Abstract
Single-cell RNA sequencing has revolutionized ocular gene expression studies. This technology has enabled researchers to identify expression signatures for rare cell types and characterize how gene expression changes across biological conditions, such as topographic region or disease status. However, sharing single-cell RNA sequencing results remains a major obstacle, particular for individuals without a computational background. To address these limitations, we developed Spectacle, an interactive web-based resource for exploring previously published single-cell RNA sequencing data from ocular studies. Spectacle is powered by a locally developed R package, cellcuratoR, which utilizes the Shiny framework in R to generate interactive visualizations for single-cell expression data. Spectacle contains five pre-processed ocular single-cell RNA sequencing data sets and is accessible via the web at OcularGeneExpression.org/singlecell. With Spectacle, users can interactively identify which cell types express a gene of interest, detect transcriptomic subpopulations within a cell type, and perform highly flexible differential expression analyses. The freely-available Spectacle system reduces the bioinformatic barrier for interacting with rich single-cell RNA sequencing studies from ocular tissues, making it easy to quickly identify cell types that express a gene of interest.
Keywords: single-cell RNA sequencing, retina, RPE, choroid, gene expression, interactive
1. Introduction:
Over the past two decades, vision researchers have employed transcriptomics to study both normal physiology and disease within ocular tissues. Early experiments using serial analysis of gene expression (SAGE) (Bowes Rickman et al., 2006; Sharon et al., 2002) and microarrays (Booij et al., 2010; Booij et al., 2009; Cai et al., 2012; Hornan et al., 2007; Radeke et al., 2007; Strunnikova et al., 2010; van Soest et al., 2007; Wagner et al., 2013; Yoshida et al., 2002) catalogued RNA expression in the human retina and retinal pigment epithelium (RPE)/choroid. As sequencing technology advanced, whole transcriptome expression analysis in these tissues was achieved with RNA sequencing (Farkas et al., 2013; Li et al., 2014; Tian et al., 2015; Whitmore et al., 2013; Whitmore et al., 2014), which captured sequence-level information including splicing and other transcriptional events. More recently, single-cell RNA sequencing has allowed for the transcriptomic profiling of individual cells, improving the ability to study gene expression in many cell types comprising complex ocular tissues (Hu et al., 2019; Lukowski et al., 2019; Orozco et al., 2020; Sridhar et al., 2020; Voigt et al., 2020a; Voigt et al., 2019a; Voigt et al., 2019b; Voigt et al., 2020b). However, these technological advancements in sequencing have been coupled with increasing data analysis complexity. Numerous bioinformatic tools aid in mapping, clustering, and visualizing single-cell data, yet accessing results can be challenging, particularly for those without a bioinformatics background (Luecken and Theis, 2019).
Several resources have been assembled to make previously generated gene expression datasets more approachable. A major example of such a resource is the Genotype-Tissue Expression (GTEx) Project, which provides a visualization platform to study the correlation between genetic variants and gene expression across a diverse collection of tissues (2013). However, ocular tissues were not included in this database, limiting the utility of this resource to vision researchers. To address this, the Ocular Tissue Database (OTDB) compiled microarray gene expression values in ten human donor eyes and summarized the results in an internet interface (Wagner et al., 2013). Subsequently, the eyeIntegration database assembled hundreds of publicly available ocular bulk RNA sequencing datasets comprising over one thousand individual cornea, retina, and RPE-choroid samples. This database provides tools for querying gene expression and offers a web-based visualization system for comparing expression profiles between different ocular tissues (Bryan et al., 2018). Likewise, the Single Cell Portal (https://portals.broadinstitute.org/single_cell) allows users to survey single-cell level gene expression across a diverse group of tissues. While this resource is impressively large (including 237 different datasets at the time of this publication), the diverse sequencing and analytic technologies used in these studies preclude interactive analysis beyond visualizing which cluster(s) express a particular gene of interest.
Each of these existing interactive gene expression resources have strengths and limitations shaped by the number of included datasets, the sequencing technology used for the experiments, and the degree of interactivity the user has with the data. We set out to develop a highly interactive, web-based visualization interface for analyzing human ocular single-cell RNA sequencing data. Our platform, Spectacle, facilitates exploring single-cell RNA sequencing expression data from the human retina and RPE/choroid. Spectacle allow users to visualize gene expression at both the cell and cluster level, analyze differentially expressed genes in interactively-selected cell populations, and identify subsets of cells exhibiting distinct gene signatures. Spectacle is freely accessible at OcularGeneExpression.org/singlecell and is powered by cellcuratoR, an open-source R package that enables users to locally interact with their own single-cell datasets.
