Abstract
Background
Panax quinquefolius and Panax notoginseng are widely used and well known for their pharmacological effects. As main pharmacological components, saponins have different distribution patterns in the root tissues of Panax plants.
Methods
In this study, the representative ginsenosides were detected and quantified by desorption electrospray ionization mass spectrometry and high-performance liquid chromatography analysis to demonstrate saponin distribution in the root tissues of P. quinquefolius and P. notoginseng, and saponin metabolite profiles were analyzed by metabolomes to obtain the biomarkers of different root tissues. Finally, the transcriptome analysis was performed to demonstrate the molecular mechanisms of saponin distribution by gene profiles.
Results
There was saponin distribution in the root tissues differed between P. quinquefolius and P. notoginseng. Eight-eight and 24 potential biomarkers were detected by metabolome analysis, and a total of 340 and 122 transcripts involved in saponin synthesis that were positively correlated with the saponin contents (R > 0.6, P < 0.05) in the root tissues of P. quinquefolius and P. notoginseng, respectively. Among them, GDPS1, CYP51, CYP64, and UGT11 were significantly correlated with the contents of Rg1, Re, Rc, Rb2, and Rd in P. quinquefolius. UGT255 was markedly related to the content of R1; CYP74, CYP89, CYP100, CYP103, CYP109, and UGT190 were markedly correlated with the Rd content in P. notoginseng.
Conclusions
These results provided the visual and quantitative profiles of and confirmed the pivotal transcripts of CYPs and UGTs regulating the saponin distribution in the root tissues of P. quinquefolius and P. notoginseng.
Keywords: Metabolome, Panax plants, Root tissues, Saponin distribution, Transcriptome
Abbreviations: UGTs, UDP-glycosyltransferases; MEP, 2-C-methyl-D-erythritol-4-phosphate; MVA, Mevalonate acid; FPS, Farnesyl pyrophosphate synthase; SS, Squalene synthase; SE, Squalene epoxidase; DS, Dammarenediol-II synthase; UPLC-MS, Ultrahigh-performance liquid chromatography quadrupole time of flight-mass spectrometry; MALDI-MS, Matrix-assisted laser desorption/ionization–mass spectrometry; HPLC-UV, High-performance liquid chromatography-ultraviolet detection; IPP, Isoprenyl diphosphate; FPP, Farnesyl diphosphate; AACT, Acetoacetyl-CoA acyltransferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MVK, Mevalonate kinase; PMK, Phosphomevalonate kinase; MVD, Mevalonate diphosphate decarboxylase; IPPI, Isopentenyl pyrophosphate isomerase; GDPS, Gerenyl diphosphatesynthase; DXPS, 1-deoxy-o-xylulose 5-phosphate synthase; DXPR, 1-deoxy-o-xylulose 5-phosphate reductoisomerase; ISPD, 2-C-methylerythritol 4-phosphatecytidyl transferase; ISPE, 4-(cytidine-5′-diphospho)-2-C-methylerythritol kinase; ORF, Open read frame; MECPS, 2-C-methylerythritol-2,4-cyclophosphate synthase; GO, Gene Ontology; HDS, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphatesynthase; ISPH, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; P450, P450-monooxygenase; NCBI Nr, NCBI Non-redundant protein; OPLS-DA, Orthogonal partial least squares-discriminant analysis; FDR, False discovery rate; WGCNA, Weighted gene coexpression network analysis
1. Introduction
Panax species mainly include Panax ginseng, Panax notoginseng, Panax quinquefolius, Panax japonicus, Panax japonicus var. major, Panax pseudoginseng, Panax vietnamensis var. fuscidicus, and Panax zingiberensis [1], [2]. P. ginseng (Asian or Korean ginseng), P. notoginseng (Chinese ginseng or sanchi ginseng), and P. quinquefolius (American ginseng) have been widely used as medicines, functional food, health-care products, and food additives [3], [4]. The therapeutic effects of Panax plants are mainly attributed to their saponin components [5], [6]. Saponins can be used to treat central nervous system diseases, cardiovascular diseases, cancer, and diabetes [7], [8], [9], [10]. Saponins have potent therapeutic effects against cardiovascular diseases (R1) [11] and induce neuroprotection activity (Rb1) [12], antiinflammatory effect (Rb2 and Rc) [13], [14], antiangiogenic activity (Rd) [15], antioxidant activity (Re) [16], and hepatoprotective effect (Rg1) [17]. The types and quantities of saponins in Panax plants change with age [18], growth environment [19], and tissue type [20]. The compositions and contents of the ginsenosides Rg1, Rc, and Rd in P. ginseng vary with cultivation age and region [18]. The ginsenoside contents of mountain-cultivated P. ginseng increase with age [19]. High-performance liquid chromatography (HPLC) analysis has indicated that the varieties and contents of saponins differ among the aerial parts (flower, stem, and leaf) and underground parts (root and fibril) of P. notoginseng [21]. A quantitative comparison of ginsenosides revealed that the ratio of Rg1/Rd, (Rg1 + Re)/Rd, and protopanaxatriol-type saponins/protopanaxadiol (PPD)-type saponins presented a substantially large difference between cultivated and wild P. quinquefolius [22]. Interestingly, the amounts of ginsenosides in the cork are higher than those in the cortex and phloem of P. ginseng as indicated by analysis using matrix-assisted laser desorption/ionization (MALDI)–mass spectrometry (MS) and ultra-high-performance liquid chromatography (UPLC)–quadrupole/time-of-flight (QTOF)–MS [23]. These results suggest that the distribution of saponin varieties and contents vary among the root tissues of Panax plants. However, visual and quantitative evidence supporting the saponin distribution in the root tissues (periderm, phloem, and xylem) of P. quinquefolius and P. notoginseng is limited.
More than 150 naturally occurring saponins, including dammarane-type and oleanane-type saponins, have been isolated from Panax species [24], and metabolomics can be applied to analyze the panorama profiles of saponins in different parts of medicinal plants [21]. The distribution of saponin contents and varieties differ among the rhizome, main root, branch root, and fibrous root of P. notoginseng, and 32 saponins are selected as potential biomarkers [25]. Nevertheless, the saponin profiles of the root tissues of P. quinquefolius and P. notoginseng have been rarely reported. As such, the saponin distribution in root tissues should be determined for the targeted breeding of Panax plants.
Saponin distribution is related to a set of putative transcripts involved in the saponin synthesis of Panax plants [26]. Triterpene saponins are synthesized by multiple synthesis transcripts in the mevalonic acid pathway, which is the main route, and the methylerythritol phosphate pathway [21], [27]. Isoprenyl diphosphate and dimethylallyl pyrophosphate, which are produced in these two pathways, are catalyzed by a series of geranyl diphosphatesynthase (GDPS), farnesyl diphosphatesynthase [28], squalene synthase [29], squalene epoxidase, and dammarenediol-II synthase (DS) [30] to form dammarenediol-II and subsequently undergo cytochrome P450 (CYP) hydroxylation and UDP-glycosyltransferase (UGT) action [31]. The analysis of these pivotal transcripts provides useful information for the genetic improvement of Panax plants. Many transcripts related to saponin synthesis and distribution have been selected via transcriptome analysis [26], [27], [28], [29], [30], [31]. In ginseng, certain transcripts, such as 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), acetoacetyl-CoA acyltransferase (AACT), dammarendiol synthases (DS), and squalene epoxidase (SE), which are involved in saponin synthesis, have been identified and found to be highly expressed in the periderm; their expression patterns are consistent with saponin distribution [32], [33]. The contents of PPD- and protopanaxatriol-type saponins in P. notoginseng are also significantly correlated with the expression levels of CYP716A47 and CYP716A53v2, respectively [27]. Thus, we hypothesized that the expression patterns of transcripts participating in saponin synthesis regulated the saponin distribution in root tissues of Panax plants.
In this study, we investigated the visualization of saponin localization and characteristic chemical markers in the root tissues of P. quinquefolius and P. notoginseng through desorption electrospray ionozation mass spectrometry (DESI–MS) and UPLC–QTOF–MS. Furthermore, the candidate transcripts related to saponin distribution were screened and verified in accordance with the transcriptome of root tissues. Our study systematically analyzed the metabolomes and transcriptomes of the root tissues of P. quinquefolius and P. notoginseng and offered relevant information for the enhanced understanding of the saponin distribution in terms of chemical and biological aspects. Results contribute to the present data for the genetic improvement and target breeding of Panax species.
2. Materials and methods
2.1. Plant materials
Three-year-old roots of P. quinquefolius and P. notoginseng were, respectively, collected from Jingyu in Liaoning Province and Wenshan in Yunnan Province at their flowering stage. The samples were divided into two parts: one part was used for DESI–MS imaging analysis, and the other part was used for metabolomic and transcriptome analyses. Each batch (15 plants) of roots was carefully washed, separated into three different parts (periderm, phloem, and xylem), and stored for chemical and molecular analyses.
2.2. DESI–MS imaging analysis
Fresh roots were cut into 1-cm cross-sections by using a blade. The sections were immediately frozen in liquid nitrogen and cut into 20-μm sections at −20°C for DESI–MS imaging. Ginsenoside distribution was detected using a DESI mass spectrometer (Xevo G2-XS; Waters Corporation Shanghai Science & Technology Co Ltd). Images were obtained through high-definition imaging (Waters Corporation). The spray solvent was composed of 90% MeOH, 10% H2O, 0.1 mM NH4Cl, and 0.1 mM leucine enkephalin. The parameters were set as follows: spray solvent speed, 1.5 μL/min; X and Y pixel sizes, 100 pm; and raster speed, 400 pm/s. The parameters of MS at negative polarity were as follows: capillary voltage, 4.5 kV; cone voltage, 80 V; and mass range, m/z 100–1200.
2.3. Metabolite analysis
All the samples were dried and crushed, and 0.1 g of the powdered sample was weighed and mixed with 1.0 mL of pure methanol containing 0.1% formic acid under vortex for 10 s. The mixture was ultrasonicated for 10 min, frozen at −20 °C for 1 h, and centrifuged at 10,000 rpm for 10 min. The upper layer was collected, filtered through a 0.22-μm filter, and transferred to a sample vial. The vial was injected into a column for UPLC–QTOF–MS analysis.
UPLC–MS analysis was performed using an UPLC system (Waters) coupled to an electrospray ionization–QTOF/MS apparatus (Waters). A C18 reversed-phase column (50 mm × 2.1 mm, 1.7 μm inner diameter, Acquity UPLC BEH; Waters, UK) was used for UPLC separation, and the sample injection volume was 10 μL. The column temperature was kept at 35°C, and the flow rate was maintained at 0.4 mL/min. The gradient was composed of water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). The linear gradient was set as follows: 0–2 min for 99%–80% A, 2–3 min for 80%–50% A, 3–7 min for 50%–20% A, 7–7.5 min for 20%–1% A, 7.5–9 min for 1% A, 9–9.1 min for 1%–99% A, and 9.1–10 min for 99% A. Multivariate data analysis was achieved using MetaboAnalyst 4.0 software (Genome, Canada) (http://www.metaboanalyst.ca/). Potential biomarkers were selected using the following threshold values: false discovery rate (FDR) ≤ 0.05 for all chemical components and FDR ≤ 1 for saponin components.
