Abstract
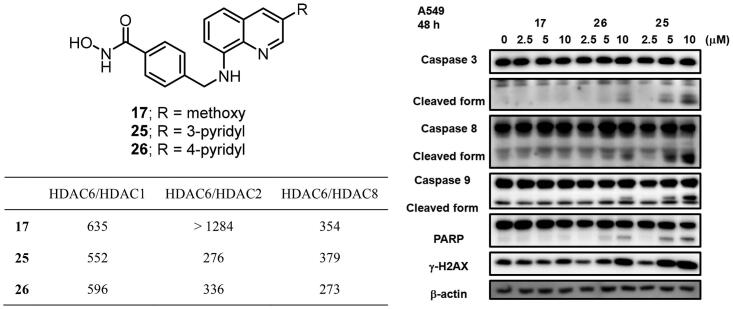
A series of 3-subsituted quinolinehydroxamic acids has been synthesised and evaluated for their effect on human lung cancer cell line (A549), human colorectal cancer cell line (HCT116) and HDAC isoforms 1, 2, 6, and 8. The results indicated that substitution at C3 of quinoline is favoured for HDAC6 selectivity. Two compounds (25 and 26) were also found to be potent anti-proliferative compounds with IC50 values ranging from 1.29 to 2.13 µM against A549 and HCT116 cells. These compounds displayed remarkable selectivity for HDAC6 over other HDAC isoforms with nanomolar IC50 values. Western blot analysis revealed that compounds of this series activate apoptotic caspase pathway as indicated by cleavage of caspase 3, 8, and 9 and also increase phosphorylated H2AX thus inducing DNA double strand fragmentation in a concentration dependent manner. Flow cytometric analysis also displayed a dose dependent increase of cell population in sub G1 phase.
Keywords: Quinoline, HDAC, lung cancer, colon cancer, hydroxamic acid, acrylamide
Graphical Abstract
1. Introduction
Over last two decades Epigenetics have emerged as a potential target for the treatment of cancer and various other physiological disorders. Gene expression regulation by epigenetic modifications play pivotal role and thus have drawn a lot interest for the development of therapeutics capable of altering gene expressions by modulating post-translational changes in histone e.g. acetylation, methylation etc. Histone post-translational modifications act as regulatory marks which are important for the control of transcription and chromatin architecture. Chromatin is a compact structure of nucleosomes and is formed from DNA-wrapped histone proteins. Two enzymes, histone acetylase (HAT) and histone deacetylase (HDAC) together control the acetylation level of lysine residues in the chromatin N-terminal region. The acetylation level is correlated with the structure of chromatin. HAT acetylates lysine residues of histone and the resultant neutralised histones loose chromatin, which subsequently results in activation of gene expression. In contrast, HDAC removes acetyl groups and the resulting positively ionised proteins lead to the condensation of chromatin, which represses gene expression. Therefore it can be stated that histone acetylation is carried away by histone transferases (HATs) and histone deacetylases (HDACs) reverse the action of HATs. Epigenetic modification regulates the genetic expression of chromatin without changing its DNA sequence, and the aberrance of this process is highly correlated with the occurrence of disease1–4,8a. Consequently, epigenetic regulation has currently become an attractive target for the treatment of a variety of diseases such as cancer5–8b, inflammation,9–17 neurological disorders18 and HIV19–20. Many HDACs have been reported to be overexpressed in various malignancies and therefore various HDAC inhibitors (HDIs) have been developed to treat these diseases. Since 2006, four HDAC inhibitors have been approved by FDA viz. Vorinostat (SAHA), Romidepsin (FK228), Belinostat (PXD101), and Panobinostat (LBH589). SAHA is the first HDI that received FDA approval in 2006 for the treatment of Cutaneous T cell Lymphoma (CTCL). Many new HDAC inhibitors, including dual/multitarget inhibitors8b, have been developed and many are now in clinical trials (Figure 1).
Figure 1.
Various examples of histone deacetylase inhibitors (1–5).
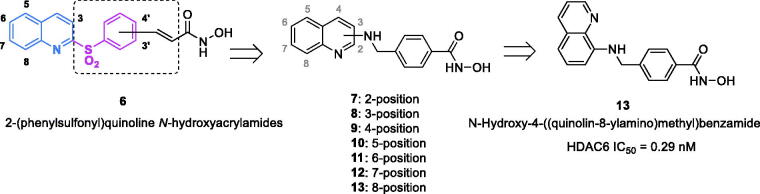
Our group has focussed on the development of HDAC inhibitors with various core scaffolds including indole21–24, indoline25,26, and quinoline27,28. In a study related of quinoline-derived compounds, we found that 2-(phenylsulfonyl)quinoline-N-hydroxyacrylamides (6) exhibit potent pan-HDAC inhibitory and antiproliferative activity27.
Recent reports brought our attention to the development of selective inhibitors and on further modifying the lead molecule (6), we reported the design and synthesis of N-hydroxy-4-((quinolin-8-ylamino)methyl)benzamide (13), a selective HDAC6 inhibitor which exhibits potent activity against multiple myeloma (Figure 2)28,29. Continuing our efforts on investigation of quinoline-containing molecules, the current study focuses on the modification of compound 13, and evaluates the effect of various substituents at C3 of quinoline to generate a series of 3-substituted quinolinehydroxamic acids with selective inhibitory and anti-proliferative activities against HDAC6 (Figure 3).
Figure 2.
Previously synthesised quinoline-containing HDAC inhibitors (6–13).
Figure 3.
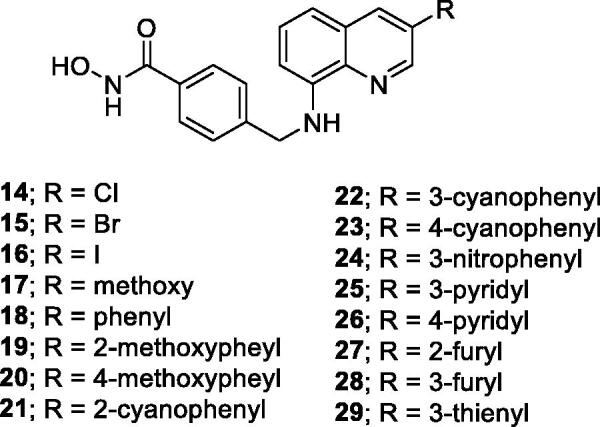
Synthetic 3-substituted quinolinehydroxamic acids (14–29).
2. Results and discussion
2.1. Chemistry
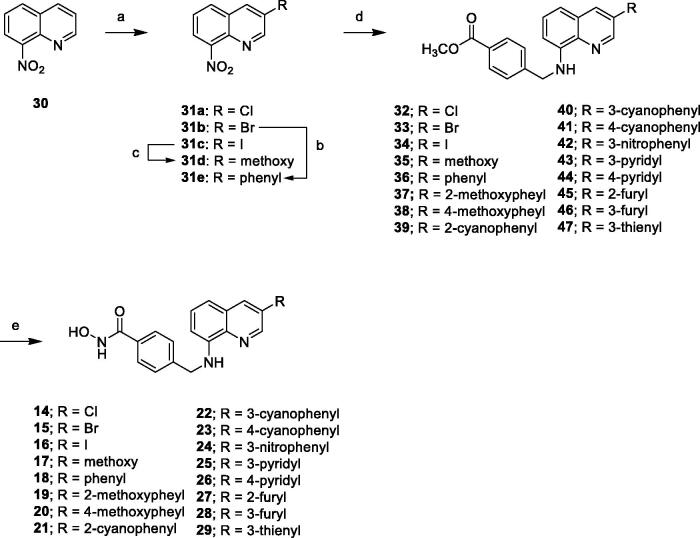
Scheme 1 describes the synthesis of designed compounds (14–29), which begins with halogenation of 8-nitroquinoline (30) with N-halosuccinimide to furnish 3-halogenated 8-nitroquinolines (31a–31c). The iodine in compound 31c was replaced by a methoxy group in the presence of CuI and 1,10-phenanthroline to generate compound 31d. Meanwhile, compound 31b underwent a Suzuki arylation with phenylboronic acid to furnish compound 31e. Subsequently, reduction of the nitro groups of 31a–31e by iron powder, followed by reductive amination with methyl 4-formylbenzoate in the presence of NaBH(OAc)3, led to compounds 32–36. Compound 33 underwent Suzuki arylation with various arylboronic acids to further afford compounds 37–47. The conversion of 32–47 to the desired hydroxamic acids (14–29) was achieved by hydrolysis of the ester by LiOH, amidation with NH2OTHP, and TFA-mediated deprotection, reactions that were conducted sequentially.
Scheme 1.
Synthetic Approaches to Compounds 14–29a. aReagents and conditions: (a) NXS, acetic acid, 110 oC, 30–61%; (b) phenylboronic acid, Pd(OAc)2, PPh3, Na2CO3, THF, H2O, MW, 90 oC, 90%; (c) CuI, 1,10-phenanthroline, Cs2CO3, MeOH, toluene, MW, 70 oC, 57%; (d) for 32–36: i. Fe powder, 1N HCl(aq), EtOH (or MeOH), reflux; ii. methyl 4-formylbenzoate, NaBH(OAc)3, DCM, 40 oC; for 37–47: 33, boronic acids, Pd(PPh3)4, TBAB, 2M K2CO3(aq), dioxane, reflux, 35–69%; (e) i. 1N LiOH(aq), dioxane, rt; ii. NH2OTHP, HBTU, DMF, TEA, rt; iii. 10% TFA(aq), MeOH, rt, 28–55%.
