012; Vigo et al., 2016). The aim of this research using animal and cellular models is to better understand the neurobiology of psychiatric disorders to develop new treatments for the individuals that experience them. In this review, we discussed models for two common overlapping psychiatric disorders: attention-deficit/hyperactivity disorder (ADHD) and autism spectrum disorder (ASD).
Attention-deficit/hyperactivity disorder and autism spectrum disorder
ADHD and ASD are two of the most widely studied neurodevelopmental disorders. According to the Diagnostic and Statistical Manual of Mental Disorders, 5th ed. (DSM-5), ADHD is a clinically heterogeneous disorder characterized by inattention, hyperactivity, and increased impulsivity (American Psychiatric Association, 2013). Approximately 2–10% of school-age children are affected by ADHD (Hawi et al., 2015), and males are more likely to be affected than females (Polanczyk et al., 2007; Nussbaum, 2012). Despite traditionally being regarded as a childhood disorder, a number of longitudinal-based and population-based epidemiological studies have identified an adult form of ADHD (Franke et al., 2012, 2018). ASD is a group of neurodevelopmental disorders characterized by impairments in learning, sensory filtering, communication, and social interactions together with restricted, repetitive, and stereotypical behaviors and interests. The prevalence of ASD in the population is estimated at ~1%, with three to four times as many males as females affected (Baird et al., 2006). There are currently no objective laboratory-based tests to diagnose ADHD or ASD. The latest version of the DSM acknowledges the high rate of comorbidity for ASD and ADHD, allowing their simultaneous diagnosis for the first time (American Psychiatric Association, 2013). The symptoms of ADHD and ASD often co-exist. Overall, 20–50% of children with ADHD also meet the criteria for ASD (Rommelse et al., 2011). Several studies have shown social deficits, peer relationship deficits, and empathy problems in children with ADHD, and the DSM-5 allows a comorbid diagnosis of both disorders. Although ADHD and ASD comorbidity has mainly been studied in children, there is also evidence that it occurs in adults (Hartman et al., 2016). The drugs most commonly used to treat ADHD are stimulants such as methylphenidate (MPH). Although effective in controlling some symptoms, MPH treatment shows a large degree of variability across patients (Wolraich and Doffing, 2004). There are currently no pharmacological treatments for the core symptoms of ASD. Behavioral therapy can improve the quality of life for patients with ASD, and pharmacological treatments may reduce secondary symptoms such as hyperactivity (LeClerc and Easley, 2015).
Genetic and environmental factors for autism spectrum disorder and attention-deficit/hyperactivity disorder
A combination of both environmental factors and genetic variants are known to influence the development and expression of ADHD and ASD (Sánchez‐Mora et al., 2015; Sjaarda et al., 2017). Meta-analysis of multiple large-scale twin studies estimated the heritability of childhood and adolescent ADHD at between 0.7 and 0.8 (Nikolas and Burt, 2010). The heritability of adult ADHD is reported to be lower, at between 0.3 and 0.4, although this may be an underestimation owing to the self-rating system commonly used in these studies (Franke et al., 2018). The true heritability could be as high as 0.7–0.8 (Brikell et al., 2015; Tick et al., 2015). ASD heritability estimates are similarly high at 0.64–0.91 (Tick et al., 2015). Familial co-aggregation of ADHD and ASD is strongly indicative of an overlap in genetic factors across disorders (Ghirardi et al., 2018). Furthermore, a number of studies have identified genes that are associated with both ADHD and ASD, including contactin-associated protein-like 2 (CNTNAP2) (Strauss et al., 2006; Elia et al., 2010; Poot et al., 2010; Rodenas-Cuadrado et al., 2014).
The genetic basis of ADHD and ASD can be explained by the common disease common variant and common disease rare variant (CDRV) hypotheses (Hawi et al., 2015). However, there is a growing recognition that most individuals with ADHD or ASD carry both common and rare variants. The common disease common variant hypothesis focuses on multiple common polymorphisms, with a frequency greater than 5%. Each polymorphism has a low level of penetrance, but when combined they can increase the risk of developing ADHD or ASD. This hypothesis has formed the basis of molecular genetic research over the past two decades starting with the investigation of preselected candidate genes in relatively small studies (Li et al., 2014). With the advent of high-throughput genetic screening, researchers could conduct hypothesis-free experiments in large samples of patients and controls. These genome-wide association studies have identified a number of polymorphisms, which confer a slightly increased risk of developing a psychiatric disorder. Common variants have been estimated to explain 22% of ADHD heritability and 32% of ASD heritability (Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017; Demontis et al., 2017). Some of the missing heritability can be explained by the CDRV hypothesis. ADHD and ASD might be caused by genetic variants that are rare in the overall population but have high penetrance in individuals. Rare variants such as monogenic or chromosomal abnormalities are a common cause of syndromic ASD, and in some cases, these abnormalities can be identified by genetic testing (Fernandez and Scherer, 2017). Nonsyndromic ASD might also be caused by monogenic mutations in genes such as Neurexin 1 (NXRN1) and SH3 and multiple ankyrin repeat domains (SHANK3) (Caglayan, 2010). Another type of rare variant that has been intensely studied is copy-number variations (CNVs). CNVs are chromosomal deletions or duplications that can span several megabases and which alter normal gene expression (Redon et al., 2006). The relevance of CNVs to a particular phenotype can be linked to their position, length, and heterozygosity. There is increasing evidence that CNVs play a role in the pathogenesis of several neurodevelopmental disorders, and it has been demonstrated that patients with ASD have a higher burden of CNVs compared with healthy controls (Shishido et al., 2013). Testing for CNVs in patients with ADHD has identified a number of genes that had previously been implicated in ASD, demonstrating the shared genetic risk for these disorders (Lionel et al., 2011). Other studies have also reported a greater burden of CNVs in patients with ADHD without reporting comorbidity with ASD (Williams et al., 2010, 2012). These CNVs include the parkin 2 (Park2) and neuropeptide Y loci (Lesch et al., 2011; Jarick et al., 2014) and genes in the metabotropic glutamate receptor family (Elia et al., 2012). There is some evidence that CNVs may contribute to adult ADHD as well (Ramos-Quiroga et al., 2014), although different genes may be affected. The growing consensus among researchers is that ASD and ADHD are caused by a number of synergistic factors including CNVs, rare variants, and common polymorphisms (Kiser et al., 2015).
Environmental factors including epigenetic modifications have also been linked to these disorders. The prenatal and perinatal stages appear to be particularly susceptible to environmental stimuli. Both premature birth and maternal smoking during pregnancy have been linked to an increased risk of developing ADHD (Halmøy et al., 2012; Zhu et al., 2014). However, when genetic and familial confounders are included, the strength of these associations decreases, calling into question any causal relationship (Sciberras et al., 2017). A further meta-analysis of 15 studies failed to find an association between maternal smoking and ASD (Rosen et al., 2014). Even within the prenatal period, the effect of environmental stimuli depends upon the trimester during which exposure occurs. Exposure to antidepressants during the second and third trimesters has been associated with an increased risk of developing ASD (Boukhris et al., 2016). However, a cohort study of more than 1.5 million Swedish children failed to find an association between maternal antidepressant use during the first trimester and risk of either disease (Sujan et al., 2017). In the perinatal period, strong associations between ASD and birth complications leading to trauma, ischemia or hypoxia have been reported (Modabbernia et al., 2017). Both prenatal and postnatal exposures to abnormal levels of metal toxicants and essential elements have been associated with ASD (Arora et al., 2017). After the antenatal period, a number of early life factors have been linked to these disorders. These include an association between maternal anxiety, depression, and ADHD in 3-year olds (Meadows et al., 2007) and increased parental age and ASD risk (Wu et al., 2016). However, whether causality between environmental factors and neurodevelopmental disorders exists can be hard to determine, and further research, including the use of animal models, is required to investigate this issue.
Cross-species models
The use of animal and cellular models to understand the pathogenesis of human disease and develop treatments has greatly contributed to our understanding of several psychiatric disorders. These models are suitable for large-scale trials that can be translated into human clinical research (Crawley, 2012; van der Voet et al., 2014; Falk et al., 2016; Kim et al., 2016; Ardhanareeswaran et al., 2017).
Mouse (Mus musculus)
The mouse is the most commonly used model organism in biomedicine. Their short generation time, small size, and social nature allow a large number of animals to be housed in a small facility. The sequencing of the mouse genome combined with the tools to manipulate it in a targeted manner has revolutionized biomedical research (Hara and Takada, 2017). As mice and humans are very similar genetically (∼99% homology), it is possible to investigate genes identified in human studies in mice. Following genetic manipulation, a combination of highly standardized behavioral and neurobiological techniques can be performed. This can provide insights into how polymorphisms or mutations can lead to the pathology of psychiatric disorders (Arguello and Gogos, 2006). Of particular relevance to psychiatric disorders, mice exhibit a range of complex behaviors including reciprocal social interactions (McCammon and Sive, 2015). Mice are often the first step in testing a new treatment for toxicity and efficacy (Vandamme, 2014).
