On May 15, 2020, prostate cancer entered the precision oncology era with accelerated approval from the US Food and Drug Administration (FDA) for the poly(ADP)-ribose polymerase (PARP) inhibitor rucaparib for treating patients with metastatic castration-resistant prostate cancer (mCRPC) who had deleterious germline or somatic BRCA1 or BRCA2 mutations and who had previously received an androgen receptor (AR)–directed therapy and a taxane-based chemotherapy. Four days later, on May 19, 2020, the FDA approved a second PARP inhibitor, olaparib, for treating patients with mCRPC who have a deleterious germline or somatic mutation in at least one of 14 homologous recombination repair (HRR) genes (BRCA1 and BRCA2 plus 12 other HRR genes: ATM, BARD1, BRIP1, CDK12, CHEK1, CHEK2, FANCL, PALB2, RAD51B, RAD51C, RAD51D, or RAD54L) who had previously received an AR-directed therapy. The FDA’s approval of olaparib in prostate cancer was based on a positive randomized phase III trial (PROfound; ClinicalTrials.gov identifier: NCT02987543) that showed an improvement in radiographic progression-free survival (rPFS) in patients with HRR-mutated mCRPC who received olaparib compared with abiraterone or enzalutamide.1 In contrast, the accelerated FDA approval of rucaparib in prostate cancer was based on an uncontrolled single-arm phase II trial (TRITON2; ClinicalTrials.gov identifier: NCT02952534) that used objective response rate (ORR) as its primary efficacy end point, as presented in this issue in an article by Abida et al.2 This accelerated regulatory approval, an unprecedented move by the FDA in the prostate cancer space, is contingent upon the confirmation of a clinical benefit with rucaparib in the ongoing randomized phase III trial (TRITON3; ClinicalTrials.gov identifier: NCT02975934), which is testing rucaparib versus physician’s choice of best systemic therapy (AR-directed therapy or taxane chemotherapy) in taxane-naïve patients with mCRPC.
As reported in Abida et al,2 the accompanying article, the TRITON2 study enrolled patients with mCRPC who had previously progressed on one to two lines of AR-directed therapy (eg, abiraterone or enzalutamide) and one line of taxane-based chemotherapy (eg, docetaxel) and who had a deleterious germline or somatic mutation in BRCA1/2 or another non-BRCA1/2 HRR gene. Patients were treated with open-label rucaparib 600 mg orally twice per day (together with ongoing medical or surgical castration), until disease progression or unmanageable toxicity. Efficacy end points were the ORR in those with measurable soft tissue metastases, a 50% or greater decrease in prostate-specific antigen (PSA50) level response rate, and rPFS. The activity of rucaparib in patients with mCRPC who had non-BRCA1/2 HRR gene mutations has been reported previously3 and was not sufficient to warrant regulatory approval for that molecular subset of patients. In the cohort of 115 patients with BRCA1 (13 patients) orBRCA2 mutations (102 patients), the ORR for rucaparib was 44% (27 of 62 evaluable patients), the PSA50 response rate was 55% (63 of 115 evaluable patients), and the median rPFS was 9.0 months (95% CI, 8.3 to 13.5 months), representing an impressive constellation of therapeutic benefits. It should also be noted that TRITON2 enrolled more patients with mCRPC who had BRCA1 and BRCA2 alterations than any other study of PARP inhibitors (including the PROfound study), an impressive feat. The safety profile of rucaparib was generally favorable but was notable for a prevalence of the following adverse events at grade 3 or greater: anemia (25%), thrombocytopenia (10%), asthenia or fatigue (9%), increases in hepatic transaminases (5%), and nausea (3%).
