Abstract
Purpose:
A prognostic model for overall survival (OS) of post-platinum patients with metastatic urothelial carcinoma (mUC) receiving PD-1/PD-L1 inhibitors is necessary since existing models were constructed in the chemotherapy setting.
Materials and methods:
Patient level data were used from phase I/II trials evaluating PD-L1 inhibitors following platinum-based chemotherapy for mUC. The derivation dataset consisted of 2 phase I/II trials evaluating atezolizumab (n=405). Two phase I/II trials that evaluated avelumab (n=242) and durvalumab (n=198) comprised the validation datasets. Cox regression analyses evaluated the association of candidate prognostic factors with OS. Stepwise selection was employed to select an optimal model using the derivation dataset. Discrimination and calibration were assessed in the avelumab and durvalumab datasets.
Results:
The 5 prognostic factors identified in the optimal model employing the atezolizumab derivation dataset were ECOG-PS (1 vs. 0; HR 1.80; 95% CI [1.36–2.36]), liver metastasis (HR 1.55; 95% CI [1.20–2.00]), platelet count (HR 2.22; 95% CI [1.54–3.18]), neutrophil-lymphocyte ratio (HR 1.94; 95% CI [1.57–2.40]) and lactate dehydrogenase (HR 1.60; 95% CI [1.28–1.99]). There was robust discrimination of survival between low, intermediate and high-risk groups. The c-statistic was 0.692 in the derivation and 0.671 and 0.773 in the avelumab and durvalumab validation datasets, respectively. A web-based interactive tool was developed to calculate the expected survival probabilities based on risk factors.
Conclusions:
A validated 5-factor model has satisfactory prognostic performance for survival across 3 PD-L1 inhibitors to treat mUC post-platinum and may assist in stratification, interpreting and designing trials incorporating PD-1/PD-L1 inhibitors in the post-platinum setting.
Keywords: Prognostic factors, Metastatic, Urothelial carcinoma, Post-platinum, PD-L1 inhibitors
Introduction
The therapeutic landscape of locally advanced unresectable or metastatic urothelial carcinoma (mUC) has changed dramatically over the last few years, with the emergence of five new PD-1/PD-L1 inhibitors for the treatment of progressive disease following platinum exposure.1–7 These agents include two PD1 inhibitors (nivolumab, pembrolizumab) and three PD-L1 inhibitors (avelumab, durvalumab, atezolizumab). Prognostic models of survival in post-platinum patients receiving chemotherapy (taxanes, vinflunine) include Eastern Cooperative Oncology Group (ECOG)-Performance status (PS), liver metastasis and hemoglobin initially proposed by Bellmunt et al, which was later enhanced by adding treatment free interval and albumin.8–10 It is unclear if these factors are applicable to post-platinum PD-1/PD-L1 inhibitors.
Other factors more reflective of the immune, inflammatory and metabolic state may confer major prognostic impact. In this context, the peripheral blood neutrophil-lymphocyte ratio (NLR), platelet count (PLT), eosinophil count and serum lactate dehydrogenase (LDH) may be relevant, given that they are universally and affordably measured factors that have been demonstrated to be prognostic in other malignancies.11–13 The different industry sponsors have employed a variety of antibodies and thresholds to define tumor PD-L1 protein expression and have inconsistently measured tumor mutation burden (TMB) and gene expression profiling using different platforms, which renders these variables difficult to apply across all of these agents.
Given the importance of accurate prognostic models to interpret data and design clinical trials, this study was undertaken to identify and validate a prognostic model for survival consisting of some the aforementioned routinely measured and available standardized clinical and analytically validated laboratory factors in the setting of PD-1/PD-L1 inhibitors administered for progressive mUC following platinum-based chemotherapy.
Patients and methods
Study design
Patient level data were used from previously reported phase I and II trials evaluating PD-L1 inhibitors following platinum-based chemotherapy for mUC to conduct this retrospective study. Individual patient level data regarding baseline patient characteristics, clinical factors and survival outcomes were collected establishing a derivation dataset that received atezolizumab and two validation datasets that received avelumab and durvalumab, respectively. The candidate prognostic factors were age, gender, body mass index (BMI), ECOG-PS, sites of metastasis (liver +/− other, non-liver visceral +/− soft tissue/ lymph node, soft tissue/lymph node only), hemoglobin, albumin, treatment-free interval from previous therapy, NLR, PLT, eosinophil count, LDH, prior platinum (cisplatin or carboplatin), primary tumor location (bladder or other), calculated creatinine clearance (Cockroft-Gault formula), clinical stage at initial diagnosis, cigarette smoking history (never or ever), prior radical cystectomy (or nephroureterectomy) and number of previous lines of treatment regimens. Laboratory values were normalized by logarithmic transformation.
Participants
The derivation dataset consisted of combining the phase I and phase II trials (PCD4989g (NCT01375842) and IMvigor210 (NCT02951767 and NCT02108652), respectively) evaluating atezolizumab (median follow-up 1.9 years; range, 1.6–2.5 years).4, 14 Patients enrolled on two separate phase I/II trials, pooled mUC cohorts from the JAVELIN Solid Tumor trial (NCT01772004) that evaluated avelumab and CD-ON-MEDI4736–1108 (NCT01693562) that evaluated durvalumab comprised the validation datasets.5, 6 These 3 trials were sponsored by different pharmaceutical companies (Genentech, EMD Serono [a business of Merck KGaA, Darmstadt, Germany] and Astrazeneca) who collaborated with the investigators of this retrospective analysis. All of these trials were approved by the respective Institutional Review Boards (IRBs), which also allowed the retrospective study of deidentified patient data.
Statistical analysis
Initial analysis to construct the prognostic model was conducted using individual patient level data from the derivation atezolizumab dataset. Cox regression analyses evaluated the association of candidate prognostic factors with survival. External validation was performed using individual patient level data from the two validation durvalumab and avelumab datasets. Forward stepwise selection was employed to create an optimal model. The prognostic index (PI), which is a continuous risk score based on the linear predictor of the Cox model, was calculated. Secondarily, a simplistic risk score based on the number of adverse risk factors using those factors identified in the Cox model was also calculated. Survival based on grouping the PI by first and third quartiles (25% and 75% quantiles) and the number of risk factors was then assessed using Kaplan-Meier method. Discrimination (separation of survival between risk groups) was assessed using concordance statistic. The c-statistic for the previously reported 3-factor prognostic model by Bellmunt (ECOG-PS > 0, hemoglobin < 100 g/L, liver metastasis) was calculated in each of the datasets separately to identify the increment in c-statistic provided by the new model.