2. Methods
2.1. Data Processing with cellcuratoR:
In order to produce consistent interactive visualization features across different single-cell RNA sequencing experiments, we developed the R package cellcuratoR. cellcuratoR provides a framework to convert single-cell RNA sequencing data analyzed in Seurat (v3.0.0 – v3.1.5) (Butler et al., 2018) into a format interpretable by the R Shiny interface. For reference, an example pipeline for creating a Seurat object from mapped FASTQ files is available as Supplementary File 1. The processed Seurat object is subsequently optimized to remove unneeded features for downstream visualizations. This serves to minimize file sizes and increase the speed of the reactive user interface. cellcuratoR is freely available as an R package from GitHub (www.github.com/drewvoigt10/cellcuratoR). The cellcuratoR package can be installed on any computer and run in a local R session, allowing users to explore data from their own experiments without requiring a webserver. Five publicly available human single-cell RNA sequencing datasets from the retina (Voigt et al., 2020a; Voigt et al., 2019b) and RPE/choroid (Voigt et al., 2019a; Voigt et al., 2020b) have been pre-processed with cellcuratoR for interactive visualizations with the web-hosted Spectacle resource (Table 1).
Table 1: Pre-processed datasets.
Five single-cell RNA sequencing datasets of human retina or RPE/choroid have been processed specifically for interactive analysis in Spectacle.
| Study | Number of analyzed cells |
Number of human donors |
Reference |
|---|---|---|---|
| Foveal vs peripheral human retina | 8,217 | 3 | (Voigt et al., 2019b) |
| Macular vs peripheral human RPE/choroid | 4,355 | 3 | (Voigt et al., 2019a) |
| Macular vs peripheral CD31-enriched RPE/choroid | 14,234 | 4 | (Voigt et al., 2019a) |
| Foveal and peripheral retina from an autoimmune retinopathy vs control donors | 23,429 | 5 | (Voigt et al., 2020a) |
| Infant vs adult CD31-enriched RPE/choroid | 37,070 | 8 | (Voigt et al., 2020b) |
2.2. Site availability:
Spectacle is freely available at OcularGeneExpression.org/singlecell or https://singlecell.ivr.uiowa.edu. The user interface is documented in a detailed “how to” guide, which is accessible from the left-hand menu of Spectacle. This guide includes an overview video and also details how to interactively load different datasets, visualize clusters of different cells, create expression heatmaps and violin plots, re-cluster cell populations of interest, and perform differential expression between different cell types across different biological conditions, as described below. In addition, the GitHub page for cellcuratoR (www.github.com/drewvoigt10/cellcuratoR) includes several animations that demonstrate salient features of the user interface.
2.3. User-interface visualizations:
Interactive visualizations of single-cell data were implemented using the Shiny (v1.3.2) framework in R (Chang et al., 2020). Several different visualization modalities are available to analyze and interpret gene expression within each dataset. Application Programming Interface (API) calls and parameters for the Seurat-based visualization functions are outlined in detail in Supplemental File 2.
Dimensionality reduction:
Visualization of graph-based clusters is achieved with a dimensionality reduction plot. By default, the dimensions are based upon the pre-computed values using uniform manifold approximation and projection (UMAP), but other methods are also supported (e.g., t-distributed Stochastic Neighbor Embedding (tSNE) and principal component analysis (PCA)). The user can zoom in on subpopulations of cells and hover over cells to determine their cluster identity. In the dimensionality reduction visualization, cells can also be shaded according to cluster identity or the originating library.
Heatmaps:
Gene expression can be visualized across clustered cells within a dimensionality reduction view in the form of heatmaps, in which each the shading of each cell is proportional to the relative expression of the specified gene. A custom legend provides reference to expression levels as transcripts per 10,000 (TP10K).
Violin Plots:
Violin plots depicting the expression of genes of interest are presented per cluster. Expression distributions are only drawn if at least 25% of cells in a cluster express the gene of interest.
Differential expression:
Differential expression can be performed between pre-defined clusters of cells or cell populations manually selected with the lasso tool. In addition, differential expression can be performed between cells that originate from different biological conditions (such as cells originating from healthy versus diseased libraries), so long as these biological conditions are dichotomous.
Reclustering:
Cell populations can be explored in a lower dimensional feature space with re-clustering. This analysis renormalizes the selected cell population and performs a dimensionality reduction on that subset. Supported dimensionality reduction algorithms include UMAP, tSNE, and PCA.
3. Results
3.1. Spectacle access and overview.
Spectacle is an interactive visualization system for ocular single-cell gene expression analysis that can be accessed at OcularGeneExpression.org/singlecell. Spectacle is a web-accessible instance of cellcuratoR, an R package available at Github (www.github.com/drewvoigt10/cellcuratoR). Spectacle currently hosts five pre-processed single-cell RNA sequencing datasets from human retina and RPE/choroid (Table 1), and additional datasets will be added in the future as they become available. A video tutorial covering salient features of Spectacle is accessible via the “How to guide” from the left-hand menu.