Zebrafish (Danio rerio)
The zebrafish is an emerging model organism for biomedical research thanks to their short generation time, small size, and genetic tractability. Zebrafish are particularly useful for developmental and live imaging studies, as they develop externally and are transparent at larval stages. Almost 70% of human genes have at least one zebrafish orthologue (Howe et al., 2013). Transient knockdown of genes in zebrafish was initially achieved by injecting morpholino oligonucleotides (Blum et al., 2015). Recently, stable mutant lines have been created using CRISPR/Cas9 genome editing (Schmid and Haass, 2013). However, modelling human disorders in zebrafish is complicated by an additional whole-genome duplication which occurred ~440 million years ago (Amores et al., 2011). Despite this issue, zebrafish are uniquely suited for high-throughput pharmacological studies, as larvae can be generated in large numbers and drugs can be applied by dilution in the embryo medium (Rihel et al., 2010; Norton, 2013; Gutiérrez et al., 2018). Zebrafish can be used to study ADHD and ASD thanks to their stereotypical behavior and social interactions. It is also possible to test cognitive deficits in this species. Spatial working memory can be measured in the habituation to novelty test, and various forms of learning can be assessed using robust behavioral paradigms (Stewart et al., 2014).
Fruit fly (Drosophila melanogaster)
The fruit fly is a widely used model system that has greatly contributed to neuroscience research (Bier, 2005). Fruit flies are small and easy to maintain with a very short generation time, which allows large-scale low-cost experiments. Although the fruit fly brain is relatively small, it is still complex enough to model some aspects of psychiatric disorders (Bellen et al., 2010). Signalling pathways and regulatory molecular networks are also well conserved. Overall, 75% of human disease genes have related sequences in the fly (Bier, 2005). Furthermore, the temporally inducible UAS-GAL4 system can be used to target specific cell types in Drosophila (Brand and Perrimon, 1993). When this system is coupled to the expression of RNA interference molecules, it allows reversible genetic manipulation. Transgenic flies can also be generated using the P-element system (Rubin and Spradling, 1982), and CRISPR/CAS9 technology now permits ubiquitous or tissue-specific genome editing (Port et al., 2014). Drosophila display a behavioral repertoire that varies from simple aggregation to more complex behaviors such as aggression and courtship (O’Kane, 2011). Therefore, even though the nervous system and social behavior of Drosophila are not that similar to vertebrates, the fruit fly presents a valuable model to characterize mechanisms underlying disease-related behaviors (van Alphen and van Swinderen, 2013; van der Voet et al., 2014).
Human induced pluripotent stem cells
The recent development and improvement of human induced pluripotent stem cell (hiPSC) technology permits the conversion of tissue into pluripotent cells that can be redifferentiated into neurons in a noninvasive and ethical manner (Takahashi et al., 2007). This technique offers the possibility to use a range of different samples including skin biopsies, urine samples, and hair follicles. A great advantage of this method is that the genetic makeup of patients is included in the experimental model rather than investigating the influence of single genes. This may be particularly important for multifactorial diseases such as ADHD and ASD. hiPSCs also represent an innovative tool for large-scale drug screening. Moreover, neurons obtained from hiPSCs can be cultured in two dimensions, for single neuron analysis, or in three-dimensional aggregates (brain organoids), allowing evaluation of network-level effects (Falk et al., 2016; Frega et al., 2017; Prytkova and Brennand, 2017). In recent years, hiPSCs have been used as a tool to model psychiatric disorders such as ASD, ADHD, bipolar disorder, Schizophrenia, and the response to lithium at a cellular level (Prilutsky et al., 2014; Ardhanareeswaran et al., 2015; Lim et al., 2015; Soliman et al., 2017; Hoffman et al., 2018; Jansch et al., 2018).
A single model cannot capture all the traits of a complex brain disorder such as ASD and ADHD. As specific behavioral and neurobiological features may be more easily studied in certain organisms, a comparative study across models should be performed whenever possible (Stewart et al., 2015; Wong and Josselyn, 2016).
Contactin-associated protein-like 2, adhesion G protein-coupled receptor L3, and parkin 2
Human genetic studies have identified a large number of genes linked to ADHD and ASD. There are currently 359 genes implicated in ADHD according to the ADHDGene database, whereas 1007 genes are included in the SFARI Gene database for ASD (Zhang et al., 2011; Abrahams et al., 2013). In this review, we have chosen to focus on three candidate genes for ADHD and ASD: CNTNAP2, adhesion G protein-coupled receptor L3 (ADGRL3), and PARK2. As well as being involved in diverse biological processes, the types of variants linking these genes to disease are also different. CNTNAP2 was initially implicated in ASD through a recessive single base pair deletion in the coding region. In contrast, the association between ADGRL3 and ADHD comes from noncoding single nucleotide polymorphisms (SNPs), which may affect gene expression levels (Strauss et al., 2006; Martinez et al., 2016). Larger scale CNVs in PARK2 are associated with ASD (Glessner et al., 2009; Scheuerle and Wilson, 2011). CNTNAP2 and PARK2 are primarily implicated in ASD but have also been linked with ADHD. ADGRL3 is linked with ADHD and substance use disorders (SUD), but there is currently no strong connection to ASD (Gau et al., 2012). Together this review provides an overview of how genes with different biological functions can be studied in model systems to understand the comorbidity of ADHD and ASD.
Contactin-associated protein-like 2
Biological and molecular description
CNTNAP2 is one of the largest human genes. It spans 2.3 Mb and is located in chromosomal region 7q35. CNTNAP2 encodes the neuronal transmembrane protein CASPR2, a member of the neurexin (NRXN) superfamily of single-pass transmembrane proteins. CASPR2 is composed of 1331 amino-acid residues organized into eight extracellular domains, two C-terminal intracellular domains, and one transmembrane domain (Poot, 2015). The intracellular domain of CASPR2 may be involved in protein–protein interactions, whereas the extracellular domains mediate cell–cell interactions and bind ligands, receptors, and extracellular matrix components (Poot, 2015; Baig et al., 2017). CNTNAP2 forms a neuron–glia adhesion complex with contactin 2 that localizes voltage-gated potassium channels to the juxtaparanodal region of myelinated axons (Poliak et al., 1999, 2003). The first link between CNTNAP2 and ASD was a recessive mutation found in an Amish family affected by a syndromic form of ASD called cortical dysplasia-focal epilepsy syndrome (Strauss et al., 2006). Several more studies have provided convergent evidence that rare and common variations in CNTNAP2 confer risk for ASD-related endophenotypes such as developmental language disorders and ASD itself (Peñagarikano and Geschwind, 2012). CNTNAP2 has also been linked to schizophrenia, epilepsy, intellectual disability, learning disability, and ADHD (Rodenas-Cuadrado et al., 2014). However, most studies have focused on the role of CNTNAP2 in ASD.
In this review, we have chosen to focus on these three candidate genes because they are linked to either ADHD (ADGRL3), ASD (CNTNAP2), or both disorders (PARK2). Each gene has been modelled in a number of different species, making these candidates ideal to demonstrate the power of comparative studies to help translate findings to human patients.
Models
Mouse
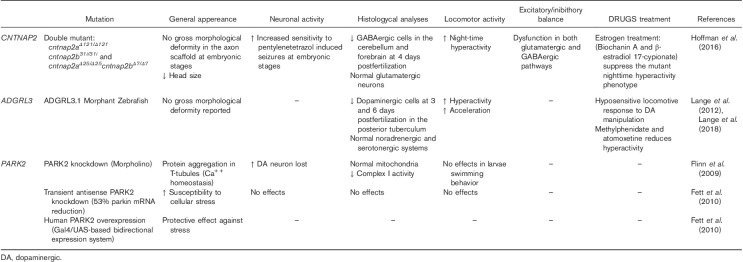
The first Cntnap2 mutant mouse was generated in 2003 (Poliak et al., 2003). After the identification of CNTNAP2 as a candidate gene for ASD, the original strain was backcrossed onto the C57BL/6J background, which is more suitable for behavioral characterization (Crawley et al., 1997; Penagarikano et al., 2011). Cntnap2 KO mice display deficits in core behavioral features of ASD: social deficits, restrictive and repetitive behaviors, and reduced vocal communication (Table 1). When compared with wild-type littermates, these mice appear normal, and there are no differences in growth rate and final size. Interestingly, after six months, Cntnap2−/− mice develop epileptic seizures and their electroencephalogram has an abnormal pattern during slow wave sleep. In addition, knockout mice are hyperactive (Penagarikano et al., 2011). Cntnap2−/− mice show neuronal migration abnormalities that reveal the fundamental role of this gene in cortical projection neuron migration (Penagarikano et al., 2011). Interestingly, it has been found that Cntnap2−/− mice display a reduced number of GABAergic interneurons (Penagarikano et al., 2011). Pharmacological treatment with risperidone rescued the stereotypic behavior of Cntnap2−/− mice, whereas social deficits were unaffected, similar to the pattern of improvements seen in human patients (Penagarikano et al., 2011). Overall, this model recapitulates certain features of patients with CNTNAP2 mutations, and this is one of the best studied genes for this disorder (Poot, 2015).
Table 1.