Statistical validation of the prognostic model was then performed following the procedures of Royston and Altman15 using the independent avelumab and durvalumab datasets. Specifically, assessment of the discrimination, calibration (survival estimate accuracy), and predictive ability was performed using concordance (c-statistics), regression on the PI, inspection of Kaplan-Meier curves and corresponding survival estimates (by PI quartile risk groups) and comparison with expected survival. Analyses using PI appeared to have better discrimination than analyses using the number of risk factors, thus, results using risk factors were omitted for simplicity. An interactive web-based application was developed to predict the survival probability based on specific risk factor values from the weighted Cox model in the atezolizumab derivation dataset using shiny, which is an R package that makes it easy to build interactive web apps straight from the open-source software R.16, 17 The overall survival probability for an individual patient i is predicted for any time t: where the baseline survival has been obtained from the prognostic model fit on the derivation data and approximated using polynomial regression S0(t) = 1 − 2.478 ⋅ 10−5 ⋅ t + 4.547 ⋅ 10−7 ⋅ t2 − 4.050 ⋅ 10−9 ⋅ t3. Averaging the PIs over all members of each risk group in the validation datasets results in the respective expected survival. The derivation of the respective confidence interval can be found the electronic supplement.
Results
Patient characteristics
The derivation atezolizumab dataset consisted of 405 patients, while the validation avelumab and durvalumab datasets consisted of 242 and 198 patients, respectively (Table 1). The patients exhibited characteristics that were typical for a post-platinum mUC population in all cohorts (Table 1). The median follow-up in the combined atezolizumab derivation dataset including IMvigor210 and PCD4989g was 1.9 years (range, 1.6–2.5 years).4, 14 The median follow-up in the avelumab JAVELIN EMR 100070–001 validation dataset was 2.7 years (range, 2.0–3.6 years) and in the durvalumab CD-ON-MEDI4736–1108 dataset was 1.5 years (range, 1.3 – 3.1 years).5, 6
Table 1.
Patient demographics and disease characteristics at baseline
| Derivation atezolizumab data (n = 405) | Validation | ||
|---|---|---|---|
| avelumab data (n = 242) | durvalumab data (n = 198) | ||
| Age (years) | |||
| Median (min; max) | 66 (32; 89) | 68 (30; 89) | 67 (34; 88) |
| Gender | |||
| Male | 313 (77.3%) | 175 (72.3%) | 142 (71.7%) |
| NLR (Neutrophil / Lymphocyte Ratio) | |||
| Missing n (%) | --- | --- | 4 (2.0%) |
| ECOG-Performance Status | |||
| ≥1 | 251 (62.0%) | 158 (65.3%) | 135 (68.2%) |
| Serum LDH (U/L) | |||
| Missing n (%) | 19 (4.7%) | 7 (2.9%) | 6 (3.0%) |
| Platelet counts (109/L) | |||
| Missing n (%) | --- | --- | 4 (2.0%) |
| Location of Metastasis* | |||
| Soft tissue/lymph node | 88 (21.7%) | 55 (22.7%) | 24 (12.1%) |
| Serum Albumin (gm/L) | |||
| Missing n (%) | --- | --- | 3 (1.5%) |
| Hemoglobin (gm/L) | |||
| Missing n (%) | --- | --- | 4 (2.0%) |
| Number of previous agents | |||
| ≥ 3 | 104 (25.7%) | 81 (33.5%) | 16 (8.1%) |
atezolizumab and avelumab datasets: Liver metastasis = liver (+/−other), visceral metastasis = visceral (non-liver, +/− soft tissue/lymph node), soft tissue/lymph node metastasis = soft tissue/lymph node (non-liver, non-visceral); durvalumab dataset: Liver metastasis = liver (+/−other), visceral metastasis = visceral (any non-lymph node metastasis including liver, soft tissue and other visceral metastasis), soft tissue/lymph node metastasis = lymph node metastasis only
Prognostic factors associated with survival in the atezolizumab derivation dataset
On univariable analysis, multiple factors were associated with survival (Supplementary Table 1). On multivariable analysis (Table 2), the 5 factors included in the optimal prognostic model in the derivation dataset were ECOG-PS (1 vs. 0; HR 1.80; 95% CI [1.36–2.36]), liver metastasis (HR 1.55; 95% CI [1.20–2.00]), log platelet counts (HR 2.22; 95% CI [1.54–3.18]), log platelet counts (HR 2.22; 95% CI [1.54–3.18]), log NLR (HR 1.94; 95% CI [1.57–2.40]) and log LDH (HR 1.60; 95% CI [1.28–1.99]). Patients were segregated into low, intermediate and high-risk groups based on the first and third quartiles of the prognostic index, and there was separation of survival curves between groups (Figure 1a). The 1-year Kaplan-Meier survival estimates and 95% CI of those in the low, intermediate and poor risk groups were 76.2% (66.1–83.6), 33.8 % (27.1–40.5) and 8.6% (3.8–15.8), respectively.
Table 2.
Multivariable analysis showing 5 significant factors associated with survival in the derivation atezolizumab dataset
| PI Coefficient | HR | 95% CI | p-value | |
|---|---|---|---|---|
| Log NLR | 0.661 | 1.937 | 1.566–2.395 | <0.001 |
| ECOG-PS | 0.583 | 1.792 | 1.362–2.358 | <0.001 |
| Log LDH | 0.467 | 1.596 | 1.277–1.994 | <0.001 |
| Log Platelet count | 0.796 | 2.217 | 1.544–3.182 | <0.001 |
| Liver metastasis | 0.437 | 1.547 | 1.199–1.997 | 0.001 |
C-index of 5-factor model is 0.692 in the derivation dataset; The risk groups are based on PI categorization: PI <7.73681 (low risk), 7.73681≤ PI <8.91837 (moderate risk), PI ≥ 8.91837 (high risk). Legend: NLR=Neutrophils-lymphocyte ratio; ECOG-PS=Eastern Cooperative Oncology Group performance status; LDH=lactate dehydrogenase; HR=hazard ratio; CI=confidence interval
Figure 1.
