Abstract
Epigenetic pharmacotherapy for CNS-related diseases is a burgeoning area of research. In particular, members of the bromodomain and extra-terminal domain (BET) family of proteins have emerged as intriguing therapeutic targets due to their putative involvement in an array of brain diseases. With their ability to bind to acetylated histones and act as a scaffold for chromatin modifying complexes, BET proteins were originally thought of as passive epigenetic ‘reader’ proteins. However, new research depicts a more complex reality where BET proteins act as key nodes in lineage-specific and signal-dependent transcriptional mechanisms to influence disease-relevant functions. Amid a recent wave of drug development efforts from basic scientists and pharmaceutical companies, BET inhibitors are currently being studied in several CNS-related disease models, but safety and tolerability remain a concern. Here we review the progress in understanding the neurobiological mechanisms of BET proteins and the therapeutic potential of targeting BET proteins for brain health and disease.
Keywords: neuroepigenetics, bromodomain, BET, epigenetic readers, BRD4, addiction, Substance use disorder, Alzheimer’s disease, neurological disorders, brain cancer, learning and memory
Introduction
Dynamic control of histone acetylation processes influences chromatin structure, DNA accessibility and transcriptional activity (Grunstein 1997). While writers (e.g., histone acetyltransferases, HATs) and erasers (e.g., histone deacetylases, HDACs) of histone acetylation have been extensively investigated in CNS-related disorders (Schneider et al., 2013; Penney and Tsai 2014; Mahgoub and Monteggia 2014), the roles for histone acetylation readers in brain health and disease have only recently emerged. In this review, we focus on a class of histone acetylation readers called bromodomain and extra-terminal (BET) proteins. We summarize the structure, function, and expression of BET proteins in the brain and review novel pharmacological approaches to selectively inhibit BET activity. We then highlight the mechanisms by which BET proteins are involved in multiple CNS-related disorders, ranging from neurodevelopmental to neurological to psychiatric disorders. Finally, we discuss challenges and future directions of BET-based therapies for brain health and disease.
BET structure and function
The bromodomain (BD) is a ~110 amino acid motif that binds to acetyl-lysine modifications on histone and non-histone proteins (Dhalluin et al., 1999). To date, 61 bromodomains have been identified in 46 diverse proteins in human cells (Filippakopoulos et al., 2012). In recent years, members of the bromodomain and extra-terminal family of proteins have attracted significant attention following the discovery of their role in multiple cancers and the development of selective, small molecule BET inhibitors (Shi and Vakoc, 2014; Devaiah et al., 2016a; Liu et al., 2017). The BET family of bromodomains is comprised of 4 proteins: bromodomain-containing protein 2 (BRD2), bromodomain-containing protein 3 (BRD3), bromodomain-containing protein 4 (BRD4), and bromodomain testis associated protein (BRDT). BRD2, BRD3, and BRD4 are expressed in most cells and tissues, while BRDT is primarily expressed in the testes (Jones et al., 1997; Taniguchi, 2016). At the structural level, BET proteins contain two tandem bromodomains (BD1 and BD2) that bind to acetyl-lysine histone residues (e.g., H3K27ac, H4K5ac, H4K12ac) and non-histone acetylated proteins (e.g., RelA subunit of NF-kappaB, Twist, and GATA1) (Huang et al., 2009; Filippakopoulos et al., 2012; Shi et al., 2014; Stonestrom et al., 2015). The affinity of BET bromodomains for specific histone acetylation marks is highly influenced by adjacent histone modifications, an indication that BET proteins recognize combinations of modifications rather than individual acetylated sites (Filippakopoulos et al., 2012). Although the amino acid sequence of BD1 and BD2 is similar across BET proteins, notable differences in amino acid sequence exist between BD1 and BD2 which may allow for selective BD1 vs. BD2 inhibition (Cheng et al., 2017). Beyond the bromodomain motif, BET proteins also contain an extra-terminal (ET) domain which is a site for multiple interactions with chromatin regulators (e.g., NSD3, JMJD6, CHD4) and viral proteins (Rahman et al., 2011; De Rijck et al., 2013; Gupta et al., 2013). Lastly, the C-terminal motif (CTM), which is unique to BRD4 and BRDT, plays an essential role in transcription elongation by modulating the activity of transcription elongation factor b (P-TEFb) (Jang et al., 2005; Yang et al., 2005; Bisgrove et al., 2007). Thus, by linking histone acetylation with transcriptional complexes, BET proteins are in a unique position to regulate disease-relevant gene expression programs.
BRD4 phosphorylation and enzymatic activity
Compared to BRD2 and BRD3, BRD4 has attracted the majority of attention within the BET family of proteins (Figure 1). Although BRD2, BRD3, and BRD4 share similar domains (e.g., BD1, BD2, ET), many of the protein-protein interaction and mechanistic studies have focused on BRD4. Figure 2 illustrates key domains within BRD4 and their known interactions with various histone and non-histone proteins. With the growing knowledge of BRD4-protein interactions, the mechanisms by which BRD4 is activated and recruited to chromatin remained an important unanswered question. Wu and colleagues revealed that BRD4 is regulated by a “phospho-switch” mechanism that influences its ability to bind to chromatin and interact with acetylated proteins (Wu et al., 2013). Casein kinase II (CK2), protein kinase A (PKA), and inhibitor of nuclear factor kappa-B kinase subunit epsilon (IKKe) were found to phosphorylate the N-terminal phosphorylation sites (NPS, amino acids 484–504) on BRD4 and protein phosphatase 2 (PP2A) was shown to remove these phosphorylation marks (Wu et al., 2013) (Figure 2). When dephosphorylated, NPS obstructs BD2 and functionally impedes BD1, which prevents BRD4 bromodomains from binding to acetylated histones and non-histone proteins. When NPS is phosphorylated, BRD4 undergoes a conformational change that uncovers the bromodomains and promotes their interaction with acetylated proteins (Chiang, 2016). Though the upstream receptor signaling mechanisms responsible for BRD4 phosphorylation are poorly understood, TrkB receptor signaling following BDNF stimulation was found to increase BRD4 phosphorylation levels in neurons in a CK2-dependent manner (Korb et al., 2015). The magnitude of BRD4 phosphorylation in neurons or in brain tissue has also been associated with neuroplasticity and drug-induced neurobehavioral effects (Korb et al., 2015; Guo et al., 2019). Thus, measuring and manipulating the phosphorylation states of BRD4 may be a unique and effective way to determine its functional activity in specific disease states.
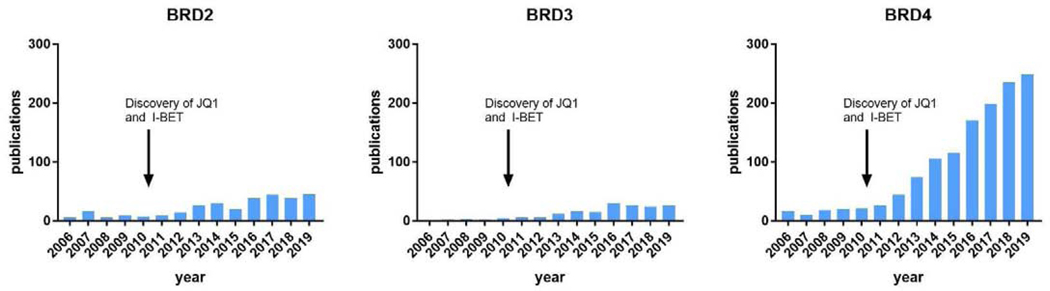
Figure 1. Yearly publications (2006–2019) for the PubMed search terms BRD2, BRD3, and BRD4.
Note the rise in publications (particularly for BRD4) following the discovery of JQ1 and I-BET.
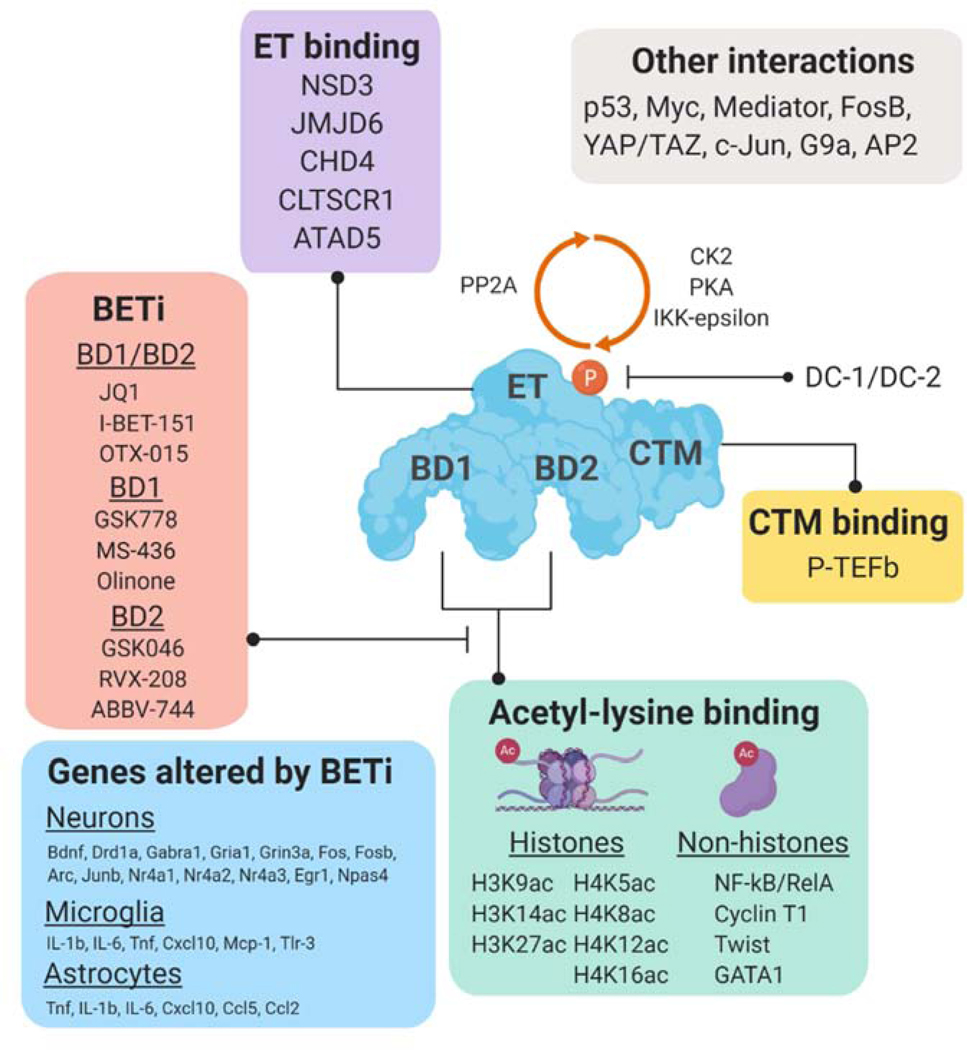
Figure 2. Interactions with BRD4 domains:
BRD4 contains tandem bromodomains (BD1 and BD2) that bind to acetylated histone and non-histone proteins, an extra-terminal domain (ET) that interacts with several chromatin regulators, and a C-terminal domain (CTM) that interacts with P-TEFb to facilitate transcription elongation. Several transcription factors have also been found to associate with BRD4-associated complexes. CK2, PKA, and IKK-epsilon phosphorylate BRD4 to alter its binding to acetylated proteins and PP2A removes the phosphorylation sites. Also listed are examples of BD1/BD2, BD1-selective, and BD2-selective BET inhibitions and key genes altered by BET inhibition in specific cell types.