Mouse models
Zebrafish
A zebrafish cntnap2 mutant model was recently created (Table 2). Cntnap2a−/− and cntnap2b−/− double mutants (cntnap2ab) harbor homozygous loss-of-function mutations in both genes resulting in a complete loss of the protein (Hoffman et al., 2016). Hoffman et al. (2016) analyzed excitatory and inhibitory neuron populations in cntnap2ab during early development, as an imbalance in inhibitory and excitatory signalling could be involved in ASD (Rubenstein and Merzenich, 2003). They observed that mutants have a smaller head than wild type. Moreover, mutants show a decrease in GABAergic cell count; however, there were no prominent deficits in glutamatergic neurons (Hoffman et al., 2016). Loss of inhibitory neurons can increase seizure susceptibility (Cobos et al., 2005). Zebrafish cntnap2ab mutants are more sensitive to drug-induced seizures and show night-time hyperactivity, another phenotype linked to the imbalance between excitatory and inhibitory neurotransmission. The night-time hyperactivity phenotype can be rescued by estrogens (Hoffman et al., 2016). This finding has reinforced the use of whole organism drug screening as an effective system to identify compounds that can be developed into pharmaceutical treatments.
Table 2.
Fish models
Fruit fly
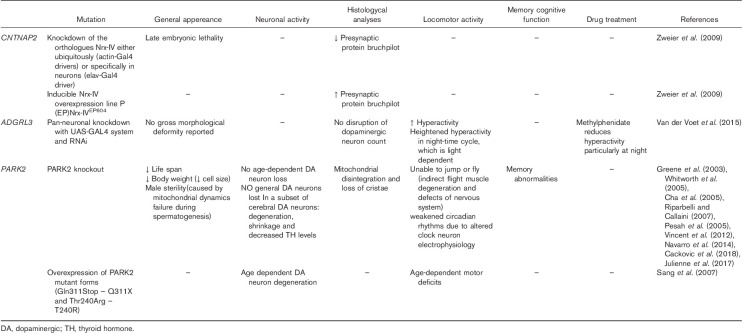
CNTNAP2 and NRXN1 are members of the neurexin superfamily. SNPs and CNVs in both genes have been associated with disorders including ASD. Zweier et al. (2009) have used Drosophila to examine a common synaptic link between these two genes and neuropsychiatric disorders (Table 3). This group studied both Nrx-I and Nrx-IV, the Drosophila orthologs of NRXN1 and CNTNAP2. Knockdown (KO) of Nrx-IV, either ubiquitously or only in neurons, led to late embryonic lethality. Immunostaining of Nrx-IV KO embryos showed an overall reduction in staining for the presynaptic protein bruchpilot that maintains the structural integrity of synaptic active zones (Wagh et al., 2006). They also found a dosage-dependent increase of bruchpilot staining when Nrx-IV was overexpressed in larval neurons. bruchpilot and Nrx-I colocalize at presynaptic active zones (Li et al., 2007). In conclusion, CNTNAP2 and NRXN1 may be involved in a common synaptic mechanism, contributing to the etiology of neurodevelopmental disorders together with bruchpilot (Zweier et al., 2009). Further studies are required to determinate the mechanism by which Nrx-I and Nrx-IV control bruchpilot levels.
Table 3.
Fruit fly
Human induced pluripotent stem cells
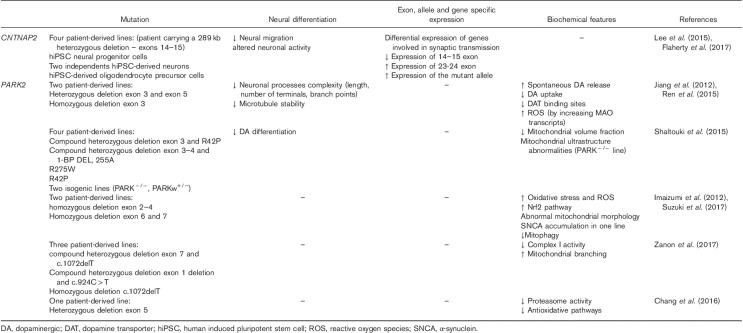
hiPSCs carrying a heterozygous deletion in CNTNAP2 have been generated to characterize the associated molecular and cellular phenotypes (Table 4). However, they were derived from a female proband who met the DSM-IV criteria for schizo-affective disorder, not ASD or ADHD. hiPSCs were also generated from both parents as well as five unrelated healthy controls (Lee et al., 2015). Neural migration was quantified using a neurosphere assay. Interestingly, hiPSC neural progenitor cells derived from the heterozygous CNTNAP2 schizo-affective patient showed significantly reduced neural migration relative to controls (Lee et al., 2015). This phenotype is consistent with the neuronal migration abnormalities observed in the Cntnap2−/− mouse model of ASD (Penagarikano et al., 2011). Global gene expression and neuronal activity were analyzed in the same cohort. HiPSC-derived neurons from individuals carrying the heterozygous CNTNAP2 deletion show altered neuronal activity and differential expression of genes involved in synaptic transmission (Flaherty et al., 2017).
Table 4.
Human induced pluripotent stem cells
Adhesion G protein-coupled receptor L3
Biological and molecular description
The latrophilin family of proteins was initially discovered owing their ability to bind α-latrotoxin, a component of black widow spider venom (Davletov et al., 1996). The ADGRL3 (previously known as LPHN3) is of particular interest in relation to neurodevelopmental disorders. Multiple polymorphisms in ADGRL3 have been linked to an increased risk of ADHD and SUD (Arcos-Burgos et al., 2010; Ribasés et al., 2010). ADGRL3 forms a trimeric complex with FLRT3 and UNC5, which supports transcellular adhesion and glutamatergic synapse development (Jackson et al., 2015). The identification of ADGRL3 in human genetic studies, coupled with its putative roles in neuronal migration and synapse development, provides strong support for ADGRL3 polymorphisms in the development of ADHD (O’Sullivan et al., 2014). The lack of naturally occurring coding variants in humans greatly limits our ability to investigate ADGRL3 (Orsini et al., 2016). Coding variants may not affect the genes that they are located in, but may rather have effects elsewhere in the genome. However, there is functional evidence that the noncoding variant rs2271338 affects ADGRL3 expression, further linking this gene to ADHD (Martinez et al., 2016).
Models
Mouse
ADGRL3−/− mice displayed hyperactivity in the open-field test, (Table 1) but no significant genotype differences were observed in the amount of time spent in the center of the open field, a common test for anxiety. ADGRL3 null mice also displayed a heightened locomotor response to cocaine. A differential response to cocaine is of interest because of the link between ADGRL3 polymorphisms and SUD (Arcos-Burgos et al., 2012). In a food reward paradigm, ADGRL3−/− mice displayed increased levels of motivation to work for food, whereas motor coordination was unaffected in the rotarod test. In the Porsolt swim test, ADGRL3−/− mice spent more time swimming and had increased latency to immobility, possibly owing to reduced depressive behavior or hyperactivity. Whole brains of ADGRL3−/− and wild-type P0 pups were used to quantify candidate gene expression by qPCR. The genes chosen were involved in neuronal differentiation and survival as well as dopaminergic (DA) and serotonergic neurotransmission. The following genes were differentially expressed in ADGRL3−/−: serotonin transporter (SLC6A4), serotonin 2C receptor (5-Htr2c), dopamine transporter (SLC6A3), dopamine receptor D4 (Drd4), neural cell adhesion molecule 1 (Ncam1), nuclear receptor subfamily 4 group A member 2 (Nr4a2), and tyrosine hydroxylase (Th). High-pressure liquid chromatography also revealed an increased level of dopamine and 5-HT in ADGRL3−/− mice (Wallis et al., 2012). Transcriptomic analysis was performed on the prefrontal cortex, striatum, and hippocampus of ADGRL3−/− and control mice at 4 days, 28 days, and 6 months. Collapsing the RNA-sequencing results across brain regions and time points identified 11 genes that are differentially expressed in ADGRL3−/− mice. Two of these genes (Pcdhgb8, upregulated and Pcdhb9, downregulated) are members of the protocadherin family of calcium-dependent cell–cell adhesion molecules (Orsini et al., 2016), which have been linked to ADHD and ASD (Rivero et al., 2015). Independent analysis of brain regions and time points revealed a large number of differentially expressed genes including overexpression of Htr2c [5-hydroxytryptamine (serotonin) receptor 2c] in the cortex and SLC6A3 (dopamine transporter) in the hippocampus of 6-month old ADGRL3−/− mice. Gene set analysis did not find a statistically significant enrichment of gene sets following adjustment for multiple testing. However, a number of the most enriched pathways were relevant to ADHD and SUD, including synaptic vesicle cycle, DA synapse, glutamatergic synapse, neurotrophin signalling pathway, and both amphetamine and nicotine addiction.
Zebrafish
Zebrafish have two paralogues of human ADGRL3: adgrl3.1 and adgrl3.2. adgrl3.1 shows a more specific expression pattern during embryonic development. adgrl3.1 morphant larvae displayed hyperactivity under both light and dark conditions at 6 days after fertilization (Table 2). The increased distance swum was because of greater acceleration and average speed throughout the trial (Lange et al., 2012). In most patients with ADHD, MPH administration results in a reduction in hyperactivity (Ramos-Quiroga et al., 2009). MPH decreased hyperactivity in adgrl3.1 morphant fish but not in controls. The total distance swam was reduced by decreasing the average speed. Application of another ADHD treatment drug, the selective noradrenaline reuptake inhibitor atomoxetine, reduced the total distance swam by adgrl3.1 morphant zebrafish by increasing the resting time. The number of DA neurons in the posterior tuberculum of adgrl3.1 morphants was reduced at 3 and 6 days after fertilization. Subregions of the posterior tuberculum were also disorganised (Lange et al., 2012). Projectome analysis has indicated a functional correlation between the posterior tuberculum and the mammalian A11 group of DA neurons (Tay et al., 2011). Administration of DA receptor agonists and antagonists showed that adgrl3.1 MO larvae are hyposensitive to DA manipulation compared with adgrl3.1 CO. A possible explanation for this hyposensitivity is that there could be a heightened level of DA in the synaptic cleft of adgrl3.1 morphants, which might desensitize both DA-1 and DA-2 like receptors. A heightened level of DA might also explain the hyperactivity observed in these animals (Lange et al., 2018). An evolutionarily conserved enhancer for adgrl3.1 has been used to express green fluorescent protein in zebrafish. This enhancer was sufficient to produce an expression pattern similar to endogenous adgrl3.1 in the fore, mid, and hindbrain but not in the telencephalon and retina (Martinez et al., 2016).
