Abstract
Background and aims
The COVID-19 pandemic has spread worldwide and poses a severe health risk. While most patients present mild symptoms, descending pneumonia can lead to severe respiratory insufficiency. Up to 50% of patients show gastrointestinal symptoms like diarrhea or nausea, intriguingly associating with prolonged symptoms and increased severity. Thus, models to understand and validate drug efficiency in the gut of COVID-19 patients are of urgent need.
Methods
Human intestinal organoids derived from pluripotent stem cells (PSC-HIOs) have led, due to their complexity in mimicking human intestinal architecture, to an unprecedented number of successful disease models including gastrointestinal infections. Here, we employed PSC-HIOs to dissect SARS-CoV-2 pathogenesis and its inhibition by remdesivir, one of the leading drugs investigated for treatment of COVID-19.
Results
Immunostaining for viral entry receptor ACE2 and SARS-CoV-2 spike protein priming protease TMPRSS2 showed broad expression in the gastrointestinal tract with highest levels in the intestine, the latter faithfully recapitulated by PSC-HIOs. Organoids could be readily infected with SARS-CoV-2 followed by viral spread across entire PSC-HIOs, subsequently leading to organoid deterioration. However, SARS-CoV-2 spared goblet cells lacking ACE2 expression. Importantly, we challenged PSC-HIOs for drug testing capacity. Specifically, remdesivir effectively inhibited SARS-CoV-2 infection dose-dependently at low micromolar concentration and rescued PSC-HIO morphology.
Conclusions
Thus, PSC-HIOs are a valuable tool to study SARS-CoV-2 infection and to identify and validate drugs especially with potential action in the gut.
Keywords: SARS-CoV-2, COVID-19, Intestinal Organoids, Remdesivir, Famotidine
Abbreviations used in this paper: 3D, 3-dimensional; ACE2, angiotensin-converting enzyme 2; CHGA, chromogranin A; COVID-19, coronavirus disease 2019; DMEM, Dulbecco’s modified Eagle’s medium; FCS, fetal calf serum; HIO, human intestinal organoid; IC50, half-maximal inhibitory concentration; LYZ, lysozyme; MUC2, mucin 2; N, nucleocapsid; PFA, paraformaldehyde; PBS, phosphate-buffered saline; PSC-HIO, pluripotent stem cell derived human intestinal organoids; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; S, spike; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TCID50, Tissue culture infectious dose 50; TMPRSS2, transmembrane serine protease 2
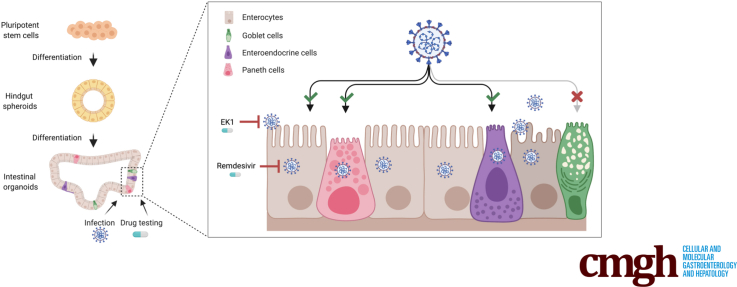
Graphical abstract
Summary.
Human pluripotent stem cell–derived intestinal organoids serve as an indefinite resource for organ-specific drug testing. SARS-CoV-2 infected and replicated within different cell types of the organoids, which was effectively inhibited by remdesivir and EK1 but not by famotidine.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects the respiratory tract with mostly mild symptoms, but up to 20% of patients develop severe pneumonia eventually followed by multiorgan failure and death.1 Intriguingly, up to 50% of patients present with gastrointestinal symptoms, associated with prolonged disease duration and increased severity.2, 3, 4, 5 Viral RNA is detected in rectal swabs and feces long after nasopharyngeal swabs tested negative.6,7 Infection of host cells with SARS-CoV-2 requires angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2). Both proteins mediate multiorgan tropism, as they are detected in esophagus, ileum, and colon.5,8, 9, 10, 11 The advent of organotypic cell culture systems provides a roadmap to successfully set up personalized medicine approaches. Organotypic cultures, also referred to as organoids, can be defined as 3-dimensional (3D) structures derived either from pluripotent or organ-restricted stem cells harboring the ability to mimic in vivo architecture and multilineage differentiation of terminally differentiated tissues.12, 13, 14, 15 Both model systems are derived from a cell source with unlimited self-renewal potentially allowing unlimited replenishments of the desired cell type or tissue such as the gut.16 While single-layered human intestinal organoids (HIOs) derived from human adult gut stem cells contain only epithelial cell types,17 pluripotent stem cells derived from HIOs (PSC-HIOs) comprise endodermal and mesodermal progeny18 resembling epithelium and fibroblasts or blood vessels of the gut, respectively.19 Thus, HIOs are less complex in architecture and lack in vivo transplantability as well as analytical access to developmental intermediate stages as compared with PSC-HIOs, while the latter are not fully mature.17, 18, 19 In turn, both models are comparable and complement each other with model-specific advantages and limitations. HIOs express ACE2 and are susceptible to SARS-CoV-2.20, 21, 22, 23 Although the exploration of new drugs is rapidly evolving, knowledge on their efficiency to inhibit intestinal infection of SARS-CoV-2 is at first unknown. However, drug testing might require a more complex organotypic culture system to reflect the true value of a given drug.24 Remdesivir is up to now the sole agent showing benefit on pulmonary phenotypes in coronavirus disease 2019 (COVID-19) patients.25 The histamine-2-blocker famotidine has been suggested to reduce severe COVID-19 course.26,27 Meanwhile, recent in silico predictions suggest that famotidine could interact with the catalytic site of 3 proteases (chymotrypsin-like, papain-like, and TMPRSS2 protease) involved in SARS-CoV-2 replication. Notably, famotidine binding affinity was low to these proteases, suggesting that additional interventions such as direct antivirals might be necessary to reach full efficiency.28 In contrast, a recent case series provided promising results by high-dose oral famotidine administration being well tolerated and associated with improved patient-reported outcomes in nonhospitalized patients with COVID-19.29 Here, we employ PSC-HIOs to study SARS-CoV-2 tropism with respect to distinct intestinal cell types and test such drug candidates in a human organotypic culture system resembling a natural 3D environment.