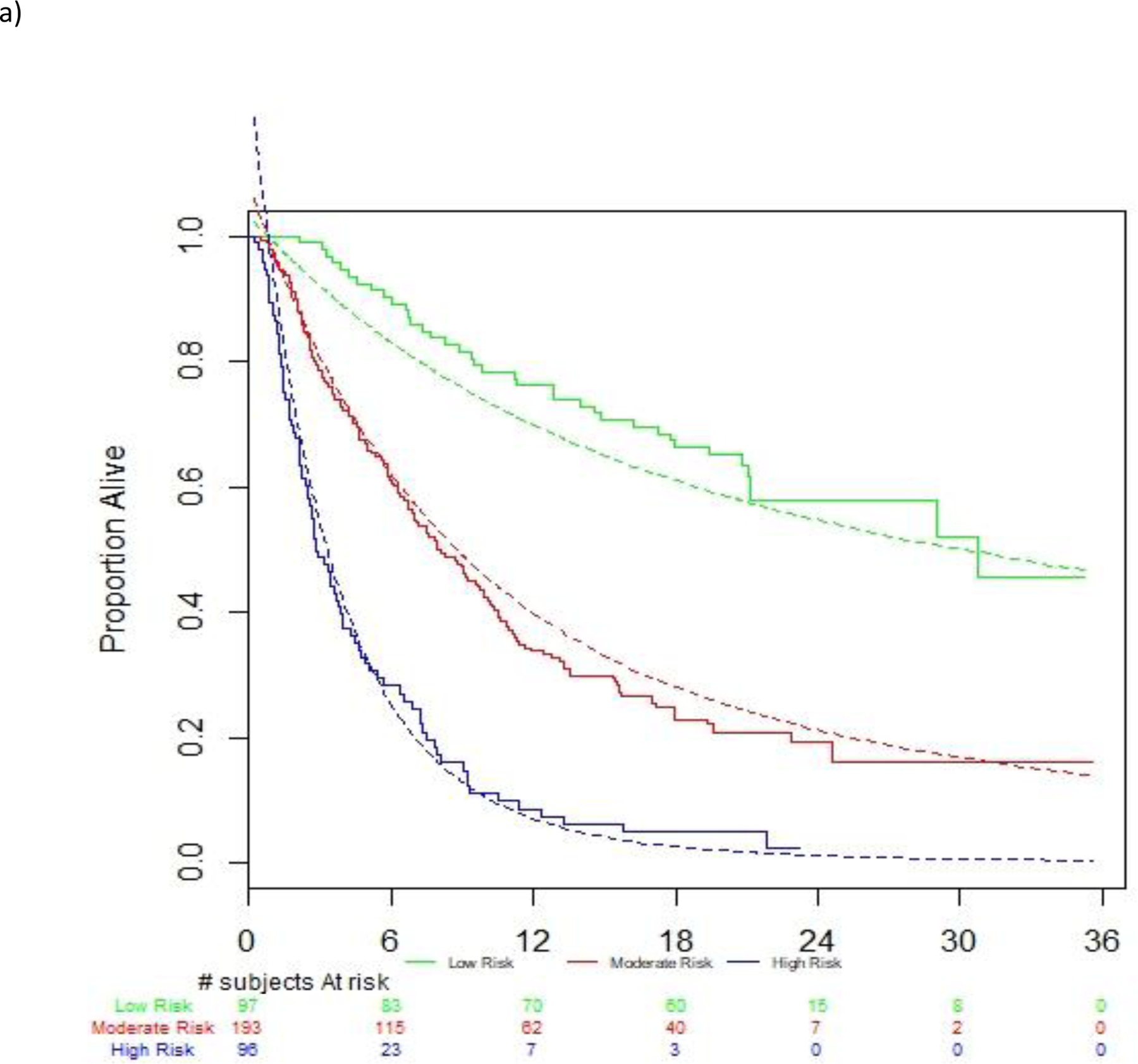
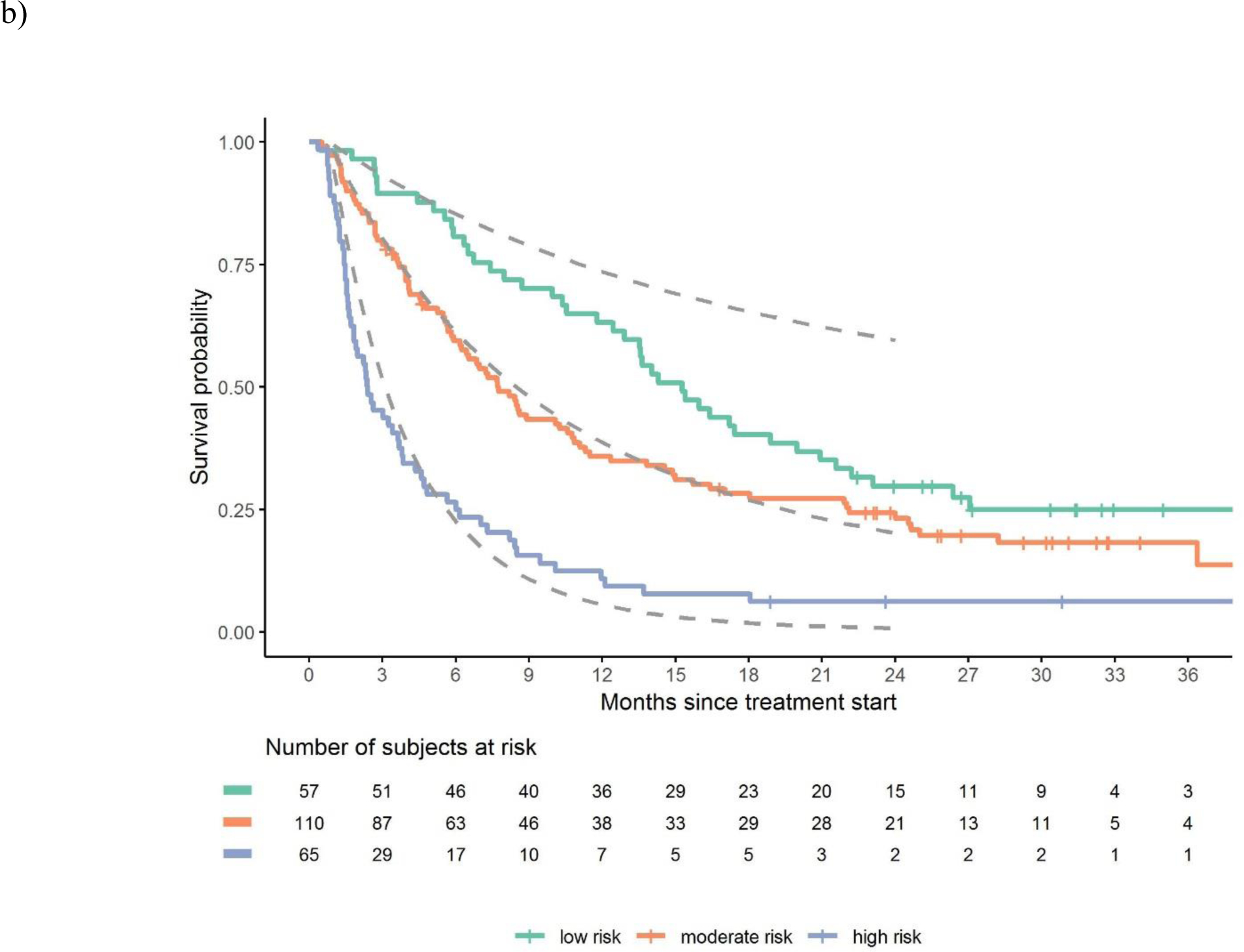
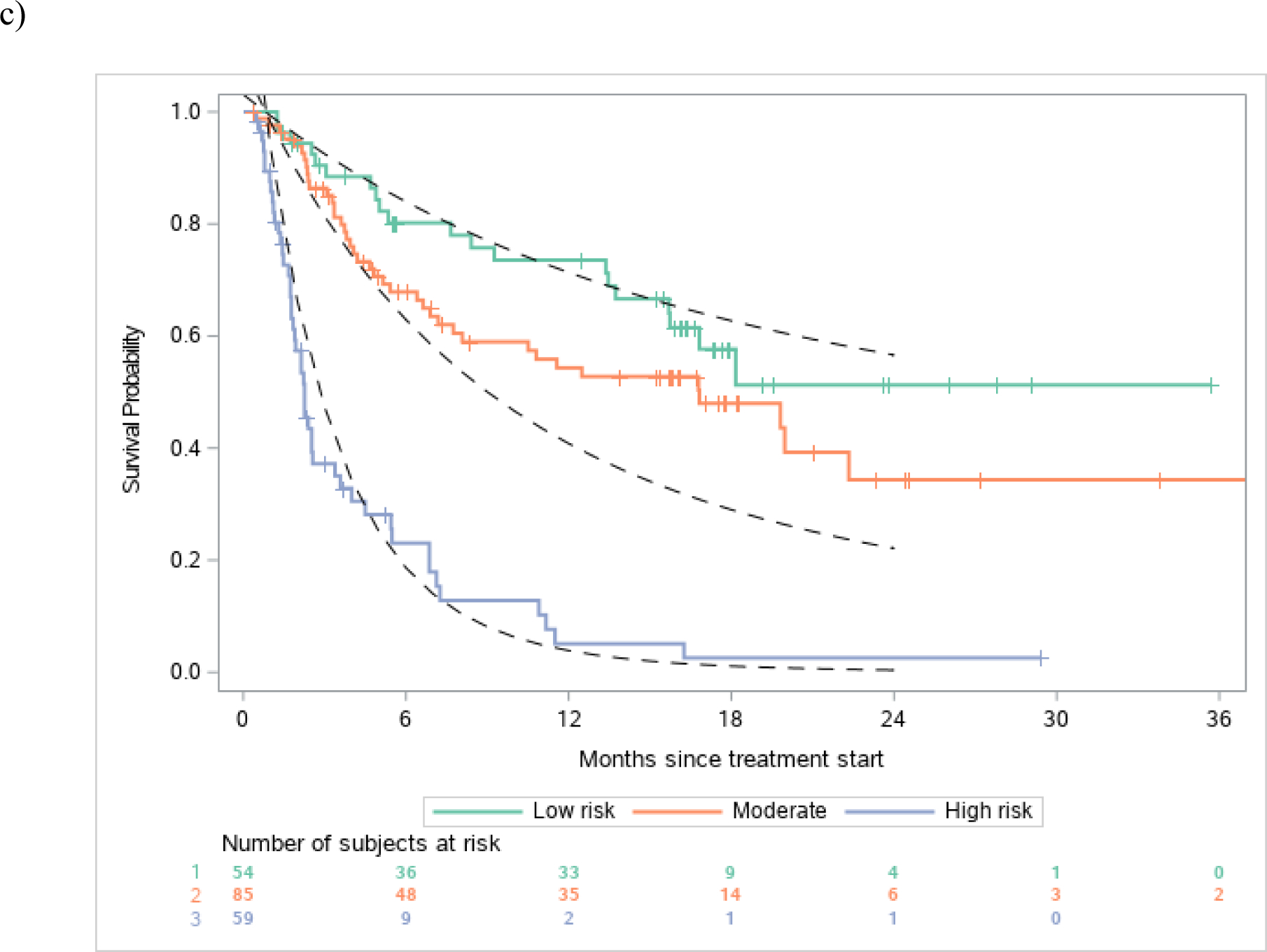
Discrimination of observed vs. estimated survival based on risk groups in the a) atezolizumab derivation dataset, b) avelumab validation dataset and c) durvalumab validation dataset
Legend: Figures show observed and estimated survival based on low, intermediate and high-risk defined by the derivation dataset. Dashed lines references observed survival in the derivation data.