2.4. HPLC–UV analysis
The standards of notoginsenoside R1 and ginsenosides Rb1, Rg1, Re, Rb2, Rd, and Rc were purchased from Shanghai Tauto Biotech Company (Shanghai, China). These standards have more than 98.0% purity, and the batch numbers of notoginsenoside R1 and ginsenosides Rb1, Rg1, Re, Rb2, Rd, and Rc were 160923, 160930, 16042724, 160907, 16060121, 160924, and 16081931, respectively.
The extracts of the samples were also used for saponin quantitative analysis. An Agilent HPLC 1260 series system (Agilent Technologies, USA) equipped with a quaternary pump, automatic sampler, column compartment, and ultraviolet detector (VWD) was used. A C18 reversed-phase column (4.6 mm × 250 mm, inner diameter of 5 μm, Eclipse XDB; Agilent) was used for separation, and the sample injection volume was set as 10 μL. The conditions for P. quinquefolius were set as follows: column temperature, 30°C; flow rate, 1.0 mL/min; and wavelength, 203 nm. The gradient was composed of acetonitrile (A) and water (B), and the linear gradient was set as follows: 0–25 min, 19% A to 20% A; 25–60 min, 20% A to 40% A; 60–90 min, 40% A to 55% A; and 90–100 min, 55% A to 60% A. The conditions for P. notoginseng were set as follows: column temperature, 25°C; flow rate, 1.0 mL/min; and wavelength, 203 nm. The gradient was composed of acetonitrile (A) and water (B), and the linear gradient was set as follows: 0–12 min for 19% A and 12–60 min for 19% A to 36% A.
2.5. RNA extraction and illumina sequencing
Total RNA was isolated from different tissues in accordance with the instructions in a plant RNA isolation kit (BioTeke, Beijing, China). The quality of RNA was evaluated on 1% agarose gel, and RNA concentrations were determined with a NanoDrop 2000 spectrophotometer (Thermo Technologies). cDNA library construction and sequencing were performed in accordance with the standards of progress. First, mRNA was enriched from the total RNA by oligo (dT) magnetic beads and broken into short fragments. A random hexamer and RNA fragments were then used to prime cDNA synthesis. After purification and connection with adapters, a cDNA library was constructed through polymerase chain reaction amplification. The length of an insert sequence was verified using an Agilent 2100 bioanalyzer, and the library was quantified using an ABI StepOnePlus Real-Time PCR System (Applied Biosystems). Finally, the qualified cDNA library was sequenced with an Illumina HiSeqTM 2000 system (Illumina Technologies). All the transcriptome sequences were submitted to the NCBI (Accession Number: SRR7764541-SRR7764549 and SRR7764553-SRR7764561).
2.6. Transcriptome analysis
Low-quality reads (more than 20% of bases with quality ≤ 10) and reads with adapters or containing more than 5% of unknown nucleotides were filtered to generate clean reads. De novo assembly and redundant sequence removal were performed in Trinity and Tgicl, respectively.
The resultant transcripts were searched against the NCBI nonredundant nucleotide (Nt) database, NCBI nonredundant protein (Nr), and SwissProt protein for functional annotation by using the BLAST algorithm with an E-value cutoff of 1e−5. The functional categories of these unique sequences were further analyzed using the Clusters of Orthologous Groups of Proteins (COG) database, Gene Ontology (GO) database, and Kyoto Encyclopedia of Genes and Genomes (KEGG) database in BLAST and Blast2GO programs.
The clean reads were mapped to the reference by using Bowtie (v2.2.6) to estimate the expression profiles of transcripts. The expression levels were calculated with the fragments per kilobase of exon per million fragments (FPKM) by using RNA-Seq by expectation maximization (RSEM) analysis. The candidate transcripts involved in saponin biosynthesis were selected in accordance with previous reports and databases with FPKM values of the transcripts converted to log10 values (FPKM ≥ 5). They were visualized in a heatmap to identify the different expression profiles among the three tissues.
2.7. Coexpression analysis
Weighted gene coexpression network analysis (WGCNA) was used to analyze the relationships between different transcripts and saponin contents with R package [34]. Before network interring, the expressing transcripts were normalized by square root transformation. All differentially expressing transcripts were clustered based on their FPKM values with the k-means method. The network construction and module detection method with default settings were used, including an unsigned type of topological overlap matrix [34]. All parameters were set as defined: “soft_power=15, deep_split=3, min_module size=30, merge Cut Height=0.2”. The P-values of 0.05 and R of 0.6 were set as the threshold for significantly high correlation. The significant and positive modules (R>0.6, P<0.05) were selected from the analysis of all transcripts with saponin contents, and then candidate transcripts involved in saponin biosynthesis were further selected in accordance with annotation information to analyze the relationship of transcripts expression profiles and saponin contents. The heatmap was constructed with R package to identify pivotal transcripts related to saponins contents.
3. Results
3.1. Distribution and contents of saponin in the root tissues of P. quinquefolius and P. notoginseng
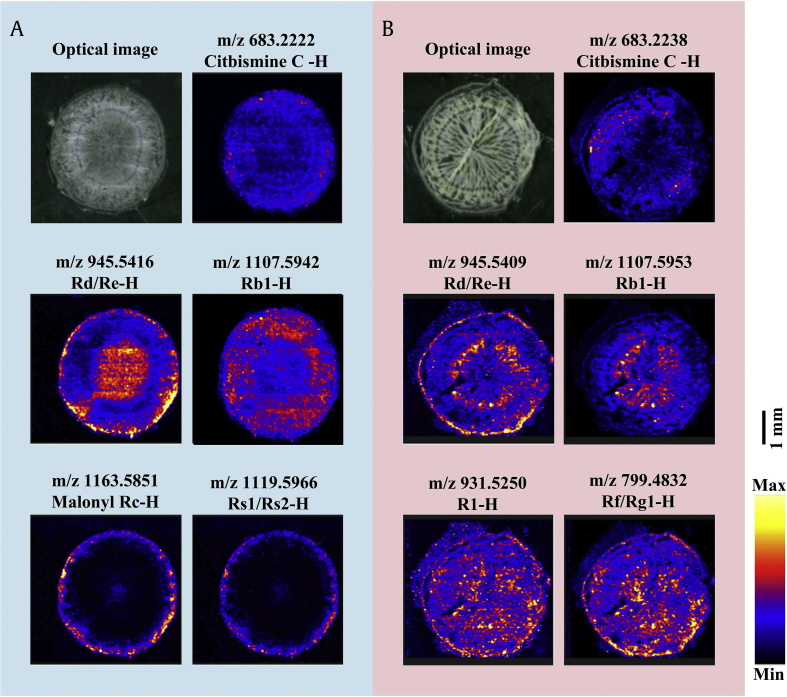
Saponin distribution in the root tissues of P. quinquefolius and P. notoginseng was visualized through DESI–MS imaging (Fig. 1). The ion images showed that the saponins had distinct localization and different relative abundances in three tissues. In P. quinquefolius, ginsenosides Rd/Re (m/z 945.5416 [M-H]) were found at high concentrations in the periderm and xylem of the root, whereas malonyl Rc and Rs1/Rs2 (m/z 1119.5966 [M-H]) were highly distributed in the periderm (Fig. 1A). In P. notoginseng, the ginsenosides Rd/Re (m/z 945.5409 [M-H]) were highly distributed in the periderm and xylem, and Rb1 (m/z 1107.5953 [M-H]) was highly concentrated in the xylem. Rg1/Rf (m/z 799.4832 [M-H]) and notoginsenoside R1 (m/z 931.5250 [M-H]) were widely distributed throughout the root sections (Fig. 1B). Thus, the saponin distribution in the root tissues differed between P. quinquefolius and P. notoginseng depending on the saponin types.
Fig. 1.
Saponin distribution in P. quinquefolius and P. notoginseng root cross sections on the basis of the DESI–MS. (A) P. quinquefolius. (B) P. notoginseng. Scale bar = 2 mm. MS, mass spectrometry.
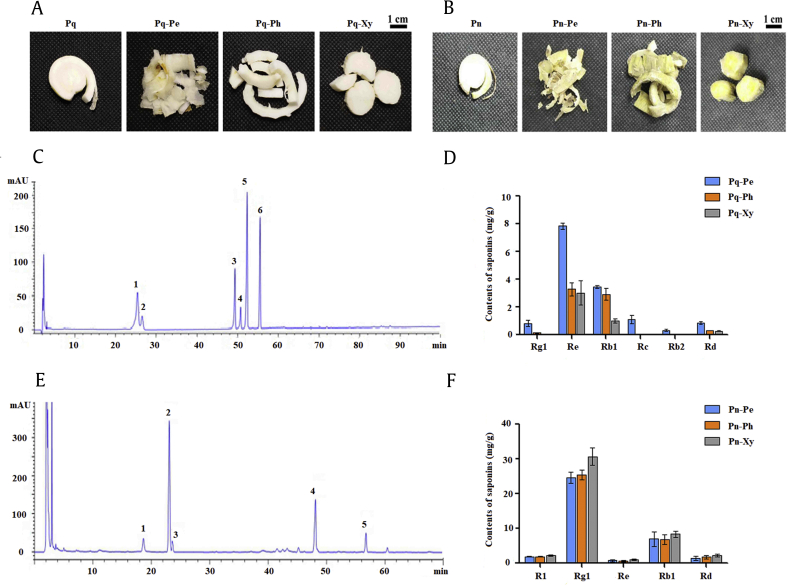
The root tissues were peeled to obtain the three tissues (periderm, phloem, and xylem) in P. quinquefolius and P. notoginseng (Fig. 2A and B). The contents of the representative saponins (R1, Rb1, Rg1, Re, Rb2, Rc, and Rd) were quantified in the root tissues of P. quinquefolius and P. notoginseng through HPLC (Fig. 2C–F). In P. quinquefolius, the highest contents of Rg1, Re, Rb1, Rc, Rb2, and Rd were detected in the periderm (0.83, 7.85, 3.46, 1.11, 0.33, and 0.86 mg/g, respectively). Their contents were significantly higher than those in the phloem (0.14, 3.29, 2.93, 0.05, 0, and 0.3 mg/g, respectively) and xylem (0.05, 3.02, 1.02, 0, 0, and 0.28 mg/g, respectively; Fig. 2D). In P. notoginseng, the contents of saponins (R1, Rb1, Re, Rb1, and Rd) were higher in the xylem (2.27, 30.69, 1.03, 8.41, and 2.30 mg/g, respectively) than in the periderm (1.93, 24.64, 0.75, 6.94, and 1.48 mg/g, respectively) and the phloem (1.91, 25.44, 0.66, 6.90, and 1.79 mg/g, respectively), but their differences were insignificant in the root tissues (Fig. 2F). These results indicated that the saponin distributions differed (P < 0.05) in the root tissues of P. quinquefolius, and the differences in the representative saponin contents were insignificant (P < 0.05) in the root tissues of P. notoginseng.
Fig. 2.