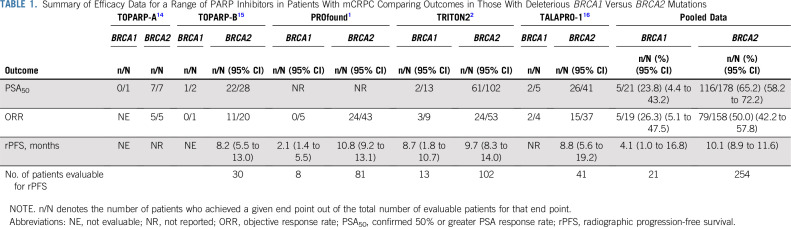
As an exploratory hypothesis-generating exercise, the authors examined the activity of rucaparib according to gene mutation (BRCA1 or BRCA2), genetic origin (germline or somatic), zygosity status (monoallelic or biallelic), and mutation type (homozygous deletion or other deleterious mutations). This is the first study to explore the clinical significance of these genomic variables in prostate cancer, so the authors should be congratulated for shedding additional light on and providing new biologic insights into the activity of PARP inhibitors in prostate cancer. Accordingly, the efficacy of rucaparib was generally greater (although not always statistically superior) in patients with germline versus somatic BRCA1/2 mutations (PSA50 response, 61% v 51%, with similar ORR estimates), in patients with biallelic versus monoallelic mutations (PSA50 response, 75% v 11%; ORR, 52% v 50%), and in patients with homozygous deletions versus other deleterious mutations (PSA50 response, 81% v 49%; ORR, 60% v 38%). However, what is perhaps most interesting is the potential difference in efficacy of rucaparib in men with BRCA1 compared with BRCA2 mutations. More specifically, the clinical activity of rucaparib seemed to be generally greater in patients with BRCA2-altered compared with BRCA1-altered mCRPC, as assessed by PSA50 response rates (60% [95% CI, 50% to 69%] v 15% [95% CI, 2% to 45%]), ORR estimates (45% [95% CI, 32% to 60%] v 33% [95% CI, 8% to 70%]), and median rPFS estimates (9.7 months [95% CI, 8.3 to 14.0 months] v 8.7 months [95% CI, 1.8 to 10.7 months]). Moreover, this apparent discrepancy in PARP inhibitor sensitivity between patients with BRCA1- and BRCA2-mutated mCRPC does not seem to be restricted to rucaparib and seems to be a class effect of PARP inhibitors in prostate cancer, as summarized in Table 1.
TABLE 1.
Summary of Efficacy Data for a Range of PARP Inhibitors in Patients With mCRPC Comparing Outcomes in Those With Deleterious BRCA1 Versus BRCA2 Mutations
What could explain this dampened clinical activity of PARP inhibitors in patients with prostate cancer who have BRCA1 compared with BRCA2 mutations? We propose four hypotheses: (1) BRCA1 alterations are less often germline lesions than BRCA2 alterations, (2) BRCA1 alterations are less often biallelic mutations than BRCA2 alterations, (3) BRCA1 mutations result in attenuated HRR deficiency compared with BRCA2 mutations, and (4) BRCA1 mutations have more genomic co-alterations (eg, in TP53 or PTEN) than BRCA2 mutations. We will examine the available evidence that may support or refute each of these claims in turn.
Hypothesis 1: Are BRCA1 alterations less often germline lesions than BRCA2 alterations? This does not seem to be the case. In the TRITON2 study,2 there were equal proportions of germline alterations affecting the two genes (38% of BRCA1 mutations [five of 13] and 38% of BRCA2 mutations [39 of 102] were of germline origin). An equal frequency of germline mutations involving BRCA1 and BRCA2 in patients with mCRPC (about 50% for both genes) was also reported in another recent study.4 However, in the largest pan-cancer analysis of BRCA1/2 alterations published to date (which included 7,185 prostate cancers) by Sokol et al,5 it was shown that germline mutations affected 35% of patients with BRCA1-altered mCRPC compared with 50% of patients with BRCA2-altered mCRPC. Thus, although it is possible that BRCA2 alterations might be slightly enriched for germline mutations compared with BRCA1 alterations, this difference is likely to be small at best.
Hypothesis 2: Are BRCA1 alterations less often biallelic mutations than BRCA2 alterations? Yes, this seems to be correct. In the TRITON2 trial, only 40% of BRCA1 mutations (two of five) that were evaluable for zygosity status were biallelic alterations, whereas 85% of evaluable BRCA2 mutations (34 of 40) were biallelic alterations.2 Similarly, in another recent study,4 the proportion of patients with germline-altered mCRPC who had somatic biallelic mutations (as evidenced by tumoral loss of heterozygosity [LOH]) was 50% in BRCA1 carriers and 60% in BRCA2 carriers. Finally, in the large pan-cancer BRCA1/2 analysis by Sokol et al,5 biallelic inactivation occurred in only 50% of BRCA1 mutations versus 90% of BRCA2 mutations; biallelic alteration was more common in both cases with germline mutation origin. Therefore, biallelic mutations in prostate cancer seem to involve BRCA2 more commonly than BRCA1.