2.2. Biological evaluation
2.2.1. In vitro cell growth inhibitory activity
All synthetic compounds (14–29) were tested for their anti-proliferative activity in A549 and HCT116 cells, with SAHA (N1-hydroxy-N8-phenyloctanediamide) or PXD101 ((E)-N-hydroxy-3–(3-(N-phenylsulfamoyl)phenyl)acrylamide) as reference compounds (Table 1)27. Cell line inhibitory results show that halogen-substituted or most aryl/heteroaryl-substituted compounds have similar cellular activity. However, the methoxyphenyl group in 19 and 20 and the 2-cyanophenyl group (21) result in diminished activity. Comparison of 21, 22, and 23 revealed that the cyano group is disfavoured at C2’ of the C3-phenyl group. Among compounds possessing a C3-heteroaryl group, compounds 25 and 26 with pyridine substituents showed improved antiproliferative activity against A549 and HCT116 cells. Compound 25 in particular, inhibits the growth of A549 and HCT116 cells with IC50 values of 1.29 and 1.61 µM, respectively.
Table 1.
Antiproliferative activity (IC50, μM) against human cancer cell lines by compounds 14–29
| Cell lines |
||
|---|---|---|
| Compd | A549 | HCT116 |
| 14 | 3.14 ± 0.31 | 4.82 ± 0.29 |
| 15 | > 10 | > 10 |
| 16 | 3.41 ± 0.9 | 4.79 ± 0.47 |
| 17 | 3.11 ± 0.24 | 4.76 ± 0.43 |
| 18 | 4.97 ± 0.06 | 4.70 ± 0.38 |
| 19 | > 10 | 7.19 ± 0.77 |
| 20 | 9.10 ± 0.30 | > 10 |
| 21 | 8.65 ± 2.35 | 6.00 ± 1.56 |
| 22 | 2.04 ± 0.35 | 3.18 ± 0.16 |
| 23 | 3.64 ± 0.66 | 3.50 ± 0.32 |
| 24 | 4.05 ± 0.42 | 2.88 ± 0.21 |
| 25 | 1.29 ± 0.41 | 1.61 ± 0.22 |
| 26 | 2.13 ± 0.13 | 2.03 ± 0.46 |
| 27 | 3.18 ± 0.19 | 2.20 ± 0.17 |
| 28 | 3.77 ± 0.28 | 3.77 ± 0.21 |
| 29 | 3.84 ± 0.40 | 3.59 ± 0.51 |
| SAHA [27] | 1.02 ± 0.15 | 0.15 ± 0.03 |
| PXD101 [27] | 0.78 ± 0.07 | 0.13 ± 0.01 |
*IC50 values higher than 10 μM are estimated based on the best curve fitting available.
2.2.2. HDAC isoform inhibitory activity
The activity of several compounds and reference compounds against trichostatin A ((R,2E,4E)-7–(4-dimethylamino)phenyl)-N-hydroxy-4.6-dimethyl-7-oxahepta-2,4-dienamide (5) were tested for HDAC isoform selectivity against HDAC1, 2, 6 and 8 (1Table 2). The results from compounds 17, 25, and 26 reveals that a polar substituent at C3 shows improvement of HDAC6 selectivity and six-membered aromatic rings at C3 are favoured for HDAC6 selectivity. Compound 25, with a 3-pyridyl group at C3 of quinoline shows remarkable HDAC6 selectivity over HDAC1, 2, and 8 with selectivity ratios of 552, 276, and 379 respectively (values under IC50 in Table 2, presented parenthetically). On the other hand, five-membered heterocycles such as furan (27, 28) and thiophene (29), which act as bioisosteres of a phenyl ring, result in decrease of HDAC6 selectivity over that of HDAC 1, 2 and 8.
Table 2.
Inhibition of the Activity (IC50a, M) of HDAC Isoforms 1, 2, 6, and 8 (selectivity ratiob is shown in brackets).
| HDAC isoforms |
||||
|---|---|---|---|---|
| Compd | HDAC1 | HDAC2 | HDAC6 | HDAC8 |
| 14 | 4.32 × 10−6 | – | 2.02 × 10−8 | 3.64 x 10−6 |
| (214) | (180) | |||
| 17 | 4.95 × 10−6 | > 10−5 | 7.79 × 10−9 | 2.76 × 10−6 |
| (635) | (> 1284) | (354) | ||
| 18 | 5.30 × 10−6 | > 10−5 | 1.94 × 10−8 | 6.88 × 10−6 |
| (273) | (> 515) | (355) | ||
| 22 | 9.11 × 10−6 | 1.98 × 10−6 | 3.62 × 10−8 | 3.83 × 10−6 |
| (252) | (55) | (106) | ||
| 25 | 2.62 × 10−6 | 1.31 × 10−6 | 4.75 × 10−9 | 1.80 × 10−6 |
| (552) | (276) | (379) | ||
| 26 | 5.13 × 10−6 | 2.89 × 10−6 | 8.61 × 10−9 | 2.35 × 10−6 |
| (596) | (336) | (273) | ||
| 27 | 3.40 × 10−6 | 1.28 × 10−6 | 2.07 × 10−8 | 3.14 × 10−6 |
| (164) | (62) | (152) | ||
| 28 | 4.59 × 10−6 | 2.10 × 10−6 | 1.34 × 10−8 | 2.91 × 10−6 |
| (343) | (157) | (216) | ||
| 29 | 6.00 × 10−6 | 3.52 × 10−6 | 3.01 × 10−8 | 4.93 × 10−6 |
| (199) | (117) | (164) | ||
| Trichostatin A | 1.00 × 10−8 | 2.00 × 10−8 | 2.22 × 10−9 | 6.34 × 10−7 |
| (5) | (9) | (286) | ||
Dashed line indicates no inhibition or compound activity that could not be fitted to an IC50 curve. IC50 value higher than 10 μM is estimated based on the best curve fitting available.
Selectivity ratio: selectivity ratio of HDAC subtypes over HDAC6.
2.2.3. Western blot analysis
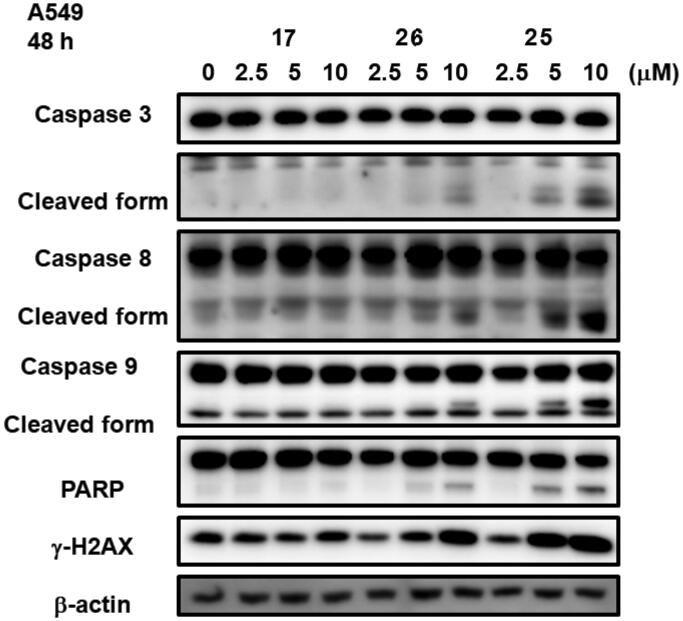
Western blot analysis indicated that compounds of this series induce apoptosis by activation of caspase and PARP pathways. Compound 25 with a 3-pyridyl substituent and compound 26 with a 4-pyridyl substitution at C3 of quinoline demonstrated dose dependent increases in caspase 3, 8, and 9 cleavage in A549 cells treated with 17, 25, and 26 for 48 h. 23 was found to be the most potent compound amongst these three tested compounds followed by 26 which was further followed by 17. The order of their efficacy in cleaving caspase isoforms also follows the same order as their in vitro cell growth inhibitory activity with 17 being the least potent amongst these three compounds. These molecules also increase dose dependent expression of gamma H2AX indicating increased DNA double strand fragmentation (Figure 4).
Figure 4.
Effect of treatment with compound 17, 25, and 26 at three different doses (0.25 µM, 5 µM, and 10 µM) on cleavage of caspase 3, 8, and 9 and PARP. Increased expression of gamma H2AX, a phosporylated form of H2AX, indicates increased DNA double strand fragmentation of A549 cells treated for 48 h in a dose- dependent manner.