Results
ACE2 and TMPRSS2 Are Expressed in the Gastrointestinal Tract and in PSC-HIOs
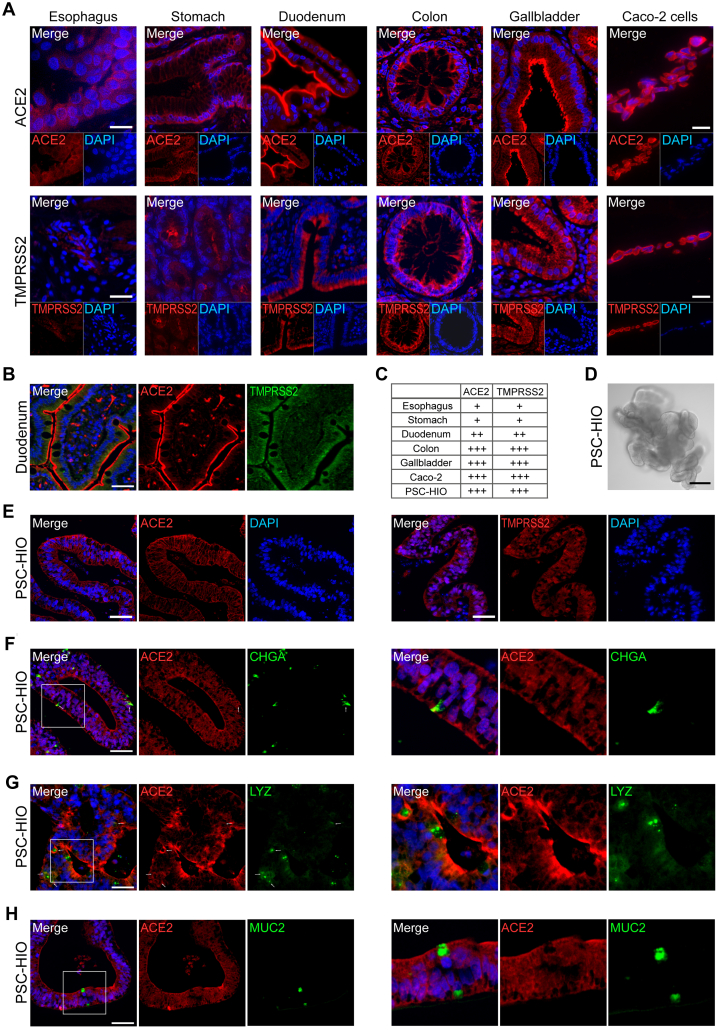
First, we studied expression of ACE2 and TMPRSS2 in organs of the gastrointestinal tract. Expression of both SARS-CoV-2 entry factors was most prominent in the epithelial lining of duodenum, gallbladder, and colon as compared with Caco-2 cells (Figure 1A–C). Duodenal cells showed apical ACE2 expression in the glycocalyx and a strong cytoplasmic TMPRSS2 signal (Figure 1A and B). Cells of gallbladder and colon presented a strong luminal expression of both proteins while sections of gastric mucosa and esophagus only revealed weak expression (Figure 1C). These findings suggest that various cell types in gastrointestinal tissues are SARS-CoV-2 target cells, though with different expression intensities of the entry factors. Next, we analyzed ACE2 and TMPRSS2 expression in in vitro differentiated PSC-HIOs (Figure 1D–H). Both proteins were readily expressed in virtually all intestinal cells present in PSC-HIOs (Figure 1E–H) mimicking expression patterns in human tissue17,18 and Caco-2 cells (Figure 1A and B). Moreover, co-staining for distinct intestinal cell type markers revealed that ACE2 is expressed on chromogranin A (CHGA)-positive enteroendocrine and lysozyme (LYZ)-positive Paneth cells (Figure 1F and G), while mucin 2 (MUC2)-positive goblet cells appeared virtually negative (Figure 1H). Thus, PSC-HIOs express cell-specifically SARS-CoV-2 entry factors and might be susceptible to infection.
Figure 1.
ACE2 and TMPRSS2 are expressed in the gastrointestinal tract and in PSC-HIOs. (A) Paraffin-embedded sections of indicated gastrointestinal tissues and embedded Caco-2 cells were stained for ACE2 and TMPRSS2 protein. Strong luminal expression is found in epithelia of duodenum, colon, and gallbladder, with weaker expression in the stomach and the mucosal lining of the esophagus. Representative images are shown. Scale bars = 20 μm. (B) Duodenum biopsy was stained for ACE2 and TMPRSS2 using primary antibodies and fluorophore-conjugated secondary antibodies and imaged by fluorescence microscopy. Scale bar = 50 μM. (C) ACE2 and TMPRSS2 expression from panel A was graded according to signal intensity (relative to the positive control, Caco-2 cells). (D) Microscopy of PSC-HIOs. Scale bar = 50 μM. (E) PSC-HIOs were stained for ACE2 (left panels) and TMPRSS2 (right panels). Nuclei are stained with DAPI in blue. (F–H) PSC-HIOs were costained for ACE2 and (F) enteroendocrine cell marker CHGA, (G) Paneth cell marker LYZ, or (H) goblet cell marker MUC2 showing coexpression with CHGA and LYZ but not with MUC2. Nuclei are stained with DAPI in blue. Arrows indicate co-expression. Left panels show representative images, right panels magnifications.
PSC-HIOs Support SARS-CoV-2 Infection and Replication
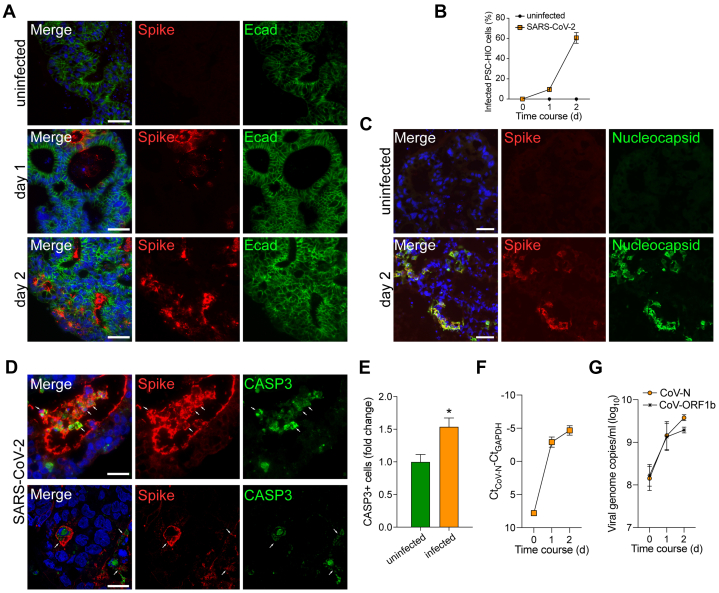
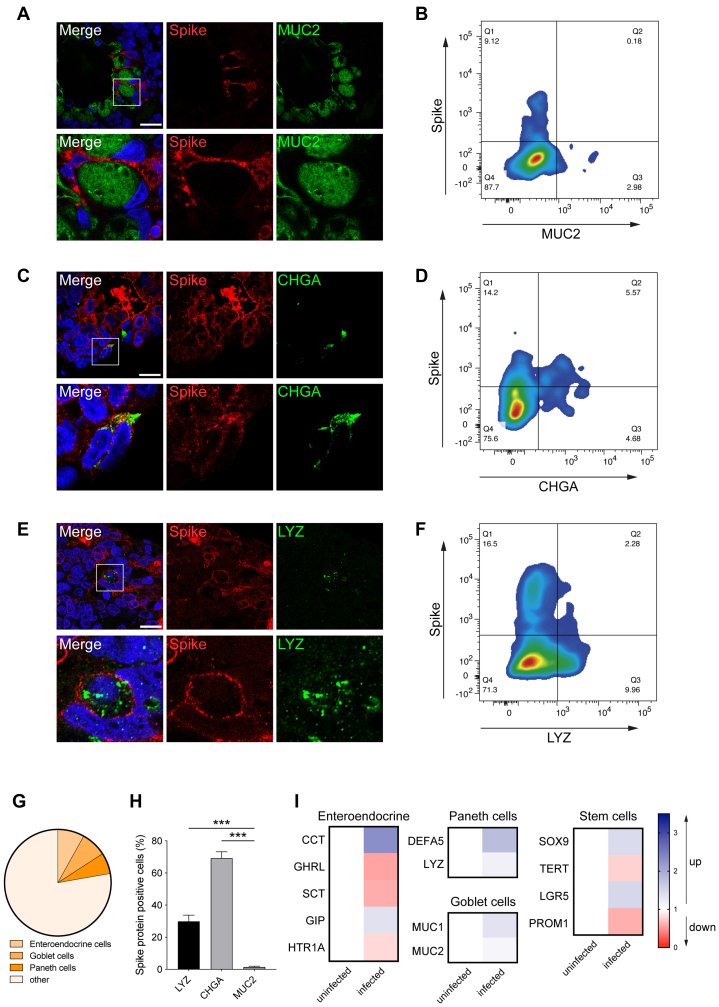
To determine susceptibility to in vitro infection, PSC-HIOs were exposed to SARS-CoV-2, and expression of viral spike (S) and nucleocapsid (N) protein together with the epithelial marker E-cadherin (Ecad) or respective labels for more specialized cell types (CHGA, LYZ, MUC2) were analyzed. Upon SARS-CoV-2 infection of PSC-HIOs, S protein was detected in 10% of the cells after 24 hours, which increased to 57% at 48 hours, suggesting viral replication and spreading infection (Figure 2A and B). Costaining for N protein confirmed this finding (Figure 2C). Notably, infected cells are positive for cleaved caspase 3 (CASP3) indicating initiated apoptosis in SARS-CoV-2-positive intestinal cells (Figure 2D and E). Increasing intra- and extracellular viral RNA levels in PSC-HIOs further corroborated progressive viral replication (Figure 2F and G). Productive viral replication in intestinal organoids was confirmed by determining infectious viral titers in the respective supernatants (Figure 2F, G). To probe cellular tropism upon SARS-CoV-2 infection of PSC-HIOs, we performed costaining of viral S protein with markers for distinct intestinal cell types. No expression of S protein was shown in goblet cells (MUC2+), despite being surrounded by S-positive cells, suggesting that they are not or only abortively infected (Figure 3A). In contrast, expression of S protein was detected in enteroendocrine (CHGA+) and Paneth cells (LYZ+) and in the vast majority of LYZ/CHGA-negative enterocytes (Figure 3C, E, and G). Results obtained by fluorescence microscopy of histological sections are matched by flow cytometric analysis of PSC-HIO cells after dissolution: no S/MUC2 double-positive cells were detected (Figure 3B), but 29% and 68% of label-positive cells (CHGA or LYZ) were also S protein positive, respectively (Figure 3D, F, and H). Surprisingly, reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analysis of whole organoid RNA revealed just a trend toward lower expression levels of most enteroendocrine hormones without a specific pattern in other cell types. Notably, MUC1/2 also remained virtually unchanged in infected PSC-HIOs (Figure 3I). Further experiments with longer follow-up postinfection need to determine cell type–specific consequences of SARS-CoV-2 infections. Taken together, these results show that SARS-CoV-2 productively infects most cell types in gut organoids, with the notable exception of goblet cells, an observation in contrast with, for example, astrovirus preferentially infecting actively secreting intestinal goblet cells.30
Figure 2.