In addition to functioning as a reader of acetyl-lysine residues and a scaffold for a variety of chromatin modifiers and transcription factors, preliminary evidence indicates that BRD4 may also have intrinsic kinase and HAT activity. Acting as an atypical kinase, BRD4 was found to promote transcription by directly and specifically phosphorylating serine 2 on the carboxyl-terminal domain (CTD) of RNA polymerase II (Devaiah et al., 2012). As a HAT, BRD4 was shown to acetylate the globular region of H3K122 residue, leading to nucleosome clearance and increased chromatin accessibility (Devaiah et al., 2016b). These intriguing studies add new layers of complexity to BRD4’s function, but further studies should be conducted in other cell types to confirm these initial observations.
Transcriptional regulation by BET proteins
BET proteins have been shown to regulate the expression of a variety of genes in brain cells and tissue, including but not limited to, immediate early genes, synaptic receptors, ion channels, cytokines, and neurotrophic factors (Korb et al., 2015; Sartor et al., 2015; Sullivan et al., 2015; Magistri et al., 2016; Benito et al., 2017; Duan et al., 2020). Interestingly, in primary neurons and brain tissue, BET inhibition was found to preferentially reduce expression of long genes (> 100 kb), likely by perturbing BET-mediated transcriptional elongation (Sullivan et al., 2015). To date, however, the vast majority of BET-mediated transcriptional studies have focused on BRD4 in cancer cell lines. Although initial experiments examined BRD4’s function at promoter regions, more recent studies indicate that BRD4-mediated gene expression is correlated with the presence of BRD4 at intergenic regions and gene bodies and not transcription start sites (TSS) (Kanno et al., 2014). Thus, although BRD4 occupies widespread genomic regions, its presence at the TSS is not always associated with active transcription.
BRD4 interactions at enhancer and super-enhancer genomic regions is another area of emerging research. Together with the Mediator complex, BRD4-associated enhancers activate P-TEFb on promoter regions via long-range enhancer-promoter interactions (Liu et al., 2013). Furthermore, BRD4 bromodomains interact with P300 and CBP to increase histone acetylation at H3K27, a modification that is often enriched at enhancer and super-enhancer regions (Wu et al., 2018). More recently, BRD4 binding at Fos enhancers was shown to be associated with activity-dependent transcription following synaptic activation (Chen et al., 2019). In addition to binding to enhancer regions, BRD4 regulates the expression of enhancer RNAs (eRNAs), and in turn, eRNA influence BRD4’s activity (Rahnamoun et al., 2018). For example, one study revealed that eRNAs dock to BRD4 bromodomains and increase BRD4 binding to genomic sites with H3K27ac and H4K16ac modifications (Rahnamoun et al., 2018). Given that the majority of eRNAs are expressed in a cell type-specific manner, future studies are needed to determine if BET inhibitors produce cell type-specific effects in neurons and other brain cells by disrupting BRD4-enhancer and/or BRD4-eRNA interactions.
BET inhibitors
In 2010, two small molecule inhibitors based on benzotriazolodiazepine (called I-BET) and thienotriazolodiazepine (called JQ1) structures were found to selectively inhibit the binding of BET bromodomains to acetylated histones (Filippakopoulos et al., 2010; Nicodeme et al., 2010). Although JQ1, I-BET151, and similar BET inhibitors are highly specific to BET bromodomains over non-BET bromodomains, these molecules bind to both BD1 and BD2 bromodomains within each BET protein and do not discriminate between BRD2, BRD3, BRD4 and BRDT. Over the years, improvements in selectivity, potency, and in vivo pharmacodynamics have led to over a dozen small-molecule BET inhibitors that are being tested in clinical trials, primarily for cancer, and many more tool compounds that are commercially available (Cochran et al., 2019). Of the published BET inhibitors, JQ1 and I-BET858 have been shown to penetrate the blood brain barrier and produce molecular and behavioral effects in rodents when injected systemically (Tables 1 and 2) (Korb et al., 2015; Sartor et al., 2015; Sullivan et al., 2015). Importantly, as shown in Table 2, BET inhibitors primarily produce disease-state dependent effects on behavior and few non-specific behavioral side effects have been reported (Korb et al., 2015; Sartor et al., 2015; Magistri et al., 2016; Benito et al., 2017; Korb et al., 2017; Duan et al., 2020). More recently, BD1 selective (GSK778, MS-436, Olinone, and BI-2536) and BD2 selective (GSK046, RVX-208, RVX-297, ABBV-744), small molecule BET inhibitors have been developed (McLure et al., 2013; Zhang et al., 2013; Gacias et al., 2014; Chen et al., 2015; Kharenko et al., 2016; Gilan et al., 2020). BD1- and BD2-selective BET inhibitors produced discrete transcriptional profiles in vitro, compared to compounds that are not selective for individual BET bromodomains (Picaud et al., 2013). However, in a more recent study, the BD1-selective inhibitor GSK778 showed similar transcriptional effects compared to pan-BET inhibitors in cancer models. Intriguingly, compared to BD1-selective and pan-BET inhibitors, the BD2-selective inhibitor GSK046 produced a unique transcriptional profile, caused limited effects on BRD4 binding to chromatin or cell proliferation, and showed an effective response in inflammatory and autoimmune disease models (Gilan et al., 2020). Thus, unlike pan-BET inhibitors, highly-selective BD2 BET inhibitors primarily modify transcription of induced genes and leave established transcriptional programs largely unchanged.
Table 1.
BET mechanisms in CNS-related disease models
| Disease model | Inhibitor | Cell line/rodent model | Genes/ targets | Mechanisms/pathways | Reference |
|---|---|---|---|---|---|
| Cerebellar ataxia | n/a | Mouse cerebellum, cerebellar granule cell progenitors | Brd4, Gli1 | Cerebellar granule cell neurogenesis, cerebellum development | (Penas et al., 2019) |
| Autism spectrum disorder | IBET858 | Primary neuron culture, young WT mice | Met, Gabra1, Cntn6, Pde4b, Npas2, Cdh10, Foxp1, Cacna1, Kcna1/4, Drd1a, Gria1–3, Fosb, Junb, Grin3a, Pde1c/4d, Ank3, Bdnf, Dscam1, Ntrk3 | Neuronal development and function | (Sullivan et al., 2015) |
| Fragile-x syndrome | JQ1 | Fmr1 KO neuronal culture, Fmr1 KO mice | Nr4a1, Shank2, Arc, Gria1 | Neuronal development and function, chromatin organization | (Korb et al., 2017) |
| Ischemic stroke | dBET | MCAO mouse model | Tnf-α, Cxcl1, Cxcl10, Ccl2, and Mmp-9 | NF-kB pathway | (DeMars et al., 2019) |
| JQ1 | MCAO mouse model | Tnf-a, Il-1b, Il-6, Il-18, Nlrp3, Asc, Caspase-1, Gsdmd | NF-kB pathway, glial activation | (Zhou et al., 2019) | |
| Spinal cord injury | JQ1 | Primary mouse macrophages, astrocytes, neurons, oligodendrocytes, microglia and mouse SCI model | Tnf-α, Il-1b, Il-6, Cxcl10, Ccl5, Ccl2, Ccl16 | NF-kB pathway, leukocyte infiltration | (Rudman et al., 2018) |
| JQ1 | HAPI microglia cells, mouse SCI model | Il-6, Il-1b, p-p65, IkBa, Iba-1, iNos, Cox-2 | NF-kB pathway, microglia activation | (Wang et al., 2018) | |
| JQ1 | Mouse SCI model | Il-6, Il-1b, Tnf-a, Ccl2, Il-4,Il-13, Il-10 | NF-kB pathway, microglia/macrophage activation | (Sanchez-Ventura et al., 2019) | |
| Multiple Sclerosis/EAE | RVX297 | U937 macrophage cell line, EAE mouse model | Il-1b, Il-6, Il-17, Tnf-α, Hist2h2be, IFN-γ, MCP-1 | NF-κB Pathway | (Jahagirdar et al., 2017) |
| Juvenile myoclonic epilepsy | n/a | Brd2+/− mice | GAD67, parvalbumin-positive neurons | GABA neurotransmission | (Velisek et al., 2011); (Chachua et al., 2014) |
| Seizures | JQ1 | Pentylenetetrazol-induced seizures in WT mice | GluA1 | Synaptic function, ion channel expression | (Korb et al., 2015) |
| Diabetes-induced cognitive impairment | JQ1 | STZ-induced diabetic rats | Il-1β, Tnf-α, p-AKT, Bax, Bcl-2 | Inflammatory cytokine pathway, Nox4-Nrf2 redox balance | (Liang et al., 2018) |
| Parkinson’s Disease | JQ1 | 6-OHDA model of L-DOPA induced dyskinesia in rats | Dab1, Esr1, Ntrk2, Arc, FosB, Nedd41, Bdnf | Corticostriatal plasticity | (Figge and Standaert, 2017) |
| Amyotrophic lateral sclerosis | JQ1, EP72, BET151 | C9/ALS fibroblasts | C9ORF72 | Epigenetic regulation of C9ORF72 | (Zeier et al., 2015) |
| Alzheimer’s disease | JQ1 | 3xTg mice | Il-b, Il-6, Ptgs2, Ccl2, Tnf-α, Nos2 | Tau phosphorylation, proinflammatory cytokine expression | (Magistri et al., 2016) |
| JQ1 | APP/PS1 mice | Egr1, Egr2, Junb, cFos, Arc, Nr4a2 | Ion channel function, DNA repair, RNA localization, synaptic transmission | (Benito et al., 2017) | |
| Substance use disorder | JQ1 | WT mice, Sprague Dawley rats | Bdnf, Il-1b, Arc, Fos, Gria1, Gria2, Creb | Cocaine-induced neuroplasticity | (Sartor et al., 2015); (Guo et al., 2019) |
| JQ | Long Evans rats | n/a | Opioid-induced chromatin conformation | (Egervari et al., 2017) | |
| Schizophrenia | JQ1 | Neurons from SZ patients, WT mice | n/a | H2A.Z and H4 acetylation mechanisms | (Farrelly et al., 2019) |
| Post-traumatic stress disorder | JQ1 | Mouse anterior cingulate cortex | IGF-2, Egr2, Grin2a, Grin2b, Sypl, Igf2 | Neuroplasticity associated with remote fear memory | (Duan et al., 2020) |
| Glioblastoma | JQ1 | T4105, T4302, T4597 primary GBM xenografts | c-MYC, p21 CIP1/WAF1, hTERT, BCL-2, BCL-XL | Cell proliferation and apoptosis | (Cheng et al., 2013) |
| JQ1 | U87, U87EGFRvIII, LN229, U373, GBM6 cell lines | SOX9 and FOXG1 | Cell proliferation and apoptosis | (Liu et al., 2015) | |
| iBET151 | U87MG, A172, SW1783 GB cell lines; UM20-patient cell lines; GBM xenografts | p21cipl, HEXIM-1 | Cell proliferation, G1/S cell cycle transition | (Pastori et al., 2014) | |