Hypothesis 3: Do BRCA1 mutations result in attenuated HRR deficiency compared with BRCA2 mutations? This is not likely. Because the efficacy of PARP inhibitors relies on impaired HRR function to induce synthetic lethality,6 a functional assessment of HRR deficiency or proficiency could theoretically predict sensitivity to PARP inhibition. With respect to the BRCA1/2 genes, however, there is no evidence of differential HRR dysfunction when comparing alterations in the two genes. In one study that computed homologous recombination deficiency scores,4 the authors showed that prostate cancers with biallelic BRCA1 and BRCA2 mutations had similar composite homologous recombination deficiency scores, and these scores were in fact slightly higher in BRCA1-altered prostate cancers. Similarly, in another study that examined genome-wide LOH (gLOH) scores as a surrogate for HRR function,5 biallelic BRCA1 and BRCA2 mutations were associated with high gLOH scores in 40% and 25% of prostate cancers, respectively; the proportion of the cancer genome under LOH (% gLOH) was slightly higher in prostate tumors with biallelic BRCA1 versus BRCA2 mutations (15% v 10%, respectively). In sum, this suggests that biallelic inactivation of BRCA1 does indeed result in significant HRR deficiency and that perhaps we need better ways to define true BRCA1 loss in prostate cancer. In other malignancies, for example, BRCA1 methylation has been associated with PARP inhibitor sensitivity,7 whereas mutations in exon 11 of BRCA1 may predict therapeutic resistance to PARP inhibition.8
Hypothesis 4: Do BRCA1-mutated cancers have more genomic co-alterations (eg, in TP53 or PTEN) than BRCA2-mutated cancers? This seems to be the case, at least for mutations in TP53. It could also be hypothesized that BRCA1/2-altered prostate cancers with concurrent mutations in other genes associated with poor prognosis (eg, TP53 or PTEN)9,10 might demonstrate attenuated sensitivity to PARP inhibitors, and we wondered whether such mutations might be distributed unequally in the two groups. In the TRITON2 study,2 for example, it was observed that patients with BRCA1 mutations had more frequent co-alterations in TP53 (62%) and PTEN (69%) compared with those who had BRCA2 mutations (TP53 alterations in 42% and PTEN alterations in 29%, respectively). To determine whether these finding were real or spurious, we analyzed publicly available genomic data from the cBioPortal repository.11 We discovered that deleterious TP53 alterations were significantly more frequent in BRCA1-mutated versus BRCA2-mutated prostate cancers (39% [19 of 49] v 23% [76 of 336], respectively; P = .02), whereas the prevalence of deleterious PTEN alterations was similar (20% [10 of 49] v 18% [59 of 336], respectively; P = .69). Intriguingly, TP53 (but not PTEN) co-alterations were also more common in BRCA1-altered versus BRCA2-altered breast cancer (66% [78 of 119] v 35% [57 of 165], respectively; P < .01) and bladder cancer (58% [14 of 24] v 34% [20 of 58]; P = .05), as well as colorectal (56% [19 of 34] v 36% [32 of 88]; P = .06), gastroesophageal (36% [29 of 81] v 26% [111 of 421]; P = .10), endometrial (55% [12 of 22] v 34% [24 of 71]; P = .13), and even cutaneous squamous cell carcinomas (42% [22 of 52] v 23% [78 of 338]; P = .01). In addition to TP53 alterations broadly portending an overall worse prognosis, recent reports suggest that TP53 mutations might be more permissive of the emergence of BRCA1/2 reversion mutations (that restore the open reading frame) in BRCA1/2-altered cancers exposed to PARP inhibitor treatment.4,12 Such reversion mutations have been associated with secondary PARP inhibitor resistance in prostate and other cancers.