PSC-HIOs support SARS-CoV-2 infection and replication. (A) PSC-HIOs were infected with SARS-CoV-2 and stained for viral S protein (red) and E-cadherin (Ecad) (green) at day 1 and day 2 postinfection. Nuclei are stained with DAPI in blue. Scale bar = 50 μM. (B) Infected cells of PSC-HIOs were manually counted according to presence of viral S protein compared with the total cell number (n = 3 independent experiments, at least 3 organoids counted per experiment). (C) Infection was further confirmed by S (red) and N (green) protein co-staining. Scale bar = 50 μM. (D) Infected cells of PSC-HIOs 2 days postinfection were stained for viral S protein (red) and CASP3 (green). Nuclei are stained with DAPI in blue. Scale bar = 50 μM. (E) Cleaved caspase 3 (CASP3)-positive cells were quantified by flow cytometry (n = 3). (F) Relative SARS-CoV-2 viral RNA copies in PSC-HIOs at different time points were quantified using primers amplifying CoV-N in RT-qPCR. For normalization, Ct values obtained for amplification of GAPDH from the same cultures were subtracted from the Ct value obtained for CoV-N (n = 2). (G) Viral genome copy number in extracellular matrix (matrigel) of infected PSC-HIOs was assessed by RT-qPCR targeting CoV-N and -ORF1b-nsp14 (n = 2). All error bars represent SEM.
Figure 3.
SARS-CoV-2 infects enteroendocrine and Paneth cells but not goblet cells. (A–F) Confocal microscopy (left column; A, C, E) and flow cytometric analysis (right column; B, D, F) of SARS-CoV-2 infected cells costained for S protein (red) and markers for goblet (green, MUC2), enteroendocrine (green, CHGA), and Paneth cells (green, LYZ). Scale bar = 20 μm. (G) Flow cytometry–based quantifications of the frequency of indicated cell types in intestinal organoids are shown. “Other” cells comprise mostly enterocytes and few stem cells.17,18 (H) Flow cytometric quantification of infected (S-positive) PSC-HIO cell types compared with total cell number (2 independent experiments with 2–3 biological replicates per experiment are shown; error bar represents SEM). (I) RT-qPCR quantification of enteroendocrine, Paneth cell–, goblet cell–, and stem cell–associated transcripts shown as fold change following infection by SARS-CoV-2 (compared with uninfected cells; 2 independent experiments with 2 biological replicates per experiments).
Drug Testing in PSC-HIOs
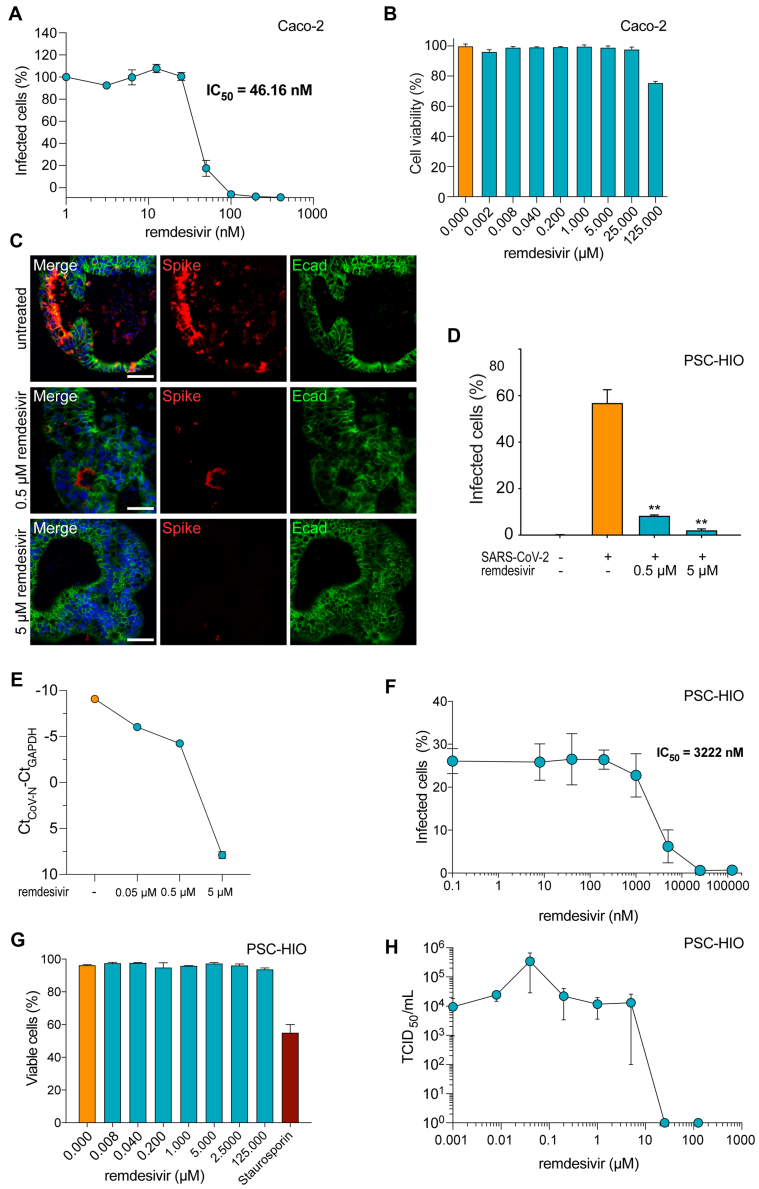
We next analyzed the effect of remdesivir on SARS-CoV-2 infection of intestinal cells. On Caco-2 cells, remdesivir inhibited viral infection in a dose-dependent manner with a half-maximal inhibitory concentration (IC50) of 46 nM (Figure 4A). Cell viability remained unaffected up to concentrations of 25 μM of the drug and was only marginally affected at 125 μM, resulting in a selectivity index value for remdesivir in colorectal Caco-2 cells of >2,717 (Figure 4A and B). Notably, remdesivir also decreased SARS-CoV-2 infection of PSC-HIOs by 86% at a concentration of 500 nM and almost completely abolished infection at 5 μM (Figure 4C and D). Accordingly, viral copies as measured via qPCR for CoV-N were strongly diminished at a dosage of 5 μM (Figure 4E). We also determined IC50 values by stepwise dose escalation of remdesivir in SARS-CoV-2–infected PSC-HIOs by flow cytometry analysis of N protein expression (Figure 4F). In contrast to the rather low IC50 value of 46 nM in monolayer cultured Caco-2 cells, the IC50 value in PSC-HIOs was ∼70-fold higher with 3.2 μM, indicating the importance of performing drug tests in a 3D culture format to avoid overestimations (Figure 4A and F). With an IC50 of 3.2 μM and a 50% cytotoxic concentration exceeding 125 μM of remdesivir on PSC-HIOs, however, the selectivity index is at least higher than 39, indicating good tolerability (Figure 4G). Tissue culture infection dose 50 (TCID50) determination of the supernatants derived from drug-treated infected PSC-HIOs confirmed a dose-dependent inhibition of SARS-CoV-2 replication (Figure 4H).