Visualizing cell clusters and gene expression:
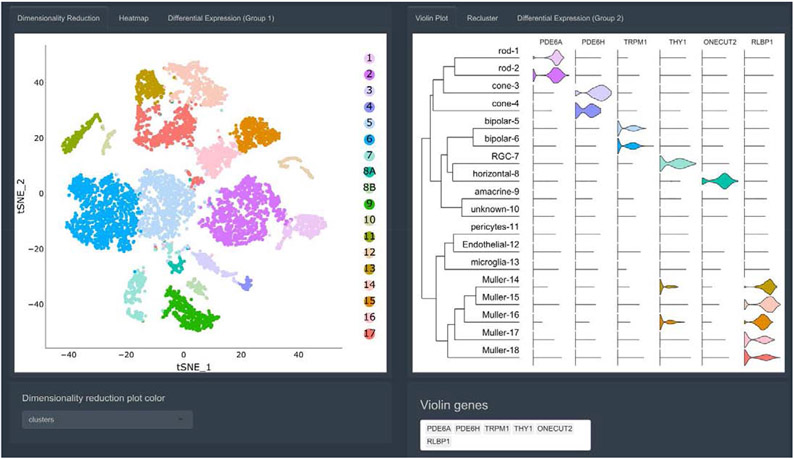
Spectacle provides several complementary visualizations for exploring gene expression across clusters. First, dimensionality reduction plots are used to visualize clustered cells (Figure 1). Second, the user can query cluster-level expression of one or more genes in the form of violin plots (Figure 1), which can be useful for classifying clusters into putative cell types or for exploring expression patterns of genes of interest. Lastly, users can color the cells in the dimensionality reduction plots by the expression of a gene of interest with heatmaps (SI Figure 1).
Figure 1: Screenshot of cluster-level expression analysis in Spectacle.
Visualization of clusters from foveal and peripheral retinal cells with tSNE-based dimensionality reduction (left). The plots are interactive, allowing zooming in on regions of interest and hovering over individual cells to identify the cell type and originating library. Violin plots (right) depict cluster-level expression for one or more genes of interest. A dendrogram is appended on the left side of the violin plots to demonstrate the relationships between all clusters.
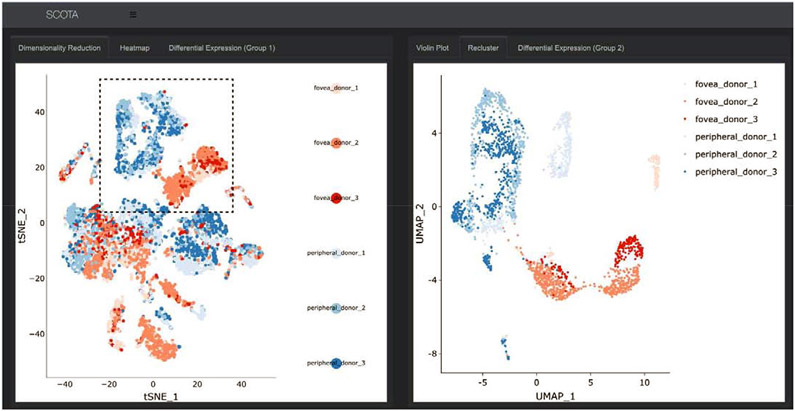
In addition to visualizing cells by cluster identity, it is often useful to analyze which libraries contribute cells to each cluster. Hence, Spectacle provides the ability to color cells by library composition, which can aid in visualizing clusters that contain a majority of cells from a specific biological condition. For example, in analyzing populations isolated from the fovea and peripheral human retina, distinct subpopulations of glial cells belonging to foveal (red) and peripheral (blue) libraries become apparent (Figure 2). Such cellular groups can further be re-clustered in attempt to identify subpopulations or to view relationships between cells in a more granular space (Figure 2). In this example, foveal and peripheral glial cells remain well segregated upon re-clustering, further suggesting that the multidimensional gene expression patterns of glial cells are influenced by the region of the retina from which they came.
Figure 2: Re-clustering of cells in Spectacle.
In addition to visualizing cluster labels, the dimensionality reduction plot can be colored by originating library (left). This is particularly useful when libraries correspond to different biological conditions, such as region of isolated tissue (with cells colored in red originating from the fovea and cells colored in blue originating from the periphery). Re-clustering of cells (right) can aid in the identification of subpopulations of cells and viewing the relationships of cells in a more granular space. In this example, after glial cell populations (dotted line) have been re-clustered, the foveal and peripheral cells remain discrete, further suggesting that gene expression in glial cells is influenced by region.