Fruit fly
The Drosophila orthologue of ADGRL3 is known as latrophilin. Locomotion activity and sleep have been assessed in latrophilin KO flies using the Drosophila Activity Monitor (Table 3). Latrophilin KO flies were found to have heightened locomotor activity and reduced sleep, particularly during the 12-h night period. Testing in constant darkness showed that this hyperactive phenotype was light dependent. As light can buffer the wake-promoting effect of DA (Shang et al., 2011), the presence of a light-sensitive phenotype hints at a disruption of the DAergic system. However, the number and position of DA neurons is normal in latrophilin flies, suggesting that impairment of the DA system may result from changes in downstream signalling. Drosophila with a pan-neuronal KO of the dopamine transporter (DAT) gene displayed a similar light-sensitive hyperactive phenotype. Furthermore, application of MPH normalized the locomotor and sleep phenotype of latrophilin flies, further linking this gene to DA signalling (van der Voet et al., 2015).
Cellular models
Although no hiPSCs-containing ADGRL3 polymorphisms are currently available, a number of conventional cellular models have been used to study this gene. A family-based genetic analysis of 838 individuals identified six evolutionary conserved regions (ECRs) in the ADGRL3 genomic region that contained SNPs or haplotypes linked to ADHD (Martinez et al., 2016). ECR 37 and 47 were the only regions found to drive luciferase expression. The ECR 37 risk variant did not affect luciferase levels whereas the ECR 47 risk haplotype reduced luciferase activity by ~40% in both B35 neuroblastoma and U87 astrocytoma cell lines. Transcription factor binding motif analysis of ECR 47 showed an overrepresentation of transcription factors associated with brain function. Electromobility shift assays showed that the rs2271338 SNP in ECR 47 prevented binding of the Yin Yang 1 (YY1) transcription factor. However, YY1 interfering small RNA molecules did not reduce ADGRL3 expression in SH-SY5Y neuroblastoma cells, perhaps because of an inability to model this complex interaction outside of its native system. Expression quantitative trait loci analysis of post-mortem human brain tissue has linked the rs2271338 AA risk genotype allele to decreased thalamic expression of ADGRL3 (Martinez et al., 2016).
PARK2
Biological and molecular descriptions
Mutations in PARK2 were first discovered in a small percentage of patients with Parkinson’s disease (Lücking et al., 2000). Subsequent whole-genome studies have linked PARK2 CNVs with Han Chinese children with ASD (Yin et al., 2016) and European populations (Glessner et al., 2009; Scheuerle and Wilson, 2011). CNVs within the PARK2 locus have also been found among ADHD patient cohorts (Jarick et al., 2012).
PARK2, also known as PRKN, codes for an E3 ubiquitin ligase previously called Parkin. One of the most studied partners of PARK2 is the serine/threonine protein kinase PINK1. Acting together, PARK2 and PINK1 constitute an internal sensor system for disparate perturbations to cellular homeostasis (Pickrell and Youle, 2015). In fact, different stimuli can lead to PINK1 accumulation on the outer mitochondrial membrane where it can recruit and activate cytosolic PARK2. PARK2 can then target several mitochondrial proteins for degradation (Hang et al., 2015). The interaction between PINK1 and PARK2 plays a pivotal role in mitochondria dynamics and quality control, regulating processes such as mitophagy, fusion and fission, biogenesis, and transport (Scarffe et al., 2014). PARK2 is also linked to apoptotic pathways and could exert a proapoptotic or antiapoptotic effect in a stressor-dependent and environmental-dependent manner (da Costa et al., 2009; Müller-Rischart et al., 2013; Hollville et al., 2014; Zhang et al., 2014).
PARK2 shows high amino-acid sequence conservation across species, suggesting that it has an evolutionarily conserved function (Pienaar et al., 2010). Orthologues of this gene have been found in most of the model organisms used in translational research, permitting the molecular function of this gene to be studied in different systems (Pienaar et al., 2010).
Models
Mouse
Surprisingly, Park2−/− mice do not represent a robust model for Parkinson’s disease (Perez and Palmiter, 2005), but phenotypic characterization of these animals can be used to investigate whether there is a link to neurodevelopmental disorders like ADHD and ASD (Table 1). Park2−/− mice did not show impairment of general activity, coordination, and gait performance (Goldberg et al., 2003; Perez and Palmiter, 2005; Rial et al., 2014) or display any olfactory changes (Kurtenbach et al., 2013). Anxiety-related behaviors were shown in one study (Zhu et al., 2007) but were not confirmed by two other groups (Perez and Palmiter, 2005; Rial et al., 2014). Similarly, Park2−/− mice did not show a strong phenotype for depression-like traits (Perez and Palmiter, 2005; Rial et al., 2014). Interestingly, most studies have shown effects on cognition and working memory. Park2−/− mice have impaired habituation and exploratory activity in a novel environment (Itier, 2003; Zhu et al., 2007) and exhibit worse object location performance in a Y maze (Rial et al., 2014). Additionally, they show less spontaneous alternation in a T-maze. There is no evidence of impaired sensorimotor gating, although one study found a reduced acoustic startle response (von Coelln et al., 2004) concomitant with a selective reduction of Th-positive neurons in the locus coeruleus and noradrenaline-positive neurons in the spinal cord and olfactory bulb. Unfortunately, biochemical and electrophysiological evaluation of these models is not clear. Although no studies have reported DA neuron degeneration, some research has found an increase of DA leakage from nerve terminals (Rial et al., 2014), higher extracellular levels of DA in the striatum, and an increase in DAT expression (Itier, 2003). Long-term potentiation was decreased in one study, and a higher rate of paired pulse facilitation and excitatory postsynaptic potentials was found after low-frequency stimulation (Itier, 2003). Proteomic studies highlighted a strong reduction in mitochondrial respiratory chain proteins and stress response proteins (Shin et al., 2011) together with mitochondrial damage (Stichel et al., 2007). Lack of Park2 also seems to affect mitochondrial morphology (Pinto et al., 2018). The great variability among Park2−/− mouse models could be related to the background line and the type of mutation introduced (Perez and Palmiter, 2005). The absence of a strong Parkinsonian phenotype in Parkin-deficient mice may be because of the presence of compensatory systems such as multiple redundant E3 ubiquitin ligases. Although Park2−/− mice have not been subjected to tests for core features of ADHD, they do show impairments in working memory, a feature shared by both patients with ADHD and those with ASD (Craig et al., 2016). Specifically, the impairments in spontaneous alternation and exploration of a novel environment (Itier, 2003) might mirror the restricted interest and cognitive rigidity that are shown by some mouse models of ASD (Kazdoba et al., 2016). A reduced acoustic startle response has been also shown in other mouse ASD models (DeLorey et al., 2011; Wurzman et al., 2015), and might be linked to the inattention that characterizes patients with ADHD. There are no published findings regarding social behavior or impulsivity, making it difficult to further speculate about the similarity of this model to ADHD or ASD phenotypes.
Zebrafish
Park2 protein is extensively expressed in ventral diencephalic DA neurons in zebrafish (Pienaar et al., 2010). No stable park2 mutants are available in this species. Transient KO zebrafish have been created and these show no motor impairments. Interestingly, one of these models showed a loss of neurons, and despite showing a normal mitochondrial subset, complex I activity was found to be reduced (Table 2; Flinn et al., 2009). Similarly, KO of park2 increased cellular susceptibility to stress, whereas overexpression of human PARK2 exerted a general protective effect against the same conditions (Fett et al., 2010).
Fruit fly
PARK2 protein is highly expressed in Drosophila and a number of studies have begun to unravel the biological functions of PARK2 in this organism. Generation of PARK2/PINK1 double mutants has been crucial to understand the PARK2 signalling pathway and identify protein substrates (Botella et al., 2009; Pienaar et al., 2010; Guo, 2012). Park−/− Drosophila have a short life span and a low body weight (Table 3). The most prominent feature of these mutants is mitochondrial disintegration (fragmented mitochondrial networks), advanced aging, and loss of inner cristae (Cackovic et al., 2018). Moreover, failure of mitochondrial dynamics leads to male sterility and most probably to muscle degeneration, leaving the flies unable to jump or fly. Variable results have been reported regarding degeneration of a small subset of DA neurons (Greene et al., 2003; Cha et al., 2005; Pesah et al., 2005; Whitworth et al., 2005; Riparbelli and Callaini, 2007; Vincent et al., 2012; Navarro et al., 2014). Overexpression of mutant isoforms of PARK2 leads to a more pronounced age-dependent loss of DA neurons (Sang et al., 2007). A recent study reported learning and memory abnormalities in park−/− and a weakening of circadian rhythms because of changes in clock neurons (Julienne et al., 2017).