| iBET151 | LN18, U87MG, A172, T98G cell lines; Postmortem GBM tissues; U87 cell lines transplanted in Crl:Nu-Foxn1 nude mice | HOTAIR, MEG3, NEAT1, DGRR5, TUG1 | Cell proliferation | (Pastori et al., 2015) | |
| OTX 051 | U87MG, T98G, UI18 cell lines, U87MG orthotopic and heterotopic mouse model | c-MYC, CDKN1A, SESN3, HEXIM-1, HIST2H2BE, HIST1H2BK, MTHFDIL, HIST2H4A, HIST1H2BJ | Cell cycle regulation, Ras/Akt/mTOR pathway, Synthesis of purines and thymidylates | (Berenguer-Daize et al., 2016) | |
| Medulloblastoma | JQ1 | D283, D425, D458, DAOY, ONS-76 cell lines | c-MYC, SOX2, Nanog, Nestin, MAP2 | Cell proliferation | (Venkataraman et al., 2014) |
| JQ1 | HD-MB3, ONS-76, UW-228, DAOY, D-341, D-283 cell lines | c-MYC, cyclin D1, E2F1 | Cell proliferation, p53 pathway, DNA replication | (Henssen et al., 2013) | |
| JQ1, iBET151 | RCMB018, RCMB025, Hh-Light2, Sufu−/− MEF cell lines | GLI1, GLI2, c-MYC, P21, CDK4 | Hedgehog signaling pathway, cell cycle | (Long et al., 2014); (Tang et al., 2014) | |
Table 2:
BET inhibitor effects on behavior
| Behavioral test | Inhibitor | Dose | Frequency | Effect | Reference |
|---|---|---|---|---|---|
| Locomotor activity | IBET858 | 30 mg/kg, i.p | Single injection | no effect | (Sullivan et al., 2015) |
| IBET858 | 30 mg/kg, i.p | 1 injection/day for 2 wks | ↓ distance traveled | (Sullivan et al., 2015) | |
| JQ1 | 25 and 50 mg/kg, i.p. | 1 injection/day for 1 wk | no effect | (Korb et al., 2017) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 1 or 3 wks | no effect | (Korb et al., 2015) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 1 wk | no effect | (Benito et al., 2017) | |
| JQ1 | 25 mg/kg, i.p. | Single injection | no effect | (Sartor et al., 2019) | |
| JQ1 | 50 mg/kg, i.p. | Single injection | no effect | (Sartor et al., 2015) | |
| JQ1 | 20 μmol/L, dorsal striatum | Single injection | no effect | (Egervari et al., 2017) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 5 days | no effect | (Duan et al., 2020) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day, 5 days/wk for 15 wks | no effect | (Magistri et al., 2016) | |
| Thigmotaxis/open field | IBET858 | 30 mg/kg, i.p | Single injection | no effect | (Sullivan et al., 2015) |
| IBET858 | 30 mg/kg, i.p | 1 injection/day for 2 wks | ↑ anxiety and stereotypic behavior | (Sullivan et al., 2015) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 1 or 3 wks | no effect | (Korb et al., 2015) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 5 days | no effect | (Duan et al., 2020) | |
| JQ1 | 25 and 50 mg/kg, i.p. | 1 injection/day for 1 wk | no effect | (Korb et al., 2017) | |
| Y-maze | JQ1 | 50 mg/kg, i.p. | 1 injection/day, 5 days/wk for 15 wks | no effect | (Magistri et al., 2016) |
| Barnes maze | JQ1 | 50 mg/kg, i.p. | 1 injection/day, 5 days/wk for 15 wks | no effect | (Magistri et al., 2016) |
| Novel object recognition | JQ1 | 50 mg/kg, i.p. | Single injection and 1 injection/day for 1 or 3 wks | ↓ novel object recognition | (Korb et al., 2015) |
| JQ1 | 25 mg/kg, i.p. | 1 injection/day for 1 wk | ↓ novel object recognition | (Korb et al., 2017) | |
| Social interaction | IBET858 | 30 mg/kg, i.p | Single injection | no effect | (Sullivan et al., 2015) |
| IBET858 | 30 mg/kg, i.p | 1 injection/day for 2 wks | ↓ social interaction | (Sullivan et al., 2015) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 1 wk | no effect in WT, ↓ in FXS mice | (Korb et al., 2017) | |
| Fear conditioning | JQ1 | 50 mg/kg, i.p. | 1 injection/day for 1 or 3 wks | no effect on acquisition, ↑ freezing in new context | (Korb et al., 2015) |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 1 wk | ↑ freezing | (Benito et al., 2017) | |
| JQ1 | 5 μg, intra-hippocampal | 1 to 4 injections | ↑ freezing | (Benito et al., 2017) | |
| JQ1 | 50 mg/kg, i.p. | 1 injection/day for 5 days | no effect on recent memory. ↓ extinction of remote memory | (Duan et al., 2020) | |
| IBET858 | 30 mg/kg, i.p | Single injection | no effect | (Sullivan et al., 2015) | |
| Marble burying | JQ1 | 25 and 50 mg/kg, i.p. | 1 injection/day for 1 wk | no effect in WT, ↓ in FXS mice | (Korb et al., 2017) |
| Morris water maze | JQ1 | 50 mg/kg, i.p. | 13 injections during training | no effect on escape latency, ↑ time in target area | (Benito et al., 2017) |
| Condition place aversion (LiCl) | JQ1 | 50 mg/kg, i.p. | 1 injection/day for 3 days | no effect | (Sartor et al., 2015) |
| Condition place preference (cocaine) | JQ1 | 25 and 50 mg/kg, i.p.; 10 μM intra-NAc | 1 injection/day for 3 days | ↓ acquisition | (Sartor et al., 2015) |
| JQ1 | 25 mg/kg, i.p. and 5–10 μM intra-NAc | Single injection | ↓ expression and reinstatement | (Guo et al., 2019) | |
| Cocaine self-administration | JQ1 | 25 mg/kg, i.p.; 10 μM intra-NAc | Single injection | ↓ cocaine intake | (Guo et al., 2019) |
| Cocaine-induced locomotor activity | JQ1 | 10 μM intra-NAc | Single injection | ↓ hyperactivity | (Guo et al., 2019) |
| Heroin self-administration | JQ1 | 20 μmol/L, dorsal striatum | 3 injections | ↓ heroin-seeking and taking behaviors | (Egervari et al., 2017) |
Recent screening experiments have revealed that some kinase inhibitors also act as potent BET inhibitors. For example, dinaciclib (CDK inhibitor), TG101209 (JAK2 inhibitor), and BI-2536 (PLK1 inhibitor) inhibit BET bromodomains at submicromolar concentrations (Ember et al., 2014; Hu et al., 2017). Thus, the effects observed in previous studies using these kinase inhibitors may be due, in part, to their inhibition of BET proteins. These findings have led to the design and development of multiple, dual BET-kinase inhibitors (Ember et al., 2017). Other dual inhibitors have also emerged for BETs and HDACs (Shao et al., 2017) as well as BETs and HATs (Picaud et al., 2015). While most of these compounds have only been studied in cancer models, future studies are needed to determine if dual BET inhibitors have utility in CNS-related disease models.
The compounds described above prevent BET bromodomain interactions with acetylated histones and non-histone proteins. Because BET proteins have bromodomain-independent functions, newer strategies have recently been developed to investigate additional molecular aspects of BET proteins. For example, in a technique known as proteolysis targeting chimera (PROTAC), a BET inhibitor, such as JQ1, is conjugated to an ubiquitin ligase recognition module (Yang et al., 2019). JQ1 displaces the BET proteins from chromatin and the ubiquitin ligase recognition site recruits E3 ubiquitin ligases, leading to polyubiquitylation of BET proteins and proteasome-dependent degradation. With the rapid and selective degradation of BET proteins, PROTAC compounds such as dBET1, ARV825 and MZ1 provide an additional opportunity to impede all functions of BET proteins (Winter et al., 2015; Zengerle et al., 2015; Sakamaki et al., 2017). Blocking the interaction between phosphorylated BRD4 and transcriptional factors is another novel therapeutic strategy to manipulate BRD4 function without blocking its bromodomain binding activity. Recently, cell-permeable peptoids (DC-1 and DC-2) were found to specifically bind to the NPS regions of human BRD4 and block BRD4 phosphorylation-specific functions (Wu et al., 2016). Thus, with the growing number of highly selective and versatile tools to manipulate BET activity, the prospect of identifying novel roles for BET proteins in CNS-related disorders is increasingly promising.
BET expression in the CNS
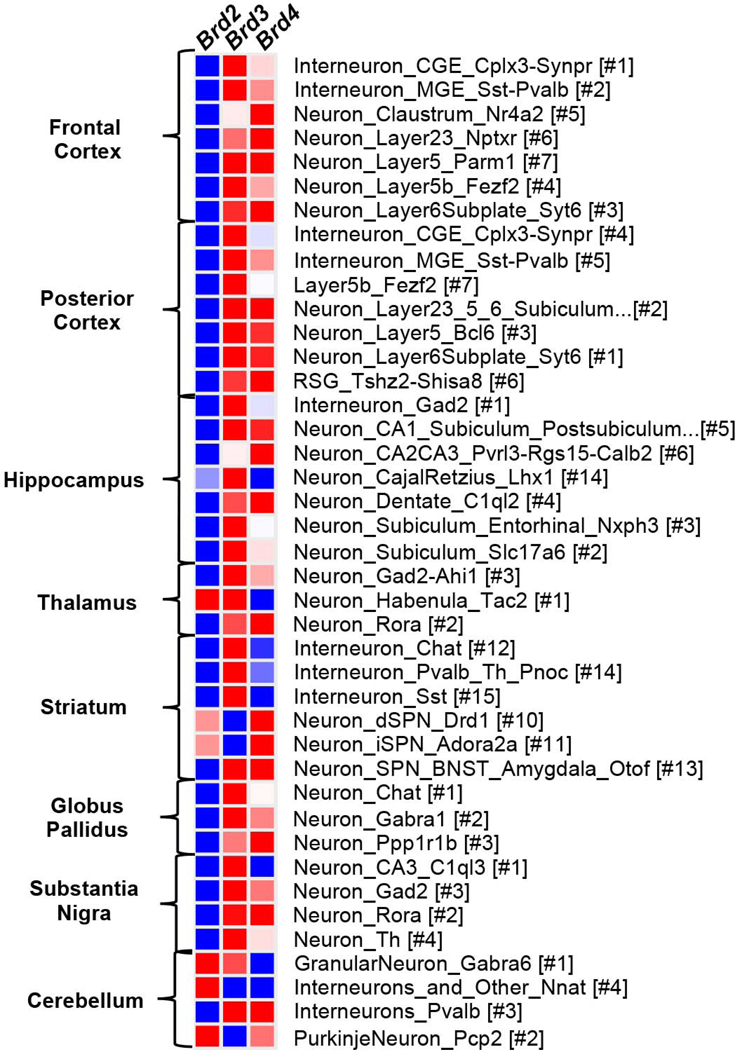
While BRD2, BRD3, and BRD4 mRNAs and proteins are thought to be ubiquitously expressed, recent studies have shown that some BETs are enriched in specific brain cells. For example, consistent with BRD4’s essential role in neuronal function, baseline BRD4 protein levels were shown to be enriched in mouse neurons compared to other brain cells (Korb et al., 2015). In the mouse nucleus accumbens (NAc), a brain region important in reward processing, baseline Brd4 mRNA levels were found to be more than 3 times higher than Brd2 and Brd3 mRNA (Guo et al., 2019), an indication that BRD4 plays an important role in reward-seeking behaviors. Consistent with a potential link between BETs and reward processing, BET proteins in the amygdala, nucleus accumbens, and midbrain were found to be expressed at higher levels compared to other brain regions using PET radiotracers in rodents and non-human primates (Bai et al., 2019). In single-cell analysis studies, differences in BET mRNA expression levels are also apparent across neuronal subtypes (Saunders et al., 2018). In the mouse brain, for example, Brd3 mRNA levels are expressed at higher levels in most neuronal subtypes compared to Brd2 and Brd4, and Brd2 levels are relatively lower compared to Brd3 and Brd4. Within the striatum, Brd4 mRNA levels are higher in dopamine 1 receptor- (D1R) and dopamine 2 receptor- (D2R) expressing neurons, whereas Brd3 is expressed at higher levels in non-D1R and -D2R neurons in the striatum (Figure 3). The enrichment of Brd4 in D1R- and D2R-expressing neurons is consistent with the findings of Guo et al., 2019, as the NAc primarily consists of D1R- and D2R-expressing neurons. Other notable differences in cell type-specific enrichment of individual BET mRNAs can be found in the prefrontal cortex (Brd4 claustrum_Nr4a2 neurons), hippocampus (Brd3 in CajalRetzius_Lhx1 and Subiculum_Entorhinal_Nxph3 neurons), and cerebellum (Brd2 in Nnat interneurons) (Figure 3). Thus, while BETs are expressed in most brain cells, expression levels vary among neuronal subtypes and may contribute to unique cell-type specific mechanisms in the brain. Future studies using cell type-specific manipulations of individual BET proteins within specific brain regions are needed to precisely understand the functional importance of BETs in brain health and disease.