Differential expression for generating hypotheses:
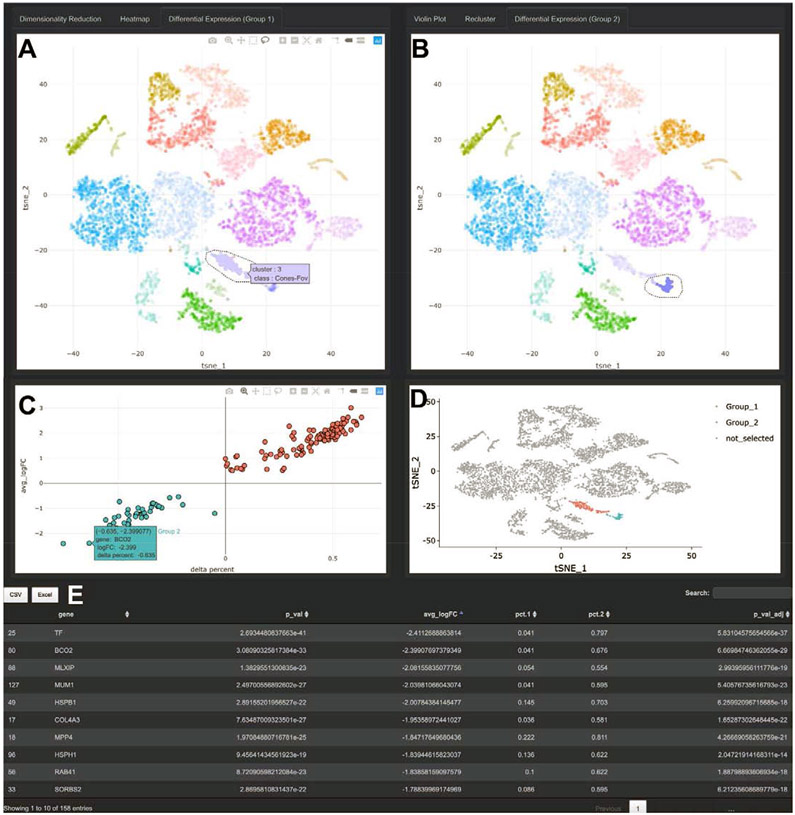
Differential expression analysis is often central to RNA-based experiments. Spectacle thus supports two modes of differential expression analysis. First, gene expression can be compared between any combination of pre-defined cellular clusters or flexibly selected cell populations with the user drawn “lasso” tool (Figure 3A). Such functionalities aid in identifying gene signatures specific for cell populations or subpopulations. Second, gene expression can be compared between biological conditions, such as anatomic region or disease state, which promotes hypothesis-generating questions about normal ocular physiology and disease pathogenesis. Spectacle displays these results as graphs (Figure 3CD) and tables (Figure 3E), which users can download as high-resolution image files and spreadsheets.
Figure 3: Differential Expression in Spectacle.
A-B. Differential expression can be performed between pre-characterized clusters of cells or interactively selected populations with the lasso tool, with cells selected on the left belonging to “Group_1” (A) and cells selected on the right belonging to “Group_2” (B). In addition to comparing expression between selected populations, differential expression can be performed between cells in the same region originating from different biological conditions, such as disease status (not shown). C. Differential expression results are displayed graphically. The y-axis depicts the log of the fold-change between cells in the Group_1 and Group_2 selections. The x-axis depicts a variable called “delta percent,” which represents the percentage of cells in Group_1 samples that express each gene minus the percent of cells in Group_2 samples that express the gene. For example, the gene BCO2 is expressed by 4.1% of cells in Group_1 and 67.6% of cells in Group_2, resulting in a delta percent of 0.041 minus 0.676 = −0.635. This visualization allows for the expression level (y-axis) and the proportion of expressing cells (x-axis) to be simultaneously evaluated. D. The cell selections are re-depicted on the standard dimensionality reduction space. E. In addition to graphical output, the differential expression results are displayed in tabular format, and can be exported to CSV or Excel files.