Main root tissue and saponin content analysis in P. quinquefolius and P. notoginseng. (A) Main root tissues in P. quinquefolius. Pq means the root cross section of P. quinquefolius; Pq-Pe means the periderm of P. quinquefolius; Pq-Ph means the periderm of P. quinquefolius; and Pq-Xy means the xylem of P. quinquefolius. (B) Main root tissues in P. notoginseng. Pn means the root cross section of P. notoginseng; Pn-Pe means the periderm of P. notoginseng; Pn-Ph means the periderm of P. notoginseng; and Pn-Xy means the xylem of P. notoginseng. (C) HPLC chromatogram profiles of P. quinquefolius plants: 1, ginsenoside Rg1; 2, ginsenoside Re; 3, ginsenoside Rb1; 4, ginsenoside Rc; 5, ginsenoside Rb2; 6, ginsenoside Rd. (D) Contents of six saponins in the roots of P. quinquefolius. (E) HPLC chromatogram profiles of P. notoginseng plants: 1, notoginsenoside R1; 2, ginsenoside Rg1; 3, ginsenoside Re; 4, ginsenoside Rb1; 5, ginsenoside Rd. (F) Contents of five saponins in the roots of P. notoginseng. HPLC, high-performance liquid chromatography.
3.2. Saponin components in the root tissues of P. quinquefolius and P. notoginseng
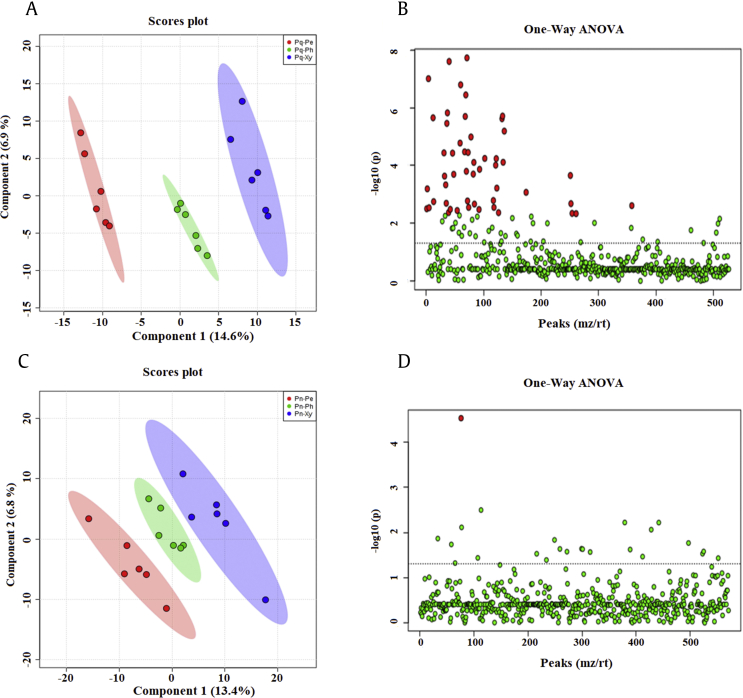
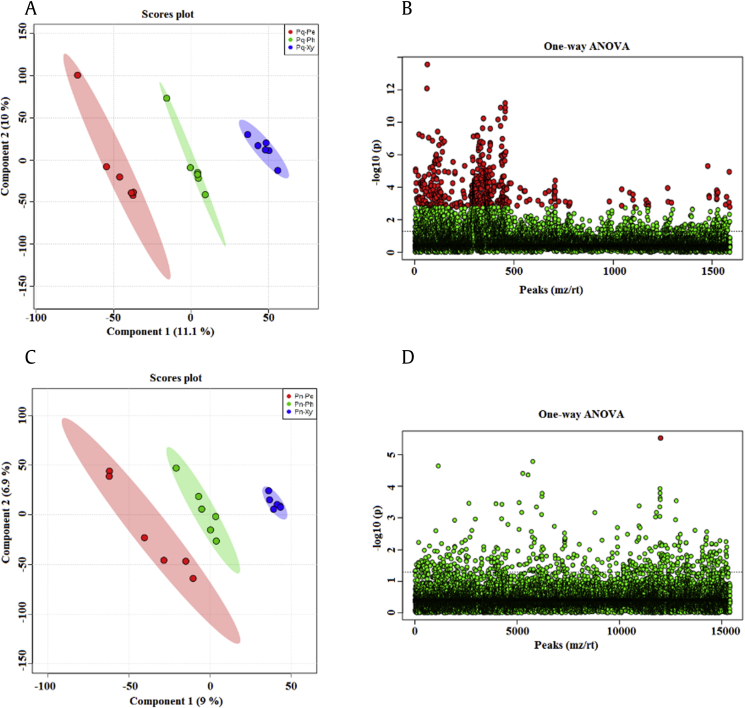
Orthogonal partial least squares-discriminant analysis (OPLS–DA) revealed the differences in the chemical components of the root tissues of P. quinquefolius and P. notoginseng (Fig. S1). In P. quinquefolius, supervised OPLS–DA results showed the clear separation of three root tissues (Fig. S1A). A total of 535 potential biomarkers were found through one-way analysis of variance (ANOVA) (FDR ≤ 0.05; Fig. S1B and Table S1). In P. notoginseng, OPLS–DA results showed that three root tissues were clearly separated (Fig. S1C). One-way ANOVA test results indicated that only one potential biomarker was found (FDR ≤ 0.05; Fig. S1D and Table S1). These data suggested that the distribution of the chemical components of the root tissues differed between P. quinquefolius and P. notoginseng, and the number of potential biomarkers was higher in the root tissues of P. quinquefolius than in those of P. notoginseng.
The saponin contents and types differed in the root tissues of P. quinquefolius and P. notoginseng (Fig. 3). OPLS–DA analysis revealed that the periderm, phloem, and xylem of P. quinquefolius clearly separated into different metabolic profiles (Fig. 3A). One-way ANOVA indicated the presence of 88 potential biomarkers in the root tissues of P. quinquefolius (FDR ≤ 1; Fig. 3B and Table S2). Furthermore, three root tissues of P. notoginseng displayed disparate metabolic profiles, and 24 potential biomarkers were detected (FDR ≤ 1; Fig. 3C and D and Table S2). These metabolite data showed that the distribution of saponins differed among the three root tissues of P. quinquefolius and P. notoginseng.
Fig. 3.
Metabolomic analysis of saponin difference among the root tissues in P. quinquefolius and P. notoginseng. (A) OPLS–DA score plots of P. quinquefolius. (B) One-way ANOVA of P. quinquefolius. (C) OPLS–DA score plots of P. notoginseng. (D) One-way ANOVA of P. notoginseng. ANOVA, analysis of variance; OPLS–DA, orthogonal partial least squares-discriminant analysis.
3.3. Transcriptome analysis of the root tissues of P. quinquefolius and P. notoginseng
Illumina Hiseq paired-end sequencing technology was used to analyze the transcriptome of root tissue samples (periderm, phloem, and xylem) of P. quinquefolius and P. notoginseng. After the low-quality reads were filtered, the averages of 6.07 G and 6.13 G clean reads were obtained from P. quinquefolius and P. notoginseng samples, respectively. After de novo assembly was conducted, the clean reads of P. quinquefolius and P. notoginseng were assembled into 84,408 and 80,472 transcripts, respectively. In P. quinquefolius, the percentage of GC was 41.68% with an average contig size of 1,197 bp and an N 50 contig size of 1,720 bp. In P. notoginseng, the percentage of GC was 41.25% with an average contig size of 1,290 bp and an N 50 contig size of 1,839 bp (Table 1).
Table 1.
Summary of the transcripts and assembly results for P. quinquefolius and P. notoginseng
| Item | P. quinquefolius | P. notoginseng |
|---|---|---|
| Average number of raw data (G) | 6.07 | 6.13 |
| Number of unigenes | 84,408 | 80,472 |
| Length of unigene (bp) | 10,11,04,546 | 10,38,55,332 |
| Average contig size (bp) | 1,197 | 1,290 |
| N50 contig size (bp) | 1,720 | 1,839 |
| GC (%) | 41.68 | 41.25 |
To investigate the function of assembled transcripts, we performed annotation via a sequence similarity search with a cutoff E-value of 10−5 against public databases, including GO, COG, KEGG, Nr, and Swiss-Prot (Table S3). In P. quinquefolius, 69,951 transcripts (82.87%) were annotated from the databases. A total of 68,720 (81.41%) transcripts showed significant matches in the Nr database, and 52,118 (61.75%), 52,176 (61.81%), 28,431 (33.68%), and 42,535 (50.39%) transcripts had significant matches with the Swiss-Prot, KEGG, COG, and GO databases, respectively. In P. notoginseng, 66,871 transcripts (83.10%) were annotated in the databases. A total of 65,889 (81.88%) transcripts exhibited significant matches in the Nr database, whereas 51,217 (63.65%), 51,130 (63.54%), 28,982 (36.02%), and 42,490 (52.80%) transcripts had significant matches with the Swiss-Prot, KEGG, COG, and GO databases, respectively.
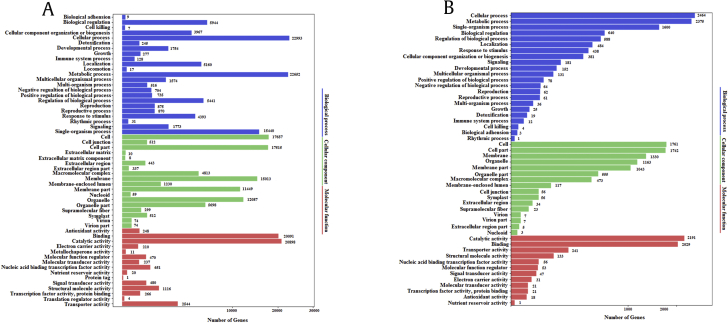
GO terms were used to assign the unigene sets and classify the gene functions. In P. quinquefolius, 230,542 transcripts were classified into three groups in GO terms, namely, biological process (95,505), cellular component (87,790), and molecular function (47,247). These terms were further categorized into 56 subcategories. Cellular process, metabolic process, and single-organism process were the most abundant terms in the biological process category. The most dominant subcategories in the cellular component were cell, cell part, and membrane. Catalytic activity and binding were the most represented terms in molecular function. In P. notoginseng, a low number of sequences were mapped to GO terms. A total of 23,016 transcripts were classified into three groups in GO terms, namely, biological process (9,799), cellular component (8,375), and molecular function (4,842). These terms were further categorized into 50 subcategories. Similar to the categories in P. quinquefolius, cellular process, metabolic process, and single-organism process were the most abundant terms in the biological process category. The most dominant subcategories in the cellular component were cell, cell part, and membrane. Catalytic activity and binding were the most represented terms in molecular function (Fig. S2).
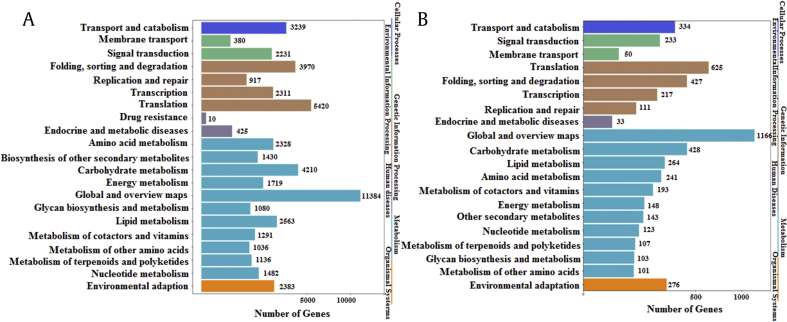
The annotated sequences were also mapped to the KEGG pathways. In P. quinquefolius, 50,945 transcripts were assigned to 21 KEGG pathways. The metabolic pathway achieved the most represented sequences (29,659), including global and overview maps (11,384), carbohydrate metabolism (4,210), and lipid metabolism (2,563). Notably, 1,136 sequences were annotated to the metabolism of terpenoids and polyketides. In P. notoginseng, 5,323 transcripts were assigned to 20 KEGG pathways. Similarly, the metabolic pathway achieved the most represented sequences (3,293), including global and overview maps (1,166), carbohydrate metabolism (428), and lipid metabolism (264). Moreover, 107 sequences were annotated to the metabolism of terpenoids and polyketides. These annotation results provided valuable information for analyzing metabolic pathways in P. quinquefolius and P. notoginseng (Fig. S3).