Figure 4.
Remdesivir inhibits SARS-CoV-2 infection of PSC-HIOs. (A) Caco-2 cells were infected with SARS-CoV-2 in the presence of increasing concentrations of remdesivir. Two days later, infection was quantified by an enzyme-based immunodetection assay against viral spike (S) protein. Values shown represent mean values derived from triplicate infections ± SEM from one representative experiment. Experiment was performed 3 times. (B) Effect of remdesivir on metabolic activity of Caco-2 cells was assessed by CellTiter-Glo assay. Shown are technical triplicates from one representative experiment. Error bars represent SEM. (C) PSC-HIOs were infected with SARS-CoV-2 and treated with indicated concentrations of remdesivir. Two days later, infection was analyzed by viral S protein and E-caherin (Ecad, green) staining. Nuclei are stained with DAPI in blue. Scale bar = 50 μM. Images are representative for at least 3 independent experiments. (D) Images from panel C were quantified according to presence of viral S protein compared with total cell number (n = 3 independent experiments, at least 3 organoids counted per experiment). (E) Two days postinfection, SARS-CoV-2 N was quantified in cellular RNA isolates of PSC-HIOs treated with either distinct dosages of remdesivir and normalized to GAPDH RNA. (n = 1, values represent means ± SEM of technical duplicates). (F) PSC-HIOs were infected with SARS-CoV-2 and treated with serial dilutions of remdesivir. Two days later, infection was analyzed by flow cytometric analysis against viral nucleocapsid protein. Data shown represent means ± SEM of 2 independent experiments, at least 2 biological replicates per experiment. (G) PSC-HIOs were treated with serial dilutions of remdesivir and relative cellular viability was assessed by annexin V staining. Data shown represent means ± SEM of 2 independent experiments, at least 2 biological replicates per experiment. (H) Infectious virus release in supernatant of PSC-HIOs treated with serial dilution of remdesivir. Supernatants were collected 2 days postinfection and infectious titer was determined by TCID50 endpoint titration (n = 2 independent experiments, at least 2 biological replicates per experiment; error bars represent SEM).
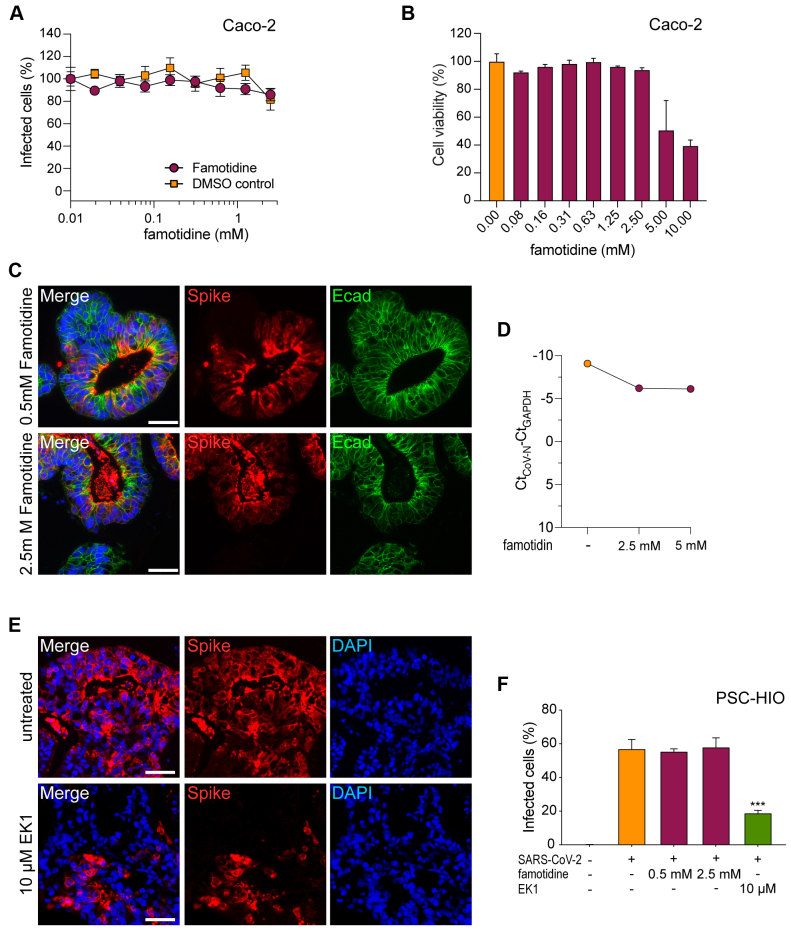
The over-the-counter histamine-2 receptor antagonist famotidine is a putative therapeutic agent for COVID-19.28,29 Thus, we tested famotidine concentrations of up to 2.5 mM but did not observe notable reduction of SARS-CoV-2 infection of Caco-2 cells (Figure 5A). In contrast to remdesivir, famotidine compromised Caco-2 cell viability starting from 5 mM upward (Figure 5B). Accordingly, PSC-HIOs did not show reduced infection rates after famotidine treatment, as assessed by immunostaining for viral S protein and CoV-N qPCR (Figure 5C, D, and F). Finally, we evaluated the antiviral activity of EK1, a recently described peptidic pan-coronavirus fusion inhibitor,31 in PSC-HIOs. In a previous study, we found EK1 to inhibit SARS-CoV-2 infection of Caco-2 with an IC50 of 303.5 nM, while not causing any cytotoxic effects up to 10 μM concentration (selectivity index > 32.9).32 Accordingly, in PSC-HIOs the presence of 10 μM EK1 during infection decreased the number of S-positive cells 48 hours after infection by 38% (Figure 5E and F). Taken together, SARS-CoV-2 productively infects most gastrointestinal cell types, with the notable exception of goblet cells, and remdesivir and the peptidic pan-coronavirus fusion inhibitor EK1 but not famotidine block SARS-CoV-2 infection of intestinal organoids.
Figure 5.
EK1 but not famotidine inhibit infection of PSC-HIOs with SARS-CoV-2. (A) Caco-2 cells were infected with SARS-CoV-2 and treated with famotidine at indicated concentrations. Two days later, infection was quantified by an enzyme-based immunodetection assay against viral S protein. Values shown represent mean values derived from triplicate infections ± SEM from one representative experiment. Experiment was performed 2 times. (B) Effect of famotidine on metabolic activity of Caco-2 cells was assessed by CellTiter-Glo assay. Values shown represent mean values derived from triplicates ± SEM from one representative experiment. (C–E) PSC-HIOs were infected with SARS-CoV-2 and treated with (C) famotidine or (E) EK1. Two days later, infection was analyzed by staining for viral S protein (red). Ecad (green) was additionally stained to visualize cells. Nuclei are stained with DAPI in blue. Scale bar = 50 μM. (D) SARS-CoV-2 N was quantified in cellular RNA isolates of PSC-HIOs treated with either distinct dosages of famotidine and normalized to GAPDH RNA (n = 1, values represent means ± SEM of technical duplicates). (F) Images from panels C and E were quantified according to the presence of viral S protein compared with total cell number (n = 2–3 independent experiments, at least 3 organoids counted per experiment; values represent mean ± SEM).