3.2. Case Study:
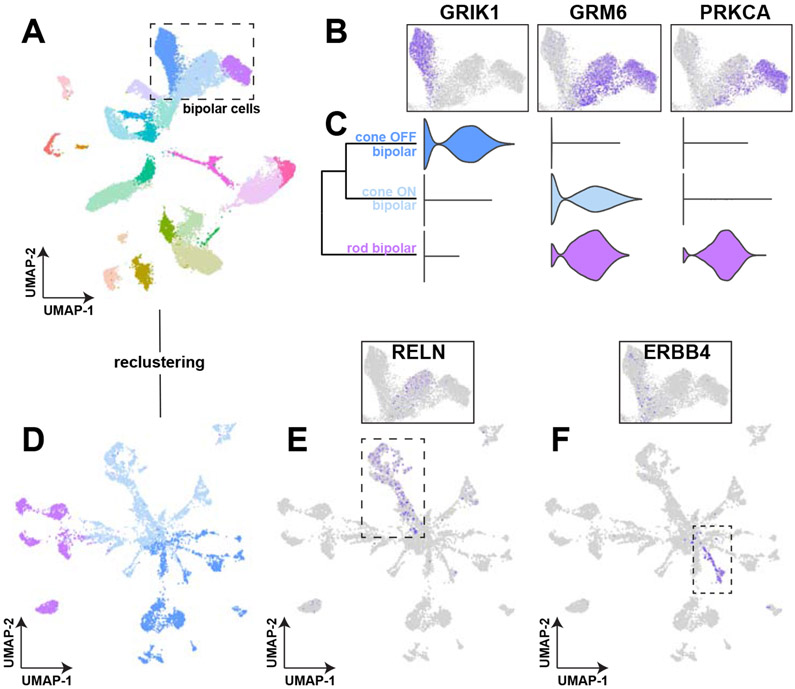
To further demonstrate the utility of Spectacle, we present a basic analysis of retina samples from five human donors: one donor with autoimmune retinopathy and four control donors (Voigt et al., 2020a). In this dataset, 23,429 cells were recovered from paired foveal and peripheral libraries. After clustering and dimensionality reduction, three distinct clusters of bipolar cells were identified (Figure 4A). These clusters were assigned to rod bipolar cell, cone OFF bipolar cell, and cone ON bipolar cell types by comparing expression of distinguishing marker genes in heatmaps (Figure 4B) and violin plots (Figure 4C). In the mouse retina, bipolar cell subsets have been extensively characterized with single-cell RNA sequencing, which resulted in the identification of 15 morphologic and transcriptomic subsets (Shekhar et al., 2016). Using the Spectacle re-clustering tool, we re-clustered the three observed bipolar cells detected in this experiment to analyze potential subpopulations (Figure 4D).
Figure 4: Spectacle investigation of bipolar cell subsets.
A. Of 23,429 total cells, three clusters were identified as bipolar cells. B. Heatmaps were used to classify the three bipolar cell clusters as cone OFF bipolar cells (by expression of GRIK1), cone ON bipolar cells (by expression of GRM6), and rod bipolar cells (by expression of PRKCA), which are supported by violin plots (C). D. In order to identify potential subpopulations of bipolar cells, re-clustering of the bipolar cell populations was performed. E. RELN expression, which is enriched in the BC7 subclass of cone ON bipolar cells, did not localize to any subpopulation within the unprocessed data (insert) but localized to a subpopulation within the re-clustered bipolar cell object (dashed line). F. ERBB4 expression, which is enriched in the BC3A subclass of cone OFF bipolar cells, did not localize to any subpopulation within the unprocessed data (insert) but did localize to a subpopulation within the re-clustered bipolar cell object (dashed line).
Next, we asked if key genes enriched in bipolar cell subsets localized to subpopulations within the re-clustered bipolar cells. RELN, an enriched gene in the BC7 subclass of cone ON bipolar cells, did not localize to any subpopulations within the unprocessed dataset (Figure 4E, insert) but demonstrated localization to subpopulation of cells within the re-clustered object (Figure 4E). Likewise, expression of ERBB4, a gene enriched in the cone OFF bipolar BC3A morphologic class, segregated to a small population of OFF bipolar cells within the re-clustered object (Figure 4F). Such re-clustering analysis highlights the utility of exploring gene expression in a reduced dimensional space for the identification and characterization of cellular subpopulations of interest.
4. Discussion
Visualizing single-cell RNA sequencing data can be complicated and usually requires a degree of bioinformatic expertise. With Spectacle, we have deployed an interactive single-cell RNA sequencing exploration resource for ocular datasets that can extend data interpretation to a broad range of vision researchers. In addition, our development of cellcuratoR provides a flexible platform for bioinformaticians to share their own single-cell RNA sequencing analyses with interdisciplinary teams. We believe that cellcuratoR will increase accessibility to and interpretability of the many information-rich, publicly available single-cell datasets.