Human induced pluripotent stem cells
At the present time, 12 different hiPSC lines with various CNVs and mutations in the PARK2 locus have been created (Table 4) (Xu et al., 2016). All the donors were diagnosed with Parkinson’s disease, and no hiPSC study has been carried out using ASD or ADHD donors. The creation of human midbrain DA neurons from these stem cells uncovered an increase of DA release from terminals accompanied by decreased DAT expression and lower reuptake (Jiang et al., 2012). The same cells show a diminished degree of neuronal process complexity, measured by different features such as length, number of terminals, and branch points, probably because of microtubule instability (Ren et al., 2015). The rate of DA differentiation is lower in hiPSCs carrying PARK2 mutations than in controls (Shaltouki et al., 2015). Most studies have found impairment in mitochondrial morphology (Imaizumi et al., 2012; Shaltouki et al., 2015; Zanon et al., 2017) and complex I activity (Zanon et al., 2017) together with an increase in the presence of reactive oxygen species and a decrease in antioxidative pathways (Chang et al., 2016; Suzuki et al., 2017). An impairment in mitophagy has also been demonstrated (Suzuki et al., 2017).
Discussion
The comparison of CNTNAP2, ADGRL3, and PARK2 across species has shown how these models can link genetic variants found in human populations to ADHD and ASD at both behavioral and neurobiological levels (Table 5). Transgenic animal models of CNTNAP2, a gene coding for a neuron–glia adhesion protein found in myelinated axons, show that this gene is essential for normal neurodevelopment. Depletion of CNTNAP2 in mice results in epileptic seizures and a reduction in the number of GABAergic interneurons (Crawley et al., 1997; Penagarikano et al., 2011). The same features are found in the equivalent zebrafish model, which shows a smaller head and increased sensitivity to drug-induced seizures (Hoffman et al., 2016). The phenotype is even stronger in Drosophila where the absence of the CNTNAP2 orthologues results in late-stage embryonic lethality (Zweier et al., 2009). Finally, neural progenitor cells derived from a heterozygous CNTNAP2 patient show significantly reduced neural migration (Lee et al., 2015), and hiPSC-derived neurons from carrier individuals show differential expression of genes involved in synaptic transmission and neuronal activity (Flaherty et al., 2017). Interneuron dysfunction has been repeatedly linked with ASD (Takano, 2015). On a behavioral level, some phenotypic features are common across species such as the hyperactivity seen in both mice and zebrafish (Penagarikano et al., 2011; Hoffman et al., 2016). This hyperactivity mirrors one of the most prominent symptoms of ADHD.
Table 5.
Overview of genes linked to attention-deficit/hyperactivity disorder and autism spectrum disorder
The biological role of ADGRL3 is also linked to transcellular adhesion and glutamatergic synapse development (Jackson et al., 2015). Multiple models of ADGRL3 have displayed alterations in their DA system including increased dopamine levels in ADGRL3 mutant mice and a reduced number of DA neurons in adgrl3.1 morphant zebrafish (Lange et al., 2012; Orsini et al., 2016). Dysregulation of dopamine has been implicated in the etiology of ADHD by both genetic and functional imaging studies (Li et al., 2006; Faraone et al., 2015), and the most effective pharmacological treatments for ADHD are stimulant medications that act on the DA and noradrenergic systems (Engert and Pruessner, 2008). Behavioral analysis of ADGRL3 transgenic animals has revealed a number of phenotypes, the most prominent of which is the cross-species hyperactivity, which is modulated by light in Drosophila. The impairment of DA signalling in ADGRL3 transgenic animals could provide insights into how genes not directly involved in this neurotransmitter pathway may confer increased risk of ADHD.
Although mutations in the PARK2 gene are historically linked with Parkinson’s disease, mouse and zebrafish do not robustly model this disease (Perez and Palmiter, 2005). Instead, knockout of Park2 in both mouse and Drosophila leads to ADHD-like impairments in working memory. Most PARK2 models show mitochondrial phenotypes including fragmented mitochondrial networks in Drosophila and disrupted mitophagy, mitochondrial morphology, and complex I activity in neurons derived from iPSCs of patients with various genetic abnormalities in PARK2 (Imaizumi et al., 2012; Shaltouki et al., 2015; Zanon et al., 2017). Different studies have underlined an association between ASD, ADHD, and mitochondrial impairment. General mitochondrial dysfunction is believed to be present in 30–40% of autistic children (Giulivi et al., 2010; Rossignol and Frye, 2011; Morris and Berk, 2015). With regards to ADHD, sporadic mtDNA mutations and increases in oxidative markers have been reported (Marazziti et al., 2011), as well as lower levels of oxygen consumption and ATP production together with increased levels of superoxide radicals (Verma et al., 2016).
PARK2 hiPSC models also indicate an impairment in the DA system. Midbrain DA neurons created from these hiPSC lines showed an increase in DA release from terminals accompanied by lower reuptake of this neurotransmitter (Jiang et al., 2012). The same cells show a diminished degree of neuronal process complexity, probably because of microtubule instability (Ren et al., 2015).
Conclusion
In this review, we highlight the use of animal and cellular models to study neuropsychiatric disorders. By focusing on three genes that are linked to one or more psychiatric disorders, we have shown that comparing phenotypes across different species can provide more convincing insights into the aetiology of these diseases. Examples of this include the link between ADGRL3.1 and DA signalling, and PARK2 and mitochondrial dysfunction. Despite the difficulties in recapitulating complex human diseases, animal models are vital to identify novel treatments. Moreover, the advent of hiPSC models may generate novel insights into pathophysiology as they can mirror the heterogeneous genetic background typical of ADHD and ASD in a manner that is difficult to achieve in animal models. The diversity of genetic variants associated with neurodevelopmental disorders including ASD may be explained by their downstream convergence on biological processes such as synapse development and neurotransmission (Sanders, 2015). In the future, additional hiPSC models and more complex animal models of ASD and ADHD coupled with stringent testing will be necessary to unravel the molecular mechanisms underlying these disorders and to develop novel treatments for them.
Acknowledgements
The project is funded by the European Union’s Horizon 2020 research and innovation program under grant agreement no 643051.
E.D.V., N.M., and V.S.P. are early-stage researchers in the MiND Marie Sklodowska-Curie who received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no 643051. S.K.S., K.P.L., A.R., A.S., and W.H.J.N. are PIs, and W.H.J.N. is leader of the animal work package in the MiND project. The authors thank Dr Olga Rivero, Dr Marta Ribases, and Prof. Barbara Franke for their supervision and support during the preparation of this review.
Conflicts of interest
There are no conflicts of interest.