2.2.4. Flow cytometry analysis
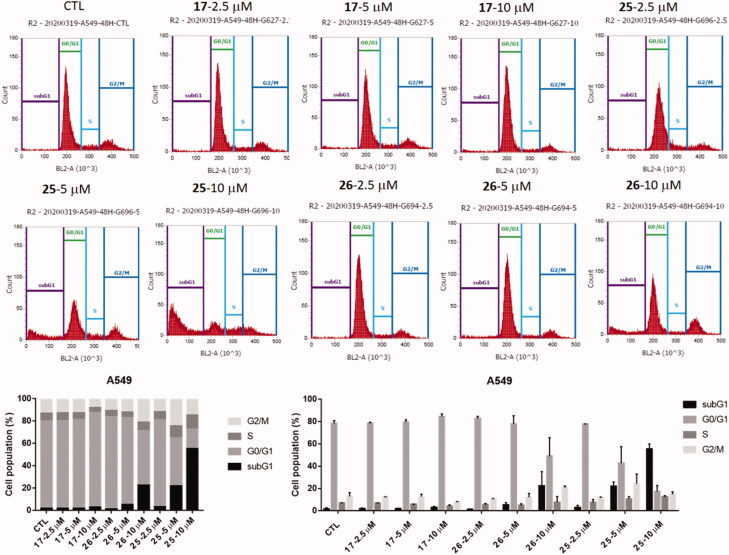
The effects of compound 17, 25, and 26 on cell cycle progression on A549 cells were examined by flow cytometry. Treatment of A549 cells with 25 and 26 resulted in concentration dependent accumulation of A549 cells in sub G1 phase with concomitant losses from the G0/G1 phase. Compounds followed the same trend in their activity as that of western blot analysis owing to same reason. Furthermore, a characteristic hypo diploid sub G0/1-peak was observed as a consequence of increased apoptosis and partial DNA loss in A549 cells treated with 25 or 26 in a dose dependent manner (Figure 5).
Figure 5.
(A) A549 cells were treated with or without 17, 25, and 26 (0.25, 0.5, 10 μM) for 48 h and were analysed by flow cytometry for cell cycle distribution. (B) After starvation for 24 h, A549 cells were then treated with compounds for the indicated time. After labelling with propidium iodide, DNA content was analysed by flow cytometry. (C) Quantification of cell population in Sub G1, G0/G1, S and G2/M phase. In A, B, and C, percent of cells = 100%.
2.2.5. Docking study of compound 17
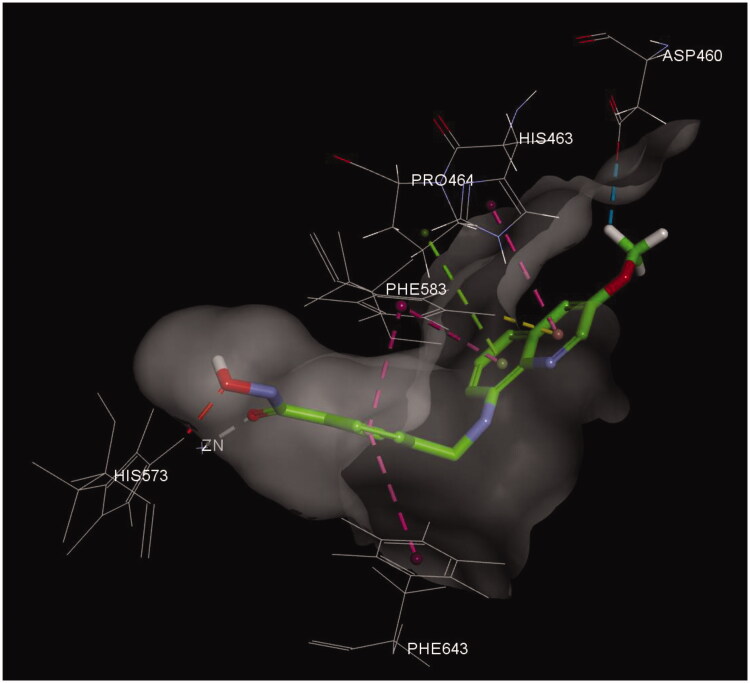
Compound 17 possesses the most HDAC6 selectivity over other isoforms; therefore, we docked 17 into the published crystal structure of HDAC6 (PDBID: 6CGP, Figure 6) in an attempt to understand the interaction of compound 17 with HDAC6, which was conducted using Discovery Studio2017R2. The hydroxamic acid of 17 interacts with a zinc ion (gray dotted line) and forms a hydrogen bond with His573 (red dotted line) in the bottom of the pocket, which is the typical interaction of hydroxamic acid-containing histone deacetylase inhibitors with HDACs. The central phenyl ring has π–π interactions with Phe583 and Phe643 (violet dotted lines) and the quinoline moiety is able to interact with the surrounding amino acids (His463, Pro464, and Phe583), including π-π (violet dotted lines), π-alkyl (green dotted line), and π-sigma interactions (yellow dotted line). Notably, the C3-methoxy group has an additional carbon hydrogen bond interaction with Asp460 (blue dotted line) in the open area, which probably contributes the higher HDAC6 selectivity.
Figure 6.
Docking of compound 17 (green) in the binding site of HDAC6 (PDB ID: 6CGP).
3. Conclusion
This study investigated the effect of C3 substitution of quinolinehydroxamic acids on related biological activities, such as antiproliferative and HDAC inhibitory activity. Synthesised compounds showed potent antitumor activities in A549 cells and HCT116 cells with single digit micromolar activities. Pyridine substitution at C3 of quinoline was found to be optimum for the activity of these compounds. The 3-pyridyl compound (25) is more potent than the 4-pyridyl isomer (26) and with IC50 = 4.75 nM, displayed better HDAC6 selectivity. These compounds exhibit their anticancer effects by induction of apoptosis and by fragmenting DNA double strands as revealed by western blot analysis. The increased population of treated A549 cells in sub G1 phase reveals cell cycle arrest in the sub G1 phase indicating DNA loss and apoptosis. These compounds (17, 25, 26) are potent and selective HDAC6 inhibitors and are crucial to development of the SAR of quinolinehydroxamic acids and to explore the possibilities of structural modifications to yield compounds with selectivity towards HDAC inhibition.
4. Experimental section
4.1. Chemistry
Nuclear magnetic resonance spectra were obtained with Bruker DRX-300 spectrometer operating at 300 MHz, with chemical shifts reported in parts per million (ppm, δ) downfield from TMS, an internal standard. High-resolution mass spectra (HRMS) were measured with a JEOL (JMS-700) electron impact (EI) mass spectrometer. Purity of the final compounds was determined using a Hitachi 2000 series HPLC system using C-18 column (Agilent ZORBAX Eclipse XDB-C18 5 µm. 4.6 mm × 150 mm) and was found to be ≥ 95% in all cases. Flash column chromatography was carried out using silica gel (Merck Kieselgel 60, No. 9385, 230–400 mesh ASTM). All reactions were done under an atmosphere of dry nitrogen.
General procedure of halogenation of 8-nitroquinoline (31a–31c): A solution of 8-nitroquinoline (1 eq) in AcOH (10 ml) was stirred at 110 °C and N-halosuccinimide (1.1–1.4 eq) was added in portions to the solution. The mixture was stirred at 110 °C for 1 h, and then cooled to room temperature (RT). The reaction mixture was poured into H2O (50 ml), and the resulting precipitate was collected by filtration and washed with H2O. The crude product was purified by flash column chromatography on silica gel eluting with EtOAc/n-Hexane to afford desired compound.
General Procedure of reductive amination (32–36): A mixture of nitro compound (1 eq), iron powder (4 eq), NH4Cl (5 eq), and 75% MeOH (10 ml) was heated to reflux. The reaction mixture was filtered and the filtrate was concentrated in vacuo, and then extracted with DCM and H2O. The combined organic layer was dried over anhydrous MgSO4, and concentrated in vacuo to afford a crude residue. A mixture of the residue (1 eq), methyl 4-formylbenzoate (1.2 eq), sodium triacetoxyborohydride (1.5 eq), AcOH (2–4 drops), and anhydrous DCM (10 ml) was stirred at RT. The mixture was poured into H2O, and then extracted with DCM. The combined organic layer was purified by flash column chromatography on silica gel with EtOAc/n-Hexane to afford corresponding ester compound 32–36.
General procedure for Suzuki arylation (37–47): A mixture of 33 (1 eq), substituted phenylboronic acid (1.2 eq), tetrakis(triphenylphosphine)palladium(0) (0.1 eq), TBAB (0.4 eq), 2 M K2CO3 (1 ml) and dioxane (10 ml) was stirred and refluxed overnight. Dioxane was removed from the reaction mixture in vacuo, and the mixture was extracted with H2O and EtOAc. The organic layer was collected and concentrated into an oily residue which was purified via column chromatography on silica gel with EtOAc/n-hexane to afford corresponding esters 37–47.
General procedure for hydroxamic acid synthesis. A mixture of 1 N LiOH (3 ml) and ester (1 mmol) was stirred at 40 °C for 2 h. The reaction was concentrated under reduced pressure and then H2O was added. The mixture was acidified with 3 N HCl to give an off-white precipitate. This off-white solid (1 mmol) was dissolved in DMF (10 ml) and EDC·HCl (1.5 mmol) was added, followed by HOBt hydrate (1.5 mmol) and TEA (3 mmol). After being stirred at RT for 30 min, NH2OTHP (1.2 mmol) was added and allowed to stir for an additional 5 h. The reaction mixture was quenched with H2O and was extracted with EtOAc (25 ml × 3). The combined organic layer was collected, dried over anhydrous MgSO4 and concentrated under reduced pressure to give a light yellow residue, which was purified by silica gel chromatography (EtOAc: n-hexane = 1:1) to give a colourless liquid. 10% TFA (5 ml) was added to the resulting product dissolved in MeOH (5 ml) and the mixture was stirred at RT for 5 h. The reaction mixture was concentrated under reduced pressure to give a white residue, which was recrystallized from MeOH to afford the desired compound.