Discussion
We report that in vitro differentiated PSC-HIOs express ACE2 and TMPRSS2 and support productive infection with SARS-CoV-2 in a cell type-specific manner, launching a valuable pathomechanistic model for SARS-CoV-2 infection of the gut. Notably, remdesivir and EK1 but not famotidine block SARS-CoV-2 in PSC-HIOs, indicating suitability to treat gastrointestinal COVID-19 and validating PSC-HIOs as a putative screening platform to test drugs that might ameliorate gastrointestinal symptoms in the clinic.
A study by Lamers et al20 recently demonstrated SARS-CoV-2 infection of enterocytes in small intestinal organoids from primary gut epithelial cells. Similarly, recent studies show infection of and replication in epithelial cells of human colon-derived intestinal organoids22 as well as human and bat small intestinal organoids.23 We extended these studies by specifically showing infection of specialized intestinal cells (e.g., enteroendocrine and Paneth cells) in addition to enterocytes, suggesting that SARS-CoV-2 infection may not only disrupt absorption and transport of metabolites, but also hamper hormone secretion and local immune defense. Similarly, infection of Paneth cells may support further disturbance of host defense by impeding secretion of antiviral and antimicrobial proteins into the gut. However, qPCR analysis of cell type–specific markers did not reveal conclusive data, potentially influenced by the sampling time point. Especially considering the recently highlighted relevance of mucosal SARS-CoV-2 defense to fight COVID-19,33 our findings of cell type–specific mucosal SARS-CoV-2 infection add information about SARS-CoV-2 tropism. Our infection pattern of SARS-CoV-2 sparing goblet cells contrasts with lung findings from deceased COVID-19 patients, in which MUC5AC-positive goblet cells as well as MUC5B-positive club-like cells have been infected.34 These tissue-specific differences should be subject to further investigation.
As COVID-19 patients suffering from gastrointestinal symptoms show prolonged disease duration and severity of disease, cells such as enteroendocrine cells, Paneth cells, and goblet cells are particularly relevant for local immune defense; thus, drugs that suppress intestinal SARS-CoV-2 infection are of high interest. Remdesivir, developed to target Ebola virus, provides the first antiviral attempt to treat COVID-19, although clinical success appears heterogeneous and benefiting subpopulations remain to be defined.25,35 Similarly, effectivity of this agent across distinct organs is completely unclear. We here show that remdesivir inhibits SARS-CoV-2 infection of and replication in PSC-HIOs, highlighting its possible use for treatment of gastrointestinal infection occurring simultaneously with or independently of respiratory manifestation of COVID-19. In addition, head-to-head comparison of IC50 values determined in a 3D vs 2-dimensional culture system underpins the relevance of screening system, as 2-dimensional cultures are likely overestimating efficiency of a given drug.
In contrast, famotidine, proposed to inhibit proteins essential for viral replication,26 did not affect SARS-CoV-2 infection and viral spread in a colon cell line and PSC-HIOs. However, our current treatment regime does not allow to determine long-term effects, which may play a role during clinical deterioration. This would be in line with low binding affinity of famotidine to SARS-CoV-2 proteases28; however, its effect on host-specific immune response as well as tissue-specific efficiency might be of relevance to ultimately judge its clinical value, particularly to treat life-limiting lung infection. In line, histamine-2 receptor antagonists are able to modulate histamine effector pathways by receptor binding, thus directing immunomodulatory effects.36 Specifically, they can boost the innate immune function and revert negative effects of histamine on production and release of cytokines such as tumor necrosis factor alpha and interferon alpha, possibly supporting immune response during viral infection.37,38 Accordingly, a recent case series suggested efficiency to treat mild-to-moderate COVID-19 with pulmonary parameters as a readout.29 Thus, the current clinical study landscape remains in support of famotidine,26 although our in vitro data appear contrary and warrant further investigation in randomized controlled trials, best with taking gastrointestinal facets of COVID-19 into account.
Conclusions
Overall, our data demonstrate that SARS-CoV-2 can infect all investigated cell types of the gut, with the exception of goblet cells, and can perturb intestinal integrity, which might lead to diarrhea among many other gastrointestinal symptoms frequently reported by COVID-19 patients. Most importantly, we tested and validated our culture platform as an indefinite resource to organ-specific drug testing, as shown by proof-of-concept studies with remdesivir and EK1, emphasizing the relevance of the system as a potential tool for drug screening. Treatment with remdesivir during in vitro infection of intestinal organoids with SARS-CoV-2 inhibited viral replication. Hence, clinical treatment with this agent might prevent SARS-CoV-2–induced gastrointestinal damage and soothe gastrointestinal symptoms.
Materials And Methods
Drugs
Remdesivir was obtained from Selleck Chemicals (#S8932; Houston, TX), famotidine was obtained from Sigma-Aldrich (#F6889-1G; St. Louis, MO), and EK1 was synthesized by the Core Facility Functional Peptidomics, Ulm University Medical Center.
Cell Culture
Caco-2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 units/mL penicillin, 100 μg/mL streptomycin (Sigma-Aldrich), 2 mM L-glutamine, 1 mM sodium pyruvate, and NEAA (#M7145; Sigma-Aldrich). Vero E6 cells were grown in the same medium but with 2.5% FCS. TMPRSS2-expressing Vero E6 cells (kindly provided by the National Institute for Biological Standards and Control, Ridge, United Kingdom; #100978) were cultivated in DMEM supplemented with 10% FCS, 2 mM L-glutamine, 100-units/mL penicillin, 100 mg/mL streptomycin, and 1 mg/mL geneticin. Cells were grown at 37°C in a 5% CO2 humidified incubator.
Stem Cell Culture and Intestinal Differentiation
Human embryonic stem cell line HUES8 (Harvard University, Cambridge, MA) was used with permission from the Robert Koch Institute according to the “79. Genehmigung nach dem Stammzellgesetz, AZ 3.04.02/0084.” Cells were cultured on human embryonic stem cell matrigel (Corning, Corning, NY) in mTeSR1 medium (STEMCELL Technologies, Vancouver, Canada) at 5% CO2, 5% O2, and 37°C. Medium was changed daily and cells were split with TrypLE Express (Invitrogen, Carlsbad, CA). For differentiation, 300,000 cells per well were seeded in 24-well plates coated with growth factor–reduced matrigel (Corning) in mTeSR1 with 10-μM Y-27632 (STEMCELL Technologies). The next day, differentiation was started at 80%-90% confluency, as reported previously.13,18,39, 40, 41
Virus Strains and Virus Propagation
Viral isolate BetaCoV/Netherlands/01/NL/2020 (#010V-03903) was obtained from the European Virus Archive global and propagated on Vero E6 cells. To this end, 70%-90% confluent cells in 75 cm2 cell culture flasks were inoculated with 100-μL SARS-CoV-2 isolate in 3.5-mL serum-free medium containing 1-μg/mL trypsin. Cells were incubated for 2 hours at 37°C, before adding 20-mL medium containing 15-mM HEPES. Cells were incubated at 37°C and supernatant harvested when a strong cytopathic effect was visible. Supernatants were centrifuged for 5 minutes at 1,000 g to remove cellular debris, and then aliquoted and stored at -80°C as virus stocks. Infectious virus titer was determined as plaque-forming units on Vero E6 cells, which was used to calculate multiplicity of infection.