Figure 3. Heatmap of Brd2, Brd3, and Brd4 mRNA expression in mouse neuronal cell types.
Each row compares relative expression of Brd2, Brd3, and Brd4 mRNA within a cell type cluster from the specified brain region. RNA expression is compared using normalized mean log values obtained from DropVis single-cell RNAseq database (http://dropviz.org/). The heatmap was generated using Morpheous tool provided by the Broad Institute (https://software.broadinstitute.org/morpheus/). Red indicates higher expression levels and blue indicates lower expression levels. Note that these are baseline expression levels and that specific disease states may augment Brd2, Brd3, Brd4 levels in a cell type-specific manner.
BETs in neurodevelopment
Arising from multifactorial genetic and environmental interactions, neurodevelopmental disorders are characterized by impairments in growth and development of the central nervous system that ultimately lead to deficits in speech, learning and memory, and/or motor skills. In embryonic developmental studies, Brd2 and Brd4 were found to be essential for survival in mice (Houzelstein et al., 2002; Shang et al., 2009). While Brd4 deficient mice result in embryonic lethality prior to implantation, conditional knockout of Brd4 in the cerebellum caused deficits in granule cell progenitor proliferation and cerebellar morphology, leading to cerebellar ataxia in mice (Penas et al., 2019). Brd2 null embryos exhibit defects in neural tube closure that most commonly appear as exencephaly of the hindbrain and die by embryonic day 13 (Gyuris et al., 2009). However, heterozygous Brd2+/− mice are viable but exhibit increased susceptibility to provoked seizures, develop unprovoked spontaneous seizures, show decreased activity in GAD67-positive neurons in the basal ganglia, and exhibit aggressive behavior (Velisek et al., 2011; Chachua et al., 2014). Similar to the findings in heterozygous Brd2+/− mice, mutations in the human BRD2 promoter region are associated with juvenile myoclonic epilepsy (JME), an adolescent onset generalized epilepsy (Durner et al., 1991; Weissbecker et al., 1991; Pal et al., 2003; Cavalleri et al., 2007) (Table 1). Thus, data from mice and humans indicate that an underlying mechanism of JME may involve a developmentally determined deficit of GABAergic neurons caused by alterations in BRD2 activity (Velisek et al., 2011; Chachua et al., 2014).
To investigate the effects of BET inhibition on neurodevelopment, Sullivan and colleagues identified and tested a novel, brain-permeable BET inhibitor called I-BET858 (Sullivan et al., 2015). In both primary neurons and young mice, I-BET858 altered the expression of genes that regulate neuronal development, synaptic formation, and synaptic function, many of which are known autism-associated genes (e.g., Met, Gabra1, Cntn6, Pde4b, Npas2, Cdh10, and Foxp1). Other genes involved in neurotransmitter receptor signaling (Grin3, Pde1c/4d), ion channels (Cacna2d1, Kcnma1, and Kcna1/4), and dendrite and axon development (Ank3, Bdnf, Camk2d, Dscaml1, Ntrk3, and Sema3a) were also altered in primary neurons and/or in the striatum of young mice treated with I-BET858 (Table 1). Interestingly, the repressive effects of I-BET858 on gene expression were significantly correlated with the gene length, with genes over 100 kb being reduced the most. In addition to changes in gene expression, multiple autism-like behaviors were observed in young mice (4 weeks old) treated with chronic but not acute I-BET858. For example, after 2 weeks of daily treatment with I-BET858 (30 mg/kg), mice displayed reduced exploration, increased anxiety-like behavior in open field, and reduced sociability and social novelty preference (Sullivan et al., 2015) (Table 2).
Although suppression of BET proteins produced autism-like behaviors in young mice, increased activity of BRD4 was shown to play an essential role in a mouse model of Fragile X syndrome (FXS). In neurons from fragile X mental retardation protein (FMRP) knockout mice, a model for FXS, BRD4 protein along with histone marks associated with active chromatin states (H3K27ac, H4K8ac, H4K16ac and H3K4me3) were significantly increased (Korb et al., 2017). When comparing the genes changed in FMRP KO neurons to wild-type neurons treated with JQ1, many of the same genes were altered. Consistent with these findings, all significantly elevated genes in FMRP KO neurons were reduced by JQ1, an indication that BRD4 plays a key role in the transcriptional dysregulation following FMRP KO. In FMRP KO mice, JQ1 reduced the elevated number of dendritic spines, decreased repetitive behaviors, and normalized social behaviors (Korb et al., 2017) (Table 2). Together these data in FXS and autism mouse models indicate that the appropriate amount of BET protein is essential for optimal neuronal function and deficits emerge from too much or too little BET activity during neurodevelopment.
BETs in neuroinflammation, myelination, and repair
Some of the earliest studies with BET inhibitors indicated that these compounds have anti-inflammatory properties by reducing Nuclear factor kappa B (NF-kB) signaling (Nicodeme et al., 2010). Initial studies revealed that the RelA subunit of NF-kB is acetylated by p300 which promotes BRD4 binding and BRD4-mediated transcriptional co-activation of NF-kB target genes (Huang et al., 2009). Since these initial studies, BET inhibitors have been shown to attenuate the expression of many genes involved in inflammation (e.g., Il-6, Il-10, Il-17, Il-1β, Tnf-α, Mcp-1/Ccl2) and prevent endotoxin-induced death in mice (Huang et al., 2009; Bandukwala et al., 2012; Belkina et al., 2013; Barrett et al., 2014; Wang et al., 2018). In chromatin immunoprecipitation studies, BRD4 was found to physically associate with the promoter regions of inflammatory cytokine genes such as Il-1β, Il-6, Cxcl8, Tnf-α in fibroblasts and macrophages, and BET inhibition reduced BRD4 binding to these promoter regions (Barrett et al., 2014; Khan et al., 2014). Consistent with other cell types, BET inhibitors also produce anti-inflammatory responses in brain cells. For example, in purified primary astrocytic and microglial cell lines, JQ1 reduced the expression of cytokines and chemokines via PA1/tPA axis and MAPK/NF-κB signaling pathways, respectively (Choi et al., 2015; Wang et al., 2018). Similarly, in primary neuronal, oligodendroglial, microglial and astrocytic cell cultures, JQ1 decreased the expression of several genes associated with inflammation (Il-6, Il-1β, Tnf-α, Ccl5, Ccl2, Cxcl10) (Rudman et al., 2018; Wang et al., 2019).
Since BET inhibition has been shown to reduce inflammatory responses in cultured brain cells, a role for BET proteins in stroke-induced neuroinflammation was recently explored. Using a middle cerebral artery occlusion model (MCAO) of ischemic stroke, BRD4 and inflammatory cytokines (Il-1b, Il-6, Il-18, and Tnf-a) were found to be significantly elevated in mouse brain tissue (Zhou et al., 2019). Functional studies using MCAO mice demonstrated that JQ1 reduced glial activation, expression of pro-inflammatory factors, pyroptosis, and infarct volume (Zhou et al., 2019). Consistent with these results, the BET degrader, dBET1, also provided neuroprotective effects in mice following ischemic stroke (DeMars et al., 2019). In other studies using a rodent model of contusive spinal cord injury (SCI), JQ1 decreased leukocyte infiltration, pro-inflammatory cytokines and chemokines, cell death, and improved functional recovery after spinal cord injury (Rudman et al., 2018; Wang et al., 2019) (Table 1). Collectively, these experiments indicate that BET inhibitors may offer neuroprotection from hyperinflammatory responses following ischemia and neural injury.
Apart from the transcriptional control of cytokines and chemokines, BET proteins have also been shown to play important role in T-cell differentiation and function, which is widely recognized as mediator of autoimmune conditions, such as multiple sclerosis (MS) and the related murine model, experimental autoimmune encephalomyelitis (EAE) (Littman and Rudensky, 2010; Jahagirdar et al., 2017). BRD4 and BRD2 have been shown to associate with the IL-17 locus in Th17 cells, control Th17 differentiation, and regulate multiple effector Th17-associated cytokines such as IL-17, IL-21, and GMCSF (Cheung et al., 2017). In the murine EAE model, BET inhibitors decreased generation and function of Th17 cells, thus protecting the EAE mice from quadriparalysis as well as decreased demyelination (Mele et al., 2013; Jahagirdar et al., 2017). While most therapeutic interventions for MS are based on immunomodulation to reduce demyelination, few studies have focused on promoting remyelination and repair. After an acute demyelinating attack, a wave of oligodendrocyte progenitor cells saturate the damaged area in an attempt to control the injury (Grade et al., 2013). However, remyelination that occurs at this stage is inadequate to control disease progression (Kuhlmann et al., 2008). The inadequacy in remyelination despite the increased presence of oligodendrocyte progenitor cells (OPCs) at the injury site is associated with increased histone acetylation in OPCs. Interestingly, the BD1-selective BET inhibitor, Olinone, was found to accelerate the progression of mouse primary oligodendrocyte progenitors towards differentiation, whereas inhibition of both bromodomains of BET proteins impeded differentiation (Gacias et al., 2014). Thus, these data demonstrate that bromodomain-selective BET inhibitors may have therapeutic advantages in lifting the OPCs differentiation block and promoting remyelination.