4.1.1. 3-Chloro-8-nitroquinoline (31a)
Using N-chlorosuccinimide (1.4 eq) and following general procedure of halogenation, the title compound (31a) was obtained in 30% yield from 30: 1H NMR (300 MHz, CDCl3) δ 7.67 (dd, J = 7.8, 8.4 Hz, 1H), 7.98 (dd, J = 1.5, 8.4 Hz, 1H), 8.04 (dd, J = 1.2, 7.5 Hz, 1H), 8.25 (d, J = 2.1 Hz, 1H), 8.97 (d, J = 2.4 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 124.0, 126.8, 129.3, 130.6, 131.3, 134.1, 137.5, 148.3, 151.9.
4.1.2. 3-Bromo-8-nitro-quinoline (31 b)
Using N-bromosuccinimide (1.2 eq) and following general procedure of halogenation, the title compound (31 b) was obtained in 40% yield from 30: 1H NMR (300 MHz, CDCl3) δ 7.67 (dd, J = 7.5, 8.1 Hz, 1H), 7.97 (dd, J = 1.5, 8.4 Hz, 1H), 8.06 (dd, J = 1.5, 7.5 Hz, 1H), 8.44 (d, J = 2.1 Hz, 1H), 9.06 (d, J = 2.1 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 119.3, 124.1, 126.7, 129.8, 131.2, 137.4, 137.6, 148.3, 153.7.
4.1.3. 3-Iodo-8-nitro-quinoline (31c)
Using N-Iodosuccinimide (1.1 eq) and following general procedure of halogenation, the title compound (31c) was obtained in 61% yield from 30: 1H NMR (300 MHz, CDCl3) δ 7.62–7.67 (m, 1H), 7.93 (dd, J = 1.5, 8.1 Hz, 1H), 8.05 (dd, J = 1.2, 7.5 Hz, 1H), 8.65 (d, J = 2.1 Hz, 1H), 9.17 (d, J = 2.1 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 92.1, 124.3, 126.5, 130.3, 131.1, 137.6, 143.8, 148.2, 157.9.
4.1.4. 3-Methoxy-8-nitro-quinoline (31d)
A mixture of 31c (0.10 g, 0.33 mmol), CuI (0.006 g, 0.03 mmol), 1,10-phenanthroline (0.011 g, 0.06 mmol), Cs2CO3 (0.16 g, 0.50 mmol), MeOH (1 ml), and toluene (0.5 ml) was placed in a 10 ml reaction vessel with a magnetic stirring bar. The vessel was sealed and placed in the microwave cavity. The reaction condition was held at 70 °C for 30 min. After it was cooled to RT, the reaction mixture was filtered and the filtrate was concentrated in vacuo, and then extracted with DCM and H2O. The combined organic layer was purified by flash column chromatography on silica gel with EtOAc/n-hexane to afford compound 31d (0.04 g, 57%): 1H NMR (300 MHz, CDCl3) δ 3.98 (s, 3H), 7.43 (d, J = 2.7 Hz, 1H), 7.55 (dd, J = 7.8, 8.1 Hz, 1H), 7.85 (dd, J = 1.5, 7.5 Hz, 1H), 7.92 (dd, J = 1.5, 8.4 Hz, 1H), 8.79 (d, J = 3.0 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 55.8, 112.1, 121.0, 126.0, 130.2, 130.9, 134.5, 147.0, 148.4, 154.3.
4.1.5. 8-Nitro-3-phenyl-quinoline (31e)
A mixture of 31 b (0.30 g, 1.19 mmol), phenylboronic acid (0.29 g, 2.38 mmol), palladium (II) acetate (0.027 g, 0.12 mmol), triphenylphosphine (0.12 g, 0.48 mmol), sodium carbonate (0.25 g, 1.78 mmol), THF (10 ml), and H2O (1 ml) was placed into 35 ml reaction vessel with a magnetic stirring bar. The vessel was sealed and placed in the microwave cavity. The temperature was held at 90 °C for 20 min. After it cooled to RT, the reaction mixture was filtered and the filtrate was concentrated in vacuo; then extracted with DCM and H2O. The combined organic layer was purified by flash column chromatography on silica gel with EtOAc/n-hexane to afford compound 31e (0.27 g, 90%): 1H NMR (300 MHz, CDCl3) δ 7.45–7.57 (m, 3H), 7.60–7.66 (m, 1H), 7.68–7.71 (m, 2H), 8.03 (d, J = 7.2 Hz, 1H), 8.09 (d, J = 8.4 Hz, 1H), 8.37 (s,1H), 9.32 (s, 1H); 13 C NMR (75 MHz, CDCl3) δ 123.7, 125.8, 127.5, 128.9, 129.0, 129.5, 132.4, 133.1, 135.7, 136.7, 138.5, 148.2, 152.3.
4.1.6. 4-[(3-Chloro-quinolin-8-ylamino)-methyl]-benzoic acid methyl ester (32)
The title compound was obtained in 51% yield from compound 31a following the general procedure for reductive amination: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.62 (d, J = 6.0 Hz, 2H), 6.54 (d, J = 7.5 Hz, 1H), 6.63 (t, J = 5.4 Hz, 1H), 6.98 (d, J = 8.4 Hz, 1H), 7.32 (t, J = 7.8 Hz, 1H), 7.48 (d, J = 8.1 Hz, 2H), 8.00 (d, J = 8.4 Hz, 2H), 8.04 (d, J = 2.4 Hz, 1H), 8.62 (d, J = 2.4 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 47.4, 52.2, 105.7, 113.7, 127.1, 129.2, 129.3, 130.1, 134.2, 136.1, 144.4, 144.5, 146.1, 167.0.
4.1.7. 4-[(3-Bromo-quinolin-8-ylamino)-methyl]-benzoic acid methyl ester (33)
The title compound was obtained in 68% yield from compound 31 b following the general procedure for reductive amination: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.62 (s, 2H), 6.56 (dd, J = 0.6, 7.5 Hz, 1H), 6.98 (dd, J = 0.9, 8.4 Hz, 1H), 7.32 (t, J = 7.8 Hz, 1H), 7.48 (d, J = 8.4 Hz, 2H), 8.01 (d, J = 8.4 Hz, 2H), 8.23 (d, J = 2.1 Hz, 1H), 8.71 (d, J = 2.1 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 47.4, 52.2, 105.8, 113.6, 117.9, 127.1, 129.1, 129.2, 129.8, 130.1, 1136.1, 137.4, 144.4, 144.5, 147.8. 167.0.
4.1.8. 4-[(3-Iodo-quinolin-8-ylamino)-methyl]-benzoic acid methyl ester (34)
The title compound was obtained in 64% yield from compound 31c following the general procedure for reductive amination: 1H NMR (300 MHz, CDCl3) δ 3.90 (s, 3H), 4.59 (s, 2H), 6.55 (dd, J = 1.2, 7.8 Hz, 1H), 6.92 (dd, J = 1.2, 8.4 Hz, 1H), 7.29 (t, J = 8.1 Hz, 1H), 7.45 (d, J = 8.4 Hz, 2H), 8.00 (dt, J = 1.8, 6.6 Hz, 2H), 8.41 (d, J = 2.1 Hz, 1H), 8.82 (d, J = 2.1 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 47.4, 52.2, 90.6, 106.0, 113.5, 127.1, 128.9, 129.2, 130.1, 130.5, 136.2, 143.8, 144.4, 144.5, 152.1, 167.0.
4.1.9. 4-[(3-Methoxy-quinolin-8-ylamino)-methyl]-benzoic acid methyl ester (35)
The title compound was obtained in 63% yield from compound 31d following the general procedure for reductive amination: 1H NMR (300 MHz, CDCl3) δ 3.90 (s, 3H), 3.93 (s, 3H), 4.61 (s, 2H), 6.43 (dd, J = 1.2, 7.8 Hz, 1H), 6.62 (brs, 1H), 6.98 (dd, J = 0.9, 8.4 Hz, 1H), 7.27 (t, J = 7.8 Hz, 2H), 7.33 (d, J = 2.7 Hz, 1H), 7.49 (dt, J = 1.8, 8.1 Hz, 2H), 8.49 (d, J = 2.7 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 47.5, 52.1, 55.6, 103.7, 113.1, 113.9, 127.2, 128.6, 129.1, 129.5, 130.0, 133.2, 140.6, 144.4, 145.0, 153.9, 167.1.
4.1.10. 4-[(3-Phenyl-quinolin-8-ylamino)-methyl]-benzoic acid methyl ester (36)
The title compound was obtained in 45% yield from compound 31e following the general procedure for reductive amination: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.65 (s, 2H), 6.57 (d, J = 7.5 Hz, 1H), 6.73 (brs, 1H), 7.14 (dd, J = 1.2, 8.1 Hz), 7.34 (t, J = 8.1 Hz, 1H), 7.40–7.47 (m, 1H), 7.50–7.55 (m, 4H), 7.70–7.73 (m, 2H), 8.01–8.06 (m, 2H), 8.23 (d, J = 2.1 Hz, 1H), 9.01 (d, J = 2.4 Hz, 1H); 13 C NMR (75 MHz, CDCl3) δ 47.5, 52.1, 105.5, 114.9, 127.2, 127.5, 128.1,128.3, 128.5, 129.1, 129.2, 130.1, 133.6, 134.3, 137.2, 138.2, 144.3, 144.9, 146.4, 167.1.