Other visualization tools for single-cell RNA sequencing data exist to interactively explore data (Hillje et al., 2019; Innes and Bader, 2018; Patel, 2018; Pont et al., 2019). We believe that Spectacle offers at least two major advantages. First, ocular tissues are excluded from many popular gene expression resources, such as GTEx. This prevents quickly determining if a gene of interest is expressed in the retina, RPE, or choroid, and instead forces a researcher to embark on a bioinformatic exercise to reprocess data from pre-published datasets. With Spectacle, one can identify the precise population(s) that express a given gene of interest within seconds, which we have found to be immensely helpful in preparing manuscripts and discussing clinical cases. Second, Spectacle is extremely interactive. Many existing single-cell visualization platforms share the basic functionalities offered by Spectacle, such as displaying gene expression in heatmaps or violin plots. But Spectacle builds upon these visualization aspects and adds several more advanced features to further hypothesis generation. For example, re-clustering of selected cell populations allows for discovery of cellular subgroups that are not discernable when analyzed with other cell types, as illustrated by our analysis of bipolar subpopulations (Figure 4). In addition, Spectacle supports highly flexible differential expression analysis to not only identify genes enriched in each cellular cluster, but also to detect genes enriched in cells across biological conditions (supporting comparisons such as foveal versus peripheral, youth versus age, health versus autoimmune retinopathy, and health versus age-related macular degeneration). Thus, we believe that Spectacle dramatically lowers the analytical barrier for vision researchers to quickly access and interact with rich ocular single-cell expression datasets.
Spectacle contains five previously published ocular datasets available for interactive visualization. We plan on updating Spectacle with future studies from our group as they become published. For other groups wishing to interactively explore their own datasets, we have made cellcuratoR, our R-package that powers Spectacle, freely available (www.github.com/drewvoigt10/cellcuratoR). After preliminary analysis in Seurat, bioinformaticians can use cellcuratoR to privately distribute results with their interdisciplinary research groups. We have found that this interactive style of sharing results is immensely helpful in generating hypotheses and drafting manuscripts. Likewise, cellcuratoR is published under the permissive GPL-3 license, allowing others to modify the codebase to their experimental needs and host their own publicly facing webservers. We will maintain both Spectacle and cellcuratoR to ensure compatibility with any future updates to the Seurat R package.
There are several limitations to this approach. First, the high degree of interactivity of the user interface requires consistent data processing of a Seurat analyzed S4 object in R. This initial processing requires a degree of bioinformatic expertise. While Seurat is a popular single-cell RNA sequencing analysis tool, several other data analysis systems exist and are used in the field (Azizi et al., 2018; Klein et al., 2015; Parekh et al., 2018; Qiu et al., 2017). In addition, the numerous visualization and analysis features of Spectacle require loading of very large expression matrices into memory. While the file sizes of such objects have been minimized where possible, loading a dataset takes several seconds. Benchmarking experiments (SI Figure 2) suggest that dataset loading times increase linearly with the number of cells in each experiment. Computationally demanding functionalities, such as differential expression, are appreciably slower with larger datasets, and we recommend adjusting thresholds to accelerate these processes when exploring larger studies. Lastly, highly interactive differential expression is powerful for identifying enriched genes across different cell types and biological conditions; however, such interactivity may permit p-hacking in data analysis (Head et al., 2015). In particular, the differential expression within Spectacle (and Seurat) treats all cells as independent observations, which inflates p-values, especially in the context of comparing expression across biological conditions. Most single-cell experiments, including those provided in Spectacle, contain limited biological replicates, and hence the statistical significance of differential expression results should be interpreted with caution.
In summary, Spectacle aids in generating publication-ready visualizations, performing basic data analysis, and interpreting results from complex ocular single-cell RNA sequencing experiments. This expedites hypothesis generation and testing to improve understanding of visual diseases.
Supplementary Material
SI Figure 1: Visualization of cellular and cluster-level expression in Spectacle. Heatmaps (left) depict the expression of a gene of interest (RLBP1) across a dimensionality reduction plot from foveal and peripheral retinal cells. A custom legend provides reference to expression levels as transcripts per 10,000. Violin plots (right) can complement heatmaps by providing a cluster-level overview of expression for multiple genes simultaneously.
SI Figure 2: Benchmarking loading and computational times within Spectacle. Six different dataset sizes (1000 cells, 10,000 cells, 25,000 cells, 50,000 cells, 100,000 cells, and 150,000 cells) were simulated before measuring the computational time of several tasks within Spectacle (accessed from oculargeneexpression.org/singlecell). Dataset loading time (red), defined as the interval between selecting a dataset and displaying the first plot, scales linearly with increasing number of cells. Differential gene expression (black) was performed under default settings between two clusters, each comprising 10% of total cells in the dataset. Differential expression on datasets with more than 50,000 cells takes noticeably longer on the web-hosted Spectacle. Lastly, 10% of cells within each dataset were reclustered under default parameters. Reclustering (green) more than 10,000 cells (for a 100,000 cell dataset) begins to scale exponentially.