BETs in learning and memory
Epigenetic regulatory mechanisms are critical for learning and memory (Alarcon et al., 2004; Korzus et al., 2004; Levenson et al., 2004; Levenson and Sweatt, 2005). BET proteins, in particular, have high affinity for histone marks (H4K5/K8/K12ac) that are associated with learning and memory (Guan et al., 2009; Peleg et al., 2010; Umehara et al., 2010b; Umehara et al., 2010a; LeRoy et al., 2012). Additionally, BET inhibitors have been shown to regulate expression of multiple genes associated with neuroplasticity and learning and memory (e.g., Bdnf, Nr4a1/2, Gria1, Fos, Arc, Egr1 and Junb) (Korb et al., 2015; Sartor et al., 2015; Benito et al., 2017; Sartor et al., 2019; Duan et al., 2020). However, little is known about the expression levels of individual BET proteins or occupancy of BET proteins at specific genomic sites following a learning event, a major impediment to understanding mechanistic roles of BETs in learning and memory. In behavioral studies, chronic administration of JQ1 attenuated long-term, but not short-term, memory in the novel object recognition task in mice (Korb et al., 2015; Korb et al., 2017). However, other types of contextual learning and memory tasks were not altered by BET inhibition (Korb et al., 2015; Sartor et al., 2015; Sullivan et al., 2015). In most of learning and memory studies, BET inhibitors were administered by intraperitoneal injections (Table 2). Thus, it is unclear if the BET inhibitor is reaching specific brain regions at sufficient concentrations to affect learning and memory. Future studies using brain region-specific injections of BET inhibitors or viral-mediated knockdown of individual BET proteins in specific brain regions are needed to accurately determine the role of BET proteins in specific aspects of learning and memory. Across multiple studies, however, other behaviors such as locomotor activity and anxiety-like behavior in an open field task were not changed by JQ1 (Table 2) (Korb et al., 2015; Sartor et al., 2015; Magistri et al., 2016; Benito et al., 2017; Korb et al., 2017; Duan et al., 2020), an indication that JQ1 is not causing wide-spread behavior changes and deficits in motor function.
Although the effects of BET inhibition on learning and memory are actively being investigated, HDAC inhibitors, in many instances, enhance learning and memory (Guan et al., 2009; Calfa et al., 2012; Graff and Tsai, 2013; Benito et al., 2015; Ganai et al., 2016; Sartor et al., 2019). Because HDAC inhibitors typically increase histone acetylation and learning and memory and because BET proteins are readers of histone acetylation, a role for BETs in HDAC inhibitor-induced enhancement of learning and memory was recently investigated. Using molecular, physiological, and behavioral techniques, BRD4 was shown to be necessary for the increased BDNF expression, long-term potentiation, and memory following HDAC3 inhibition (Sartor et al., 2019). Thus, the neuroprotective and cognition-enhancing properties of some HDAC inhibitors are mediated, in part, by BET proteins.
BETs in neurodegenerative disorders
Targeting histone acetylation factors is a promising therapeutic strategy for the treatment of neurodegenerative disorders (Berson et al., 2018). In particular, HDAC inhibitors appear to show beneficial effects in animal models of Alzheimer’s disease (AD) (Govindarajan et al., 2011; Graff et al., 2012; Volmar et al., 2017; Janczura et al., 2018), but the effectiveness of BET inhibitors in AD models remains unclear. For example, in the 3xTg mouse model of AD, repeated exposure to JQ1 did not affect learning and memory in the Barnes maze and Y-maze tasks, but did reduce neuroinflammation and Tau phosphorylation (Magistri et al., 2016). Contrary to the findings above, Benito and colleagues found that JQ1 enhanced learning and memory and hippocampal long-term potentiation in the APP/PS1 mouse model for AD (Benito et al., 2017) (Table 2). Methodological differences such as mouse strain and age, treatment regimen, dose, vehicle used for JQ1, and behavior training protocols may explain these divergent results. Clearly, more testing is needed to determine if BET inhibitors have potential therapeutic value for AD. Additionally, as little is known about the functional importance of BETs in aging, future studies should focus on BETs in both pathological and non-pathological brain function associated with the aging.
Although mixed results have been reported in animal models of AD, some evidence indicates that selective BET inhibitors may improve cognitive function and AD by augmenting biomarkers typically associated with cardiovascular disease. For example, the BD2-selective BET inhibitor, RVX-208, has been shown to increase apolipoprotein A1 (ApoA1) and high-density lipoprotein (HDL) (McNeill 2010), two factors that are often decreased in patients with AD and cognitive impairments (Merched et al., 2000; Button et al., 2019). Consistent with the potential benefits of BD2-selective BET inhibitors in AD, RVX-208 was found to increase plasma levels of Aβ40 by 12–14% in AD patients (Rvx 208 2011). Although these initial studies are promising, large-scale clinical trials are needed to determine if RVX-208-induced changes ApoA1, HDL and Aβ40 levels leads to improved cognitive function in AD patients.
Apart from Alzheimer’s disease, BET proteins have also been shown to play important roles in other models of neurodegenerative disorders. In the 6-OHDA rodent model of levodopa-induced dyskinesia, an animal model for Parkinson’s disease, increased levels of Brd2 mRNA were observed in the dorsal striatum, along with increased binding of BRD2 and BRD4 proteins to the promoter and enhancer regions of the genes dysregulated during dyskinesia induction (e.g., Dab1, Esr1, Ntrk2, Arc, FosB, and Nedd41). Consistent with these molecular results, JQ1 was shown to reduce levodopa-induced dyskinesia in rats (Figge and Standaert, 2017), an indication that BET inhibition may improve symptoms assoicated with Parkinson’s disease. In in vitro models of amyotrophic lateral sclerosis (ALS), BET inhibitors and BRD3 siRNA elevated the expression of unexpanded C9ORF72 mRNA and pre-mRNA without altering repressive epigenetic signatures of expanded C9ORF72 alleles (Zeier et al., 2015). Interestingly, BET inhibition did not alter expression of expanded alleles in fibroblasts from Fragile X syndrome or Freiderich’s ataxia patients, indicating a C9ORF72-specific effect (Table 1). Thus, BET and/or BRD3 inhibition may be effective at rescuing C9ORF72 haploinsufficiency and reducing symptoms associated with ALS. Future experiments in animal models of ALS are needed to corroborate these initial in vitro studies.
BETs in psychiatric disorders
Alterations in histone acetylation mechanisms are associated with many psychiatric conditions (Tremolizzo et al., 2002; Kumar et al., 2005; Renthal et al., 2007; Malvaez et al., 2010; Kurita et al., 2012; Stafford et al., 2012; Moonat et al., 2013). In animal models of substance use disorder (SUD), for example, chronic administration of cocaine elevates global histone acetylation levels in reward-related brain regions, such as the nucleus accumbens (Renthal et al., 2009), and manipulation of histone acetyltransferases and histone deacetylases drastically affects behavioral and molecular responses to psychostimulants in rodents (Kumar et al., 2005; Renthal et al., 2007; Sun et al., 2008; Renthal et al., 2009; Im et al., 2010; Malvaez et al., 2010; Wang et al., 2010; Malvaez et al., 2011; Kennedy et al., 2013; Malvaez et al., 2013; Rogge et al., 2013). Given that histone acetylation factors impact drug-induced neuroadaptations and behaviors, a role for BET proteins was recently examined in animal models of SUD. BRD4, but not BRD2 or BRD3, protein was found to be significantly elevated in the NAc of rats following cocaine self-administration (Sartor et al., 2015). In behavioral studies, systemic and intra-NAc administration of JQ1 attenuated cocaine conditioned place preference (CPP), an animal model measuring contextual reward associations, but did not affect locomotor activity or other types of contextual learning (Sartor et al., 2015) (Table 2). Investigating the underlying mechanisms, repeated cocaine administration was found to increase binding of BRD4 to the promoter region of Bdnf in the NAc, while systemic injection of JQ1 reduced cocaine-induced expression of Bdnf in the NAc. These data are consistent with other results showing BET-mediated regulation of Bdnf (Sullivan et al., 2015; Zeier et al., 2015; Guo et al., 2019; Sartor et al., 2019). In unpublished results from our laboratory, JQ1 also attenuated nicotine and amphetamine CPP, indicating that BET proteins play a role in multiple psychostimulant-induced behaviors.
More recently, Guo and colleagues found that phospho-BRD4 levels were elevated in the NAc following cocaine-seeking behaviors, and JQ1 attenuated expression and reinstatement of cocaine CPP and cocaine self-administration in mice (Guo et al., 2019). In other SUD-related studies, JQ1 injections in the dorsal striatum reduced heroin self-administration and cue-induced heroin-seeking behavior in rats (Egervari et al., 2017) (Table 2). Though no studies have examined a functional role for BETs in alcohol-induced behaviors, Barbier and colleagues found that Brd3 and Brd4 mRNA levels were reduced in the dorsal medial prefrontal cortex of alcohol-dependent rats (Barbier et al., 2017). Together, these observations support the view of BET proteins as novel regulators of drug-induced neuroadaptations and that modulation of BET activity may have therapeutic efficacy in SUD-related behaviors.
Although still in relatively early stages, multiple reports have shown a potential link between BET activity and other psychiatric disorders. For example, single nucleotide polymorphisms (SNPs) associated with schizophrenia were found to be highly enriched within BRD4 binding sites, potentially linking BRD4 to an increased susceptibility to schizophrenia (Zuber et al., 2017). In neurons from patients with schizophrenia, binding sites of BRD4 (H2A.ZK4K7K11ac and H4K5K8K12ac) were significantly increased, and in mechanistic studies, JQ1 partially ameliorated transcriptional and behavioral deficits associated with schizophrenia in rodents (Farrelly et al., 2019). In fear conditioning, an animal model for post-traumatic stress disorder (PTSD), alternative splicing changes in Brd2 and Brd3 were observed in the hippocampus following retrieval of a fear memory (Poplawski et al., 2016). However, the behavioral effects of pharmacological inhibition of BET proteins with respect to fear conditioning have been inconsistent (Korb et al., 2015; Sullivan et al., 2015; Benito et al., 2017; Duan et al., 2020) (Table 2). For example, acquisition of fear conditioning was increased by JQ1 in one study (Benito et al., 2017), but unchanged in multiple other studies (Korb et al., 2015; Sullivan et al., 2015; Duan et al., 2020). In the Korb et al., 2015 study where acquisition of fear conditioning was unchanged, JQ1-treated mice showed increased freezing behavior when exposed to a new context, an effect the authors contribute to BET’s involvement in hippocampus-dependent test of context discrimination (Korb et al., 2015). In a more recent study, JQ1 was shown to impair extinction of remote fear memory, but not a recent fear memory (Duan et al., 2020). In mechanistic experiments, insulin like growth factor 2 (IGF-2) was shown to be upregulated in the anterior cingulate cortex following extinction of the remote fear memory, and JQ1 reduced the rate of extinction by blocking this increase of IGF-2. Thus, while BET inhibitors show promising effects in some animal models of psychiatric disorders, they do not appear to be a favorable therapeutic approach in animal models of fear conditioning.