4.1.11. Methyl 4-(((3–(2-methoxyphenyl)quinolin-8-yl)amino)methyl)benzoate (37)
The title compound was obtained in 35% yield from compound 33 followed the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.85 (s, 3H), 3.92 (s, 3H), 4.65 (s, 2H), 6.58 (dd, J = 0.9, 7.8 Hz, 1H), 7.05 (d, J = 8.1 Hz, 1H), 7.13 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 7.8 Hz, 1H), 7.41–7.46 (m, 2H), 7.52 (d, J = 8.4 Hz, 2H), 8.04 (d, J = 8.4 Hz, 2H), 8.20 (d, J = 2.1 Hz, 1H), 8.98 (d, J = 2.1 Hz, 1H).
4.1.12. Methyl 4-(((3–(4-methoxyphenyl)quinolin-8-yl)amino)methyl)benzoate (38)
The title compound was obtained in 38% yield from compound 33 followed the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.90 (s, 6H), 4.71 (s, 2H), 6.66 (d, J = 7.8 Hz, 1H), 7.08 (d, J = 8.7 Hz, 2H), 7.20 (d, J = 8.1 Hz, 1H), 7.43 (t, J = 7.8 Hz, 1H), 7.53 (d, J = 8.1 Hz, 2H), 7.66 (d, J = 8.7 Hz, 2H), 8.01 (d, J = 8.4 Hz, 2H), 8.49 (s, 1H), 9.03 (d, J = 2.1 Hz, 1H).
4.1.13. Methyl 4-(((3–(2-cyanophenyl)quinolin-8-yl)amino)methyl)benzoate (39)
The title compound was obtained in 45% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.66 (s, 2H), 6.62 (dd, J = 0.9, 7.5 Hz, 1H), 7.16 (dd, J = 0.9, 8.4 Hz, 1H), 7.38 (t, J = 7.8 Hz, 1H), 7.50–7.56 (m, 3H), 7.62 (d, J = 7.2 Hz, 1H), 7.73 (td, J = 1.5, 7.7 Hz, 1H), 7.85 (td, J = 1.2, 7.8 Hz, 1H), 8.02 (d, J = 8.4 Hz, 2H), 8.31 (d, J = 2.1 Hz, 1H), 8.90 (d, J = 2.4 Hz, 1H).
4.1.14. Methyl 4-(((3–(3-cyanophenyl)quinolin-8-yl)amino)methyl)benzoate (40)
The title compound was obtained in 59% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.66 (s, 2H), 6.62 (d, J = 7.2 Hz, 1H), 7.16 (d, J = 8.1 Hz, 1H), 7.38 (t, J = 7.8 Hz, 1H), 7.51 (d, J = 8.4 Hz, 2H), 7.64 (t, J = 7.5 Hz, 1H), 7.73 (d, J = 7.8 Hz, 1H), 7.93 (d, J = 7.8 Hz, 1H), 7.98–8.03 (m, 3H), 8.26 (d, J = 2.1 Hz, 1H), 8.94 (d, J = 2.1 Hz, 1H).
4.1.15. Methyl 4-(((3–(4-cyanophenyl)quinolin-8-yl)amino)methyl)benzoate (41)
The title compound was obtained in 51% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.69 (s, 2H), 6.67 (d, J = 7.8 Hz, 1H), 7.19 (d, J = 8.4 Hz, 1H), 7.43 (t, J = 8.1 Hz, 1H), 7.52 (d, J = 8.4 Hz, 2H), 7.83 (m, 1H), 8.02 (d, J = 8.1 Hz, 2H), 8.39 (s, 1H), 9.00 (d, J = 2.4 Hz, 1H).
4.1.16. Methyl 4-(((3–(3-nitrophenyl)quinolin-8-yl)amino)methyl)benzoate (42)
The title compound was obtained in 61% yield from compound 33 followed the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.90 (s, 3H), 4.70 (s, 2H), 6.70 (d, J = 7.8 Hz, 1H), 7.22 (s, 1H), 7.46 (t, J = 8.1 Hz, 1H), 7.52 (d, J = 8.4 Hz, 2H), 7.75 (t, J = 8.1 Hz, 1H), 8.00–8.06 (m, 3H), 8.34 (d, J = 8.1 Hz, 1H), 8.51 (s, 1H), 8.59 (s, 1H), 9.05 (d, J = 2.4 Hz, 1H).
4.1.17. Methyl 4-(((3-(pyridin-3-yl)quinolin-8-yl)amino)methyl)benzoate (43)
The title compound was obtained in 69% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ3.91 (s, 3H), 4.67 (s, 2H), 6.64 (d, J = 7.5 Hz, 1H), 7.17 (d, J = 7.8 Hz, 1H), 7.40 (t, J = 8.1 Hz, 1H), 7.51 (d, J = 8.4 Hz, 2H), 7.61 (m, 1H), 8.02 (d, J = 8.1 Hz, 2H), 8.19 (d, J = 6.6 Hz, 1H), 8.31 (s, 1H), 8.73 (s, 1H), 8.98 (s, 1H), 9.02 (s, 1H).
4.1.18. Methyl 4-(((3-(pyridin-3-yl)quinolin-8-yl)amino)methyl)benzoate (44)
The title compound was obtained in 62% yield from compound 33 following the general procedure for Suzuki arylation: 1HNMR (300 MHz, CDCl3) δ3.91 (s, 3H), 4.68 (s, 2H), 6.69 (d, J = 7.8 Hz, 1H), 7.20 (d, J = 7.8 Hz, 1H), 7.44 (t, J = 7.8 Hz, 1H), 7.51 (d, J = 8.1 Hz, 2H), 8.03 (m, 3H), 8.44 (s, 1H), 8.83 (s, 2H), 9.05 (s, 1H).
4.1.19. Methyl 4-(((3-(furan-2-yl)quinolin-8-yl)amino)methyl)benzoate (45)
The title compound was obtained in 40% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.90 (s, 3H), 4.63 (s, 2H), 6.53 (d, J = 0.6 Hz, 1H), 6.55 (dd, J = 1.8, 3.3 Hz, 1H), 6.85 (d, J = 3.3 Hz, 1H), 7.10 (d, J = 8.1 Hz, 1H), 7.31 (t, J = 7.8 Hz, 1H), 7.50 (d, J = 8.1 Hz, 2H), 7.57 (d, J = 1.5 Hz, 1H), 8.01 (d, J = 8.1 Hz, 2H), 8.30 (d, J = 2.1 Hz, 1H), 9.04 (d, J = 2.1 Hz, 1H).
4.1.20. Methyl 4-(((3-(furan-3-yl)quinolin-8-yl)amino)methyl)benzoate (46)
The title compound was obtained in 37% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.65 (s, 2H), 6.56 (d, J = 7.8 Hz, 1H), 6.83 (dd, J = 0.9, 1.8 Hz, 1H), 7.09 (d, J = 8.1 Hz, 1H), 7.33 (t, J = 8.1 Hz, 1H), 7.51 (d, J = 8.4 Hz, 2H), 7.57 (t, J = 1.8 Hz, 1H), 7.91 (s, 1H), 8.01 (d, J = 8.4 Hz, 2H), 8.15 (d, J = 1.8 Hz, 1H), 8.91 (d, J = 2.1 Hz, 1H).
4.1.21. Methyl 4-(((3-(thiophen-3-yl)quinolin-8-yl)amino)methyl)benzoate (47)
The title compound was obtained in 36% yield from compound 33 following the general procedure for Suzuki arylation: 1H NMR (300 MHz, CDCl3) δ 3.91 (s, 3H), 4.63 (s, 2H), 6.54 (d, J = 7.5 Hz, 1H), 6.72 (s, 1H), 7.10 (d, J = 8.1 Hz, 1H), 7.32 (t, J = 7.8 Hz, 1H), 7.46–7.51 (m, 4H), 7.62 (s, 1H), 8.02 (d, J = 8.4 Hz, 2H), 8.20 (d, J = 1.8 Hz, 1H), 9.01 (d, J = 2.1 Hz, 1H).
4.1.2. Synthesis of hydroxamic acids 14–29
4.1.2.1. 4-[(3-Chloro-quinolin-8-ylamino)-methyl]-N-hydroxy-benzamide (14)
The title compound was obtained in 33% overall yield from compound 32 following the general procedure for hydroxamic acid synthesis: mp 188.3–188.9 °C; 1H NMR (300 MHz, DMSO-d6) δ 4.58 (d, J = 6.3 Hz, 2H), 6.54 (d, J = 7.5 Hz, 1H), 7.03 (d, J = 7.8 Hz, 1H), 7.29–7.34 (m, 2H), 7.45 (d, J = 8.1 Hz, 2H), 7.70 (d, J = 8.1 Hz, 2H), 8.38 (d, J = 2.4 Hz, 1H), 8.73 (d, J = 2.4 Hz, 1H), 8.99 (s, 1H), 11.14 (s, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.7, 105.4, 112.7, 126.9, 127.0, 128.0, 128.9, 129.2, 131.4, 134.1, 135.6, 143.1, 144.3, 145.4, 164.2; HRMS (ESI) for C17H15ClN3O2 (M + H+) calcd 328.0853, found 328.0849; HPLC purity of 100.00% (retention time = 29.63).