Performance of the prognostic model in the avelumab and durvalumab validation datasets
There was robust discrimination of survival in the validation datasets between low, intermediate and high-risk defined by the derivation dataset (Figure 1a–c). The c-statistics was 0.692 in the derivation dataset and 0.671 and 0.773 in the avelumab and durvalumab validation datasets, respectively. The c-statistic for the 3-factor Bellmunt model was 0.635 in the derivation dataset, and 0.657 and 0.688 in the Avelumab and durvalumab datasets, respectively. Thus, the increments in c-statistic in the 3 datasets when using the new 5-factor model compared to the old 3-factor model were 0.057, 0.014 and 0.085, respectively. Acceptable or good calibration of expected 6-month and 1-year survival rates was observed (Supplementary Table 2).
Construction of online tool to estimate expected survival with post-platinum PD-L1 inhibitor
The supplementary online tool (https://ucprognosis.apps.hcie.io/) constructed using the derivation dataset provided estimates for survival using the suggested prognostic model (Figure 2). The tool enabled the calculation of the probability of 6- and 12-month survival based on specific values for the 5 baseline prognostic factors.
Figure 2.
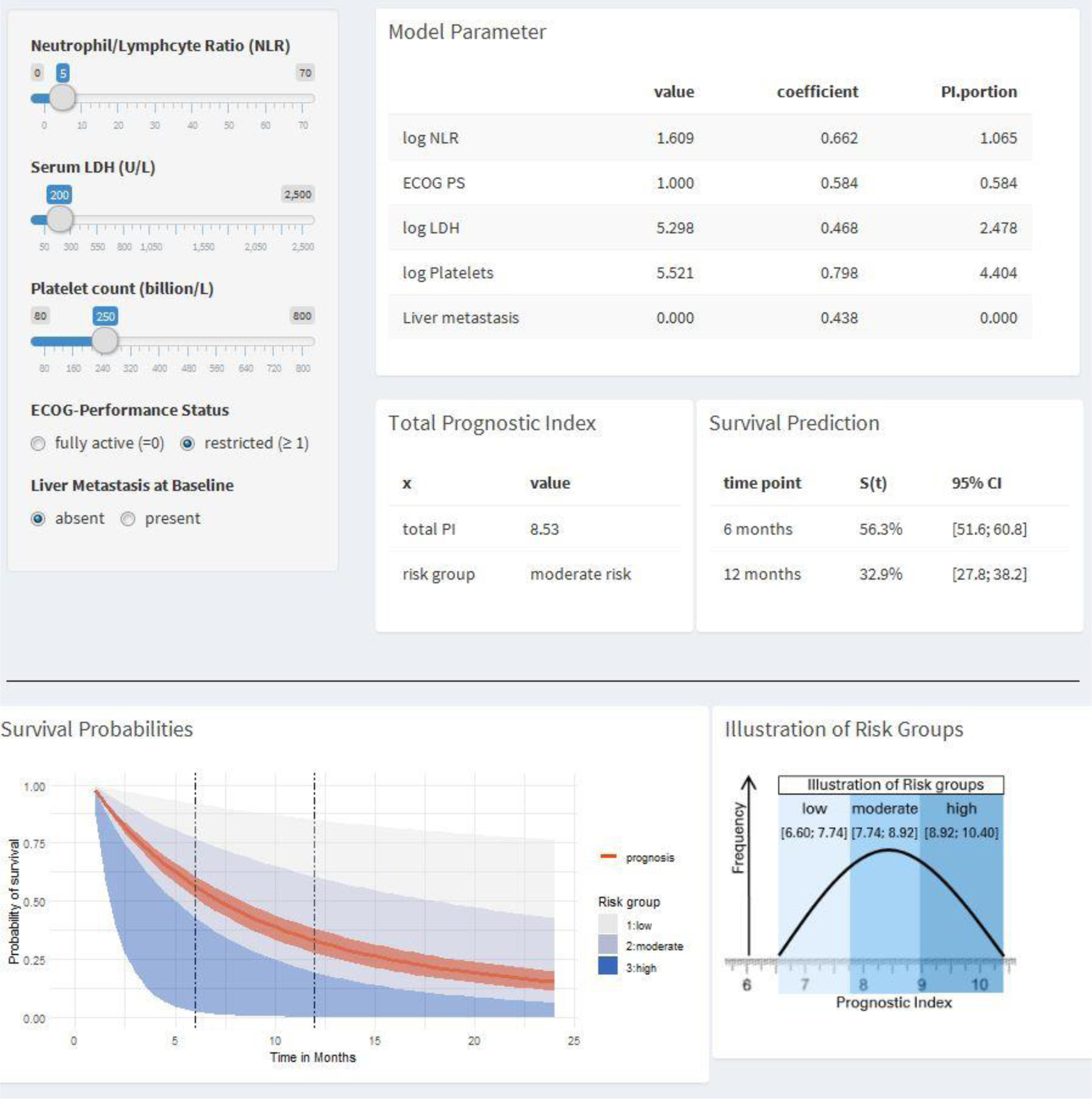
Interactive nomogram tool to calculate expected survival from the prognostic model
Legend: An interactive online tool (https://ucprognosis.apps.hcie.io/) calculates and visualizes the expected survival probabilities based on risk factors
Discussion
A 5-factor validated prognostic model for survival is proposed using data from a total of 845 mUC post-platinum patients treated with PD-L1 inhibitors. The model was developed using a combined analysis of two studies of 405 patients receiving atezolizumab. Validation was performed using two independent datasets of 242 patients receiving avelumab and 198 patients receiving durvalumab. The model calibration and concordance appeared acceptable overall and in a priori defined risk groups showed good discrimination and usability. This model may assist in prognostic stratification and interpreting nonrandomized trials of post-platinum PD-1/PD-L1 inhibitors. Further validation of this model in additional datasets of patients treated with different PD-1 inhibitors, e.g. pembrolizumab and nivolumab, is warranted.
An interactive tool was constructed to predict expected survival probabilities based on specific risk factor values and the prognostic model. This tool can be useful to compare observed versus expected survival in non-randomized phase I/II trials evaluating combinations of PD-1/PD-L1 inhibitors and novel agents in the post-platinum setting. Indeed, misinterpretation of nonrandomized data has led to large randomized phase III trials, which did not confirm initially ‘promising’ phase I/II results, e.g. combinations of PD-1/PD-L1 inhibitors and indole-deoxygenase (IDO)-1 inhibitors.18, 19 Thus, the availability of such a tool may assist rational and cost-effective drug development.