Footnotes
*Elisa Dalla Vecchia, Niall Mortimer and Viola S. Palladino contributed equally to the writing of this article.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. (2013). SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. (2011). Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188:799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. (2010). A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry 15:1053–1066. [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Vélez JI, Solomon BD, Muenke M. (2012). A common genetic network underlies substance use disorders and disruptive or externalizing disorders. Hum Genet 131:917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardhanareeswaran K, Coppola G, Vaccarino F. (2015). The use of stem cells to study autism spectrum disorder. Yale J Biol Med 88:5–16. [PMC free article] [PubMed] [Google Scholar]
- Ardhanareeswaran K, Mariani J, Coppola G, Abyzov A, Vaccarino FM. (2017). Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat Rev Neurol 13:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello PA, Gogos JA. (2006). Modeling madness in mice: one piece at a time. Neuron 52:179–196. [DOI] [PubMed] [Google Scholar]
- Arora M, Reichenberg A, Willfors C, Austin C, Gennings C, Berggren S, et al. (2017). Fetal and postnatal metal dysregulation in autism. Nat Commun 8:15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig DN, Yanagawa T, Tabuchi K. (2017). Distortion of the normal function of synaptic cell adhesion molecules by genetic variants as a risk for autism spectrum disorders. Brain Res Bull 129:82–90. [DOI] [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D, Charman T. (2006). Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP). Lancet 368:210–215. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Tong C, Tsuda H. (2010). 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci 11:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E. (2005). Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet 6:9–23. [DOI] [PubMed] [Google Scholar]
- Bloom DE, Cafiero ET, Jané-Llopis E, Abrahams-Gessel S, Bloom LR, Fathima S, et al. (2012). The global economic burden of noncommunicable diseases. PGDA Working Papers 8712, Program on the Global Demography of Aging. [Google Scholar]
- Blum M, De Robertis EM, Wallingford JB, Niehrs C. (2015). Morpholinos: antisense and sensibility. Dev Cell 35:145–149. [DOI] [PubMed] [Google Scholar]
- Botella JA, Bayersdorfer F, Gmeiner F, Schneuwly S. (2009). Modelling Parkinson’s disease in Drosophila. Neuromolecular Med 11:268–280. [DOI] [PubMed] [Google Scholar]
- Boukhris T, Sheehy O, Mottron L, Bérard A. (2016). Antidepressant use during pregnancy and the risk of autism spectrum disorder in children. JAMA Pediatr 170:117–124. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
- Brikell I, Kuja-Halkola R, Larsson H. (2015). Heritability of attention-deficit hyperactivity disorder in adults. Am J Med Genet B Neuropsychiatr Genet 168:406–413. [DOI] [PubMed] [Google Scholar]
- Cackovic J, Gutierrez-Luke S, Call GB, Juba A, O'Brien S, Jun CH, et al. (2018). Vulnerable parkin loss-of-function Drosophila dopaminergic neurons have advanced mitochondrial aging, mitochondrial network loss and transiently reduced autophagosome recruitment. Front Cell Neurosci 12:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan AO. (2010). Genetic causes of syndromic and non-syndromic autism. Dev Med Child Neurol 52:130–138. [DOI] [PubMed] [Google Scholar]
- Cha GH, Kim S, Park J, Lee E, Kim M, Lee SB, et al. (2005). Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc Natl Acad Sci USA 102:10345–10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K-H, Lee-Chen GJ, Wu YR, Chen YJ, Lin JL, Li M, et al. (2016). Impairment of proteasome and anti-oxidative pathways in the induced pluripotent stem cell model for sporadic Parkinson’s disease. Parkinsonism Relat Disord 24:81–88. [DOI] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, et al. (2005). Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci 8:1059–1068. [DOI] [PubMed] [Google Scholar]
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium (2017). Meta-analysis of GWAS of over 16 000 individuals with autism spectrum disorder highlights a novel locus at 10q24. 32 and a significant overlap with schizophrenia. Mol Autism 8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig F, Margari F, Legrottaglie AR, Palumbi R, de Giambattista C, Margari L. (2016). A review of executive function deficits in autism spectrum disorder and attention-deficit/hyperactivity disorder. Neuropsychiatr Dis Treat 12:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. (2012). Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin Neurosci 14:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Belknap JK, Collins A, Crabbe JC, Frankel W, Henderson N, et al. (1997). Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology (Berl) 132:107–124. [DOI] [PubMed] [Google Scholar]
- da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, et al. (2009). Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson's disease. Nat Cell Biol 11:1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletov BA, Shamotienko OG, Lelianova VG, Grishin EV, Ushkaryov YA. (1996). Isolation and biochemical characterization of a Ca2+-independent alpha-latrotoxin-binding protein. J Biol Chem 271:23239–23245. [DOI] [PubMed] [Google Scholar]
- DeLorey TM, Sahbaie P, Hashemi E, Li WW, Salehi A, Clark DJ. (2011). Somatosensory and sensorimotor consequences associated with the heterozygous disruption of the autism candidate gene, Gabrb3. Behav Brain Res 216:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. (2017). Discovery of the first genome-wide significant risk loci for ADHD. bioRxiv. doi: dx.doi.org/10.1101/145581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (2013). Diagnostic and statistical manual of mental disorders. USA: American Psychiatric Association. [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, et al. (2010). Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry 15:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, et al. (2012). Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 44:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert V, Pruessner JC. (2008). Dopaminergic and noradrenergic contributions to functionality in ADHD: the role of methylphenidate. Curr Neuropharmacol 6:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Heine VM, Harwood AJ, Sullivan PF, Peitz M, Brüstle O, et al. (2016). Modeling psychiatric disorders: from genomic findings to cellular phenotypes. Mol Psychiatry 21:1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Asherson P, Banaschewski T, Biederman J, Buitelaar JK, Ramos-Quiroga JA, et al. (2015). Attention-deficit/hyperactivity disorder. Nat Rev Dis Primers 1:15020. [DOI] [PubMed] [Google Scholar]
- Fernandez BA, Scherer SW. (2017). Syndromic autism spectrum disorders: moving from a clinically defined to a molecularly defined approach. Dialogues Clin Neurosci 19:353–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett ME, Pilsl A, Paquet D, van Bebber F, Haass C, Tatzelt J, et al. (2010). Parkin is protective against proteotoxic stress in a transgenic zebrafish model. PLoS ONE 5:11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty E, Deranieh RM, Artimovich E, Lee IS, Siegel AJ, Levy DL, et al. (2017). Patient-derived hiPSC neurons with heterozygous CNTNAP2 deletions display altered neuronal gene expression and network activity. NPJ Schizophr 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn L, Mortiboys H, Volkmann K, Köster RW, Ingham PW, Bandmann O. (2009). Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio). Brain 132:1613–1623. [DOI] [PubMed] [Google Scholar]
- Franke B, Faraone SV, Asherson P, Buitelaar J, Bau CH, Ramos-Quiroga JA, et al. (2012). The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol Psychiatry 17:960–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke B, Michelini G, Asherson P, Banaschewski T, Bilbow A, Buitelaar JK, et al. (2018). Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur Neuropsychopharmacol 28:1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frega M, van Gestel SH, Linda K, van der Raadt J, Keller J, Van Rhijn JR, et al. (2017). Rapid neuronal differentiation of induced pluripotent stem cells for measuring network activity on micro-electrode arrays. J Vis Exp. doi: 10.3791/54900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau JM, Stice E, Rohde P, Seeley JR. (2012). Negative life events and substance use moderate cognitive behavioral adolescent depression prevention intervention. Cogn Behav Ther 41:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi L, Brikell I, Kuja-Halkola R, Freitag CM, Franke B, Asherson P, et al. (2018). The familial co-aggregation of ASD and ADHD: a register-based cohort study. Mol Psychiatry 23:257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulivi C, Zhang YF, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, et al. (2010). Mitochondrial dysfunction in autism. JAMA 304:2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. (2009). Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, et al. (2003). Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem 278:43628–43635. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. (2003). Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA 100:4078–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. (2012). Drosophila as a model to study mitochondrial dysfunction in Parkinson’s disease. Cold Spring Harb Perspect Med 2:a009944–a009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez HC, Vacca I, Pons AI, Norton W. (2018). Automatic quantification of juvenile zebrafish aggression. J Neurosci Methods 296:23–31. [DOI] [PubMed] [Google Scholar]
- Halmøy A, Klungsøyr K, Skjærven R, Haavik J. (2012). Pre- and perinatal risk factors in adults with attention-deficit/hyperactivity disorder. Biol Psychiatry 71:474–481. [DOI] [PubMed] [Google Scholar]
- Hang L, Thundyil J, Lim KL. (2015). Mitochondrial dysfunction and Parkinson disease: a Parkin-AMPK alliance in neuroprotection. Ann N Y Acad Sci 1350:37–47. [DOI] [PubMed] [Google Scholar]
- Hara S, Takada S. (2017). Genome editing for the reproduction and remedy of human diseases in mice. J Hum Genet 63:107–113. [DOI] [PubMed] [Google Scholar]
- Hartman CA, Geurts HM, Franke B, Buitelaar JK, Rommelse N. (2016). Changing ASD-ADHD symptom co-occurrence across the lifespan with adolescence as crucial time window: illustrating the need to go beyond childhood. Neurosci Biobehav Rev 71:529–541. [DOI] [PubMed] [Google Scholar]
- Hawi Z, Cummins TD, Tong J, Johnson B, Lau R, Samarrai W, et al. (2015). The molecular genetic architecture of attention deficit hyperactivity disorder. Mol Psychiatry 20:289–297. [DOI] [PubMed] [Google Scholar]
- Hoffman EJ, Turner KJ, Fernandez JM, Cifuentes D, Ghosh M, Ijaz S, et al. (2016). Estrogens suppress a behavioral phenotype in zebrafish mutants of the autism risk gene, CNTNAP2. Neuron 89:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Schrode N, Flaherty E, Brennand KJ. (2018). New considerations for hiPSC-based models of neuropsychiatric disorders. Mol Psychiatry. doi: 10.1038/s41380-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollville E, Carroll RG, Cullen SP, Martin SJ. (2014). Bcl-2 Family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell 55:451–466. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, et al. (2012). Mitochondrial dysfunction associated with increased oxidative stress and α-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, et al. (2003). Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet 12:2277–2291. [DOI] [PubMed] [Google Scholar]