BETs in brain cancer
With its role as an epigenetic regulator of the cell cycle, BRD4 has emerged as a promising target for cancer treatment. In neuro-oncology, several preclinical studies have implicated a role for BET proteins in glioblastoma (GBM), the most common, aggressive primary malignant brain cancer. For example, BRD2 and BRD4 mRNAs were found to be significantly elevated in GBM tumors, and pharmacological inhibition of BETs reduced cell proliferation and promoted apoptosis in ex vivo cultures derived from primary GBM xenograft lines and orthotopic GBM tumors (Cheng et al., 2013; Pastori et al., 2014). BET inhibition also resulted in significant changes in expression of important GBM-associated genes (c-MYC, p21CIPI/WAFI, hTERT, Bcl-2 and Bcl-xL) and GBM-associated long noncoding RNA, HOTAIR (Cheng et al., 2013; Pastori et al., 2015). In similar studies, Liu and colleagues demonstrated that JQ1 suppressed aggressive tumor growth of GBM cells with oncogenic EGFR mutations (Liu et al., 2015). In medulloblastoma studies, JQ1 significantly decreased c-MYC expression and inhibited c-MYC-induced gene expression and cell proliferation. Survival of mice harboring medulloblastoma xenografts was also prolonged by JQ1 treatment (Henssen et al., 2013). Consistent with these results, JQ1 was found to produce anti-proliferative, pro-senescence effects in athymic mice harboring medulloblastoma xenografts (Venkataraman et al., 2014). BET inhibition was also shown to be effective in non-MYC driven medulloblastoma tumors by regulating hedgehog-targeted genes (Long et al., 2014; Tang et al., 2014). Taken together, these studies indicate that BET inhibitors may be effective in treating multiple types of aggressive brain cancer.
As cancers are known to develop resistance to monotherapies, recent studies have shown that combination therapy of BET inhibitors (JQ1 or OTX015) with HDAC inhibitor (panabinostat) or with kinase inhibitor (sorafenib) markedly reduced cell proliferation and induced apoptosis in GBM cells (Berenguer-Daize et al., 2016; Meng et al., 2018; Zhang et al., 2018). Additionally, the BET inhibitor OTX015 in combination with conventional therapies in glioblastoma models, such as SN38, temozolomide or everolimus have shown synergistic anti-tumor effects in vivo and in vitro (Berenguer-Daize et al., 2016). While combination therapies have provided enhanced efficacy compared to single agent therapies, effective treatment of GBM and other brain cancers are limited by the drug’s ability to cross the blood-brain barrier (BBB). To address this limitation, Lam and colleagues developed a transferrin functionalized nanoparticle (Tf-NP) that traverses the BBB in mice and delivers dual drug combination therapies to the tumor (Lam et al., 2018). In this experiment, Tf-NPs were loaded with temozolomide and JQ1 and systemically injected in GBM tumor-bearing mice. This treatment increased DNA damage and apoptosis in GBM cells, decreased tumor size, and increased survival compared to free-drug dosing (Lam et al., 2018). These studies support the potential therapeutic use of BET inhibitors in combination with other therapies for the treatment of brain cancers and the use of nanoparticle systems for improved brain delivery of BET therapeutics.
Summary and future directions
BET proteins have emerged as an integral component of gene regulatory networks associated with brain health and disease. With BET proteins playing fundamental roles in transcriptional responses, tolerability and side effects of BET inhibitors are an undeniable concern. In the limited clinical data available on BET inhibitors, major side effects such as thrombocytopenia and gastrointestinal toxicities have been reported (Amorim et al., 2016; Berthon et al., 2016). These side effects were shown to be dose-dependent and reversible upon removal of the BET inhibitor. To date, however, there have been no clinical studies using a BET inhibitor for the treatment of a brain disease, and there are believed to be limited clinical studies using a BET inhibitor that effectively crosses the BBB. Thus, CNS-related side effects of BET inhibitors in humans remains a major unknown.
Although BET manipulation during early brain development resulted in neurobehavioral side effects in rodents (Sullivan et al., 2015), BET inhibition in adult rodents has had mixed consequences on behavior, learning and memory, and neuronal function, with many of the effects observed being disease-state dependent (Table 2). The fewer observed side effects in adult rodents may be due to the short half-life of JQ1 in the brain (Matzuk et al., 2012). Thus, utilizing BET inhibitors with a relatively short half-life may be a preferred approach to normalize disease-related transcriptional responses while limiting non-specific side effects. Nonetheless, an objective for future pre-clinical and clinical studies will be to determine whether more selective manipulations of BET protein activity will have similar therapeutic efficacy with fewer side effects. Compounds selective for BET BD1 verses BD2 bromodomains have been developed and have yielded promising results in preclinical and clinical studies (Gacias et al., 2014; Tsujikawa et al., 2019; Faivre et al., 2020). Other tools to inhibit bromodomain-independent functions of BET proteins have also recently emerged (Cai et al., 2011; Yang et al., 2019), but their effects in CNS-related disease models have yet to be tested.
Despite the tremendous number of advancements in BET therapeutics, many unanswered questions remain. For example, what role, if any, does BRD2 and BRD3 play in CNS-related diseases? BRD3 is expressed at relatively high levels in the brain (www.proteinatlas.org) and BRD2 appears to play a role in seizure susceptibility (Velisek et al., 2011), yet most research has focused on BRD4. Because functions of individual BET proteins are not completely over-lapping (Anders et al., 2014) and because current BET inhibitors are not selective for BRD4, more studies are needed to elucidate the mechanisms of action of individual BET proteins within the brain. Additional understanding of upstream signals (e.g., receptor signaling cascades, post-translational modifications to individual BET proteins), downstream interactions (e.g., histone, non-histone protein, eRNA, and genomic binding regions), and cell type-specific mechanisms of individual BET proteins in the brain are required for therapeutic advancement. Finally, the treatment regimen for BET inhibitors remains unclear for many brain-related diseases other than cancer. For example, if a patient suffering from substance use disorder (SUD) was prescribed a BET inhibitor, how long would the patient take the medication? Because of the potential side effects of first-generation BET inhibitors, it is unlikely to be a long-term treatment. Would a SUD patient be administered a BET inhibitor during a relatively short period to reduce early withdrawal symptoms, drug craving, and relapse and then transition to more traditional medications? Could a BET inhibitor be administered in conjunction with FDA-approved medications to potentiate the effects of the approved medication? Will delivery systems (e.g., nanoparticles) be required to effectively deliver BET therapies to the brain or specific to brain cells? Continued efforts to address these questions will reveal additional therapeutic opportunities for targeting BET proteins in brain health and disease.
Highlights:
This review describes the progress in understanding the neurobiological mechanisms of BET proteins, and the therapeutic potential of targeting BET proteins for brain health and disease.
Acknowledgements:
This research was supported by National Institute on Drug Abuse grant R00DA040744.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A (2004) Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42:947–959. [DOI] [PubMed] [Google Scholar]
- Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K, Herait P, Kahatt C, Lokiec F, Salles G, Facon T, Palumbo A, Cunningham D, Zucca E, Thieblemont C (2016) Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 3:e196–204. [DOI] [PubMed] [Google Scholar]
- Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Loven J, Sigova AA, Smith WB, Lee TI, Bradner JE, Young RA (2014) Genome-wide localization of small molecules. Nat Biotechnol 32:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Wey HY, Patnaik D, Lu X, Lan Y, Rokka J, Stephanie F, Haggarty SJ, Wang C (2019) Positron emission tomography probes targeting bromodomain and extra-terminal (BET) domains to enable in vivo neuroepigenetic imaging. Chem Commun (Camb) 55:12932–12935. [DOI] [PubMed] [Google Scholar]
- Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, Molesworth AM, Smithers N, Lee K, Witherington J, Tough DF, Prinjha RK, Peters B, Rao A (2012) Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci U S A 109:14532–14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Johnstone AL, Khomtchouk BB, Tapocik JD, Pitcairn C, Rehman F, Augier E, Borich A, Schank JR, Rienas CA, Van Booven DJ, Sun H, Natt D, Wahlestedt C, Heilig M (2017) Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol Psychiatry 22:1746–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E, Brothers S, Wahlestedt C, Beurel E (2014) I-BET151 selectively regulates IL-6 production. Biochim Biophys Acta 1842:1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Nikolajczyk BS, Denis GV (2013) BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol 190:3670–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Ramachandran B, Schroeder H, Schmidt G, Urbanke H, Burkhardt S, Capece V, Dean C, Fischer A (2017) The BET/BRD inhibitor JQ1 improves brain plasticity in WT and APP mice. Transl Psychiatry 7:e1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Urbanke H, Ramachandran B, Barth J, Halder R, Awasthi A, Jain G, Capece V, Burkhardt S, Navarro-Sala M, Nagarajan S, Schutz AL, Johnsen SA, Bonn S, Luhrmann R, Dean C, Fischer A (2015) HDAC inhibitor-dependent transcriptome and memory reinstatement in cognitive decline models. J Clin Invest 125:3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer-Daize C, Astorgues-Xerri L, Odore E, Cayol M, Cvitkovic E, Noel K, Bekradda M, MacKenzie S, Rezai K, Lokiec F, Riveiro ME, Ouafik L (2016) OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int J Cancer 139:2047–2055. [DOI] [PubMed] [Google Scholar]
- Berson A, Nativio R, Berger SL, Bonini NM. Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci. 2018;41(9):587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, Kahatt C, Quesnel B, Michallet M, Recher C, Lokiec F, Preudhomme C, Dombret H (2016) Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol 3:e186–195. [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E (2007) Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A 104:13690–13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button EB, Robert J, Caffrey TM, Fan J, Zhao W, Wellington CL. HDL from an Alzheimer’s disease perspective. Curr Opin Lipidol. 2019;30(3):224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Lee AY, Chiang CM, Kodadek T (2011) Peptoid ligands that bind selectively to phosphoproteins. Bioorg Med Chem Lett 21:4960–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfa G, Chapleau CA, Campbell S, Inoue T, Morse SJ, Lubin FD, Pozzo-Miller L (2012) HDAC activity is required for BDNF to increase quantal neurotransmitter release and dendritic spine density in CA1 pyramidal neurons. Hippocampus 22:1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalleri GL et al. (2007) A multicenter study of BRD2 as a risk factor for juvenile myoclonic epilepsy. Epilepsia 48:706–712. [DOI] [PubMed] [Google Scholar]
- Chachua T, Goletiani C, Maglakelidze G, Sidyelyeva G, Daniel M, Morris E, Miller J, Shang E, Wolgemuth DJ, Greenberg DA, Veliskova J, Velisek L (2014) Sex-specific behavioral traits in the Brd2 mouse model of juvenile myoclonic epilepsy. Genes Brain Behav 13:702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yap JL, Yoshioka M, Lanning ME, Fountain RN, Raje M, Scheenstra JA, Strovel JW, Fletcher S (2015) BRD4 Structure-Activity Relationships of Dual PLK1 Kinase/BRD4 Bromodomain Inhibitor BI-2536. ACS Med Chem Lett 6:764–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Lin YT, Gallegos DA, et al. Enhancer Histone Acetylation Modulates Transcriptional Bursting Dynamics of Neuronal Activity-Inducible Genes. Cell Rep. 2019;26(5):1174–1188.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Diao H, Zhang F, Wang Y, Wang K, Wu R (2017) Deciphering the mechanisms of selective inhibition for the tandem BD1/BD2 in the BET-bromodomain family. Phys Chem Chem Phys 19:23934–23941. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, Wang J (2013) Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res 19:1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CM (2016) Phospho-BRD4: transcription plasticity and drug targeting. Drug Discov Today Technol 19:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, Hong SH, Sim S, Cho KS, Kim JW, Yang SM, Jeon SJ, You JS, Shin CY (2015) The Epigenetic Reader BRD2 as a Specific Modulator of PAI-1 Expression in Lipopolysaccharide-Stimulated Mouse Primary Astrocytes. Neurochem Res 40:2211–2219. [DOI] [PubMed] [Google Scholar]