3.4. Analysis of differential gene expression in the root tissues of P. quinquefolius and P. notoginseng
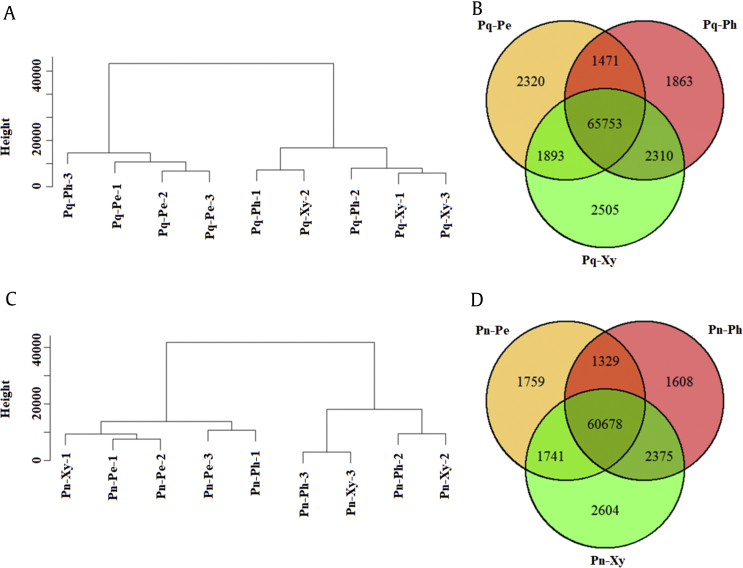
Cluster dendrograms and Venn profiles were constructed to investigate the transcription distinction among the root tissues of P. quinquefolius and P. notoginseng on the basis of the FPKM value (Fig. 4). Cluster dendrogram results showed that the periderm and xylem were finely assembled into each group in P. quinquefolius, and three root tissues of P. notoginseng were not clearly separated into independent branches. The Venn results revealed that 78,115 transcripts were obtained in P. quinquefolius, and 65,753 of these transcripts were shared among the three root tissues. Furthermore, 2,320, 1,863, and 2,505 transcripts were specifically expressed in the periderm, phloem, and xylem, respectively (Fig. 4A). In P. notoginseng, 72,094 transcripts were obtained, and 60,678 of these transcripts were shared among the three root tissues. Moreover, 1,759, 1,608, and 2,604 transcripts were specifically expressed in the periderm, phloem, and xylem, respectively (Fig. 4B). These results showed that the transcripts of the three root tissues of P. quinquefolius and P. notoginseng were differentially expressed.
Fig. 4.
Clustering and Venn profiles of gene expression in the three tissues of roots in P. quinquefolius and P. notoginseng. (A) Clustering tree of gene expression in three tissues of P. quinquefolius roots. (B) Venn profiles of gene expression in the three tissues of P. notoginseng roots. (C) Clustering tree of gene expression in the three tissues of P. quinquefolius roots. (D) Venn profiles of gene expression in the three tissues of P. notoginseng roots.
3.5. Coexpression analysis of all transcripts and saponin contents
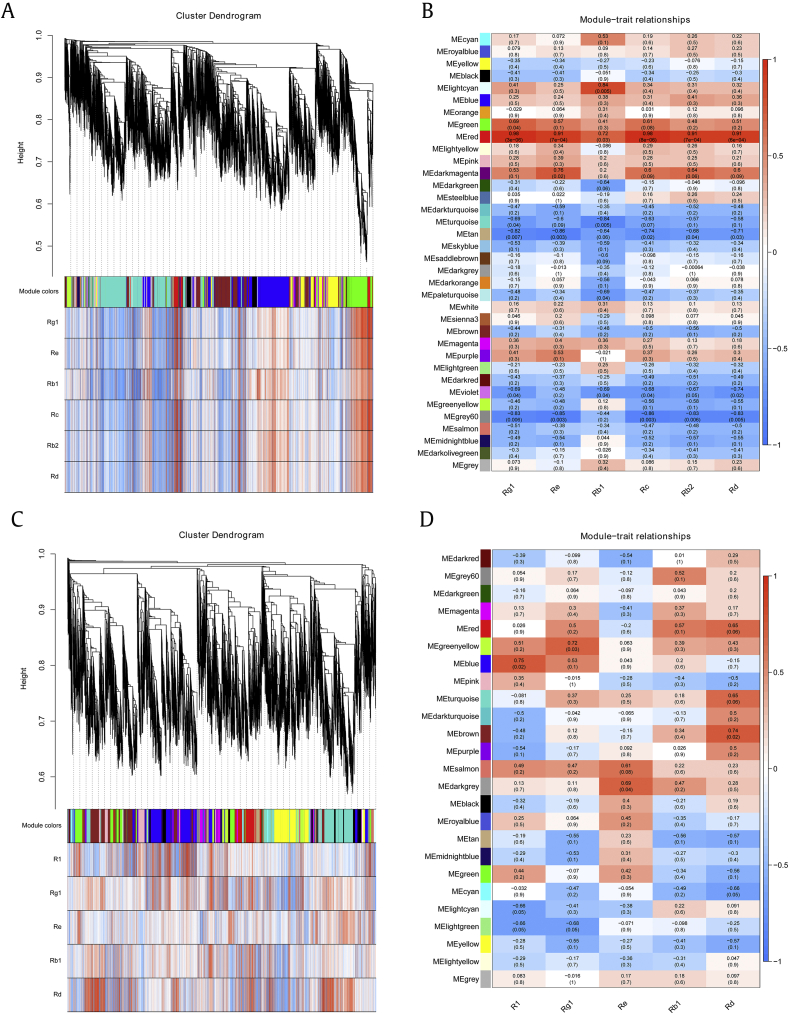
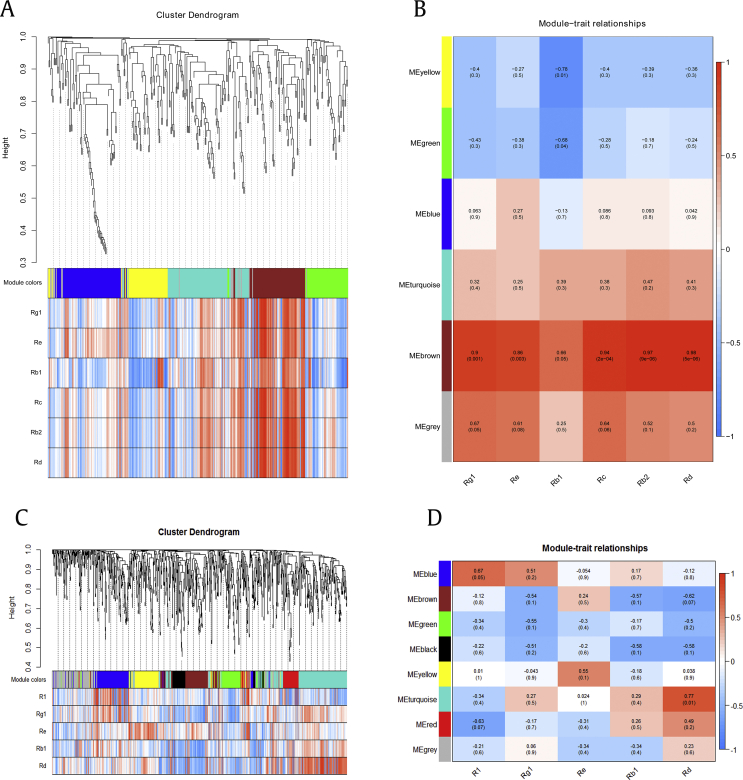
The hierarchical cluster tree was constructed with transcript expression and saponin contents through WGCNA. In P. quinquefolius, all transcripts were grouped into 35 unique modules, of which 44,067 transcripts were positively associated with saponin contents. Among these transcripts, four modules were positively correlated with saponin contents, namely, MElightcyan, MEgreen, MEred, and MEdarkmagenta (R>0.6, P<0.05; Fig. 5A). Further results showed that 73 transcripts in the MElightcyan module were positively correlated with the content of ginsenoside Rb1 (R=0.84, P<0.05). A total of 604 transcripts in the MEgreen module were positively associated with the content of ginsenoside Rg1 (R=0.69, P<0.05). By contrast, 449, 427, 179, 443, 404, and 412 transcripts in the MEred module were strongly positively correlated with the contents of ginsenosides Rg1 (R=0.98, P<0.05), Re (R=0.91, P<0.05), Rb1(R=0.72, P<0.05), Rc (R=0.98, P<0.05), Rb2 (R=0.91, P<0.05), and Rd (R=0.91, P<0.05). Thirteen transcripts in the MEdarkmagenta module were positively correlated with the content of ginsenoside Re (R=0.76, P<0.05; Fig. 5B).
Fig. 5.
Coexpression profiles of all transcripts and saponin contents in root tissues of P. quinquefolius and P. notoginseng. (A) Hierarchical cluster tree showing coexpression modules in P. quinquefolius. (B) Module–saponin association in P. quinquefolius. (C) Hierarchical cluster tree showing coexpression modules in P. notoginseng. (D) Module–saponin association in P. notoginseng.
In P. notoginseng, 25 unique modules were identified, of which 59,503 transcripts were positively associated with saponin contents, and four coexpression modules, namely, MEgreenyellow, MEblue, MEbrown, and MEdarkgrey, were highly positively correlated with saponin contents (R>0.6, P<0.05; Fig. 5C). For example, 115 transcripts in the MEgreenyellow module were positively correlated with the content of ginsenoside Rg1 (R=0.72, P<0.05). A total of 660 transcripts in the MEblue module were positively associated with the content of notoginsenoside R1 (R=0.75, P<0.05). Approximately 331 transcripts in the MEbrown module were positively correlated with the content of ginsenoside Rd (R=0.74, P<0.05). One transcript in the MEdarkgrey module was positively correlated with the content of ginsenoside Re (R=0.69, P<0.05; Fig. 5D). These results suggested the involvement of complex mechanisms of saponin synthesis in P. quinquefolius and P. notoginseng.