4.1.2.2. 4-[(3-Bromo-quinolin-8-ylamino)-methyl]-N-hydroxy-benzamide (15)
The title compound was obtained in 30% overall yield from compound 33 following the general procedure for hydroxamic acid synthesis: mp 188.5–189.2 °C; 1H NMR (300 MHz, DMSO-d6) δ 4.58 (d, J = 6.3 Hz, 2H), 6.55 (d, J = 7.8 Hz, 1H), 7.02 (dd, J = 0.6, 8.1 Hz, 1H), 7.29–7.34 (m, 2H), 7.44 (d, J = 8.1 Hz, 2H), 7.68 (d, J = 8.4 Hz, 2H), 8.55 (d, J = 2.4 Hz, 1H), 8.79 (d, J = 2.4 Hz, 1H), 8.97 (s, 1H), 11.12 (s, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.7, 105.5, 112.6, 117.2, 126.9, 127.0, 129.2, 129.6, 131.4, 135.6, 137.3, 143.1, 144.3, 147.2, 164.2; HRMS (ESI) for C17H15BrN3O2 (M + H+) calcd 372.0348, found 372.0343; HPLC purity of 98.75% (retention time = 30.55).
4.1.2.3. N-Hydroxy-4-[(3-iodo-quinolin-8-ylamino)-methyl]-benzamide (16)
The title compound was obtained in 29% overall yield from compound 34 following the general procedure for hydroxamic acid synthesis: mp 193.8–194.7 °C; 1H NMR (300 MHz, DMSO-d6) δ 4.57 (d, J = 6.3 Hz, 2H), 6.53 (dd, J = 1.2, 7.8 Hz, 1H), 6.98 (dd, J = 0.9, 8.1 Hz, 1H), 7.25–7.31 (m, 2H), 7.44 (d, J = 8.1 Hz, 2H), 7.67 (d, J = 8.1 Hz, 2H), 8.69 (d, J = 2.1 Hz, 1H), 8.88 (d, J = 2.1 Hz, 1H), 8.97 (s, 1H), 11.12 (s, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.7, 91.4, 105.5, 112.5, 126.9, 127.0, 128.8, 130.2, 131.4, 135.7, 143.2, 143.4, 144.3, 151.7, 164.2; HRMS (ESI) for C17H15IN3O2 (M + H+) calcd 420.0209, found 420.0203; HPLC purity of 95.67% (retention time = 31.33).
4.1.2.4. N-Hydroxy-4-[(3-methoxy-quinolin-8-ylamino)-methyl]-benzamide (17)
The title compound was obtained in 30% overall yield from compound 35 following the general procedure for hydroxamic acid synthesis: mp 171.8–172.9 °C; 1H NMR (300 MHz, DMSO-d6) δ 3.91 (s, 3H), 4.55 (d, J = 6.3 Hz, 2H), 6.37 (d, J = 7.2 Hz, 1H), 7.14 (t, J = 6.3 Hz, 1H), 7.21 (t, J = 8.1 Hz, 1H), 7.45 (d, J = 8.1 Hz, 2H), 7.61 (d, J = 2.7 Hz, 1H), 7.68 (d, J = 8.1 Hz, 2H), 8.49 (d, J = 2.7 Hz, 1H), 8.98 (s, 1H), 11.13 (s, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.9, 55.5, 103.0, 112.8, 113.2, 126.9, 127.0, 128.4, 129.2, 131.3, 132.5, 139.9, 143.5, 144.2, 153.4, 164.2; HRMS (ESI) for C18H18N3O3 (M + H+) calcd 324.1348, found 324.1349; HPLC purity of 97.57% (retention time = 25.47).
4.1.2.5. N-Hydroxy-4-[(3-phenyl-quinolin-8-ylamino)-methyl]-benzamide (18)
The title compound was obtained in 40% overall yield from compound 36 following the general procedure for hydroxamic acid synthesis: mp 172.7–173.2 °C; 1H NMR (300 MHz, DMSO-d6) δ 4.61 (d, J = 6.3 Hz, 2H), 6.53 (d, J = 7.5 Hz, 1H), 7.14 (d, J = 7.8 Hz, 1H), 7.27–7.33 (m, 2H), 7.40–7.60 (m, 5H), 7.70 (d, J = 8.1 Hz, 2H), 7.87 (d, J = 7.2 Hz, 2H), 8.48 (d, J = 2.1 Hz, 1H), 8.99 (s, 1H), 9.09 (d, J = 2.1 Hz, 1H), 11.15 (s, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.8, 105.0, 113.9, 126.9, 127.0, 127.2, 128.1, 129.2, 131.4, 133.0, 133.2, 136.7, 137.2, 143.4, 144.1, 145.8, 164.2; HRMS (ESI) for C23H20N3O2 (M + H+) calcd 370.1556, found 370.1557; HPLC purity of 97.59% (retention time = 33.67).
4.1.2.6. N-Hydroxy-4-(((3–(2-methoxyphenyl)quinolin-8-yl)amino)methyl)benzamide (19)
The title compound was obtained in 45% overall yield from compound 37 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 3.84 (s, 3H), 4.64 (s, 2H), 6.57 (d, J = 0.9 Hz, 1H), 6.60–7.11 (m, 2H), 7.14 (d, J = 7.8 Hz, 1H), 7.27 (t, J = 8.1 Hz, 1H), 7.42 (d, J = 7.5 Hz, 2H), 7.52 (d, J = 8.1 Hz, 2H), 7.71 (d, J = 8.4 Hz, 2H), 8.18 (d, J = 2.4 Hz, 1H), 8.86 (d, J = 2.1 Hz, 1H); 13C NMR (75 MHz, DMSO-d6) δ 45.9, 55.6, 105.0, 111.9, 113.7, 121.1, 126.6, 126.9, 127.0, 128.0, 129.8, 130.7, 131.4, 131.7, 135.3, 136.1, 143.4, 144.0, 148.0, 156.4, 164.2; HRMS (EI) for C24H22N3O3(M + H+) calcd 400.1661, found 400.1662; HPLC purity of 93.21% (retention time = 32.91).
4.1.2.7. N-Hydroxy-4-(((3–(4-methoxyphenyl)quinolin-8-yl)amino)methyl)benzamide (20)
The title compound was obtained in 37% overall yield from compound 38 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 3.83 (s, 3H), 4.61 (s, 2H), 6.51 (dd, J = 0.9 Hz, 7.5 Hz, 1H), 7.10–7.13 (m, 3H), 7.28 (t, J = 8.1 Hz, 1H), 7.47 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 8.1 Hz, 2H), 7.81–7.85 (m, 2H), 8.41 (d, J = 2.1 Hz, 1H), 9.06 (d, J = 2.4 Hz, 1H); 13C NMR (75 MHz, DMSO-d6) δ 46.0, 55.3, 105.1, 113.9, 114.7, 127.0, 127.1, 128.2, 128.3, 128.3, 129.4, 131.4, 132.2, 133.3, 136.1, 143.3, 143.9, 145.6, 159.5, 164.2; HRMS (EI) for C24H22N3O3 (M + H+) calcd 400.1661, found 400.1659; HPLC purity of 99.02% (retention time = 32.49).
4.1.2.8. 4-(((3–(2-Cyanophenyl)quinolin-8-yl)amino)methyl)-N-hydroxybenzamide amide (21)
The title compound was obtained in 38% overall yield from compound 39 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.68 (s, 2H), 4.68 (s, 2H), 6.65 (d, J = 7.5 Hz, 1H), 7.17 (d, J = 8.1 Hz, 1H), 7.35 (d, J = 8.1 Hz, 1H), 7.54 (d, J = 8.1 Hz, 2H), 7.63 (td, J = 0.9, 1.8 Hz, 7.5 Hz, 1H), 7.72 (d, J = 8.1 Hz, 2H), 7.75 (d, J = 7.5 Hz, 1H), 7.83 (d, J = 7.8 Hz, 1H), 7.93 (d, J = 7.5 Hz, 1H), 8.36 (d, J = 2.4 Hz, 1H), 8.91 (d, J = 2.1 Hz, 1H); 13C NMR (75 MHz, DMSO-d6) δ 52.0, 97.9, 103.0, 106.2, 109.9, 118.8, 118.9, 119.7, 120.1, 120.2, 122.1, 122.7, 123.4, 125.1, 125.5, 127.6, 129.3, 133.8, 135.7, 136.1, 138.1, 158.5; HRMS (EI) for C24H19N4O2 (M + H+) calcd 395.1508, found 395.1512; HPLC purity of 97.06% (retention time = 29.41).
4.1.2.9. 4-(((3–(3-Cyanophenyl)quinolin-8-yl)amino)methyl)-N-hydroxybenzamide (22)
The title compound was obtained in 55% overall yield from compound 40 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.62 (d, J = 6.0 Hz, 2H), 6.56 (d, J = 7.8 Hz, 1H), 7.13 (d, J = 7.5 Hz, 1H), 7.30–7.35 (m, 2H), 7.47 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 8.4 Hz, 2H), 7.76 (t, J = 8.1 Hz, 1H), 7.92 (d, J = 8.1 Hz, 1H), 8.25 (d, J = 8.4 Hz, 1H), 8.41 (d, J = 1.8 Hz, 1H), 8.62 (d, J = 2.1 Hz, 1H), 9.15 (d, J = 2.4 Hz, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.8, 105.5, 112.3, 113.9, 118.7, 126.9, 127.1, 128.0, 128.4, 130.3, 130.7, 131.2, 131.4, 131.6, 131.9, 133.8, 137.0, 138.5, 143.3, 144.1,145.5, 164.2; HRMS (EI) for C24H18N4O2 (M + H+) calcd 395.1508, found 395.1511; HPLC purity of 93.59% (retention time = 30.47).