The prognostic model does not include molecular factors. A decision was made to exclude tumor PD-L1 expression since different assays (Ventana SP142 in atezolizumab trials, Dako 73–10 in avelumab trials, Ventana SP263 in durvalumab trials) and different cutoffs were employed for these agents in the phase I/II trials. Moreover, the assays measured either the immune cell (atezolizumab trials) or tumor cell (avelumab trials) or a combination of immune and tumor cells (durvalumab trials). Other molecular factors such as TMB, DNA damage repair gene alterations and gene expression profile (to derive intrinsic subtype or the interferon- γ signature) may be prognostic but were variably measured using different methodologies or unavailable for most patients on these trials to incorporate them in our study.20–23 Moreover, the platform employed to measure these molecular factors may also impact their interpretability. Indeed, PD-L1 protein expression, TMB and intrinsic subtype may all complement each other and confer prognostic impact, but were considered challenging to incorporate in our nomogram for universal applicability.4, 14 PD-L1 expression has been approved by regulatory agencies to select cisplatin-ineligible patients for first-line pembrolizumab or atezolizumab, but the utility of other molecular markers and our prognostic model to develop precision medicine is unclear.
Thus, the 5 factors are proposed as components of a prognostic model and cannot be employed to in an algorithm to select patients for therapy. For example, patients in the poor risk group demonstrated poor survival, but it is unclear if these patients would exhibit worse outcomes when using a different agent. The examination of the differential prognostic impact of this model in phase III post-platinum trials comparing a PD-1/PD-L1 inhibitor with conventional chemotherapy such as the KEYNOTE-045 and IMvigor211 trials may shed insights, but the datasets of these trials were not available.3, 24 Indeed, our model could not be validated in two PD-1 inhibitors approved for post-platinum patients, pembrolizumab and nivolumab, since these datasets were also not available. However, we hypothesize that the model would very likely be applicable to patients who received any PD-1/PD-L1 inhibitor for post-platinum patients with mUC, given that these agents have similar activity when examining the data across trials.7 The proposed model exhibits varying performance in both validation datasets. In comparison to the derivation data, the discrimination of the risk groups is poorer in the avelumab and better in the durvalumab validation dataset. Indeed, it is unusual that the model performance seems to be better in the durvalumab validation dataset (c-index 0.773, calibration slope 1.14) than in the derivation dataset (c-index 0.692). The increments in c-statistic in the 3 datasets when employing the new 5-factor model compared to the 3-factor model (0.014 to 0.085) may be characterized as modest. However, the delta for improvement in concordance-statistics between the models is >0.005, which is considered adequate to enhance the prognostic information according to previously published recommendations.25 The model exhibits varying calibration accuracy in the different risk groups. In particular, the survival in the low risk group might be overestimated. The frequency of number of previous agents shows some imbalances in the derivation and the avelumab validation data set (Table 1). However, it is noteworthy that the number of prior agents was not significantly associated with survival in the univariable analysis (Supplementary Table 1). Although prognostic models are generally not optimal to predict outcomes for individual patients, the interactive tool may be useful to estimate expected survival and facilitate interpretability of observed survival in nonrandomized datasets.15
Conclusion
In conclusion, we propose the first statistically validated prognostic model for survival in the setting of post-platinum mUC receiving 3 different PD-L1 inhibitors. The model uses readily available clinical and laboratory factors and provides a meaningful improvement over the previously reported 3-factor Bellmunt model. Thus, the model may be universally applicable and enhances the ability to interpret non-randomized trials as well as stratify trials employing PD-1/PD-L1 inhibitor backbone in the post-platinum setting. Moreover, patients in the poor risk tertile could potentially undergo evaluation of new agents and combinations using a nonrandomized registration trial design. Similar efforts in other settings using checkpoint inhibitors need to be pursued to enable the capitalization of prognostic models in these setting, e.g. the first-line cisplatin-ineligible and platinum-ineligible settings. The routine incorporation of uniformly measured and analytically validated molecular (genomic, transcriptomic, proteomic, epigenetic, immune, metabolomic) factors in phase III trials may assist in further refining this prognostic model and develop its potential utility in precision medicine.
Supplementary Material
Previous presentations.
Presented in part at the Genitourinary Cancer Symposium, San Francisco, CA February 2018 as an oral presentation and poster presentation at the Genitourinary Cancer Symposium, San Francisco, CA, February 2019, and poster presentation at the annual American Society of Clinical Oncology (ASCO) conference, Chicago, IL, June 2019.
Acknowledgments
We gratefully acknowledge the provision of access to clinical trial data by Genentech, Astrazeneca, and EMD Serono, a business of Merck KGaA, Darmstadt, Germany, and is part of an alliance between Merck KGaA, Darmstadt, Germany and Pfizer.
Funding support
The research was funded in part by National Cancer Institute Cancer Center Support grant P30 CA008748.
Role of the funder
The research was supported in part by National Cancer Institute Cancer Center Support grant P30 CA008748. The funding source had no role in the writing of the manuscript or the decision to submit it for publication. All authors had full access to the data analysis results presented in this manuscript. The corresponding author had final responsibility for the decision to submit for publication.