In Vitro Infection and Cell-Based SARS-CoV-2 Immunodetection Assay
To determine SARS-CoV-2 infection, 30,000 Caco-2 cells were seeded in 96-well plates. The next day, the compound of interest was added and the cells were infected with a multiplicity of infection of 0.001 in a total volume of 180 μL. Two days later immunodetection assay was performed as previously described32; cells were fixed in 4% paraformaldehyde (PFA), permeabilized using 0.1% Triton-X, and stained with 1:5000 diluted anti-SARS-CoV-2 S protein antibody 1A9 (Biozol GTX-GTX632604) in antibody buffer (10% FCS and 0.3% Tween 20 in phosphate-buffered saline [PBS]) for 1 hour at 37°C. After 3 washes, the secondary horseradish peroxidase–conjugated antibody (#A16066; Thermo Fisher Scientific, Waltham, MA) (1:15,000) was incubated for 1 hour at 37°C. Following 4 times of washing, the TMB peroxidase substrate (#52-00-04; Medac, North Augusta, SC) was added for 5 minutes and the reaction stopped using 0.5 M H2SO4. The optical density was recorded at 450–620 nm using the Asys Expert 96 UV microplate reader (Biochrom, Cambridge, United Kingdom). Values were corrected for the background signal derived from uninfected cells and untreated controls were set to 100% infection.
In Vitro Cell Viability Assay
The effect of analyzed compounds on the metabolic activity of Caco-2 cells was analyzed using the CellTiter-Glo Luminescent Cell Viability Assay (#G7571; Promega, Madison, WI) as previously described.
Infection of Organoids and Drug Testing
To prepare in vitro differentiated organoids for infection, matrigel was dissolved in Collagenase/Dispase (Roche, Basel, Switzerland) for 2 hours at 37°C and stopped by cold neutralization solution (DMEM, 1% bovine serum albumin, and 1% penicillin-streptomycin). Organoids were transferred into 1.5-mL tubes and infected in 350-μL virus inoculum containing 3 × 105 plaque-forming units SARS-CoV-2 with or without drugs for 1 hour at 37°C. Organoids were then resuspended in 35-μL cold growth factor–reduced matrigel to generate cell-matrigel domes in 48-well plates. After 10 minutes at 37°C, intestinal growth medium (DMEM F12 [Gibco, Gaithersburg, MD], 1× B27 supplement [Thermo Fisher Scientific], 2-mM L-glutamine, 1% penicillin-streptomycin, 40 mM HEPES [Sigma-Aldrich], 3 μM CHIR99021, 200 nM LDN-193189 [Sigma-Aldrich], 100 ng/mL hEGF [Novoprotein, Summit, NJ], and 10 μM Y-27632 [STEMCELL Technologies]) was added and organoids were incubated at 37°C.
Flow Cytometry Analysis of Organoids
After infection and drug treatment, matrigel was dissolved in Collagenase/Dispase (Roche) for 2 hours at 37°C and stopped by cold neutralisation solution (DMEM, 1% bovine serum albumin, and 1% penicillin-streptomycin). Single cells were generated by incubation with accutase (STEMCELL Technologies) for 30 minutes at 37°C. Cells were fixed in 4% PFA for 1 hour. For flow cytometry analysis, cells were blocked in 5% normal donkey serum in 0.1% Triton-X in PBS for 30 minutes on ice. Cells were incubated with primary antibodies diluted in blocking solution over night at 4°C. After washing twice with 2% normal donkey serum in 0.1% Triton-X in PBS, cells were incubated with secondary antibodies (Alexa Fluor IgG H+L [Invitrogen]; 1:500) in blocking solution for 1.5 hours on ice in the dark. Cells were washed twice and filtered, and fluorescence was measured using a BD LSR II flow cytometer (BD Biosciences, San Jose, CA).
Annexin V Viability Assay
Organoids were treated with drugs for 48 hours. Matrigel was dissolved in Collagenase/Dispase (Roche) for 2 hours at 37°C and stopped by cold neutralization solution (DMEM, 1% bovine serum albumin, and 1% penicillin-streptomycin). Single cells were generated by incubation with accutase (STEMCELL Technologies) for 30 minutes at 37°C. Cells were stained using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to manufacturer’s instructions and analyzed using a BD LSR II Flow cytometer (BD Biosciences).
Isolation of RNA From Matrigel and Organoids for RT-qPCR
Viral RNA from matrigel of infected organoids or organoids themselves was isolated using the Qiagen Viral RNA Mini Kit (#52906; Qiagen, Hilden, Germany) or Qiagen RNeasy Plus Mini Kit (#74136; Qiagen), respectively, both as described by the manufacturer. For this purpose, matrigel was digested as described previously, mixed with AVL buffer and incubated at room temperature for 20 minutes before freezing at -20°C. Similarly, matrigel was dissolved and removed and organoids were lysed in 600-μL RLT Plus buffer containing 1% β-mercaptoethanol, vortexed for 30 seconds and then frozen at -20°C until analysis. Organoid lysates were homogenized using QIAshredder (#79656; Qiagen).
Reverse-Transcription qPCR
RT-qPCR from matrigel was performed with primer sets targeting N (nucleoprotein) and ORF1b-nsp14 (ORF1b)42 as previously described43 using TaqMan Fast Virus 1-Step Master Mix (#4444436; Thermo Fisher Scientific) and a StepOnePlus Real-Time PCR System (96-well format, fast mode; Thermo Fisher Scientific). Synthetic SARS-CoV-2 RNA (#102024 [Twist Bioscience, San Francisco, CA] or #VR-3276SD [American Type Culture Collection, Manassas, VA]) was used as a standard to obtain viral copy numbers. For RNA isolated from organoids, GAPDH was analyzed as an endogenous control (#4310884E; Applied Biosystems, Foster City, CA) and resulting Ct values subtracted from those obtained for CoV-N reactions for the same samples. All reactions were run in duplicates.
For RT-qPCR of intestinal cell type transcripts, RNA was transcribed into cDNA using the iScript cDNA synthesis Kit (170-8891; Bio-Rad, Hercules, CA). RT-qPCR was performed using Qiagen QuantiTect Primer Assays and SensiMix SYBR No Rox Kit (QT650-02; Bioline, London, United Kingdom) in 96-well format. Expression was normalized to hydroxymethylbilane synthase.
TCID50 Endpoint Titration
To determine the TCID50, supernatant samples were taken at day 0 and day 2. Samples were serially diluted and used to inoculate target cells. To this end, 20,000 Vero E6 or TMPRSS2-expressing Vero E6 cells were seeded per well in 96 flat-bottom well plates in 100 μL of medium and incubated over night before 62 μL of fresh medium was added. Next, 18 μL of 5-fold titrated sample was used for inoculation in triplicates. Cells were then incubated for 6 days and monitored for cytopathic effect. TCID50/mL was calculated according to Reed and Muench.44
Histology of Organoids and Tissue Sections
Sections of human gastrointestinal tissues were provided by the pathology department of Ulm University. Experiments were conducted in accordance with guidelines of the Ethics Committee of the Federal General Medical Council and approved by the Ethics Committee of Ulm University. For histological examination of organoids, they were fixed in 4% PFA over night at 4°C, washed with PBS, and pre-embedded in 2% agarose (Sigma-Aldrich) in PBS. After serial dehydration, intestinal organoids were embedded in paraffin, sectioned at 4 μm, deparaffinized, rehydrated, and subjected to heat-mediated antigen retrieval in tris buffer (pH 9) or citrate buffer (pH 6). Tissue was permeabilized with 0.5% Triton-X for 30 minutes at room temperature and stained overnight with primary antibodies (Table 1) in antibody diluent (ZYTOMED Systems, Berlin, Germany) in a wet chamber at 4°C. After washing with PBS with Tween 20, slides were incubated with secondary antibodies (Alexa Fluor IgG H+L; 1:500) and 500-ng/mL DAPI in Antibody Diluent for 90 minutes in a wet chamber at room temperature. After washing with PBS with Tween 20 and water, slides were mounted with Fluoromount-G (Southern Biotech, Birmingham, AL). Negative controls were performed using IgG controls or irrelevant polyclonal serum (anti-mycobacterium tuberculosis) for polyclonal antibodies, respectively. Absence of background staining confirmed specificity of the primary antibodies. Images were acquired with Keyence BZ 9000 microscope (Keyence, Osaka, Japan) and quantified with ImageJ (Version 2.0.0, National Institutes of Health, Bethesda, MD). Laser Scanning confocal images were acquired using the Zeiss LSM710. Z-stacks were deconvoluted and edited in Huygens (Scientific Volume Imaging, Hilversum, the Netherlands) and Fiji (ImageJ).