4.1.2.10. 4-(((3–(4-Cyanophenyl)quinolin-8-yl)amino)methyl)-N-hydroxybenzamide (23)
The title compound was obtained in 34% overall yield from compound 41 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.61 (d, J = 6.3 Hz, 2H), 6.57 (d, J = 7.2 Hz, 1H), 7.15 (d, J = 8.1 Hz, 1H), 7.33 (t, J = 7.8 Hz, 2H), 7.46 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 8.1 Hz, 2H), 8.02 (d, J = 8.4 Hz, 2H), 8.12 (d, J = 8.4 Hz, 2H), 8.62 (d, J = 2.1 Hz, 1H), 9.14 (d, J = 2.1 Hz,1H); 13 C NMR (75 MHz, DMSO-d6) δ 46.2, 106.1, 111.1, 114.5, 119.3, 127.4, 127.5, 128.4, 128.5, 128.9, 131.6, 133.5, 134.6, 137.5, 142.3, 143.7, 144.5, 146.0, 164.7; HRMS (EI) for C24H19N4O2 (M + H+) calcd 395.1508, found 395.1505; HPLC purity of 96.50% (retention time = 30.24).
4.1.2.11. N-Hydroxy-4-(((3–(3-nitrophenyl)quinolin-8-yl)amino)methyl)benzamide (24)
The title compound was obtained in 50% overall yield from compound 42 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.62 (d, J = 6.0 Hz, 2H), 6.57 (d, J = 7.8 Hz, 1H), 7.18 (d, J = 7.5 Hz, 1H), 7.31–7.36 (m, 2H), 7.47 (d, J = 7.8 Hz, 2H), 7.69 (d, J = 8.1 Hz, 2H), 7.85 (t, J = 7.8 Hz, 1H), 8.31 (d, J = 7.5 Hz, 1H), 8.37 (d, J = 8.1 Hz, 1H), 8.67 (s, 2H), 9.17 (d, J = 1.8 Hz, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 46.3, 106.0, 114.5, 122.1, 123.2, 127.4, 127.5, 128.4, 128.9, 131.2, 131.5, 131.9, 134.2, 134.5, 137.5, 139.5, 143.7, 144.6, 146.0,149.1; HRMS (EI) for C23H19N4O4 (M + H+) calcd 415.1406, found 415.1408; HPLC purity of 93.65% (retention time = 32.15).
4.1.2.12. N-Hydroxy-4-(((3-(pyridin-3-yl)quinolin-8-yl)amino)methyl)benzamide (25)
The title compound was obtained in 41% overall yield from compound 43 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.62 (d, J = 6.3 Hz, 2H), 6.56 (t, J = 7.5 Hz, 1H), 7.14 (d, J = 7.5 Hz, 1H), 7.32 (d, J = 8.1 Hz, 2H), 7.47 (d, J = 8.4 Hz, 2H), 7.58 (dd, J = 4.5 Hz, 7.9 Hz, 1H), 7.69 (d, J = 8.1 Hz, 2H), 8.30 (td, J = 1.8 Hz, 4.2 Hz, 1H), 8.59 (d, J = 2.4 Hz, 1H), 8.66 (dd, J = 1.8 Hz, 4.8 Hz, 1H), 9.10 (d, J = 1.8 Hz, 1H), 9.14 (d, J = 2.4 Hz, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.7, 105.3, 113.7, 124.0, 126.8, 127.0, 127.9, 128.2, 130.2, 131.3, 132.8, 133.5, 134.5, 136.8, 143.2, 144.0, 145.5, 147.9, 149.0, 164.1; HRMS (EI) for C22H19N4O2 (M + H+) calcd 371.1508, found 371.1507; HPLC purity of 98.11% (retention time = 19.77).
4.1.2.13. N-Hydroxy-4-(((3-(pyridin-4-yl)quinolin-8-yl)amino)methyl)benzamide (26)
The title compound was obtained in 40% overall yield from compound 44 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.63 (s, 2H), 6.62 (d, J = 7.5 Hz, 1H), 7.19 (d, J = 7.8 Hz, 1H), 7.37 (t, J = 8.1 Hz, 1H), 7.47 (d, J = 8.1 Hz, 2H), 7.69(d, J = 8.1 Hz, 2H), 8.27 (d, J = 6.0 Hz, 2H), 8.81 (d, J = 2.1 Hz, 1H), 8.89 (d, J = 5.7 Hz, 2H), 9.26 (d, J = 2.1 Hz, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.8, 106.5, 114.2, 123.1, 126.9, 127.0, 127.7, 128.8, 131.4, 135.4, 137.7, 143.2, 144.1, 145.2, 145.8, 149.5, 164.2, 179.2; HRMS (EI) for C22H19N4O2 (M + H+) calcd 371.1508, found 371.1505; HPLC purity of 94.78% (retention time = 16.94).
4.1.2.14. 4-(((3-(Furan-2-yl)quinolin-8-yl)amino)methyl)-N-hydroxybenzamide (27)
The title compound was obtained in 32% overall yield from compound 45 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.59 (d, J = 6.3 Hz, 2H), 6.51 (d, J = 6.9 Hz, 1H), 6.70 (dd, J = 1.8 Hz, 3.4 Hz, 1H), 7.11 (d, J = 8.1 Hz, 1H), 7.25 (d, J = 4.2 Hz, 2H), 7.29 (d, J = 8.1 Hz, 1H), 7.46 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 8.1 Hz, 2H), 7.88 (dd, J = 0.6 Hz, 1.8 Hz, 1H), 8.43 (d, J = 2.4 Hz, 1H), 9.14 (d, J = 2.4 Hz, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.8, 105.1, 107.5, 112.3, 113.7, 123.9, 126.8, 126.9, 127.9, 128.4, 128.8, 131.4, 136.2, 143.1, 143.8, 144.1, 150.7, 164.0; HRMS (EI) for C21H18N3O3 (M + H+) calcd 360.1348, found 360.1350; HPLC purity of 96.16% (retention time = 30.57).
4.1.2.15. 4-(((3-(Furan-3-yl)quinolin-8-yl)amino)methyl)-N-hydroxybenzamide (28)
The title compound was obtained in 32% overall yield from compound 46 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.59 (d, J = 6.3 Hz, 2H), 6.49 (d, J = 8.1 Hz, 1H), 7.04 (d, J = 7.5 Hz, 1H), 7.18 (s, 1H), 7.23 (d, J = 6.9 Hz, 1H), 7.28 (d, J = 7.8 Hz, 1H), 7.46 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 8.4 Hz, 2H), 7.84 (s, 1H), 8.39 (d, J = 2.1 Hz, 1H), 8.45 (s, 1H), 9.07 (d, J = 2.1 Hz, 1H);13C NMR (75 MHz, DMSO-d6) δ 45.8, 104.7, 108.6, 113.4, 123.0, 125.6, 126.8, 127.0, 128.1, 128.2, 130.9, 131.3, 136.3, 140.2, 143.4, 144.1, 144.6, 145.2, 164.2; HRMS (EI) for C21H18N3O3 (M + H+) calcd 360.1348, found 360.1348; HPLC purity of 98.61% (retention time = 29.15).
4.1.2.16. N-Hydroxy-4-(((3-(thiophen-3-yl)quinolin-8-yl)amino)methyl)benzamide (29)
The title compound was obtained in 28% overall yield from compound 47 following the general procedure for hydroxamic acid synthesis: 1H NMR (300 MHz, DMSO-d6) δ 4.60 (s, 2H), 6.50 (dd, J = 0.9 Hz, 7.8 Hz, 1H), 7.08 (dd, J = 0.9 Hz, 8.1 Hz, 1H), 7.28 (t, J = 7.8 Hz, 1H), 7.47 (d, J = 8.1 Hz, 2H), 7.69 (d, J = 8.4 Hz, 2H), 7.74–7.80 (m, 2H), 8.18 (dd, J = 1.5 Hz, 2.7 Hz, 1H), 8.51 (d, J = 2.4 Hz, 1H), 9.17 (d, J = 2.1 Hz, 1H); 13 C NMR (75 MHz, DMSO-d6) δ 45.9, 105.0, 113.7, 122.4, 126.3, 126.9, 127.1, 127.6, 128.2, 128.2, 128.5, 131.4, 131.8, 136.3, 138.4, 143.4, 144.1, 145.6, 164.2; HRMS (EI) for C21H18N3O2S (M + H+) calcd 376.1120, found 376.1122; HPLC purity of 97.78% (retention time = 31.49).
4.2. Biology
4.2.1. Tumour cell culture
All human cancer cells were maintained in RPMI 1640 medium supplemented with 10% FBS and penicillin (100 units/ml)/streptomycin (100 µg/ml)/amphotericin B (0.25 µg/ml). All cells were maintained in humidified air containing 5% CO2 at 37 °C and cultured every 2–3 days. All cells were cultured in tissue culture flasks in humidified air containing 5% CO2 at 37 °C and cultured every 2–3 days.