Highlights.
Spectacle is a resource for exploring ocular single-cell RNA sequencing datasets
Spectacle includes five datasets from the retina, RPE, and choroid
Spectacle facilitates interactive expression analyses on pre-classified cell types
Spectacle is freely available at OcularGeneExpression.org/singlecell
Acknowledgements
This work was supported by National Institutes of Health grants EY027038 and EY025580, the MSTP training grant T32 GM007337, the Roy J. Carver, Jr. Chair in Bioinformatics and Computational Biology (TES), and support from Research to Prevent Blindness.
Footnotes
Declarations of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 2013. The Genotype-Tissue Expression (GTEx) project. Nature genetics 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, Choi K, Fromme RM, Dao P, McKenney PT, Wasti RC, Kadaveru K, Mazutis L, Rudensky AY, Pe'er D, 2018. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij JC, ten Brink JB, Swagemakers SM, Verkerk AJ, Essing AH, van der Spek PJ, Bergen AA, 2010. A new strategy to identify and annotate human RPE-specific gene expression. PloS one 5, e9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij JC, van Soest S, Swagemakers SM, Essing AH, Verkerk AJ, van der Spek PJ, Gorgels TG, Bergen AA, 2009. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics 10, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes Rickman C, Ebright JN, Zavodni ZJ, Yu L, Wang T, Daiger SP, Wistow G, Boon K, Hauser MA, 2006. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Investigative ophthalmology & visual science 47, 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan JM, Fufa TD, Bharti K, Brooks BP, Hufnagel RB, McGaughey DM, 2018. Identifying core biological processes distinguishing human eye tissues with precise systems-level gene expression analyses and weighted correlation networks. Human molecular genetics 27, 3325–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Fields MA, Hoshino R, Priore LV, 2012. Effects of aging and anatomic location on gene expression in human retina. Front Aging Neurosci 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire JJ, Xie Y, McPherson J, 2020. shiny: Web Application Framework for R; R Foundation for Statistical Computing. [Google Scholar]
- Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA, 2013. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics 14, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head ML, Holman L, Lanfear R, Kahn AT, Jennions MD, 2015. The extent and consequences of p-hacking in science. PLoS Biol 13, e1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillje R, Pelicci PG, Luzi L, 2019. Cerebro: Interactive visualization of scRNA-seq data. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornan DM, Peirson SN, Hardcastle AJ, Molday RS, Cheetham ME, Webster AR, 2007. Novel retinal and cone photoreceptor transcripts revealed by human macular expression profiling. Investigative ophthalmology & visual science 48, 5388–5396. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang X, Hu B, Mao Y, Chen Y, Yan L, Yong J, Dong J, Wei Y, Wang W, Wen L, Qiao J, Tang F, 2019. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol 17, e3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes BT, Bader GD, 2018. scClustViz - Single-cell RNAseq cluster assessment and visualization. F1000Res 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jia C, Kazmierkiewicz KL, Bowman AS, Tian L, Liu Y, Gupta NA, Gudiseva HV, Yee SS, Kim M, Dentchev T, Kimble JA, Parker JS, Messinger JD, Hakonarson H, Curcio CA, Stambolian D, 2014. Comprehensive analysis of gene expression in human retina and supporting tissues. Human molecular genetics 23, 4001–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken MD, Theis FJ, 2019. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 15, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski SW, Lo CY, Sharov AA, Nguyen Q, Fang L, Hung SS, Zhu L, Zhang T, Grunert U, Nguyen T, Senabouth A, Jabbari JS, Welby E, Sowden JC, Waugh HS, Mackey A, Pollock G, Lamb TD, Wang PY, Hewitt AW, Gillies MC, Powell JE, Wong RC, 2019. A single-cell transcriptome atlas of the adult human retina. Embo j 38, e100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Chen HH, Cox C, Katschke KJ Jr., Arceo R, Espiritu C, Caplazi P, Nghiem SS, Chen YJ, Modrusan Z, Dressen A, Goldstein LD, Clarke C, Bhangale T, Yaspan B, Jeanne M, Townsend MJ, van Lookeren Campagne M, Hackney JA, 2020. Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration. Cell Rep 30, 1246–1259.e1246. [DOI] [PubMed] [Google Scholar]
- Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I, 2018. zUMIs - A fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MV, 2018. iS-CellR: a user-friendly tool for analyzing and visualizing single-cell RNA sequencing data. Bioinformatics 34, 4305–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont F, Tosolini M, Fournie JJ, 2019. Single-Cell Signature Explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res 47, e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C, 2017. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeke MJ, Peterson KE, Johnson LV, Anderson DH, 2007. Disease susceptibility of the human macula: differential gene transcription in the retinal pigmented epithelium/choroid. Experimental eye research 85, 366–380. [DOI] [PubMed] [Google Scholar]