3.6. Analysis of transcripts involved in saponin synthesis in the root tissues of P. quinquefolius and P. notoginseng
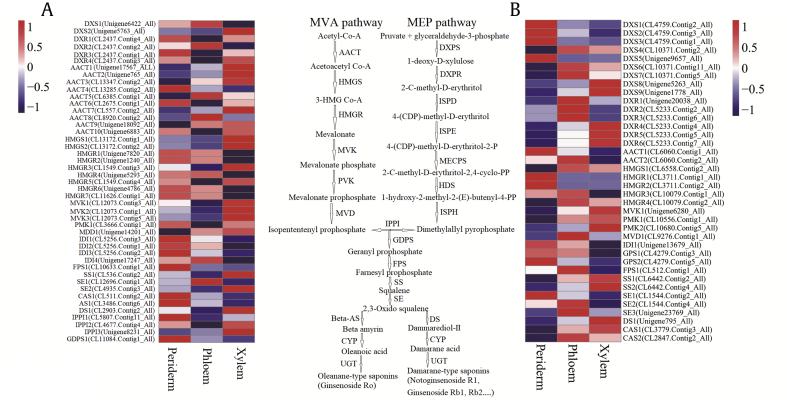
The annotation results showed that 19 (406 transcripts) and 18 genes (1174 transcripts) encoding the enzymes involved in saponin biosynthesis in the mevalonic acid and methylerythritol phosphate pathways were identified in P. quinquefolius and P. notoginseng, respectively. Multiple copies of the key transcripts and enzymes resulted in a widened variety of regulatory controllers in saponin biosynthesis.
A total of 158 and 426 transcripts were obtained in the root tissues of P. quinquefolius and P. notoginseng (FPKM ≥ 5), respectively, and the expression of most transcripts significantly differed (Table S4). In the upstream pathway of P. quinquefolius, 45 transcripts were selected to analyze expression profiles, and 21 (46.7%), 5 (11.1%), and 19 (42.2%) transcripts were expressed at the highest levels in the periderm, phloem, and xylem, respectively (Table S4 and Fig. 6A). For example, four 1-Deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) transcripts (CL2437. Contig 1_All, Contig 2_All, Contig 3_All, and Contig 4_All), two AACT transcripts (CL13285. Contig2_All and CL2675. Contig1_All), three Isopentenyl diphosphate isomerase (IDI) transcripts (CL5256. Contig1_All, Contig2_All, and Contig3_All), one AS transcript (CL3486. Contig6_All), one isopentenyl pyrophosphate isomerase transcript (CL5807. Contig11_All), and one GDPS transcript (CL11084. Contig1_All) showed the highest expression levels in the periderm. Five AACT transcripts (Unigene17567_All, Unigene765_All, CL13347. Contig2_All, CL557. Contig2_All, and Unigene18092_All) and three mevalonate kinase transcripts (CL12073. Contig1_All, Contig2_All, and Contig3_All) exhibited the highest expression levels in the xylem. In the upstream pathway of P. notoginseng, 38 transcripts with FPKM ≥ 5 were selected to analyze expression profiles, and 11 (28.9%), 15 (39.5%), and 12 (31.6%) transcripts yielded the highest expression levels in the periderm, phloem, and xylem, respectively (Table S4 and Fig. 6B). The expression levels of three 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) (CL4759. Contig1_All, Contig2_All, and Contig3_All), one AACT (CL6060. Contig1_All), two HMGR (CL3711. Contig1_All and Contig2_All), one IDI (Unigene13679_All), and two squalene epoxidase (CL1544. Contig2_All and Contig4_All) transcripts were high in the periderm. The most predominant expression levels of two DXR (CL5233. Contig4_All and Contig5_All) transcripts, one mevalonate kinase (Unigene6280_All) transcript, and one DS (Unigene795_All) transcript were observed in the xylem.
Fig. 6.
Expression of MVA pathway transcripts involved in saponin biosynthesis in P. quinquefolius and P. notoginseng. (A) Heatmap of gene expression in P. quinquefolius. (B) Heatmap of gene expression in P. notoginseng. AACT, acetoacetyl-CoA acyltransferase; DS, dammarenediol-II synthase; DXPR, 1-deoxy-o-xylulose 5-phosphate reductoisomerase; DXPS, 1-deoxy-o-xylulose 5-phosphate synthase; FPS, farnesyl pyrophosphate synthase; GDPS, geranyl diphosphatesynthase; HDS, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphatesynthase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; IPPI, isopentenyl pyrophosphate isomerase; ISPD, 2-C-methylerythritol 4-phosphatecytidyl transferase; ISPE, 4-(cytidine-5′-diphospho)-2-C-methylerythritol kinase; ISPH, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; MECPS, 2-C-methylerythritol-2,4-cyclophosphate synthase; MEP, methylerythritol phosphate; MVA, mevalonic acid; MVK, mevalonate kinase; MVD, mevalonate diphosphate decarboxylase; SE, squalene epoxidase; SS, squalene synthase. PVK, Phosphomevalonate kinase.
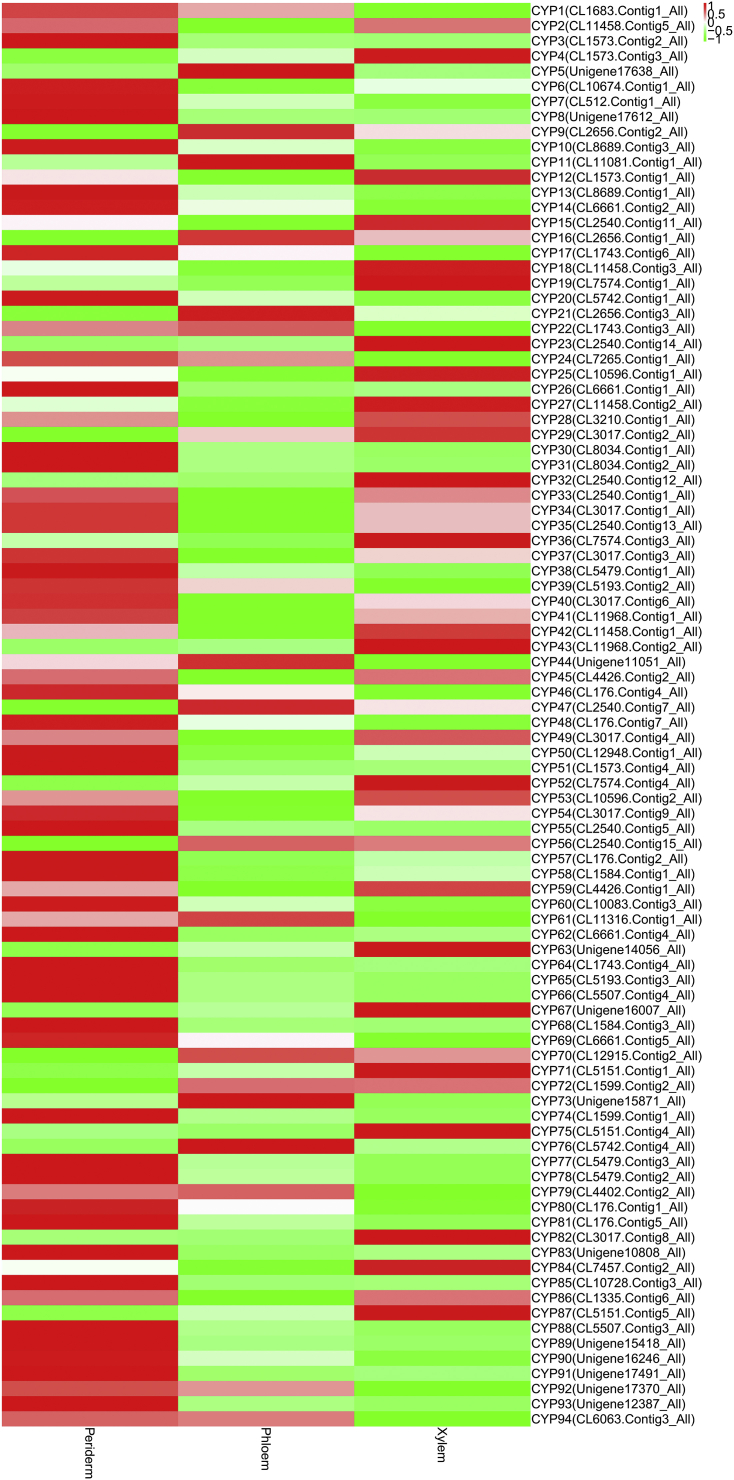
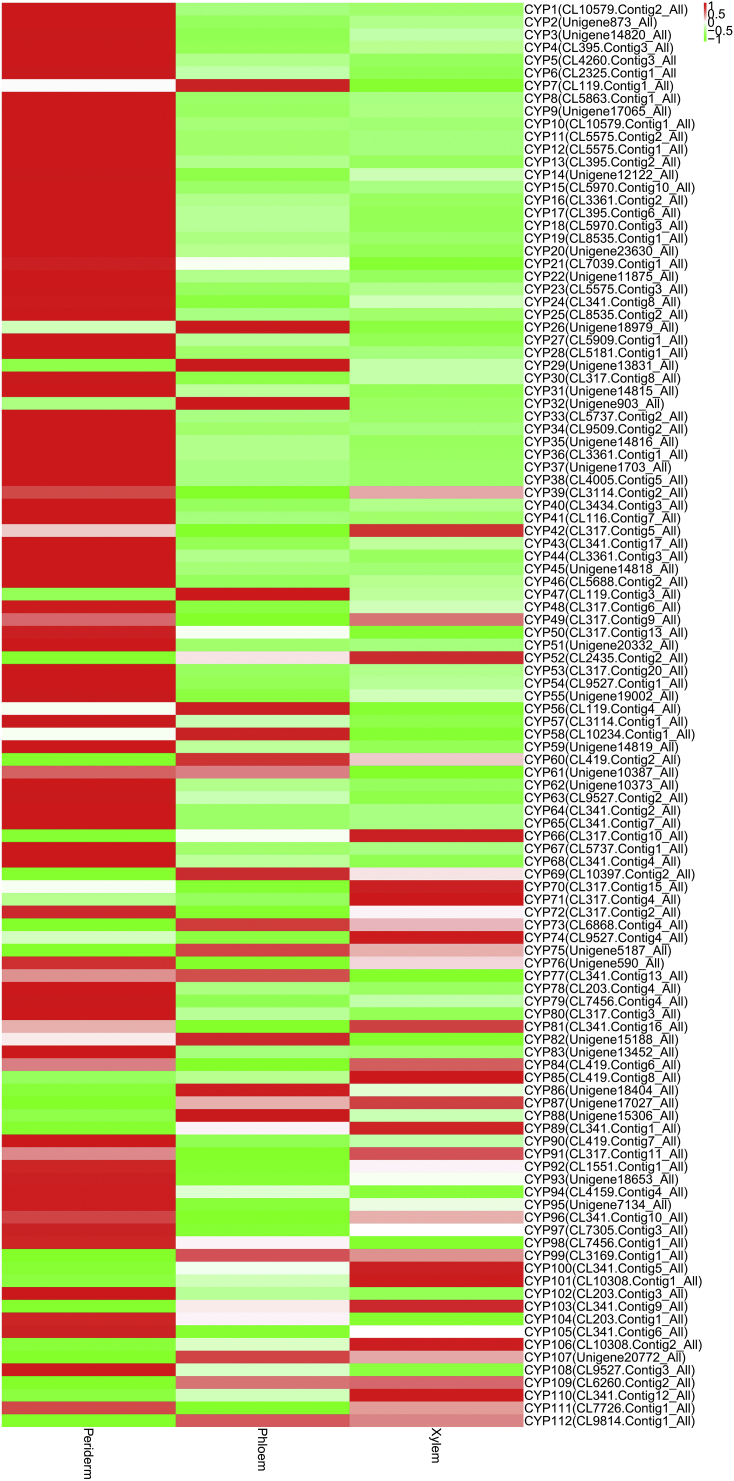
CYP families had different expression profiles in the root tissues of P. quinquefolius and P. notoginseng. A total of 94 CYP transcripts were identified in P. quinquefolius; 54, 15, and 25 of these CYP transcripts (57.4%, 16.0%, and 26.6%) showed the highest expression levels in the periderm, phloem, and xylem, respectively (Table S4 and Fig. S4). A total of 112 CYP transcripts were identified in P. notoginseng. Of these CYP transcripts, 75 (67.0%) and 17 (15.2%) were upregulated in the periderm and phloem, respectively, and 20 (17.8%) were upregulated in the xylem (Table S4 and Fig. S5). These results indicated that the CYP transcripts were more highly expressed in the periderm than in the other root tissues of P. quinquefolius and P. notoginseng.