Abbreviation list
- mUC
Metastatic urothelial carcinoma
- PD-1
Programmed Death-1
- PD-L1
Programmed Death-Ligand-1
- ECOG-PS
Eastern Cooperative Oncology Group-Performance status
- NLR
Neutrophil-lymphocyte ratio
- PLT
platelet count
- LDH
Lactate dehydrogenase
- TMB
Tumor mutation burden
- BMI
Body mass index
- PI
Prognostic index
- IDO-1
Indole-deoxygenase-1
Footnotes
Declaration of interests
Guru Sonpavde: Advisory board for BMS, Exelixis, Bayer, Sanofi, Pfizer, Novartis, Eisai, Janssen, Amgen, Astrazeneca, Merck, Genentech, EMD Serono, Astellas/Agensys; Research support to institution from Astrazeneca, Bayer, Amgen, Boehringer-Ingelheim, Janssen, Merck, Sanofi, Pfizer; Author for Uptodate; Steering committee for Astrazeneca, BMS, Astellas, Debiopharm, Bavarian Nordic; Speaker for Onclive; Research to Practice; Physician Education Resource (PER)
Juliane Manitz: Employed by EMD Serono
Chen Gao: Employed by AstraZeneca
Daniel Hennessy: Employed by EMD Serono
Doris Makari: Employed by AstraZeneca
Guenter Niegisch: lectures: Pfizer Pharma GmbH, Pierre Fabre Pharma GmbH, Roche Pharma AG, medac GmbH; advisory board/advisory role: Roche Parma AG, IMS Health AG, BMS AG, Sanofi-Aventis
Jonathan Rosenberg: Stock and Other Ownership Interests: Merck, Illumina. Honoraria: UpToDate, Bristol-Myers Squibb, AstraZeneca, Medscape, Vindico, Peerview, Chugai Pharma, Gilmore Philips, Research to Practice, Clinical Care Options, Clinical Mind, Intellisphere. Consulting or Advisory Role: Eli Lilly, Merck, Agensys, Genentech, Sanofi, AstraZeneca, MedImmune, Bristol-Myers Squibb, EMD Serono, Seattle Genetics, Bayer, Inovio Pharmaceuticals, BioClin Therapeutics, QED Therapeutics, Adicet Bio, Sensei Biotherapeutics, Fortress Biotech, Pharmacyclics, Western Oncolytics, GlaxoSmithKline, Astellas, Janssen.
Research Funding: Genentech (Inst), Oncogenex (Inst), Agensys (Inst), Mirati Therapeutics (Inst), Novartis (Inst), Viralytics (Inst), Genentech (Inst), Incyte (Inst), Seattle Genetics (Inst), Bayer (Inst), AstraZeneca (Inst), QED (Inst). Patents, Royalties, Other Intellectual Property: Predictor of platinum sensitivity (Inst). Travel, Accommodations, Expenses: Genentech, Bristol-Myers Squibb
Dean Bajorin: Bristol-Myers Squibb, Novartis, Roche/Genentech, Merck, Genentech, Roche, Lilly, Fidia Farmaceutici S. p. A., Eisai, Urogen Pharma, Pfizer, EMD Serono, Merck Sharp & Dohme, Dendreon, Amgen, Astellas Pharma, Seattle Genetics/Astellas
Petros Grivas: Consulting/honoraria (within 3 years) with Merck & Co., Bristol-Myers Squibb, AstraZeneca, Pfizer, EMD Serono, Clovis Oncology, QED Therapeutics, Driver Inc., Heron Therapeutics, Janssen, Foundation Medicine, Seattle Genetics, Genentech, Exelixis, Dendreon, Bayer, Biocept, Mirati Therapeutics, Genzyme Corporation. Prior (not current; with direct input in content) educational program with Genentech and Bristol-Myers Squibb. Clinical research (funding to institution, within 3 years): Merck & Co., Genentech, Oncogenex, Mirati Therapeutics, AstraZeneca, Pfizer, Bayer, Bavarian Nordic, Clovis Oncology, Immunomedics, Debiopharm, Bristol-Myers Squibb.
Andrea Apolo: None
Robert Dreicer: Astellas Pharma, Genentech/Roche, EMD Serono, Incyte, Pfizer, Eisai, Genentech, Seattle Genetics, BioClin Therapeutics, Janssen Oncology, Merck
Noah Hahn: Consultant: Merck, Genentech, BMS, AstraZeneca, Seattle Genetics, Incyte, Ferring, TARIS, TransMed, Inovio, Pieris. Research Funding to Institution: Incyte, AstraZeneca, BMS, Genentech, Merck, Seattle Genetics, Pieris, Inovio, Blue Buffalo. Honoraria: Bladder Cancer Academy, Advanced Health
Matthew Galsky: Consulting Fees: BioMotiv, Janssen, Dendreon, Merck, GlaxoSmithKline, Astellas, Genentech, BMS, Novartis, Pfizer, EMD Serono, AZ, Seattle Genetics, Incyte, Aileron, Dracen, Inovio, NuMab, Dragonfly Therapeutics
Contracted Research: Janssen, Denderon, Novartis BMS, Merck, Astra Zeneca, Genetech
Ownership Interest:Rappta Therapeutics
Andrea Necchi: consultant for Merck, Astra Zeneca, Janssen, Incyte, Roche, Rainier Therapeutics, Clovis Oncology, Bayer, and Astellas/Seattle Genetics. Grant/Research support from: Merck and Astra Zeneca. Travel expenses/Honoraria from: Roche, Merck, Astra Zeneca, and Janssen.
Sandy Srinivas: Genentech/Roche, Genentech, Bristol-Myers Squibb, Merck, Exelixis
Thomas Powles: honoraria - Roche AZ BMS Merck MSD Pfizer Seattle Genetics Exelexis; research funding - aZ Roche
Toni K. Choueiri: Leadership (no compensation) - Miscellaneous present or past leadership roles: Director of GU Oncology Division at Dana-Farber and past President of medical Staff at Dana-Farber), member of NCCN Kidney panel and the GU Steering Committee, past chairman of the Kidney cancer Association Medical and Scientific Steering Committee);
Stock or Other Ownership - Pionry, Tempest; Honoraria - AstraZeneca, Alexion, Sanofi/Aventis, Bayer, BMS, Cerulean, Eisai, Foundation Medicine Inc., Heron Therapeutics, Exelixis, Genentech, Roche, Lilly, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, EMD Serono, Prometheus Labs, Corvus, Ipsen, Up-to-Date, NCCN, Analysis Group, NCCN, Michael J. Hennessy (MJH) Associates, Inc (Healthcare Communications Company with several brands such as OnClive and PER), L-path, Kidney Cancer Journal, Clinical Care Options, Platform Q, Navinata Healthcare, Harborside Press, American Society of Medical Oncology, NEJM, Lancet Oncology; Consulting or Advisory Role - AstraZeneca, Alexion, Sanofi/Aventis, Bayer, BMS, Cerulean, Eisai, Foundation Medicine Inc., Exelixis, Heron therapeutics, Genentech, Roche, GlaxoSmithKline, Lilly, Merck, Novartis, Peloton, Pfizer, EMD Serono, Prometheus Labs, Corvus, Ipsen, Up-to-Date, NCCN, Analysis Group; Research Funding (Institutional and Personal) - Research (Institutional and personal): AstraZeneca, Bayer, BMS, Cerulean, Eisai, Foundation; Patents, Royalties, and other Intellectual Property - International Patent Application No. PCT/US2018/12209, entitled “PBRM1 Biomarkers Predictive of Anti-Immune Checkpoint Response,” filed January 3, 2018, claiming priority to U.S. Provisional Patent Application No. 62/445,094, filed January 11, 2017 --International Patent Application No. PCT/US2018/058430, entitled “Biomarkers of Clinical Response and Benefit to Immune Checkpoint Inhibitor Therapy,” filed October 31, 2018, claiming priority to U.S. Provisional Patent Application No. 62/581,175, filed November 3, 2017; Travel, Accommodations, Expenses - Part of Consulting, Advisory role and Honoraria; Other Relationship - The institution (Dana-Farber Cancer Institute) may have received additional independent funding/royalties from drug companies potentially involved in research around the subject matter.