- Sharon D, Blackshaw S, Cepko CL, Dryja TP, 2002. Profile of the genes expressed in the human peripheral retina, macula, and retinal pigment epithelium determined through serial analysis of gene expression (SAGE). Proceedings of the National Academy of Sciences of the United States of America 99, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR, 2016. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar A, Hoshino A, Finkbeiner CR, Chitsazan A, Dai L, Haugan AK, Eschenbacher KM, Jackson DL, Trapnell C, Bermingham-McDonogh O, Glass I, Reh TA, 2020. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep 30, 1644–1659.e1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, Sergeev Y, Chen W, Edwards AO, Stambolian D, Abecasis G, Swaroop A, Munson PJ, Miller SS, 2010. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Human molecular genetics 19, 2468–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Kazmierkiewicz KL, Bowman AS, Li M, Curcio CA, Stambolian DE, 2015. Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics 105, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soest SS, de Wit GM, Essing AH, ten Brink JB, Kamphuis W, de Jong PT, Bergen AA 2007. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Molecular vision 13, 1608–1617. [PubMed] [Google Scholar]
- Voigt AP, Binkley E, Flamme-Wiese MJ, Zeng S, DeLuca AP, Scheetz TE, Tucker BA, Mullins RF, Stone EM, 2020a. Single-Cell RNA Sequencing in Human Retinal Degeneration Reveals Distinct Glial Cell Populations. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt AP, Mulfaul K, Mullin NK, Flamme-Wiese MJ, Giacalone JC, Stone EM, Tucker BA, Scheetz TE, Mullins RF, 2019a. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proceedings of the National Academy of Sciences of the United States of America 116, 24100–24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt AP, Whitmore SS, Flamme-Wiese MJ, Riker MJ, Wiley LA, Tucker BA, Stone EM, Mullins RF, Scheetz TE, 2019b. Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Experimental eye research 184, 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt AP, Whitmore SS, Mulfaul K, Chirco KR, Giacalone JC, Flamme-Wiese MJ, Stockman A, Stone EM, Tucker BA, Scheetz TE, Mullins RF, 2020b. Bulk and single-cell gene expression analyses reveal aging human choriocapillaris has pro-inflammatory phenotype. Microvasc Res, 104031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Anand VN, Wang WH, Chatterton JE, Sun D, Shepard AR, Jacobson N, Pang IH, Deluca AP, Casavant TL, Scheetz TE, Mullins RF, Braun TA, Clark AF, 2013. Exon-level expression profiling of ocular tissues. Experimental eye research 111, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore SS, Braun TA, Skeie JM, Haas CM, Sohn EH, Stone EM, Scheetz TE, Mullins RF, 2013. Altered gene expression in dry age-related macular degeneration suggests early loss of choroidal endothelial cells. Molecular vision 19, 2274–2297. [PMC free article] [PubMed] [Google Scholar]
- Whitmore SS, Wagner AH, DeLuca AP, Drack AV, Stone EM, Tucker BA, Zeng S, Braun TA, Mullins RF, Scheetz TE, 2014. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Experimental eye research 129, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Yashar BM, Hiriyanna S, Swaroop A, 2002. Microarray analysis of gene expression in the aging human retina. Investigative ophthalmology & visual science 43, 2554–2560. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI Figure 1: Visualization of cellular and cluster-level expression in Spectacle. Heatmaps (left) depict the expression of a gene of interest (RLBP1) across a dimensionality reduction plot from foveal and peripheral retinal cells. A custom legend provides reference to expression levels as transcripts per 10,000. Violin plots (right) can complement heatmaps by providing a cluster-level overview of expression for multiple genes simultaneously.
SI Figure 2: Benchmarking loading and computational times within Spectacle. Six different dataset sizes (1000 cells, 10,000 cells, 25,000 cells, 50,000 cells, 100,000 cells, and 150,000 cells) were simulated before measuring the computational time of several tasks within Spectacle (accessed from oculargeneexpression.org/singlecell). Dataset loading time (red), defined as the interval between selecting a dataset and displaying the first plot, scales linearly with increasing number of cells. Differential gene expression (black) was performed under default settings between two clusters, each comprising 10% of total cells in the dataset. Differential expression on datasets with more than 50,000 cells takes noticeably longer on the web-hosted Spectacle. Lastly, 10% of cells within each dataset were reclustered under default parameters. Reclustering (green) more than 10,000 cells (for a 100,000 cell dataset) begins to scale exponentially.