- Cheung KL, Zhang F, Jaganathan A, Sharma R, Zhang Q, Konuma T, Shen T, Lee JY, Ren C, Chen CH, Lu G, Olson MR, Zhang W, Kaplan MH, Littman DR, Walsh MJ, Xiong H, Zeng L, Zhou MM (2017) Distinct Roles of Brd2 and Brd4 in Potentiating the Transcriptional Program for Th17 Cell Differentiation. Mol Cell 65:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran AG, Conery AR, Sims RJ 3rd, (2019) Bromodomains: a new target class for drug development. Nat Rev Drug Discov 18:609–628. [DOI] [PubMed] [Google Scholar]
- Figge DA, Standaert DG (2017) Dysregulation of BET proteins in levodopa-induced dyskinesia. Neurobiol Dis 102:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K, Gijsbers R, Debyser Z (2013) The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep 5:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMars KM, Yang C, Candelario-Jalil E (2019) Neuroprotective effects of targeting BET proteins for degradation with dBET1 in aged mice subjected to ischemic stroke. Neurochem Int 127:94–102. [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Gegonne A, Singer DS (2016a) Bromodomain 4: a cellular Swiss army knife. J Leukoc Biol 100:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ 3rd, Singer DS (2012) BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 109:6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, Singer DS (2016b) BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 23:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491–496. [DOI] [PubMed] [Google Scholar]
- Duan Q, Huang FL, Li SJ, et al. BET proteins inhibitor JQ-1 impaired the extinction of remote auditory fear memory: An effect mediated by insulin like growth factor 2 [published online ahead of print, 2020 Jul 28]. Neuropharmacology. 2020;177:108255. [DOI] [PubMed] [Google Scholar]
- Durner M, Sander T, Greenberg DA, Johnson K, Beck-Mannagetta G, Janz D (1991) Localization of idiopathic generalized epilepsy on chromosome 6p in families of juvenile myoclonic epilepsy patients. Neurology 41:1651–1655. [DOI] [PubMed] [Google Scholar]
- Egervari G, Landry J, Callens J, Fullard JF, Roussos P, Keller E, Hurd YL (2017) Striatal H3K27 Acetylation Linked to Glutamatergic Gene Dysregulation in Human Heroin Abusers Holds Promise as Therapeutic Target. Biol Psychiatry 81:585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ember SW, Zhu JY, Olesen SH, Martin MP, Becker A, Berndt N, Georg GI, Schonbrunn E (2014) Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem Biol 9:1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ember SW, Lambert QT, Berndt N, Gunawan S, Ayaz M, Tauro M, Zhu JY, Cranfill PJ, Greninger P, Lynch CC, Benes CH, Lawrence HR, Reuther GW, Lawrence NJ, Schonbrunn E (2017) Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics. Mol Cancer Ther 16:1054–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre EJ et al. (2020) Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature 578:306–310. [DOI] [PubMed] [Google Scholar]
- Farrelly L, Zhang S, Topol A, Bastle RM, Flaherty E, Bhanu N, Garcia B, Li H, Brennand K, Maze I (2019) Chromatin profiling in human neurons reveals aberrant roles for histone H2A.Z acetylation and its associated interactions with BRD4 in schizophrenia. Program no. 736.08. 2019 Neuroscience meeting planner Chicago, IL. Society for neuroscience, 2019. [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P et al. (2010) Selective inhibition of BET bromodomains. Nature 468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, Vincek A, Joshua J, Casaccia P, Zhou MM (2014) Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem Biol 21:841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai SA, Ramadoss M, Mahadevan V (2016) Histone Deacetylase (HDAC) Inhibitors - emerging roles in neuronal memory, learning, synaptic plasticity and neural regeneration. Curr Neuropharmacol 14:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilan O, Rioja I, Knezevic K, et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science. 2020;368(6489):387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26(1):187–197. [DOI] [PubMed] [Google Scholar]
- Grade S, Bernardino L, Malva JO (2013) Oligodendrogenesis from neural stem cells: perspectives for remyelinating strategies. Int J Dev Neurosci 31:692–700. [DOI] [PubMed] [Google Scholar]
- Gräff J, Rei D, Guan JS, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483(7388):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Tsai LH (2013) The potential of HDAC inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol 53:311–330. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Long H, Bu Q, Zhao Y, Wang H, Tian J, Cen X (2019) Role of BRD4 phosphorylation in the nucleus accumbens in relapse to cocaine-seeking behavior in mice. Addict Biol:e12808. [DOI] [PubMed] [Google Scholar]
- Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF (2013) Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J Virol 87:12721–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris A, Donovan DJ, Seymour KA, Lovasco LA, Smilowitz NR, Halperin AL, Klysik JE, Freiman RN (2009) The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim Biophys Acta 1789:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen A, Thor T, Odersky A, Heukamp L, El-Hindy N, Beckers A, Speleman F, Althoff K, Schafers S, Schramm A, Sure U, Fleischhack G, Eggert A, Schulte JH (2013) BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget 4:2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS (2002) Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol 22:3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Li Y, Xu L, Cao D, Song S, Damaneh MS, Wang X, Meng T, Chen YL, Shen J, Miao Z, Xiong B (2017) Discovery of a series of dihydroquinoxalin-2(1H)-ones as selective BET inhibitors from a dual PLK1-BRD4 inhibitor. Eur J Med Chem 137:176–195. [DOI] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF (2009) Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol 29:1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ (2010) MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci 13:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahagirdar R, Attwell S, Marusic S, Bendele A, Shenoy N, McLure KG, Gilham D, Norek K, Hansen HC, Yu R, Tobin J, Wagner GS, Young PR, Wong NCW, Kulikowski E (2017) RVX-297, a BET Bromodomain Inhibitor, Has Therapeutic Effects in Preclinical Models of Acute Inflammation and Autoimmune Disease. Mol Pharmacol 92:694–706. [DOI] [PubMed] [Google Scholar]
- Janczura KJ, Volmar CH, Sartor GC, Ricciardi NR, Lambert G, Brothers SP, Wahlestedt C. Inhibition of Histone Deacetylase 3 Activity with RGFP-966 Reverses Alzheimer’s Disease (AD)-related Pathological Hallmarks and Alleviates Memory Impairment in the Triple Transgenic AD Mouse Model. (2018) Proceedings of the National Academy of Sciences 115(47):E11148-E11157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–534. [DOI] [PubMed] [Google Scholar]
- Jones MH, Numata M, Shimane M (1997) Identification and characterization of BRDT: A testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics 45:529–534. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, Siebenlist U, O’Shea JJ, Ozato K (2014) BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 21:1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han MH, Bassel-Duby R, Olson EN, Nestler EJ (2013) Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci 16:434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan YM, Kirkham P, Barnes PJ, Adcock IM (2014) Brd4 is essential for IL-1beta-induced inflammation in human airway epithelial cells. PLoS One 9:e95051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharenko OA, Gesner EM, Patel RG, Norek K, White A, Fontano E, Suto RK, Young PR, McLure KG, Hansen HC (2016) RVX-297- a novel BD2 selective inhibitor of BET bromodomains. Biochem Biophys Res Commun 477:62–67. [DOI] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD (2015) BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci 18:1464–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Gresack J, Allis CD, Darnell RB (2017) Excess Translation of Epigenetic Regulators Contributes to Fragile X Syndrome and Is Alleviated by Brd4 Inhibition. Cell 170:1209–1223 e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M (2004) CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42:961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann T, Miron V, Cui Q, Wegner C, Antel J, Bruck W (2008) Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain 131:1749–1758. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ (2005) Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48:303–314. [DOI] [PubMed] [Google Scholar]
- Kurita M et al. (2012) HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neurosci 15:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FC, Morton SW, Wyckoff J, Vu Han TL, Hwang MK, Maffa A, Balkanska-Sinclair E, Yaffe MB, Floyd SR, Hammond PT (2018) Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat Commun 9:1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Chepelev I, DiMaggio PA, Blanco MA, Zee BM, Zhao K, Garcia BA (2012) Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol 13:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD (2005) Epigenetic mechanisms in memory formation. Nat Rev Neurosci 6:108–118. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD (2004) Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem 279:40545–40559. [DOI] [PubMed] [Google Scholar]
- Liang E, Ma M, Wang L, Liu X, Xu J, Zhang M, Yang R, Zhao Y (2018) The BET/BRD inhibitor JQ1 attenuates diabetes-induced cognitive impairment in rats by targeting Nox4-Nrf2 redox imbalance. Biochem Biophys Res Commun 495:204–211. [DOI] [PubMed] [Google Scholar]
- Littman DR, Rudensky AY (2010) Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140:845–858. [DOI] [PubMed] [Google Scholar]
- Liu F et al. (2015) EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell 60:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal A, Rosenfeld MG (2013) Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang P, Chen H, Wold EA, Tian B, Brasier AR, Zhou J (2017) Drug Discovery Targeting Bromodomain-Containing Protein 4. J Med Chem 60:4533–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Li B, Rodriguez-Blanco J, Pastori C, Volmar CH, Wahlestedt C, Capobianco A, Bai F, Pei XH, Ayad NG, Robbins DJ (2014) The BET bromodomain inhibitor I-BET151 acts downstream of smoothened protein to abrogate the growth of hedgehog protein-driven cancers. J Biol Chem 289:35494–35502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M, Velmeshev D, Makhmutova M, Patel P, Sartor GC, Volmar CH, Wahlestedt C, Faghihi MA (2016) The BET-Bromodomain Inhibitor JQ1 Reduces Inflammation and Tau Phosphorylation at Ser396 in the Brain of the 3xTg Model of Alzheimer’s Disease. Curr Alzheimer Res 13:985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahgoub M, Monteggia LM. A role for histone deacetylases in the cellular and behavioral mechanisms underlying learning and memory. Learn Mem. 2014;21(10):564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA (2010) Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry 67:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA (2011) CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J Neurosci 31:16941–16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA (2013) HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci U S A 110:2647–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE (2012) Small-molecule inhibition of BRDT for male contraception. Cell 150:673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLure KG, Gesner EM, Tsujikawa L, Kharenko OA, Attwell S, Campeau E, Wasiak S, Stein A, White A, Fontano E, Suto RK, Wong NC, Wagner GS, Hansen HC, Young PR (2013) RVX-208, an inducer of ApoA-I in humans, is a BET bromodomain antagonist. PLoS One 8:e83190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E. RVX-208, a stimulator of apolipoprotein AI gene expression for the treatment of cardiovascular diseases. Curr Opin Investig Drugs. 2010;11(3):357–364. [PubMed] [Google Scholar]
- Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, Lora JM (2013) BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med 210:2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Wang B, Mao W, Wang J, Zhao Y, Li Q, Zhang C, Tang Y, Ma J (2018) Enhanced efficacy of histone deacetylase inhibitor combined with bromodomain inhibitor in glioblastoma. J Exp Clin Cancer Res 37:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merched A, Xia Y, Visvikis S, Serot JM, Siest G. Decreased high-density lipoprotein cholesterol and serum apolipoprotein AI concentrations are highly correlated with the severity of Alzheimer’s disease. Neurobiol Aging. 2000;21(1):27–30. [DOI] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC (2013) Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol Psychiatry 73:763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A (2010) Suppression of inflammation by a synthetic histone mimic. Nature 468:1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal DK, Evgrafov OV, Tabares P, Zhang F, Durner M, Greenberg DA (2003) BRD2 (RING3) is a probable major susceptibility gene for common juvenile myoclonic epilepsy. Am J Hum Genet 73:261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad NG, Wahlestedt C (2015) The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A 112:8326–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Daniel M, Penas C, Volmar CH, Johnstone AL, Brothers SP, Graham RM, Allen B, Sarkaria JN, Komotar RJ, Wahlestedt C, Ayad NG (2014) BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics 9:611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A (2010) Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328:753–756. [DOI] [PubMed] [Google Scholar]
- Penas C, Maloof ME, Stathias V, Long J, Tan SK, Mier J, Fang Y, Valdes C, Rodriguez-Blanco J, Chiang CM, Robbins DJ, Liebl DJ, Lee JK, Hatten ME, Clarke J, Ayad NG (2019) Time series modeling of cell cycle exit identifies Brd4 dependent regulation of cerebellar neurogenesis. Nat Commun 10:3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney J, Tsai LH. Histone deacetylases in memory and cognition. Sci Signal. 2014;7(355):re12. [DOI] [PubMed] [Google Scholar]
- Picaud S, Wells C, Felletar I, Brotherton D, Martin S, Savitsky P, Diez-Dacal B, Philpott M, Bountra C, Lingard H, Fedorov O, Muller S, Brennan PE, Knapp S, Filippakopoulos P (2013) RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A 110:19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picaud S et al. (2015) Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer Res 75:5106–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawski SG, Peixoto L, Porcari GS, Wimmer ME, McNally AG, Mizuno K, Giese KP, Chatterjee S, Koberstein JN, Risso D, Speed TP, Abel T (2016) Contextual fear conditioning induces differential alternative splicing. Neurobiol Learn Mem 134 Pt B:221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM (2011) The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31:2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, Lauberth SM (2018) RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol 25:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ (2007) Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56:517–529. [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE 3rd, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ (2009) Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Singh H, Dang R, Wood MA (2013) HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci 33:6623–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudman MD, Choi JS, Lee HE, Tan SK, Ayad NG, Lee JK (2018) Bromodomain and extraterminal domain-containing protein inhibition attenuates acute inflammation after spinal cord injury. Exp Neurol 309:181–192. Rvx 208. Drugs R D. 2011;11(2):207–213. [DOI] [PubMed] [Google Scholar]
- Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O’Prey J, Clark W, Hedley A, Nixon C, Long JS, New M, Van Acker T, Tooze SA, Lowe SW, Dikic I, Ryan KM (2017) Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol Cell 66:517–532 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ventura J, Amo-Aparicio J, Navarro X, Penas C (2019) BET protein inhibition regulates cytokine production and promotes neuroprotection after spinal cord injury. J Neuroinflammation 16:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, Powell SK, Brothers SP, Wahlestedt C (2015) Epigenetic Readers of Lysine Acetylation Regulate Cocaine-Induced Plasticity. J Neurosci 35:15062–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, Malvezzi AM, Kumar A, Andrade NS, Wiedner HJ, Vilca SJ, Janczura KJ, Bagheri A, Al-Ali H, Powell SK, Brown PT, Volmar CH, Foster TC, Zeier Z, Wahlestedt C (2019) Enhancement of BDNF Expression and Memory by HDAC Inhibition Requires BET Bromodomain Reader Proteins. J Neurosci 39:612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, McCarroll SA (2018) Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174:1015–1030 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Chatterjee S, Bousiges O, et al. Acetyltransferases (HATs) as targets for neurological therapeutics. Neurotherapeutics. 2013;10(4):568–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ (2009) Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn 238:908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, He L, Zheng L, Huang L, Zhou Y, Wang T, Chen Y, Shen M, Wang F, Yang Z, Chen L (2017) Structure-based design, synthesis and in vitro antiproliferative effects studies of novel dual BRD4/HDAC inhibitors. Bioorg Med Chem Lett 27:4051–4055. [DOI] [PubMed] [Google Scholar]
- Shi J, Vakoc CR (2014) The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 54:728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, Rusinova E, Zhang G, Wang C, Zhu H, Yao J, Zeng YX, Evers BM, Zhou MM, Zhou BP (2014) Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25:210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM (2012) Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol Psychiatry 72:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonestrom AJ, Hsu SC, Jahn KS, Huang P, Keller CA, Giardine BM, Kadauke S, Campbell AE, Evans P, Hardison RC, Blobel GA (2015) Functions of BET proteins in erythroid gene expression. Blood 125:2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Badimon A, Schaefer U, Ayata P, Gray J, Chung CW, von Schimmelmann M, Zhang F, Garton N, Smithers N, Lewis H, Tarakhovsky A, Prinjha RK, Schaefer A (2015) Autism-like syndrome is induced by pharmacological suppression of BET proteins in young mice. J Exp Med 212:1771–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wang L, Jiang B, Hui B, Lv Z, Ma L (2008) The effects of sodium butyrate, an inhibitor of histone deacetylase, on the cocaine- and sucrose-maintained self-administration in rats. Neurosci Lett 441:72–76. [DOI] [PubMed] [Google Scholar]
- Tang Y et al. (2014) Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 20:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y (2016) The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. Int J Mol Sci 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremolizzo L, Carboni G, Ruzicka WB, Mitchell CP, Sugaya I, Tueting P, Sharma R, Grayson DR, Costa E, Guidotti A (2002) An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci U S A 99:17095–17100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa LM, Fu L, Das S, Halliday C, Rakai BD, Stotz SC, Sarsons CD, Gilham D, Daze E, Wasiak S, Studer D, Rinker KD, Sweeney M, Johansson JO, Wong NCW, Kulikowski E (2019) Apabetalone (RVX-208) reduces vascular inflammation in vitro and in CVD patients by a BET-dependent epigenetic mechanism. Clin Epigenetics 11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T, Nakamura Y, Wakamori M, Ozato K, Yokoyama S, Padmanabhan B (2010a) Structural implications for K5/K12-di-acetylated histone H4 recognition by the second bromodomain of BRD2. FEBS Lett 584:3901–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T, Nakamura Y, Jang MK, Nakano K, Tanaka A, Ozato K, Padmanabhan B, Yokoyama S (2010b) Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J Biol Chem 285:7610–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velisek L, Shang E, Veliskova J, Chachua T, Macchiarulo S, Maglakelidze G, Wolgemuth DJ, Greenberg DA (2011) GABAergic neuron deficit as an idiopathic generalized epilepsy mechanism: the role of BRD2 haploinsufficiency in juvenile myoclonic epilepsy. PLoS One 6:e23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S, Alimova I, Balakrishnan I, Harris P, Birks DK, Griesinger A, Amani V, Cristiano B, Remke M, Taylor MD, Handler M, Foreman NK, Vibhakar R (2014) Inhibition of BRD4 attenuates tumor cell self-renewal and suppresses stem cell signaling in MYC driven medulloblastoma. Oncotarget 5:2355–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmar CH, Salah-Uddin H, Janczura JK, Halley P, Lambert G, Wodrich A, Manoah S, Patel N, Mehta M, Sartor GC, Mehta N, Miles N, Desse S, Dorcius D, Wahlestedt C. (2017) M344 promotes non-amyloidogenic amyloid precursor protein processing while normalizing Alzheimer’s disease genes and improving memory. Proceedings of the National Academy of Sciences, 114(43): E9135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang W, Liang M, Shi Y, Zhang C, Li Q, Liu M, Shou Y, Yin H, Zhu X, Sun X, Hu Y, Shen Z (2018) (+)-JQ1 attenuated LPS-induced microglial inflammation via MAPK/NFkappaB signaling. Cell Biosci 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen J, Jin H, Lin D, Chen Y, Chen X, Wang B, Hu S, Wu Y, Wu Y, Zhou Y, Tian N, Gao W, Wang X, Zhang X (2019) BRD4 inhibition attenuates inflammatory response in microglia and facilitates recovery after spinal cord injury in rats. J Cell Mol Med 23:3214–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, Ma L (2010) Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology 35:913–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbecker KA, Durner M, Janz D, Scaramelli A, Sparkes RS, Spence MA (1991) Confirmation of linkage between juvenile myoclonic epilepsy locus and the HLA region of chromosome 6. Am J Med Genet 38:32–36. [DOI] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE (2015) DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348:1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM (2013) Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell 49:843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Nin DS, Lee AY, Simanski S, Kodadek T, Chiang CM (2016) BRD4 Phosphorylation Regulates HPV E2-Mediated Viral Transcription, Origin Replication, and Cellular MMP-9 Expression. Cell Rep 16:1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Kamikawa YF, Donohoe ME (2018) Brd4’s Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep 25:1756–1771. [DOI] [PubMed] [Google Scholar]
- Yang CY, Qin C, Bai L, Wang S (2019) Small-molecule PROTAC degraders of the Bromodomain and Extra Terminal (BET) proteins - A review. Drug Discov Today Technol 31:43–51. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19:535–545. [DOI] [PubMed] [Google Scholar]
- Zeier Z, Esanov R, Belle KC, Volmar CH, Johnstone AL, Halley P, DeRosa BA, Khoury N, van Blitterswijk M, Rademakers R, Albert J, Brothers SP, Wuu J, Dykxhoorn DM, Benatar M, Wahlestedt C (2015) Bromodomain inhibitors regulate the C9ORF72 locus in ALS. Exp Neurol 271:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengerle M, Chan KH, Ciulli A (2015) Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol 10:1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Plotnikov AN, Rusinova E, Shen T, Morohashi K, Joshua J, Zeng L, Mujtaba S, Ohlmeyer M, Zhou MM (2013) Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J Med Chem 56:9251–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ishida CT, Ishida W, Lo SL, Zhao J, Shu C, Bianchetti E, Kleiner G, Sanchez-Quintero MJ, Quinzii CM, Westhoff MA, Karpel-Massler G, Canoll P, Siegelin MD (2018) Combined HDAC and Bromodomain Protein Inhibition Reprograms Tumor Cell Metabolism and Elicits Synthetic Lethality in Glioblastoma. Clin Cancer Res 24:3941–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gu Y, Liu J (2019) BRD4 suppression alleviates cerebral ischemia-induced brain injury by blocking glial activation via the inhibition of inflammatory response and pyroptosis. Biochem Biophys Res Commun 519:481–488. [DOI] [PubMed] [Google Scholar]
- Zuber V, Bettella F, Witoelar A, Consortium P, Cruk G, Consortium B, Consortium T, Andreassen OA, Mills IG, Urbanucci A (2017) Bromodomain protein 4 discriminates tissue-specific super-enhancers containing disease-specific susceptibility loci in prostate and breast cancer. BMC Genomics 18:270. [DOI] [PMC free article] [PubMed] [Google Scholar]