4.2.2. The sulforhodamine B assays
Cells were seeded at the density of 5000 cells/well into 96-plate overnight. Basal cells were fixed with 10% trichloroacetic acid (TCA) to represent the cell population at the time of compound addition (T0). After additional incubation of DMSO (C) or different doses of test compounds (Tx) for 48 h, cells were fixed with 10% TCA and stained with SRB at 0.4% (w/v) in 1% AcOH. Unbound SRB was washed out using 1% AcOH and SRB bound cells were solubilised with 10 mM Trizma base. The absorbance was read at a wavelength of 515 nm. The 50% growth inhibition (GI50) was calculated by 100 − [(Tx – T0)/(C – T0)] × 100.
4.2.3. HDAC biochemical assays
The HDACs in vitro activities of human recombinant HDAC 1, 2, 4, 6, and 8 were conducted by EurofinPanlabs (Taipei, Taiwan). In brief, indicated compounds were incubated with specific HDAC enzyme and Fluor-de-Lys deacetylase substrate. Fluor-de-Lys deacetyl substrates were spectrofluorimetrically quantitated compared to control.
4.2.4. Western blot analysis
Cells were incubated with indicated compounds for 24 h and lysed with ice-cold lysis buffer (20 mMTris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 2.5 mM β-glycerylphosphate, 1 mM Na4P2O7, 5 mM NaF, 1 mM Na3VO4 and protease inhibitor cocktail from Millipore) on ice for 30 min following by centrifugation at 13000 rpm for 30 min. Protein concentrations were determined and equal amounts of protein were separated by 8–15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to poly(vinylidene difluoride) (PVDF) membranes. Membranes were immunoblotted with specific antibodies overnight at 4 °C and then applied to appropriate horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG secondary antibodies for 1 h at RT. Signals were detected using an enhanced chemiluminescence (Amersham, Buckinghamshire, UK).
4.2.5. Flow cytometry
Briefly, A549 cells were starved with EBM-2 medium overnight and were subsequently replenished with EGM-2 medium with or without 17, 25, and 26 (0.25, 0.5, 10 µM) for 48 h. After being trypsinized and fixed in ice-cold 75% MeOH for 1 h at −20 °C, A549 cells were washed with PBS and resuspended in 0.2 ml DNA extraction buffer (0.2 M Na2HPO4, 0.1 M citric acid; pH 7.8) for 30 min. Then the cells were stained with propidium iodide solution (PI; 100 µg/ml RNase, 80 µg/ml propidium iodide, 0.1% Triton X-100) in PBS. FACScan flow cytometry was utilised to determine cell cycle distribution, and data analysis was performed with CellQuest software (BD Biosciences).
4.2.8. Statistical and graphical analyses
Each experiment was performed independently at least three times and the data are presented as mean ± SEM for the indicated number of separate experiments. Student’s t-test was used to compare the mean of each group with that of the control group in experiments and one-way ANOVA was used in animal study. p Values <0.05 were considered significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplementary Material
Funding Statement
This research was supported by the Ministry of Science and Technology, Taiwan [grant no. MOST108-2320-B-038–042-MY3].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Barnes P, Adcock IM, Ito K.. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Resp J 2005;25:552–63. [DOI] [PubMed] [Google Scholar]
- 2.Dokmanovic M, Clark C, Marks PA.. Histone deacetylase inhibitors: overview andperspectives. Mol Cancer Res 2007;5:981–9. [DOI] [PubMed] [Google Scholar]
- 3.Adcock IM. HDAC inhibitors as anti‐inflammatory agents. Br J Pharmacol 2007;150:829–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adcock IM, Ito K, Barnes PJ.. Histone deacetylation: an important mechanism in inflammatory lung diseases. COPD 2005;2:445–55. [DOI] [PubMed] [Google Scholar]
- 5.(a) Mehndiratta S, Pan SL, Kumar S, et al. Indole-3-ethylsulfamoylphenylacrylamides with potent anti-proliferative and anti-angiogenic activities. Anticancer Agents Med Chem 2016;16:907–13. [DOI] [PubMed] [Google Scholar]; (b) Huang YC, Huang FI, Mehndiratta S, et al. Anticancer activity of MPT0G157, a derivative of indolylbenzenesulfonamide, inhibits tumor growth and angiogenesis. Oncotarget 2015;6:18590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehndiratta S, Lin MH, Wu YW, et al. N-alkyl-hydroxybenzoyl anilide hydroxamates as dual inhibitors of HDAC and HSP90, downregulating IFN-γ induced PD-L1 expression. Eur J Med Chem 2020;185:111725. [DOI] [PubMed] [Google Scholar]
- 7.Ceccacci E, Minucci S.. Inhibition of histone deacetylases in cancer therapy: lessons from leukaemia. Br J Cancer 2016;114:605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Mehndiratta S, Liou JP.. Histone lysine specific demethylase 1 inhibitors. RSC Med Chem 2020;11:969–81. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Quintás-Cardama A, Santos FPS, Garcia-Manero G.. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia 2011;25:226–35. [DOI] [PubMed] [Google Scholar]
- 9.Mehndiratta S, Hsieh YL, Liu YM, et al. Indole-3-ethylsulfamoylphenylacrylamides: potent histone deacetylase inhibitors with anti-inflammatory activity. Eur J Med Chem 2014;85:468–79. [DOI] [PubMed] [Google Scholar]
- 10.Leoni F, Zaliani A, Bertolini G, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci 2002;99:2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinarello CA. Anti-inflammatory agents: present and future. Cell 2010;140:935–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi JH, Oh SW, Kang MS, et al. Trichostatin A attenuates airway inflammation in mouse asthma model. Clin Exper Allergy 2005;35:89–96. [DOI] [PubMed] [Google Scholar]
- 13.Nasu Y, Nishida K, Miyazawa S, et al. Trichostatin A, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthr Cartil 2008;16:723–32. [DOI] [PubMed] [Google Scholar]
- 14.Choo QY, Ho PC, Tanaka Y, et al. Histone deacetylase inhibitors MS-275 and SAHA induced growth arrest and suppressed lipopolysaccharide-stimulated NF-κB p65 nuclear accumulation in human rheumatoid arthritis synovial fibroblastic E11 cells. Rheumatology 2010;49:1447–60. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Zhang Z, Schluesener H.. MS-275 a histone deacetylase inhibitor, reduces the inflammatory reaction in rat experimental autoimmune neuritis. Neuroscience 2010;169:370–7. [DOI] [PubMed] [Google Scholar]
- 16.Leoni F, Fossati G, Lewis EC, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med 2005;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joosten LA, Leoni F, Meghji S, et al. Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Mol Med 2011;17:391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganai SA, Abdullah E, Rashid R, et al. Combinatorial in silico strategy towards identifying potential hotspots during inhibition of structurally identical HDAC1 and HDAC2 enzymes for effective chemotherapy against neurological disorders. Front Mol Neurosci 2017;10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Archin NM, Espeseth A, Parker D, et al. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 2009;25:207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Routy JP. Valproic acid: a potential role in treating latent HIV infection. Lancet 2005;366:523–4. [DOI] [PubMed] [Google Scholar]
- 21.Mehndiratta S, Wang RS, Huang HL, et al. 4-Indolyl-N-hydroxyphenylacrylamides as potent HDAC Class I and IIB inhibitors in vitro and in vivo. Eur J Med Chem 2017;134:13–23. [DOI] [PubMed] [Google Scholar]
- 22.Nepali K, Lee HY, Lai MJ, et al. Ring-opened tetrahydro-γ-carbolines display cytotoxicity and selectivity with histone deacetylase isoforms. Eur J Med Chem 2017; 127:115–27. [DOI] [PubMed] [Google Scholar]
- 23.Lee HY, Lee JF, Kumar S, et al. 3-Aroylindoles display antitumor activity in vitro and in vivo: Effects of N1-substituents on biological activity. Eur J Med Chem 2017;125:1268–78. [DOI] [PubMed] [Google Scholar]
- 24.Lee HY, Fan SJ, Huang FI, et al. 5-Aroylindoles act as selective histone deacetylase 6 inhibitors ameliorating Alzheimer’s disease phenotypes. J Med Chem 2018;61:7087–102. [DOI] [PubMed] [Google Scholar]
- 25.Huang HL, Lee HY, Tsai AC, et al. Anticancer activity of MPT0E028, a novel potent histone deacetylase inhibitor, in human colorectal cancer HCT116 cells in vitro and in vivo. PLoS ONE 2012;8:e43645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HY, Yang CR, Lai MJ, et al. 1-Arylsulfonyl-5-(N-hydroxyacrylamide) indolines histone deacetylase inhibitors are potent cytokine release suppressors. ChemBioChem 2013;14:1248–54. [DOI] [PubMed] [Google Scholar]
- 27.Lee H-Y, Chang C-Y, Su C-J, et al. 2-(Phenylsulfonyl)quinoline N-hydroxyacrylamides as potent anticancer agents inhibiting histone deacetylase. Eur J Med Chem 2016;122:92–101. [DOI] [PubMed] [Google Scholar]
- 28.Lee HY, Nepali K, Huang FI, et al. (N-Hydroxycarbonylbenylamino)quinolines as selective histone deacetylase 6 inhibitors suppress growth of multiple myeloma in vitro and in vivo. J Med Chem 2018;61:905–17. [DOI] [PubMed] [Google Scholar]
- 29.Tu HJ, Lin YJ, Chao MW, et al. The anticancer effects of MPT0G211, a novel HDAC6 inhibitor, combined with chemotherapeutic agents in human acute leukemia cells. Clin Epigenetics 2018;10:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.