Constanze Kaiser: Employed by Genentech
Darren Tayama: Employed by Genentech
Ashok Gupta: Employed by AstraZeneca
Shaad Essa Abdullah: Employed by AstraZeneca
Gregory R. Pond: Family member works for Roche Canada and owns stock with Roche Canada; Received consulting fees for Data Safety Monitoring Board Membership from Takeda
References
- 1.Sonpavde G: PD-1 and PD-L1 Inhibitors as Salvage Therapy for Urothelial Carcinoma. N Engl J Med, 376: 1073, 2017 [DOI] [PubMed] [Google Scholar]
- 2.Sharma P, Retz M, Siefker-Radtke A et al. : Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol, 18: 312, 2017 [DOI] [PubMed] [Google Scholar]
- 3.Bellmunt J, de Wit R, Vaughn DJ et al. : Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. New England Journal of Medicine, 376: 1015, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg JE, Hoffman-Censits J, Powles T et al. : Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet, 387: 1909, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powles T, O’Donnell PH, Massard C et al. : Efficacy and Safety of Durvalumab in Locally Advanced or Metastatic Urothelial Carcinoma: Updated Results From a Phase 1/2 Open-label Study. JAMA Oncol: e172411, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel MR, Ellerton J, Infante JR et al. : Avelumab in metastatic urothelial carcinoma after platinum failure (JAVELIN Solid Tumor): pooled results from two expansion cohorts of an open-label, phase 1 trial. Lancet Oncol, 19: 51, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niglio SA, Jia R, Ji J et al. : Programmed Death-1 or Programmed Death Ligand-1 Blockade in Patients with Platinum-resistant Metastatic Urothelial Cancer: A Systematic Review and Meta-analysis. Eur Urol, 2019 [DOI] [PubMed] [Google Scholar]
- 8.Bellmunt J, Choueiri TK, Fougeray R et al. : Prognostic factors in patients with advanced transitional cell carcinoma of the urothelial tract experiencing treatment failure with platinum-containing regimens. J Clin Oncol, 28: 1850, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Sonpavde G, Pond GR, Fougeray R et al. : Time from prior chemotherapy enhances prognostic risk grouping in the second-line setting of advanced urothelial carcinoma: a retrospective analysis of pooled, prospective phase 2 trials. Eur Urol, 63: 717, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonpavde G, Pond GR, Rosenberg JE et al. : Improved 5-Factor Prognostic Classification of Patients Receiving Salvage Systemic Therapy for Advanced Urothelial Carcinoma. J Urol, 195: 277, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Templeton AJ, McNamara MG, Seruga B et al. : Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst, 106: dju124, 2014 [DOI] [PubMed] [Google Scholar]
- 12.Heng DY, Xie W, Regan MM et al. : External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: a population-based study. Lancet Oncol, 14: 141, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNeel DG, Gardner TA, Higano CS et al. : A transient increase in eosinophils is associated with prolonged survival in men with metastatic castration-resistant prostate cancer who receive sipuleucel-T. Cancer Immunol Res, 2: 988, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powles T, Eder JP, Fine GD et al. : MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature, 515: 558, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Altman DG, Royston P: What do we mean by validating a prognostic model? Stat Med, 19: 453, 2000 [DOI] [PubMed] [Google Scholar]
- 16.Chang W, C. J, Allaire JJ, Xie Y, McPherson J shiny: Web Application Framework for R. R package version 1.3.2. https://CRAN.R-project.org/package=shiny, 2019 [Google Scholar]
- 17.Team RC: R: A language and environment for statistical computing.. R Foundation for Statistical Computing, Vienna, Austria, 2019 [Google Scholar]
- 18.Long GV, Dummer R, Hamid O et al. : Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol, 20: 1083, 2019 [DOI] [PubMed] [Google Scholar]
- 19.Mitchell TC, Hamid O, Smith DC et al. : Epacadostat Plus Pembrolizumab in Patients With Advanced Solid Tumors: Phase I Results From a Multicenter, Open-Label Phase I/II Trial (ECHO-202/KEYNOTE-037). J Clin Oncol: JCO2018789602, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teo MY, Seier K, Ostrovnaya I et al. : Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From PD-1/PD-L1 Blockade in Advanced Urothelial Cancers. J Clin Oncol, 36: 1685, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legrand FA, Gandara DR, Mariathasan S et al. : Association of high tissue TMB and atezolizumab efficacy across multiple tumor types. Journal of Clinical Oncology, 36: 12000, 2018 [Google Scholar]
- 22.Robertson AG, Kim J, Al-Ahmadie H et al. : Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell, 171: 540, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayers M, Lunceford J, Nebozhyn M et al. : IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest, 127: 2930, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powles T, Duran I, van der Heijden MS et al. : Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet, 391: 748, 2018 [DOI] [PubMed] [Google Scholar]
- 25.Nguyen CT, Kattan MW: How to tell if a new marker improves prediction. Eur Urol, 60: 226, 2011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