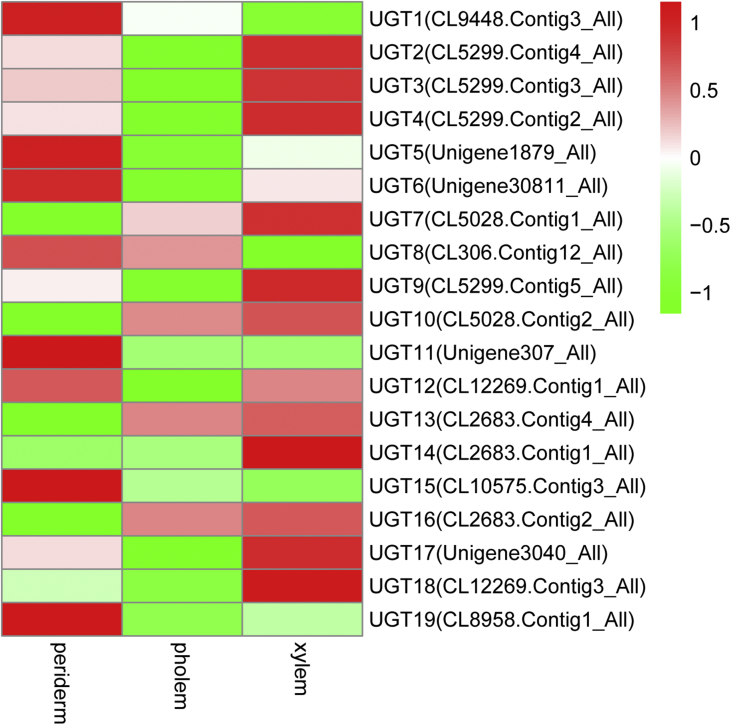
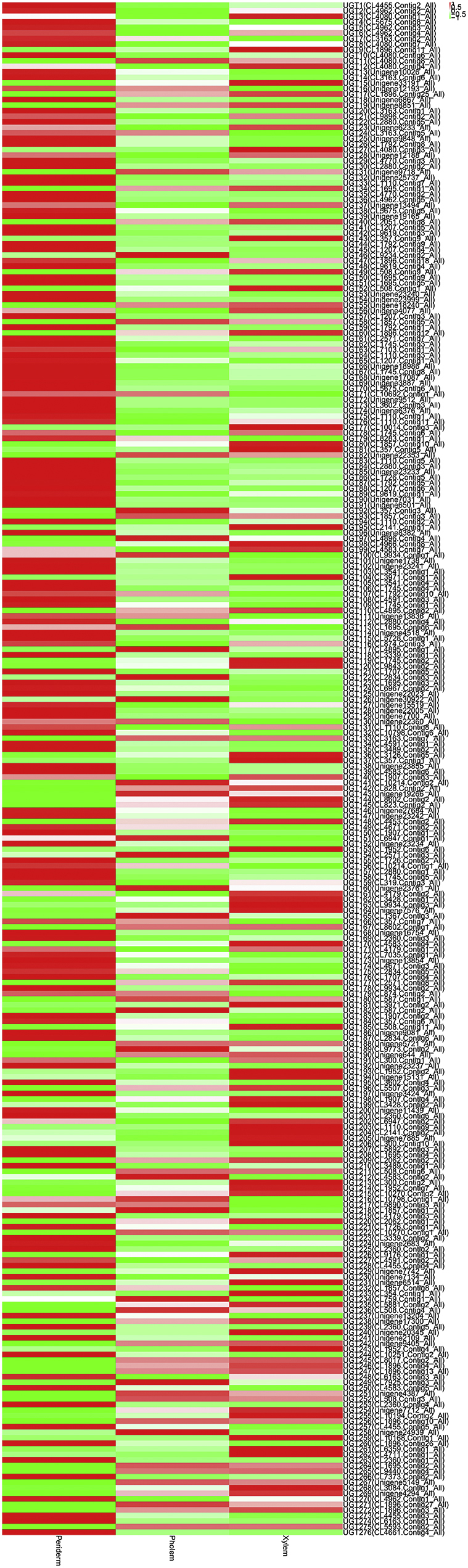
The UGT families also had different expression profiles in the root tissues of P. quinquefolius and P. notoginseng. A total of 19 UGT transcripts were identified in P. quinquefolius. Among these UGT transcripts, 8 (42.1%) had the highest expression in the periderm and 10 (52.6%) had the highest expression in the xylem (Table S4 and Fig. S6). In P. notoginseng, 276 UGT transcripts were identified; the expression levels of 152 (55.1%), 42 (15.2%), and 82 UGT transcripts (29.7%) were the highest in the periderm, phloem, and xylem, respectively (Table S4 and Fig. S7). These results indicated that the UGT transcripts were highly expressed in the periderm and xylem of P. quinquefolius and P. notoginseng.
3.7. Coexpression analysis of saponin contents and transcripts involved in saponin synthesis
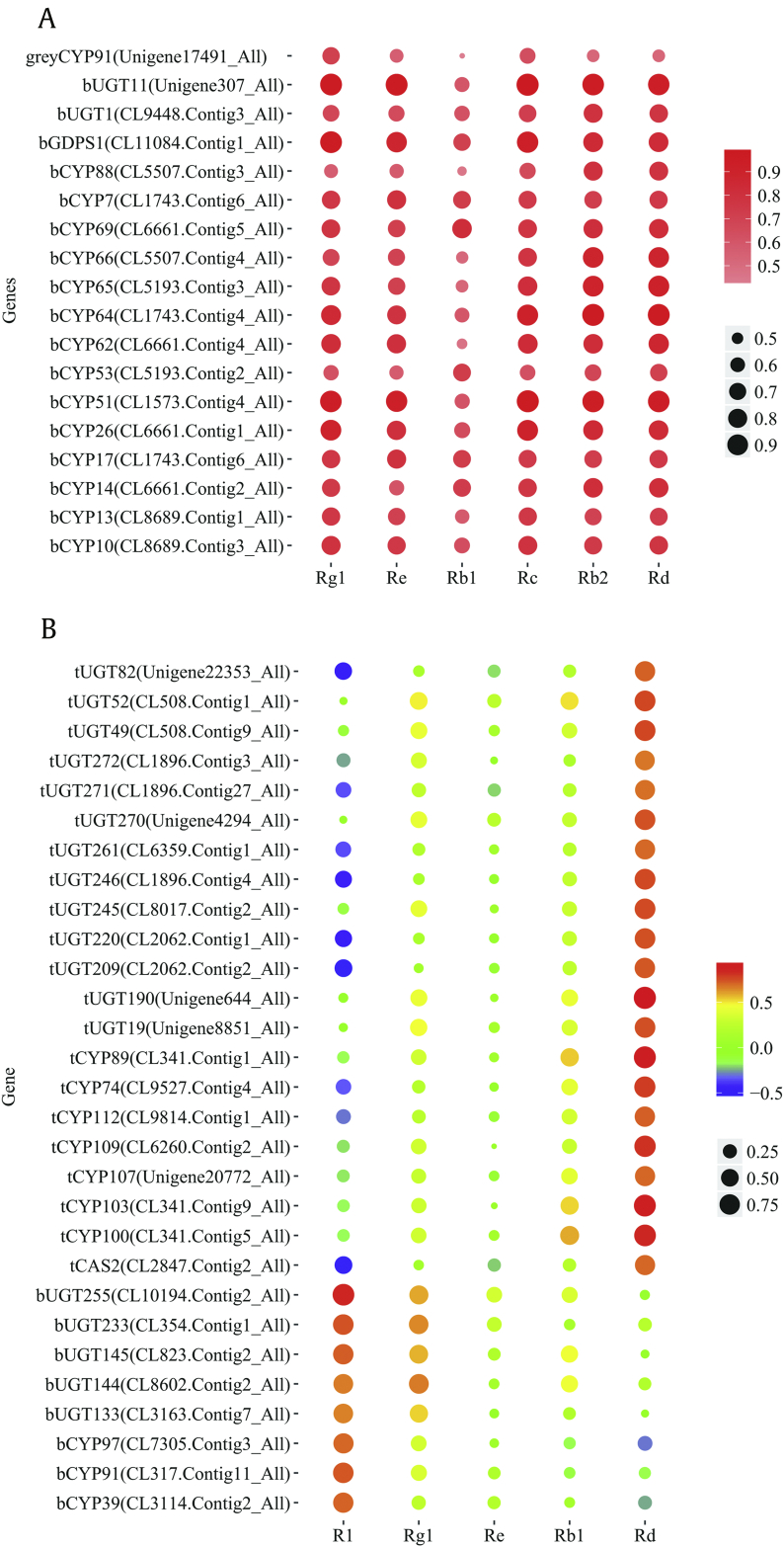
Transcripts involving unique modules that were positively associated with saponin contents were selected from the WGCNA (Fig. S8). Transcripts involving saponin synthesis were further selected according to annotation information to analyze the relationship of transcript expression profiles and saponin contents. A total of 340 transcripts were positively associated with saponin contents, and MEbrown and MEgrey were highly positively correlated with saponin contents (R>0.6, P<0.05; Fig. S8A). A total of 32, 25, 4, 3, 30, and 25 transcripts in the MEbrown module were significantly positively correlated with the contents of ginsenosides Rg1, Re, Rb1, Rc, Rb2, and Rd, respectively. Unigene17491_All in the MEgrey module was markedly positively associated with the ginsenoside Rg1 content (Fig. S8B). Highly expressed transcripts (FPKM ≥ 5) were selected and annotated as GDPS (1 transcript), CYP (15 transcripts), and UGT (2 transcripts), among which GDPS1 (CL11084.Contig1_All), CYP51 (CL1573.Contig4_All), CYP64 (CL1743.Contig4_All), and UGT11 (Unigene307_All) had significantly high positive correlations with Rg1, Re, Rc, Rb2, and Rd (R>0.8, P<0.05; Fig. 7A).
Fig. 7.
Pearson correlation bubble chart of annotation transcripts and saponin contents in P. quinquefolius and P. notoginseng. (A) P. quinquefolius: b represents brown; g represents grey. (B) P. quinquefolius: b represents blue; t represents turquoise.