Ultimately, because of the relative rarity of both germline and somatic BRCA1 mutations compared with BRCA2 mutations in prostate cancer10,13 (unlike the situation in breast or ovarian cancers, where the prevalence of two genes is roughly equal), our conclusions related to PARP inhibitor sensitivity should be interpreted with caution. As presented in Table 1, the total number of patients with BRCA1-altered mCRPC included in the publicly available global literature is a mere 21 patients, which limits making robust conclusions. Additional studies and meta-analyses will be required to gain clearer insights on the potential differential efficacy of PARP inhibitors in prostate cancers with BRCA1 versus BRCA2 mutations and to understand the biologic mechanisms underpinning this phenomenon. At this time, both rucaparib and olaparib are indicated, and should be considered, for the treatment of mCRPC patients with either BRCA1 or BRCA2 mutations. Indeed, the availability of genomically selected therapies for our patients with BRCA1/2-altered advanced prostate cancer represents a welcome addition to our therapeutic armamentarium and is a giant step forward in the management of this disease.
ACKNOWLEDGMENT
Supported by National Institutes of Health Cancer Center Support Grant P30CA006973 (E.S.A.) and a Prostate Cancer Foundation Young Investigator Award (M.C.M).
The authors thank Alexander Baras for assistance with analysis and interpretation of the cBioPortal database and Su Jin Lim and Hao Wang for assistance with pooled biostatistical analyses
Footnotes
See accompanying article on page 3763
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Administrative support: Emmanuel S. Antonarakis
Collection and assembly of data: Emmanuel S. Antonarakis
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
BRCA1 Versus BRCA2 and PARP Inhibitor Sensitivity in Prostate Cancer: More Different Than Alike?
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Mark C. Markowski
Honoraria: Clovis Oncology, Exelixis
Emmanuel S. Antonarakis
Honoraria: Sanofi, Dendreon, Medivation, Janssen Biotech, ESSA, Astellas Pharma, Merck, AstraZeneca, Clovis Oncology
Consulting or Advisory Role: Sanofi, Dendreon, Janssen Biotech, ESSA, Merck, AstraZeneca, Clovis Oncology, Eli Lilly, Bayer
Research Funding: Janssen Biotech (Inst), Johnson & Johnson (Inst), Sanofi (Inst), Dendreon (Inst), Aragon Pharmaceuticals (Inst), Exelixis (Inst), Millennium Pharmaceuticals (Inst), Genentech (Inst), Novartis (Inst), Astellas Pharma (Inst), Tokai Pharmaceuticals (Inst), Merck (Inst), AstraZeneca (Inst), Clovis Oncology (Inst), Constellation Pharmaceuticals (Inst)
Patents, Royalties, Other Intellectual Property: Co-inventor of a biomarker technology that has been licensed to QIAGEN
Travel, Accommodations, Expenses: Sanofi, Dendreon, Medivation
No other potential conflicts of interest were reported.
REFERENCES
- 1.de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 2.Abida W, Patnaik A, Campbell D, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 38:3763-3772, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abida W, Campbell D, Patnaik A, et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: Analysis from the phase II TRITON2 study. Clin Cancer Res. 2020;26:2487–2496. doi: 10.1158/1078-0432.CCR-20-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson P, Bandlamudi C, Cheng ML, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokol ES, Pavlick D, Khiabanian H, et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol. doi: 10.1200/PO.19.00345. 4:442-465, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Kondrashova O, Topp M, Nesic K, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9:3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Bernhardy AJ, Cruz C, et al. The BRCA1-Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76:2778–2790. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maughan BL, Guedes LB, Boucher K, et al. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2018;21:260–268. doi: 10.1038/s41391-017-0027-4. [DOI] [PubMed] [Google Scholar]
- 10.Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.cBioPortal for Cancer Genomics www.cbioportal.org
- 12.Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 2017;7:999–1005. doi: 10.1158/2159-8290.CD-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateo J, Porta N, Bianchini D, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Bono JS, Mehra N, Higano CS, et al. TALAPRO-1: Phase II study of talazoparib in men with DNA damage repair alterations (DDRmut) and metastatic castration-resistant prostate cancer (mCRPC): Updated interim analysis (IA) J Clin Oncol. 2020;38(suppl; abstr 119) [Google Scholar]