- Jackson VA, del Toro D, Carrasquero M, Roversi P, Harlos K, Klein R, et al. (2015). Structural basis of latrophilin–FLRT interaction. Structure 23:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansch C, Günther K, Waider J, Ziegler GC, Forero A, Kollert S, et al. (2018). Generation of a human induced pluripotent stem cell (iPSC) line from a 51-year-old female with attention-deficit/hyperactivity disorder (ADHD) carrying a duplication of SLC2A3. Stem Cell Res 28:136–140. [DOI] [PubMed] [Google Scholar]
- Jarick I, Volckmar AL, Pütter C, Pechlivanis S, Nguyen TT, Dauvermann MR, et al. (2014). Genome-wide analysis of rare copy number variations reveals PARK2 as a candidate gene for attention-deficit/hyperactivity disorder. Mol Psychiatry 19:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarick I, Volckmar AL, Pütter C, Pechlivanis S, Nguyen TT, Dauvermann MR, et al. (2014). Genome-wide analysis of rare copy number variations reveals PARK2 as a candidate gene for attention-deficit/hyperactivity disorder. Mol Psychiatry 19:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ren Y, Yuen EY, Zhong P, Ghaedi M, Hu Z, et al. (2012). Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat Commun 3:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julienne H, Buhl E, Leslie DS, Hodge J. (2017). Drosophila PINK1 and parkin loss-of-function mutants display a range of non-motor Parkinson’s disease phenotypes. Neurobiol Dis 104:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgensen S, Castillo E, Leslie PE. (2015). Selective Dysregulation of Hippocampal Inhibition in the Mouse Lacking Autism Candidate Gene CNTNAP2. J Neurosci 35:14681–14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazdoba T, Leach P, Crawley JN. (2016). Behavioral phenotypes of genetic mouse models of autism. Genes Brain Behav 15:7–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Rim YA, Yi H, Park N, Park SH, Ju JH. (2016). The generation of human induced pluripotent stem cells from blood cells: an efficient protocol using serial plating of reprogrammed cells by centrifugation. Stem Cells Int 2016:1329459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser DP, Rivero O, Lesch KP. (2015). Annual research review: the (epi)genetics of neurodevelopmental disorders in the era of whole-genome sequencing – unveiling the dark matter. J Child Psychol Psychiatry 56:278–295. [DOI] [PubMed] [Google Scholar]
- Kurtenbach S, Wewering S, Hatt H, Neuhaus EM, Lübbert H. (2013). Olfaction in three genetic and two MPTP-induced Parkinson’s disease mouse models. PLoS One 8:77509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, et al. (2012). The ADHD-susceptibility gene lphn3. 1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry 17:946–954. [DOI] [PubMed] [Google Scholar]
- Lange M, Froc C, Grunwald H, Norton W, Bally-Cuif L. (2018). Pharmacological analysis of zebrafish lphn3. 1 morphant larvae suggests that saturated dopaminergic signaling could underlie the ADHD-like locomotor hyperactivity. Prog Neuropsychopharmacol Biol Psychiatry 84:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc S, Easley D. (2015). Pharmacological therapies for autism spectrum disorder: a review. P T 40:389–397. [PMC free article] [PubMed] [Google Scholar]
- Lee IS, Carvalho CM, Douvaras P, Ho SM, Hartley BJ, Zuccherato LW, et al. (2015). Characterization of molecular and cellular phenotypes associated with a heterozygous CNTNAP2 deletion using patient-derived hiPSC neural cells. NPJ Schizophr 1:15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch K-P, Selch S, Renner TJ, Jacob C, Nguyen TT, Hahn T, et al. (2011). Genome-wide copy number variation analysis in attention-deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Mol Psychiatry 16:491–503. [DOI] [PubMed] [Google Scholar]
- Li D, Sham PC, Owen MJ, He L. (2006). Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum Mol Genet 15:2276–2284. [DOI] [PubMed] [Google Scholar]
- Li J, Ashley J, Budnik V, Bhat MA. (2007). Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron 55:741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chang SH, Zhang LY, Gao L, Wang J. (2014). Molecular genetic studies of ADHD and its candidate genes: a review. Psychiatry Res 219:10–24. [DOI] [PubMed] [Google Scholar]
- Lim C-S, Yang JE, Lee YK, Lee K, Lee JA, Kaang BK. (2015). Understanding the molecular basis of autism in a dish using hiPSCs-derived neurons from ASD patients. Mol Brain 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, et al. (2011). Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med 3:95ra75. [DOI] [PubMed] [Google Scholar]
- Lücking CB, Dürr A, Bonifati V, Vaughan J, De Michele G, Gasser T, et al. (2000). Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med 342:1560–1567. [DOI] [PubMed] [Google Scholar]
- Marazziti D, Baroni S, Picchetti M, Landi P, Silvestri S, Vatteroni E, et al. (2011). Mitochondrial alterations and neuropsychiatric disorders. Curr Med Chem 18:4715–4721. [DOI] [PubMed] [Google Scholar]
- Martinez AF, Abe Y, Hong S, Molyneux K, Yarnell D, Löhr H, et al. (2016). An Ultraconserved brain-specific enhancer within ADGRL3 (LPHN3) underpins attention-deficit/hyperactivity disorder susceptibility. Biol Psychiatry 80:943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon JM, Sive H. (2015). Addressing the genetics of human mental health disorders in model organisms. Annu Rev Genomics Hum Genet 16:173–197. [DOI] [PubMed] [Google Scholar]
- Meadows SO, McLanahan S, Brooks-Gunn J. (2007). Parental depression and anxiety and early childhood behavior problems across family types. J Marriage Fam 69:1162–1177. [Google Scholar]
- Modabbernia A, Velthorst E, Reichenberg A. (2017). Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol Autism 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G, Berk M. (2015). The many roads to mitochondrial dysfunction in neuroimmune and neuropsychiatric disorders. BMC Med 13:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, et al. (2013). The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell 49:908–921. [DOI] [PubMed] [Google Scholar]
- Navarro JA, Heßner S, Yenisetti SC, Bayersdorfer F, Zhang L, Voigt A, et al. (2014). Analysis of dopaminergic neuronal dysfunction in genetic and toxin-induced models of Parkinson’s disease in Drosophila. J Neurochem 131:369–382. [DOI] [PubMed] [Google Scholar]
- Nikolas MA, Burt SA. (2010). Genetic and environmental influences on ADHD symptom dimensions of inattention and hyperactivity: a meta-analysis. J Abnorm Psychol 119:1–17. [DOI] [PubMed] [Google Scholar]
- Norton WHJ. (2013). Toward developmental models of psychiatric disorders in zebrafish. Front Neural Circuits 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum NL. (2012). ADHD and female specific concerns: a review of the literature and clinical implications. J Atten Disord 16:87–100. [DOI] [PubMed] [Google Scholar]
- Orsini CA, Setlow B, DeJesus M, Galaviz S, Loesch K, Ioerger T, Wallis D. (2016). Behavioral and transcriptomic profiling of mice null forLphn3, a gene implicated in ADHD and addiction. Mol Genet Genomic Med 4:322–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane CJ. (2011). Drosophila as a model organism for the study of neuropsychiatric disorders Curr Top Behav Neurosci 7:37–60. [DOI] [PubMed] [Google Scholar]
- O’sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. (2014). LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Develop 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, et al. (2011). Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez FA, Palmiter RD. (2005). Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA 102:2174–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesah Y, Burgess H, Middlebrooks B, Ronningen K, Prosser J, Tirunagaru V, et al. (2005). Whole-mount analysis reveals normal numbers of dopaminergic neurons following misexpression of alpha-synuclein in Drosophila. Genesis 41:154–159. [DOI] [PubMed] [Google Scholar]
- Peñagarikano O, Geschwind DH. (2012). What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol Med 18:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell MA, Youle JR. (2015). The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85:257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienaar IS, Götz J, Feany MB. (2010). Parkinson’s disease: insights from non-traditional model organisms. Prog Neurobiol 92:558–571. [DOI] [PubMed] [Google Scholar]
- Pinto M, Nissanka N, Moraes CT. (2018). Lack of parkin anticipates the phenotype and affects mitochondrial morphology and mtDNA levels in a mouse model of Parkinson’s disease. J Neurosci 38:1042–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. (2007). The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am J Psychiatry 164:942–948. [DOI] [PubMed] [Google Scholar]
- Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, et al. (1999). Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron 24:1037–1047. [DOI] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, et al. (2003). Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol 162:1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M. (2015). Connecting the CNTNAP2 networks with neurodevelopmental disorders. Mol Syndromol 6:7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Beyer V, Schwaab I, Damatova N, Van't Slot R, Prothero J, et al. (2010). Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics 11:81–89. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA 111:2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilutsky D, Palmer NP, Smedemark-Margulies N, Schlaeger TM, Margulies DM, Kohane IS. (2014). iPSC-derived neurons as a higher-throughput readout for autism: promises and pitfalls. Trends Mol Med 20:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prytkova I, Brennand KJ. (2017). Prospects for modeling abnormal neuronal function in schizophrenia using human induced pluripotent stem cells. Front Cell Neurosci 11:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Quiroga JA, Corominas M, Castells X, Bosch R, Casas M. (2009). OROS methylphenidate for the treatment of adults with attention-deficit/hyperactivity disorder. Expert Rev Neurother 9:1121–1131. [DOI] [PubMed] [Google Scholar]
- Ramos-Quiroga J-A, Sánchez-Mora C, Casas M, Garcia-Martínez I, Bosch R, Nogueira M, et al. (2014). Genome-wide copy number variation analysis in adult attention-deficit and hyperactivity disorder. J Psychiatr Res 49:60–67. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. (2006). Global variation in copy number in the human genome. Nature 444:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Jiang H, Hu Z, Fan K, Wang J, Janoschka S, et al. (2015). Parkin mutations reduce the complexity of neuronal processes in iPSC-derived human neurons. Stem Cells 33:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial D, Castro AA, Machado N, Garção P, Gonçalves FQ, Silva HB, et al. (2014). Behavioral phenotyping of Parkin-deficient mice: looking for early preclinical features of Parkinson’s disease. PLoS One 9:114216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribasés M, Ramos-Quiroga JA, Sánchez-Mora C, Bosch R, Richarte V, Palomar G, et al. (2011). Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav 10:149–157. [DOI] [PubMed] [Google Scholar]