Eight unique modules were identified in P. notoginseng, of which 122 transcripts were positively correlated with saponin contents, and modules of MEblue and MEturquoise were highly positively correlated with saponin contents (R>0.6, P<0.05; Fig. S8C). Investigating the MEblue module and saponin correlation revealed that 23 transcripts were slightly positively correlated with the content of notoginsenoside R1 (R=0.67, P<0.05). A total of 76 transcripts in the MEturquoise module were highly positively correlated with the content of ginsenoside Rd (R=0.77, P<0.05; Fig. S8D). The selected highly expressed transcripts (FPKM ≥ 5) were annotated as CAS (1 transcript), CYP (10 transcripts), and UGT (18 transcripts). Among these transcripts, the relative abundance of UGT255 (CL10194.Contig2_All) had significantly high correlation with R1 (R=0.88, P<0.05) and CYP74 (CL9527.Contig4_All), whereas the relative abundance of CYP89 (CL341.Contig1_All), CYP100 (CL341.Contig5_All), CYP103 (CL341.Contig9_All), CYP109 (CL6260.Contig2_All), and UGT190 (Unigene644_All) had significantly strong positive correlation with Rd (R>0.8, P<0.05; Fig. 7B).
4. Discussion
In this study, the saponin distribution was dependent on the types of saponins and demonstrated tissue specificity in P. quinquefolius and P. notoginseng roots. Transcript expression profiles were tissue specific in the roots of P. quinquefolius and P. notoginseng. In addition, WGCNA further confirmed that the pivotal CYP and UGT transcripts regulated the saponin distribution in the root tissues. This study could offer useful information for investigating the genetic and biochemical mechanisms of saponin synthesis.
Visual and quantitative analyses revealed that the saponin content (Rd, Re, and Rc) was significantly higher in the periderm of P. quinquefolius than those in the phloem and xylem. By contrast, the saponin (R1, Rg1, Re, Rb1, and Rd) distribution was not significantly different (P < 0.05) throughout the root sections of P. notoginseng. Laser microdissection analysis demonstrated that the ginsenosides were more highly accumulated in the periderm than in the medulla of P. ginseng [23]. Researchers also found that ginsenosides are highly located in the outer core and poorly located in the central parts of P. ginseng [33]. In the present study, ginsenosides (e.g., Rd, Re, malonyl Rc, and Rs1/Rs2) were highly concentrated in the periderm than in the phloem and xylem of P. quinquefolius. These highly accumulating ginsenosides in the periderm of P. quinquefolius and P. ginseng can provide protection against animal and insect attacks [35]. The saponins (R1 and Rg1/Rf) in P. notoginseng were widely distributed throughout the root sections and with a higher concentration in the xylem than in other plant parts. Rd/Re had a higher distribution in the periderm and xylem than in other plant parts, and these findings were similar to those of previous results [36], suggesting that the saponins exhibited similar distribution profiles in the root tissues of P. ginseng and P. quinquefolius but differed from those of P. notoginseng. HPLC results showed that the contents of saponins (Rg1, Re, Rb1, Rc, Rb2, and Rd) were abundantly distributed in the periderm but scarcely distributed in the xylem of P. quinquefolius; these findings were consistent with those of previous studies [37]. The contents of saponins (R1, Rg1, Re, Rb1, and Rd) of P. notoginseng were widely abundant throughout the root sections and slightly higher in the xylem, but they did not significantly vary; these findings were also consistent with those of previous studies [38]. The distribution of saponins depending on types demonstrated tissue specificity in P. quinquefolius and P. notoginseng roots. Although Panax plants are morphologically similar, their saponin types and contents in root tissues differ among species. These differences in saponin distribution and accumulation among Panax plants are reflected by genetic diversity and influenced by their growth environments [26]. P. ginseng and P. quinquefolius grow in north China, whereas P. notoginseng thrives in south China. The large environmental difference between the north and south regions could contribute to the variation in saponin distribution.
Metabolite profiles revealed that the saponin components showed tissue specificity in the root tissues of P. quinquefolius and P. notoginseng. A total of 88 and 24 biomarkers were detected in the root tissues of P. quinquefolius and P. notoginseng, respectively. The saponins differed in the aerial parts (flower, stem, and leaf) and underground parts (root and fibril) of three-year-old P. notoginseng, and 21 potential markers were found [21]. Furthermore, five different parts (root, leaf, flower bud, berry, and seed) of P. ginseng showed chemical differentiation, and 11 robust biomarkers were discovered [39]. These studies confirmed that the saponin components in different organs varied substantially, and our work evidentially reported the profiles of saponin components in the root tissues of Panax plants.
The transcript expression in P. quinquefolius and P. notoginseng was tissue specific, and 158 and 426 transcripts (FPKM ≥ 5) were identified in saponin synthesis in P. quinquefolius and P. notoginseng, respectively. Numerous transcripts involved in saponin synthesis were identified, and they show distinct expression profiles in different parts or tissues of P. ginseng [32], [33], [40]. HMGR, DXS, IDI, and DS transcripts are more highly expressed in the periderm than in the phloem and xylem of P. ginseng [32]. In our study, transcripts encoding DXR (4), AACT (2), IDI (3), AS (1), isopentenyl pyrophosphate isomerase (1), GDPS (1), CYP (54), and UGT (8) from P. quinquefolius were more highly expressed in the periderm than in the phloem and xylem; this trend was similar to the results in P. ginseng [32]. In addition, CYPs (57.4%) and UGTs (42.1%) were highly expressed in the periderm of P. quinquefolius, and CYPs and UGTs were 67.0% and 55.1% in the periderm of P. notoginseng, respectively. WGCNA further showed that 340 and 122 transcripts related to saponin biosynthesis were positively correlated with the saponin contents (R>0.6, P<0.05) in root tissues of P. quinquefolius and P. notoginseng, respectively. In P. quinquefolius, the abundance of GDPS1, CYP51, CYP64, and UGT11 was significantly correlated with the contents of Rg1, Re, Rc, Rb2, and Rd (R>0.8, P<0.05). In P. notoginseng, the abundance of CYP74, CYP89, CYP100, CYP103, CYP109, and UGT190 was correlated with the Rd content (R>0.8, P<0.05). Meanwhile, coexpression data of P. ginseng tissue showed that relative transcripts were positively correlated with the contents of ginsenosides Rg1, Re, and Rb1 [32]. Previous studies showed that UGTPg45 selectively transferred a glucose moiety to the C3 hydroxyl of PPD to form ginsenoside Rh2; UGTPg29 selectively transferred a glucose moiety to the C3 hydroxyl Rh2 to form ginsenoside Rg3 [41]. CYP716A94 was β-amyrin 28-oxidase involved in oleanolic acid production from β-amyrin, and CYP72A397 was oleanolic acid 23-hydroxylase involved in hederagenin production from oleanolic acid [42]. UGRdGT from P. notoginseng and UGRh2GT from P. ginseng were shown to be responsible for the synthesis of ginsenoside Rb1 from Rd and Rg3 from Rh2, respectively [43], [44]. Jung et al. [45] have characterized that PgUGT74AE2 catalyzed the transfer of a glucose moiety from UDP-glucose to the C3 hydroxyl groups of PPD and compound K, yielding Rh2 and F2, respectively; PgUGT94Q2 could transfer a glucose moiety from UDP-glucose to Rh2 and F2 to generate Rg3 and Rd, respectively. These studies revealed that CYP and UGT transcripts involved in saponin synthesis. CYP and UGT transcripts correlated with the contents of saponins in our study could regulate the saponin distribution in the root tissues of Panax plants. These results offered molecular evidence for analysis of saponin distribution and targeted transcripts for genetic improvement. In addition, integrated analysis of metabolomes and transcriptomes could reveal the correlated genes regulating the accumulation of active compounds and provide useful information for understanding the molecular mechanism of biosynthesis [46], [48]. Our study could offer useful data for investigating the molecular and chemical information of saponin distribution in Panax plants.
5. Conclusion
In summary, the distribution and contents of saponins demonstrated tissue specificity in P. quinquefolius and P. notoginseng roots. Gene expression profiles showed tissue specificity in the roots of P. quinquefolius and P. notoginseng. The metabolomes and transcriptomes systematically confirmed the pivotal transcripts of CYPs and UGTs regulating the saponin distribution in the root tissues of P. quinquefolius and P. notoginseng. These results served as a basis for genetically improving and breeding medicinal plants.
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
L.D. and S.C. conceived and designed the experiments. G.W., J.X., F.Y., F.W., L.Z., Y.G., J.Q., Y.W., Z.J., and H.S. performed the experiments and analyzed the data. L.D., G.W., J.X., and Z.C. wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by grants from the National Nature Science Foundation of China (No. 81603238), the Beijing Nova Program (No. Z181100006218020), Yunnan Provincial Science and Technology Department Science and Technology Talent and Platform Program Project (No. 2017IC038), and the Fundamental Research Funds for the Central public welfare research institutes (No. ZXKT17049).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2019.05.009.
Contributor Information
Guangfei Wei, Email: 15098855936@163.com.
Feng Yang, Email: yangfenggoodluck@163.com.
Fugang Wei, Email: weifugang@live.com.
Lianjuan Zhang, Email: 913872290@qq.com.
Ying Gao, Email: yinggao@caf.ac.cn.
Jun Qian, Email: qianjun@biozeron.com.
Zhongjian Chen, Email: 18687656337@126.com.
Zhengwei Jia, Email: zhengwei_jia@waters.com.
Yong Wang, Email: ws-wangyong37@163.com.
He Su, Email: 824414677@qq.com.
Linlin Dong, Email: lldong@icmm.ac.cn.
Jiang Xu, Email: jxu@icmm.ac.cn.
Shilin Chen, Email: slchen@icmm.ac.cn.
Appendix A. Supplementary data
The following are the supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
figs8.