Table 1.
Primary Antibodies
| Antigen | Species | Cat. No. | Company | Dilution |
|---|---|---|---|---|
| ACE2 | rabbit | ab15348 | Abcam | 1:1500 |
| ACE2 (used for costainings) | mouse | sc390851 | Santa Cruz | 1:100 |
| SARS-CoV-2 spike | mouse | GTX632604 | Biozol | 1:500 |
| TMPRSS2 | rabbit | ab92323 | Abcam | 1:250 |
| CASP3 | rabbit | 9664 | Cell Signaling | 1:1000 |
| CHGA | rabbit | A0430 | DAKO | 1:500 |
| MUC2 | rabbit | sc-7314 | Santa Cruz | 1:200 |
| LYZ | rabbit | ab108508 | Abcam | 1:1000 |
| Ecad | rabbit | 24E10 | Cell Signaling | 1:200 |
| SARS-CoV-2 nucleocapsid | mouse | 40143 | Sino Biologicals | 1:500 |
ACE2, angiotensin-converting enzyme 2; CASP3, caspase 3; CHGA, chromogranin A; Ecad, E-cadherin; LYZ, lysozyme; MUC2, mucin 2; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane serine protease 2.
Statistics
Statistics were performed using GraphPad Prism (GraphPad Software, San Diego, CA). IC50 was determined by nonlinear regression (inhibitor) vs normalized response. For Figure 3G, 4D, and 5D 1-way analysis of variance and Dunnett’s multiple comparison test were performed. Data of Figure 2E were analyzed with a paired t test. P values <.05 were classified as significant (∗P < .05, ∗∗P < .01, ∗∗∗ P < .001, ∗∗∗∗P < .0001).
Acknowledgments
The authors thank Katrin Köhn, Aref Saed, Daniela Krnavek, Nicola Schrott, and Juliane Nell for their excellent technical assistance and Kanishka Tiwary and Karolin Walther for providing resources. They also thank Markus Breunig, Michael Melzer, Jessica Merkle, and Meike Hohwieler for helpful discussions. The graphical abstract was created with biorender.com.
CRediT Authorship Contributions
Jana Krüger (Data curation: Equal; Formal analysis: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Rüdiger Groß (Data curation: Equal; Formal analysis: Equal; Writing – original draft: Supporting; Writing – review & editing: Equal)
Carina Conzelmann (Data curation: Equal; Formal analysis: Equal; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Janis A. Müller (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Lennart Koepke (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting)
Konstantin M. J. Sparrer (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Equal)
Tatjana Weil (Data curation: Supporting)
Desiree Schütz (Data curation: Supporting)
Thomas Seufferlein (Writing – review & editing: Supporting)
Thomas F.E. Barth (Formal analysis: Equal; Resources: Supporting; Writing – review & editing: Supporting)
Steffen Stenger (Resources: Supporting) Sandra Heller, Dr. (Supervision: Supporting; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Jan Münch (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Lead)
Alexander Kleger (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding Rüdiger Groß, Carina Conzelmann, Lennart Koepke, Tatjana Weil, and Jana Krüger are part of and Rüdiger Groß is funded by a scholarship from the International Graduate School in Molecular Medicine Ulm. This work was supported by the European Union’s Horizon 2020 research and innovation programme (Fight-nCoV, 101003555 to Jan Münch), by grants from the MWK Baden-Württemberg (to Janis A. Müller, Jan Münch, and Alexander Kleger), and by the DFG (CRC1279 to Steffen Stenger and Jan Münch). The project also received relevant funding within the 'Fokus-Förderung COVID-19' to Alexander Kleger (KL 2544/8-1) and Jan Münch (MU 3115/14-1). Additional funding came from the Deutsche Forschungsgemeinschaft (DFG) 'Sachbeihilfe' (KL 2544/5-1,7-1) and 'Heisenberg-Programm' (KL 2544/6-1). Alexander Kleger is a Principal Investigator of the HEIST RTG funded by the DFG GRK 2254/1 and a Else-Kröner-Fresenius Excellence fellow.
Contributor Information
Jan Münch, Email: jan.muench@uni-ulm.de.
Alexander Kleger, Email: alexander.kleger@uni-ulm.de.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei X.-S., Wang X., Niu Y.-R., Ye L.-L., Peng W.-B., Wang Z.-H., Yang W.-B., Yang B.-H., Zhang J.-C., Ma W.-L. Diarrhea is associated with prolonged symptoms and viral carriage in COVID-19. Clin Gastroenterol Hepatol. 2020;18:1753–1759.e2. doi: 10.1016/j.cgh.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo S., Zhang X., Xu H. Don't overlook digestive symptoms in patients with 2019 novel coronavirus disease (COVID-19) Clin Gastroenterol Hepatol. 2020;18:1636–1637. doi: 10.1016/j.cgh.2020.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F., Wang H., Fan J., Zhang Y., Wang H., Zhao Q. Pancreatic injury patterns in patients with coronavirus disease 19 pneumonia. Gastroenterology. 2020;159:367–370. doi: 10.1053/j.gastro.2020.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai C.-C., Ko W.-C., Lee P.-I., Jean S.-S., Hsueh P.-R. Extra-respiratory manifestations of COVID-19. Int J Antimicrob Agents. 2020;56:106024. doi: 10.1016/j.ijantimicag.2020.106024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y., Freedberg D.E., Kirtane A.J., Parikh S.A., Maurer M.S., Nordvig A.S., Accili D., Bathon J.M., Mohan S., Bauer K.A., Leon M.B., Krumholz H.M., Uriel N., Mehra M.R., Elkind M.S.V., Stone G.W., Schwartz A., Ho D.D., Bilezikian J.P., Landry D.W. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., Braun F., Lu S., Pfefferle S., Schroder A.S., Edler C., Gross O., Glatzel M., Wichmann D., Wiech T., Kluge S., Pueschel K., Aepfelbacher M., Huber T.B. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liebau S., Russ H.A., Kleger A. Stem cell derived organoids in human disease and development. Stem Cells Int. 2019;2019:7919427. doi: 10.1155/2019/7919427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohwieler M., Renz S., Liebau S., Lin Q., Lechel A., Klaus J., Perkhofer L., Zenke M., Seufferlein T., Illing A. “Miniguts” from plucked human hair meet Crohn’s disease. Z Gastroenterol. 2016;54:748–759. doi: 10.1055/s-0042-105520. [DOI] [PubMed] [Google Scholar]
- 14.Huang L., Bockorny B., Paul I., Akshinthala D., Frappart P.O., Gandarilla O., Bose A., Sanchez-Gonzalez V., Rouse E., Lehoux S., Pandell N., Lim C., Clohessy J.G., Grossman J.E., Gonzalez R.S., Perea S., Daaboul G., Sawhney M., Freedman S.D., Kleger A., Cummings R.D., Emili A., Muthuswamy L., Hidalgo M., Muthuswamy S. PDX-derived organoids model in vivo drug response and secrete biomarkers. JCI Insight. 2020;5:135544. doi: 10.1172/jci.insight.135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkhofer L., Frappart P.O., Müller M., Kleger A. Importance of organoids for personalized medicine. Person Med. 2018;15:461–465. doi: 10.2217/pme-2018-0071. [DOI] [PubMed] [Google Scholar]
- 16.Clevers H. COVID-19: organoids go viral. Nat Rev Mol Cell Biol. 2020;21:355–356. doi: 10.1038/s41580-020-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 18.Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., Shroyer N.F., Wells J.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holloway E.M., Wu J.H., Czerwinski M., Sweet C.W., Wu A., Tsai Y.H., Huang S., Stoddard A.E., Capeling M.M., Glass I., Spence J.R. Differentiation of human intestinal organoids with endogenous vascular endothelial cells. Dev Cell. 2020;54:516–528.e7. doi: 10.1016/j.devcel.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B., van Schayck J.P., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanifer M.L., Kee C., Cortese M., Zumaran C.M., Triana S., Mukenhirn M., Kraeusslich H.-G., Alexandrov T., Bartenschlager R., Boulant S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 24.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 25.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., ACTT-1 Study Group Members Remdesivir for the treatment of Covid-19 - preliminary report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freedberg D.E., Conigliaro J., Wang T.C., Tracey K.J., Callahan M.V., Abrams J.A., Sobieszczyk M.E., Markowitz D.D., Gupta A., O’Donnell M.R. Famotidine use is associated with improved clinical outcomes in hospitalized COVID-19 patients: a propensity score matched retrospective cohort study. Gastroenterology. 2020;159:1129–1131.e3. doi: 10.1053/j.gastro.2020.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sethia R., Prasad M., Jagannath S., Nischal N., Soneja M., Garg P., Shalimar Efficacy of famotidine for COVID-19: a systematic review and meta-analysis. medRxiv. https://www.medrxiv.org/content/10.1101/2020.09.28.20203463v1 Available at: Accessed October 22, 2020.