- Rihel J, Prober DA, Schier AF. (2010). Monitoring sleep and arousal in zebrafish. Methods Cell Biol 281–294. [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G. (2007). The Drosophila parkin homologue is required for normal mitochondrial dynamics during spermiogenesis. Dev Biol 303:108–120. [DOI] [PubMed] [Google Scholar]
- Rivero O, Selten MM, Sich S, Popp S, Bacmeister L, Amendola E, et al. (2015). Cadherin-13, a risk gene for ADHD and comorbid disorders, impacts GABAergic function in hippocampus and cognition. Transl Psychiatry 5:655–e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas-Cuadrado P, Ho J, Vernes SC. (2014). Shining a light on CNTNAP2: complex functions to complex disorders. Eur J Hum Genet 22:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelse NN, Geurts HM, Franke B, Buitelaar JK, Hartman CA. (2011). A review on cognitive and brain endophenotypes that may be common in autism spectrum disorder and attention-deficit/hyperactivity disorder and facilitate the search for pleiotropic genes. Neurosci Biobehav Rev 35:1363–1396. [DOI] [PubMed] [Google Scholar]
- Rosen BN, Lee BK, Lee NL, Yang Y, Burstyn I. (2014). Maternal smoking and autism spectrum disorder: a meta-analysis. J Autism Dev Disord 45:1689–1698. [DOI] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. (2012). Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry 17:290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. (2003). Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218:348–353. [DOI] [PubMed] [Google Scholar]
- Sanders SJ. (2015). First glimpses of the neurobiology of autism spectrum disorder. Curr Opin Genet Dev 33:80–92. [DOI] [PubMed] [Google Scholar]
- Sang TK, Chang HY, Lawless GM, Ratnaparkhi A, Mee L, Ackerson LC, et al. (2007). A Drosophila model of mutant human parkin-induced toxicity demonstrates selective loss of dopaminergic neurons and dependence on cellular dopamine. J Neurosci 27:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe LA, Stevens DA, Dawson VL, Dawson TM. (2014). Parkin and PINK1: much more than mitophagy. Trends Neurosci 37:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerle A, Wilson K. (2011). PARK2 copy number aberrations in two children presenting with autism spectrum disorder: further support of an association and possible evidence for a new microdeletion/microduplication syndrome. Am J Med Genet B Neuropsychiatr Genet 156:413–420. [DOI] [PubMed] [Google Scholar]
- Schmid B, Haass C. (2013). Genomic editing opens new avenues for zebrafish as a model for neurodegeneration. J Neurochem 127:461–470. [DOI] [PubMed] [Google Scholar]
- Sciberras E, Mulraney M, Silva D, Coghill D. (2017). Prenatal risk factors and the etiology of ADHD: review of existing evidence. Curr Psychiatry Rep 19:1. [DOI] [PubMed] [Google Scholar]
- Shaltouki A, Sivapatham R, Pei Y, Gerencser AA, Momčilović O, Rao MS, et al. (2015). Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 Patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Reports 4:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Haynes P, Pírez N, Harrington KI, Guo F, Pollack J, et al. (2011). Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-H, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. (2011). PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 144:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E, Aleksic B, Ozaki N. (2014). Copy-number variation in the pathogenesis of autism spectrum disorder. Psychiatry Clin Neurosci 68:85–95. [DOI] [PubMed] [Google Scholar]
- Sjaarda CP, Hecht P, McNaughton A, Zhou A, Hudson ML, Will MJ, et al. (2017). Interplay between maternal Slc6a4 mutation and prenatal stress: a possible mechanism for autistic behavior development. Sci Rep 7:8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman MA, Aboharb F, Zeltner N, Studer L. (2017). Pluripotent stem cells in neuropsychiatric disorders. Mol Psychiatry 22:1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AM, Nguyen M, Wong K, Poudel MK, Kalueff AV. (2014). Developing zebrafish models of autism spectrum disorder (ASD). Prog Neuropsychopharmacol Biol Psychiatry 50:27–36. [DOI] [PubMed] [Google Scholar]
- Stewart AM, Ullmann JF, Norton WH, Parker MO, Brennan CH, Gerlai R, et al. (2015). Molecular psychiatry of zebrafish. Mol Psychiatry 20:2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichel CC, Zhu XR, Bader V, Linnartz B, Schmidt S, Lübbert H. (2007). Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum Mol Genet 16:2377–2393. [DOI] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. (2006). Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med 354:1370–1377. [DOI] [PubMed] [Google Scholar]
- Sujan AC, Rickert ME, Öberg AS, Quinn PD, Hernández-Díaz S, Almqvist C, et al. (2017). Associations of maternal antidepressant use during the first trimester of pregnancy with preterm birth, small for gestational age, autism spectrum disorder, and attention-deficit/hyperactivity disorder in offspring. JAMA 317:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Akamatsu W, Kisa F, Sone T, Ishikawa KI, Kuzumaki N, et al. (2017). Efficient induction of dopaminergic neuron differentiation from induced pluripotent stem cells reveals impaired mitophagy in PARK2 neurons. Biochem Biophys Res Commun 483:88–93. [DOI] [PubMed] [Google Scholar]
- Sánchez-Mora C, Richarte V, Garcia-Martínez I, Pagerols M, Corrales M, Bosch R, et al. (2015). Dopamine receptor DRD4 gene and stressful life events in persistent attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 168:480–491. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. [DOI] [PubMed] [Google Scholar]
- Takano T. (2015). Interneuron dysfunction in syndromic autism: recent advances. Dev Neurosci 37:467–475. [DOI] [PubMed] [Google Scholar]
- Tay TL, Ronneberger O, Ryu S, Nitschke R, Driever W. (2011). Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nat Commun 2:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tick B, Bolton P, Happé F, Rutter M, Rijsdijk F. (2016). Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry 57:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen B, van Swinderen B. (2013). Drosophila strategies to study psychiatric disorders. Brain Res Bull 92:1–11. [DOI] [PubMed] [Google Scholar]
- van der Voet M, Nijhof B, Oortveld MA, Schenck A. (2014). Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci Biobehav Rev 46 Pt 2:326–342. [DOI] [PubMed] [Google Scholar]
- van der Voet M, Harich B, Franke B, Schenck A. (2016). ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry 21:565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme TF. (2014). Use of rodents as models of human diseases. J Pharm Bioallied Sci 6:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Singh A, Nthenge-Ngumbau DN, Rajamma U, Sinha S, Mukhopadhyay K, et al. (2016). Attention deficit-hyperactivity disorder suffers from mitochondrial dysfunction. BBA Clin 6:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigo D, Thornicroft G, Atun R. (2016). Estimating the true global burden of mental illness. Lancet Psychiatry 3:171–178. [DOI] [PubMed] [Google Scholar]
- Vincent A, Briggs L, Chatwin GF, Emery E, Tomlins R, Oswald M, et al. (2012). Parkin-induced defects in neurophysiology and locomotion are generated by metabolic dysfunction and not oxidative stress. Hum Mol Genet 21:1760–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, et al. (2004). Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci USA 101:10744–10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, et al. (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49:833–844. [DOI] [PubMed] [Google Scholar]
- Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, Wellman PJ, et al. (2012). Initial characterization of mice null for Lphn3, a gene implicated in ADHD and addiction. Brain Res 1463:85–92. [DOI] [PubMed] [Google Scholar]
- Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. (2005). Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci 102:8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, et al. (2010). Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 376:1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Franke B, Mick E, Anney RJ, Freitag CM, Gill M, et al. (2012). Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13. 3. Am J Psychiatry 169:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolraich ML, Doffing MA. (2004). Pharmacokinetic considerations in the treatment of attention-deficit hyperactivity disorder with methylphenidate. CNS Drugs 18:243–250. [DOI] [PubMed] [Google Scholar]
- Wong AH, Josselyn SA. (2016). Caution when diagnosing your mouse with schizophrenia: the use and misuse of model animals for understanding psychiatric disorders. Biol Psychiatry 79:32–38. [DOI] [PubMed] [Google Scholar]
- Wu S, Wu F, Ding Y, Hou J, Bi J, Zhang Z. (2017). Advanced parental age and autism risk in children: a systematic review and meta-analysis. Acta Psychiatr Scand 135:29–41. [DOI] [PubMed] [Google Scholar]
- Wurzman R, Forcelli PA, Griffey CJ, Kromer LF. (2015). Repetitive grooming and sensorimotor abnormalities in an ephrin-A knockout model for autism spectrum disorders. Behav Brain Res 278:115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Huang J, Li J, Liu L, Han C, Shen Y, et al. (2016). Induced pluripotent stem cells and Parkinson's disease: modelling and treatment. Cell Prolif 49:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C-L, Chen HI, Li LH, Chien YL, Liao HM, Chou MC, et al. (2016). Genome-wide analysis of copy number variations identifies PARK2 as a candidate gene for autism spectrum disorder. Mol Autism 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanon A, Kalvakuri S, Rakovic A, Foco L, Guida M, Schwienbacher C, et al. (2017). SLP-2 interacts with Parkin in mitochondria and prevents mitochondrial dysfunction in Parkin-deficient human iPSC-derived neurons and Drosophila. Hum Mol Genet 26:2412–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chang S, Li Z, Zhang K, Du Y, Ott J, et al. (2012). ADHDgene: a genetic database for attention deficit hyperactivity disorder. Nucleic Acids Res 40:1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lee S, Peng Y, Bunker E, Giaime E, Shen J, et al. (2014). PINK1 triggers autocatalytic activation of parkin to specify cell fate decisions. Curr Biol 24:1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-R, Maskri L, Herold C, Bader V, Stichel CC, Güntürkün O, et al. (2007). Non-motor behavioural impairments in parkin-deficient mice. Eur J Neurosci 26:1902–1911. [DOI] [PubMed] [Google Scholar]
- Zhu JL, Olsen J, Liew Z, Li J, Niclasen J, Obel C. (2014). Parental smoking during pregnancy and ADHD in children: the Danish National Birth Cohort. Pediatrics 134:382–388. [DOI] [PubMed] [Google Scholar]
- Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, et al. (2009). CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet 85:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]