References
- 1.Zhang G.H., Ma C.H., Zhang J.J., Chen J.W., Tang Q.Y., He M.H., Xu X.Z., Jiang N.H., Yang S.C. Transcriptome analysis of Panax vietnamensis var. fuscidicus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genomics. 2015;16:159. doi: 10.1186/s12864-015-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong L.L., Xu J., Li Y., Fang H.L., Niu W.H., Li X.W., Zhang Y.J., Ding W.L., Chen S.L. Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol Biochem. 2018;12:64–74. [Google Scholar]
- 3.Sharma S.K., Pandit M.K. A new species of Panax L. (Araliaceae) from Sikkim Himalaya, India. Syst Bot. 2009;34:434–438. [Google Scholar]
- 4.Dong L.L., Xu J., Feng G.Q., Li X.W., Chen S.L. Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Sci Rep. 2016;6:31802. doi: 10.1038/srep31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D.H. Chemical diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng. J Ginseng Res. 2012;36:1–15. doi: 10.5142/jgr.2012.36.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong L.L., Xu J., Zhang L.J., Cheng R.Y., Wei G.F., Su H., Yang J., Qian J., Xu R., Chen S.L. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm Sin B. 2018;8:272–282. doi: 10.1016/j.apsb.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H.J., Kim P., Chan Y.S. A comprehensive review of the therapeutic and pharmacological effects of ginseng and ginsenosides in central nervous system. J Ginseng Res. 2013;37:8–29. doi: 10.5142/jgr.2013.37.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C.H., Kim J.H. A review on the medicinal potentials of ginseng and ginsenosides on cardiovascular diseases. J Ginseng Res. 2014;38:161–166. doi: 10.1016/j.jgr.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong A.S., Che C.M., Leung K.W. Recent advances in ginseng as cancer therapeutics: a functional and mechanistic overview. Nat Prod Rep. 2015;32:256–272. doi: 10.1039/c4np00080c. [DOI] [PubMed] [Google Scholar]
- 10.Li W., Zhang M., Gu J., Meng Z.J., Zhao L.C., Zheng Y.N., Chen L., Yang G.L. Hypoglycemic effect of protopanaxadiol-type ginsenosides and compound K on type 2 diabetes mice induced by high-fat diet combining with streptozotocin via suppression of hepatic gluconeogenesis. Fitoterapia. 2012;83:192–198. doi: 10.1016/j.fitote.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Fang H.H., Yang S.L., Luo Y.Y., Zhang C., Rao Y., Liu R., Feng Y., Yu J. Notoginsenoside R1 inhibits vascular smooth muscle cell proliferation, migration and neointimal hyperplasia through PI3K/Akt signaling. Sci Rep. 2018;8:7595. doi: 10.1038/s41598-018-25874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y.B., Tang J.P., Khatibi N.H., Zhu M., Chen D., Tu L., Chen L., Wang S. Treatment with ginsenoside Rb1, a component of Panax ginseng, provides neuroprotection in rats subjected to subarachnoid hemorrhage-induced brain injury. Acta Neurochir. 2011;110:75–79. doi: 10.1007/978-3-7091-0356-2_14. [DOI] [PubMed] [Google Scholar]
- 13.Huang Q., Wang T., Wang H.Y. Ginsenoside Rb2 enhances the anti-inflammatory effect of ω-3 fatty acid in LPS-stimulated RAW264.7 macrophages by upregulating GPR120 expression. Acta Pharm Sin B. 2017;38:192–200. doi: 10.1038/aps.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma L.J., Liu F., Zhong Z.F., Wan J.B. Comparative study on chemical components and anti-inflammatory effects of Panax notoginseng flower extracted by water and methanol. J Sep Sci. 2017;40:4730–4739. doi: 10.1002/jssc.201700641. [DOI] [PubMed] [Google Scholar]
- 15.Zhang E.Y., Shi H.L., Yang L., Wu X.J., Wang Z.T. Ginsenoside Rd regulates the Akt/mTOR/p70S6K signaling cascade and suppresses angiogenesis and breast tumor growth. Curr Oncol Rep. 2017;38:359–367. doi: 10.3892/or.2017.5652. [DOI] [PubMed] [Google Scholar]
- 16.Cho W.C., Chung W.S., Lee S.K., Leung A.W., Cheng C.H., Yue K.K. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur J Pharmacol. 2006;550:173–179. doi: 10.1016/j.ejphar.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 17.Ning C.Q., Gao X.G., Wang C.Y., Huo X.K., Liu Z.H., Sun H.J., Yang X.B., Sun P.Y., Ma X.D., Meng Q. Hepatoprotective effect of ginsenoside Rg1 from Panax ginseng on carbon tetrachloride-induced acute liver injury by activating Nrf2 signaling pathway in mice. Environ Toxicol. 2018;33:1050–1060. doi: 10.1002/tox.22616. [DOI] [PubMed] [Google Scholar]
- 18.Xiao D., Yue H., Xiu Y., Sun X., Wang Y., Liu S.Y. Accumulation characteristics and correlation analysis of five ginsenosides with different cultivation ages from different regions. J Ginseng Res. 2015;39:338–344. doi: 10.1016/j.jgr.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi Y.E., Kim Y.S., Yi M.J., Park W.G., Yi J.S., Chun S.R., Han S.S., Lee S.J. Physiological and chemical characteristics of field- and mountain-cultivated ginseng roots. J Plant Biol. 2007;50:198–205. [Google Scholar]
- 20.Wei G.F., Wei F.G., Yuan C., Chen Z.J., Wang Y., Xu J., Zhang Y.Q., Dong L.L., Chen S.L. Integrated chemical and transcriptome analysis reveals the distribution of protopanaxadiol- and protopanaxatriol- type saponins in Panax notoginseng. Molecules. 2018;23:1773–1787. doi: 10.3390/molecules23071773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei G.F., Dong L.L., Yang J., Zhang L.J., Xu J., Yang F., Cheng R.Y., Xu R., Chen S.L. Integrated metabolomic and transcriptomic analyses revealed the distribution of saponins in Panax notoginseng. Acta Pharm Sin B. 2018;8:458–465. doi: 10.1016/j.apsb.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J.R., Leung C.Y., Ho H.M., Chai S., Yau L.F., Zhao Z.Z., Jiang Z.H. Quantitative comparison of ginsenosides and polyacetylenes in wild and cultivated American ginseng. Chem Biodiversity. 2010;7:975–983. doi: 10.1002/cbdv.200900264. [DOI] [PubMed] [Google Scholar]
- 23.Liang Z.T., Chen Y.J., Xu L., Qin M.J., Yi T., Chen H.B., Zhao Z.Z. Localization of ginsenosides in the rhizome and root of Panax ginseng by laser microdissection and liquid chromatography-quadrupole/time of flight-mass spectrometry. J Pharm Biomed Anal. 2015;105:121–133. doi: 10.1016/j.jpba.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Chen C.F., Chiou W.F., Zhang J.T. Comparison of the pharmacological effects of Panax ginseng and Panax quinquefolium. Acta Pharm Sin B. 2008;291:1103–1108. doi: 10.1111/j.1745-7254.2008.00868.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang J.R., Yau L.F., Gao W.N., Liu Y., Yick P.W., Liu L., Jiang Z.H. Quantitative comparison and metabolite profiling of saponins in different parts of the root of Panax notoginseng. J Agric Food Chem. 2014;62:9024–9034. doi: 10.1021/jf502214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y.J., Zhang D.B., Yang D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33:717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Luo H.M., Sun C., Sun Y.Z., Wu Q., Li Y., Song J.Y., Niu Y.Y., Cheng X.L., Xu H.X., Li C.Y. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthesis genes and genetic markers. BMC Genomics. 2011;12:S5. doi: 10.1186/1471-2164-12-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustin J.M., Kuzina V., Andersen S.B., Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72:435–457. doi: 10.1016/j.phytochem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Haralampidis K., Bryan G., Qi X., Papadopoulou K., Bakht S., Melton R., Osbourn A. A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc Natl Acad Sci USA. 2001;98:13431–13436. doi: 10.1073/pnas.231324698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincken J., Heng L., Groot A., Gruppen H. Saponin, classification and occurrence in the plant Kingdom. Phytochemistry. 2007;68:275–297. doi: 10.1016/j.phytochem.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Wang J., Gao W., Zhang J., Zuo B.M., Zhang L.M., Huang L.Q. Advances in study of ginsenoside biosynthesis pathway in Panax ginseng C. A. Meyer. Acta Physiol Plant. 2012;34:397–403. [Google Scholar]
- 32.Zhang J.J., Su H., Zhang L., Liao B.S., Xiao S.M., Dong L.L., Hu Z.G., Wang P., Li X.W., Huang Z.H. Comprehensive characterization for ginsenosides biosynthesis in ginseng root by integration analysis of chemical and transcriptome. Molecules. 2017;22:889–901. doi: 10.3390/molecules22060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J., Chu Y., Liao B.S., Xiao S.M., Yin Q.G., Bai R., Su H., Dong L.L., Li X.W., Qian J. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience. 2017;6:1–15. doi: 10.1093/gigascience/gix093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallavadhani U.V., Mahapatra A., Raja S.S., Manjula C. Antifeedant activity of some pentacyclic triterpene acids and their fatty acid ester analogues. J Agric Food Chem. 2003;51:1952–1955. doi: 10.1021/jf020691d. [DOI] [PubMed] [Google Scholar]
- 36.Wang S.J., Bai H.R., Cai Z.W., Gao D., Jiang Y.Y., Liu J., Liu H.X. MALDI imaging for the localization of saponins in root tissues and rapid differentiation of three Panax herbs. Electrophoresis. 2016;37:1956–1966. doi: 10.1002/elps.201600027. [DOI] [PubMed] [Google Scholar]
- 37.Wang K.Y., Liu W.C., Zhang M.P., Zhao M.Z., Wang Y.F., Li L., Sun C.Y., Hu K.X., Cong Y.Y., Wang Y. Expression of saponin biosynthesis related genes in different tissues of Panax quinquefolius. China J Clin Mater Med. 2018;43:65–71. doi: 10.19540/j.cnki.cjcmm.20171030.006. [DOI] [PubMed] [Google Scholar]
- 38.Sha M.C., Zhou Y.F., Zhang H.Z., Li Y., Qin Q. Study on difference of chemical components in different parts of Panax notoginseng. Mod Chin Med. 2018;20:832–836. [Google Scholar]
- 39.Qiu S., Yang W.Z., Yao C.L., Qiu Z.D., Shi X.J., Zhang J.X., Hou J.J., Wang Q.R., Wu W.Y., Guo D.A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J Chromatogr A. 2016;1453:78–87. doi: 10.1016/j.chroma.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 40.Wang K.Y., Jiang S.C., Sun C.Y., Lin Y.P., Yin R., Wang Y., Zhang M.P. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C.A. Meyer. Sci Rep. 2015;5:18283. doi: 10.1038/srep18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P.P., Wei Y.J., Fan Y., Liu Q.F., Wei W., Yang C.S., Zhang L., Zhao G.P., Yue J.M., Yan X. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Han J.Y., Chun J.H., Oh S.A., Park S.B., Hwang H.S., Lee H.L., Choi Y.E. Transcriptomic analysis of Kalopanax septemlobus and characterization of KsBAS, CYP716A94 and CYP72A397 genes involved in hederagenin saponin biosynthesis. Plant Cell Physiol. 2018;59(2):319–330. doi: 10.1093/pcp/pcx188. [DOI] [PubMed] [Google Scholar]
- 43.He Y.P., Yue C.J. Establishment of measurement system of ginsenoside Rh2, glycosyltransfer-ase activity. Med Plant. 2010;1(4):58–60. [Google Scholar]
- 44.Yue C.J., Zhong J.J. Purification and characterization of UDPG: ginsenoside Rd glucosyltransferase from suspended cells of Panax notoginseng. Process Biochem. 2005;40:3742–3748. [Google Scholar]
- 45.Jung S.C., Kim W., Park S.C., Jeong J., Park M.K., Lim S., Lee Y., Im W.T, Lee J.H., Choi G. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 2014;55(12):2177–2188. doi: 10.1093/pcp/pcu147. [DOI] [PubMed] [Google Scholar]
- 46.Jeon J., Kim J.K., Kim H., Kim Y.J., Park Y.J., Kim S.J., Kim C., Park S.U. Transcriptome analysis and metabolite profiling of green and red kale (Brassica oleracea var. acephala) seedings. Food Chem. 2018;241:7–13. doi: 10.1016/j.foodchem.2017.08.067. [DOI] [PubMed] [Google Scholar]
- 48.Dong T.T., Han R.P., Yu J.W., Zhu M.K., Zhang Y., Gong Y., Li Z.Y. Anthocyanins accumulation and molecular analysis of correlated genes by metabolome and transcriptome in green and purple asparaguses (Asparagus officinalis, L.) Food Chem. 2019;271:18–28. doi: 10.1016/j.foodchem.2018.07.120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.