- 28.Ortega J.T., Serrano M.L., Jastrzebska B. Class A G protein-coupled receptor antagonist famotidine as a therapeutic alternative against SARS-CoV2: an in silico analysis. Biomolecules. 2020;10:954. doi: 10.3390/biom10060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janowitz T., Gablenz E., Pattinson D., Wang T.C., Conigliaro J., Tracey K., Tuveson D. Famotidine use and quantitative symptom tracking for COVID-19 in non-hospitalised patients: a case series. Gut. 2020;69:1592–1597. doi: 10.1136/gutjnl-2020-321852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortez V., Boyd D.F., Crawford J.C., Sharp B., Livingston B., Rowe H.M., Davis A., Alsallaq R., Robinson C.G., Vogel P., Rosch J.W., Margolis E., Thomas P.G., Schultz-Cherry S. Astrovirus infects actively secreting goblet cells and alters the gut mucus barrier. Nature Communications. 2020;11:2097. doi: 10.1038/s41467-020-15999-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conzelmann C., Gilg A., Groß R., Schütz D., Preising N., Ständker L., Jahrsdörfer B., Schrezenmeier H., Sparrer K.M., Stamminger T. An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antiviral Res. 2020;181:104882. doi: 10.1016/j.antiviral.2020.104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., Kashentseva E., McCune B.T., Bailey A.L., Zhao H., VanBlargan L.A., Dai Y.N., Ma M., Adams L.J., Shrihari S., Danis J.E., Gralinski L.E., Hou Y.J., Schafer A., Kim A.S., Keeler S.P., Weiskopf D., Baric R.S., Holtzman M.J., Fremont D.H., Curiel D.T., Diamond M.S. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos da Silva S., Ju E., Meng W., Paniz Mondolfi A.E., Dacic S., Green A., Bryce C., Grimes Z., Fowkes M.E., Sordillo E.M., Cordon-Cardo C., Guo H., Gao S.-J. Broad SARS-CoV-2 cell tropism and immunopathology in lung tissues from fatal COVID-19. medRxiv. https://www.medrxiv.org/content/10.1101/2020.09.25.20195818v1 Available at: Accessed October 22, 2020. [DOI] [PMC free article] [PubMed]
- 35.WHO Solidarity Consortium. Pan H., Peto R., Karim Q.A., Alejandria M., Restrepo A.M.H., Garcia C.H., Kieny M.P., Malekzadeh R., Murthy S., Preziosi M.-P., Reddy S., Periago M.R., Sathiyamoorthy V., Røttingen J.A., Swaminathan S. Repurposed antiviral drugs for COVID-19; interim WHO SOLIDARITY trial results. medRxiv. https://www.medrxiv.org/content/10.1101/2020.10.15.20209817v1 Available at: Accessed October 22, 2020.
- 36.Hahm K., Kim W.H., Lee S., Rang J., Park I. Comparison of immunomodulative effects of the histamine-2 receptor antagonists cimetidine, ranitidine, and famotidine on peripheral blood mononuclear cells in gastric cancer patients. Scand J Gastroenterol. 1995;30:265–271. doi: 10.3109/00365529509093275. [DOI] [PubMed] [Google Scholar]
- 37.Morichika T., Takahashi H.K., Iwagaki H., Yoshino T., Tamura R., Yokoyama M., Mori S., Akagi T., Nishibori M., Tanaka N. Histamine inhibits lipopolysaccharide-induced tumor necrosis factor-α production in an intercellular adhesion molecule-1-and B7. 1-dependent manner. J Pharmacol Exp Ther. 2003;304:624–633. doi: 10.1124/jpet.102.042515. [DOI] [PubMed] [Google Scholar]
- 38.Smolinska S., Groeger D., Perez N.R., Schiavi E., Ferstl R., Frei R., Konieczna P., Akdis C.A., Jutel M., O'Mahony L. Histamine receptor 2 is required to suppress innate immune responses to bacterial ligands in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2016;22:1575–1586. doi: 10.1097/MIB.0000000000000825. [DOI] [PubMed] [Google Scholar]
- 39.Fischer S., Uckert A.K., Landenberger M., Papatheodorou P., Hoffmann-Richter C., Mittler A.K., Ziener U., Hagele M., Schwan C., Muller M., Kleger A., Benz R., Popoff M.R., Aktories K., Barth H. Human peptide alpha-defensin-1 interferes with Clostridioides difficile toxins TcdA, TcdB, and CDT. FASEB J. 2020;34:6244–6261. doi: 10.1096/fj.201902816R. [DOI] [PubMed] [Google Scholar]
- 40.di Masi A., Leboffe L., Polticelli F., Tonon F., Zennaro C., Caterino M., Stano P., Fischer S., Hagele M., Muller M., Kleger A., Papatheodorou P., Nocca G., Arcovito A., Gori A., Ruoppolo M., Barth H., Petrosillo N., Ascenzi P., Di Bella S. Human serum albumin is an essential component of the host defense mechanism against Clostridium difficile intoxication. J Infect Dis. 2018;218:1424–1435. doi: 10.1093/infdis/jiy338. [DOI] [PubMed] [Google Scholar]
- 41.Ernst K., Schmid J., Beck M., Hagele M., Hohwieler M., Hauff P., Uckert A.K., Anastasia A., Fauler M., Jank T., Aktories K., Popoff M.R., Schiene-Fischer C., Kleger A., Muller M., Frick M., Barth H. Hsp70 facilitates trans-membrane transport of bacterial ADP-ribosylating toxins into the cytosol of mammalian cells. Sci Rep. 2017;7:2724. doi: 10.1038/s41598-017-02882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu D.K., Pan Y., Cheng S.M., Hui K.P., Krishnan P., Liu Y., Ng D.Y., Wan C.K., Yang P., Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groß R., Conzelmann C., Müller J., Stenger S., Steinhart K., Kirchhoff F., Münch J. Detection of SARS-CoV-2 in human breast milk. Lancet. 2020;395:1757–1758. doi: 10.1016/S0140-6736(20)31181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints12. Am J Epidemiol. 1938;27:493–497. [Google Scholar]