Abstract
Ongoing research using cell transplantation and viral-mediated gene therapy has been making progress to restore vision by retinal repair, but targeted delivery and complete cellular integration remain challenging. An alternative approach is to induce endogenous Müller glia (MG) to regenerate lost neurons and photoreceptors, as occurs spontaneously in teleost fish and amphibians. Extracellular vesicles (EVs) can transfer protein and RNA cargo between cells serving as a novel means of cell-cell communication. We conducted an in vivo screen in zebrafish to identify sources of EVs that could induce MG to dedifferentiate and generate proliferating progenitor cells after intravitreal injection into otherwise undamaged zebrafish eyes. Small EVs (sEVs) from C6 glioma cells were the most consistent at inducing MG-derived proliferating cells. Ascl1a expression increased after intravitreal injection of C6 sEVs and knockdown of ascl1a inhibited the induction of proliferation. Proteomic and RNAseq analyses of EV cargo content were performed to begin to identify key factors that might target EVs to MG and initiate retina regeneration.
Keywords: Retina, Müller glia, Extracellular Vesicle, Regeneration
1. Introduction
In mammals and humans, the extent of spontaneous repair after retina injury or disease is either nonexistent or extremely limited (Karl and Reh, 2010). Rather than regenerate, damaged mammalian retinas commonly undergo reactive gliosis and scar formation (Bringmann et al., 2006). Numerous strategies are being tested to treat a variety of human retinal disorders, including gene therapy approaches and transplantation of stem cell-derived progenitor cells (Cehajic-Kapetanovic et al., 2015; MacLaren et al., 2006; Pearson et al., 2012; Roska and Sahel, 2018; Stern et al., 2018). An attractive alternative would be to induce endogenous regeneration from a resident pluripotent adult retinal stem cell (Müller glia; MG) (Ahmad et al., 2011). The adult zebrafish retina contains two classes of cells with regenerative capacity derived from MG. After retina damage in zebrafish, MG spontaneously dedifferentiate and then undergo asymmetric division for self-renewal and the production of a pool of proliferating progenitor cells that can regenerate all lost or damaged retinal cell types (Bernardos et al., 2007; Wan and Goldman, 2016). Therapeutic strategies designed to induce endogenous mammalian MG to regenerate lost retinal neurons and photoreceptors would be a powerful approach to restore vision.
Extracellular vesicles (EVs) are secreted nanoparticle sized, membrane bound vesicles containing lipid, protein, and RNA cargo (Maas et al., 2017; Thery et al., 2018; Tkach and Thery, 2016; van Niel et al., 2018). All cells secrete a diverse array of heterogeneous extracellular vesicles that can mediate cell-cell communication through the delivery of cargo and/or induction of recipient cell signaling cascades in numerous biological contexts (Maia et al., 2018; McGough and Vincent, 2016; Raposo and Stahl, 2019; Robbins and Morelli, 2014). EVs are classified both by size and biogenesis pathway (Colombo et al., 2014). Larger microvesicles (greater than 150nm) and a heterogeneous mixture of other vesicles are released by direct budding from the plasma membrane (Booth et al., 2006; Kowal et al., 2016). Smaller vesicles of endosomal origin (exosomes) are secreted when multivesicular bodies (MVBs) fuse with the plasma membrane, thereby releasing their intraluminal contents (Kalluri, 2016). These different classes of vesicles utilize distinct mechanisms controlling cargo selection in cell- and disease-specific contexts (Maas et al., 2017; Shifrin et al., 2013; Simons and Raposo, 2009). As purification strategies have been refined, protein markers and other cargo content found within the different classes of vesicles are being reassessed (Jeppesen et al., 2019). Heterogeneous mixtures of EVs can be readily purified by differential ultracentrifugation, but high resolution density gradient fractionation is now increasingly being utilized to enable separation and purification of small EVs (sEVs), large EVs, and non-vesicular fractions (Jeppesen et al., 2019; Kowal et al., 2016; Willms et al., 2018).
Once in the extracellular space, EVs can act both locally and distant in an autocrine or paracrine fashion, (Cha et al., 2015; Gutierrez-Vazquez et al., 2013; Tkach and Thery, 2016; Wortzel et al., 2019). The precise mechanisms mediating selective EV uptake remain largely unknown, but because of their ability to transfer cargo between cell types and across membrane barriers, EVs have emerged as potentially potent therapeutic agents (Farber and Katsman, 2016; Kalluri, 2016; Murphy et al., 2019; Silva et al., 2015; Wassmer et al., 2017; Wiklander et al., 2019). Here, we report the results of a screen for sources of EVs capable of inducing proliferation that can mimic the early stages of retina regeneration. We were prompted to test EVs based on papers demonstrating EV release from multiple retinal cell types and their ability to induce changes in gene expression in immortalized MG (Katsman et al., 2012; Peng et al., 2018), retinal ganglion cells (Mead and Tomarev, 2017), and retinal progenitor cells (Zhou et al., 2018). We sought to systematically test sources of EVs that could induce a proliferative response after intravitreal injection in otherwise undamaged retinas and identified 12 different cell lines that secrete small EVs capable of inducing proliferation in zebrafish.
2. Methods
2.1. Zebrafish
Wild-type AB or Tg(1016tuba1a:gfp) (Fausett and Goldman, 2006) zebrafish, 5–7 months old, were used for all experiments. All zebrafish lines were maintained at 28.5°C on a 14/10-hour light/dark cycle. Following retinal injections, zebrafish were maintained at 30°C for 72 hours before analysis. All procedures were approved by the Vanderbilt University Institutional Animal Care and Use Committee (IACUC).
2.2. Isolation of EVs
EVs were isolated from culture media. For standard lines, EVs were isolated after final culture for 48 hours in the absence of serum; stem cell culture media lacks serum. For C6 cells, T175 flasks (Corning) were seeded between 6–7×106 cells per flask and grown in the presence of serum to 80% confluency (~48 hours), washed three times with 1x Dulbecco’s PBS (DPBS; Gibco), and then grown for 48 hours in serum free media. Media was collected and subjected to differential centrifugation in three steps: 1000rpm for 10 minutes (room temperature), 2000xg for 25 minutes (4°C), and then 10,000xg for 30 minutes (4°C). These steps produce cell pellets, cell debris and large EVs, and microvesicles, respectively. P100 pellets (crude sEVs) were obtained by centrifuging conditioned media (CM) through the three steps above, followed by an additional 17 hr at 100,000xg (4°C). Pellets were suspended in 1xDPBS and washed by centrifugation at 100,000xg twice for 70 minutes each (4°C). Final sEV pellets were resuspended in 20μL 1xDPBS. This level of purity was used for the EV screen shown in Fig 2. For density gradient preparations, CM subject to the three steps above was concentrated using a 100K concentrator (MilliPore) to ~5mL and then layered onto 1mL 60% iodixanol cushions (Optiprep), and centrifuged at 100,000xg for 17 hours (4°C). The bottom 3mL were collected and then layered on top of an iodixanol discontinuous gradient consisting of 3 ml layers of 40%, 20% (CM layer), 10%, and 5% iodixanol. After centrifugation at 100,000xg for 17 hours (4°C), 1mL fractions (12) were collected (top to bottom). Each fraction was diluted in 12mL 1xDPBS and then centrifuged at 100,000xg for 3 hours (4°C). Final pellets were resuspended in 10–30μL 1xDPBS.
Fig 2. Knockdown of Ascl1a blocks C6 EV induced proliferation.
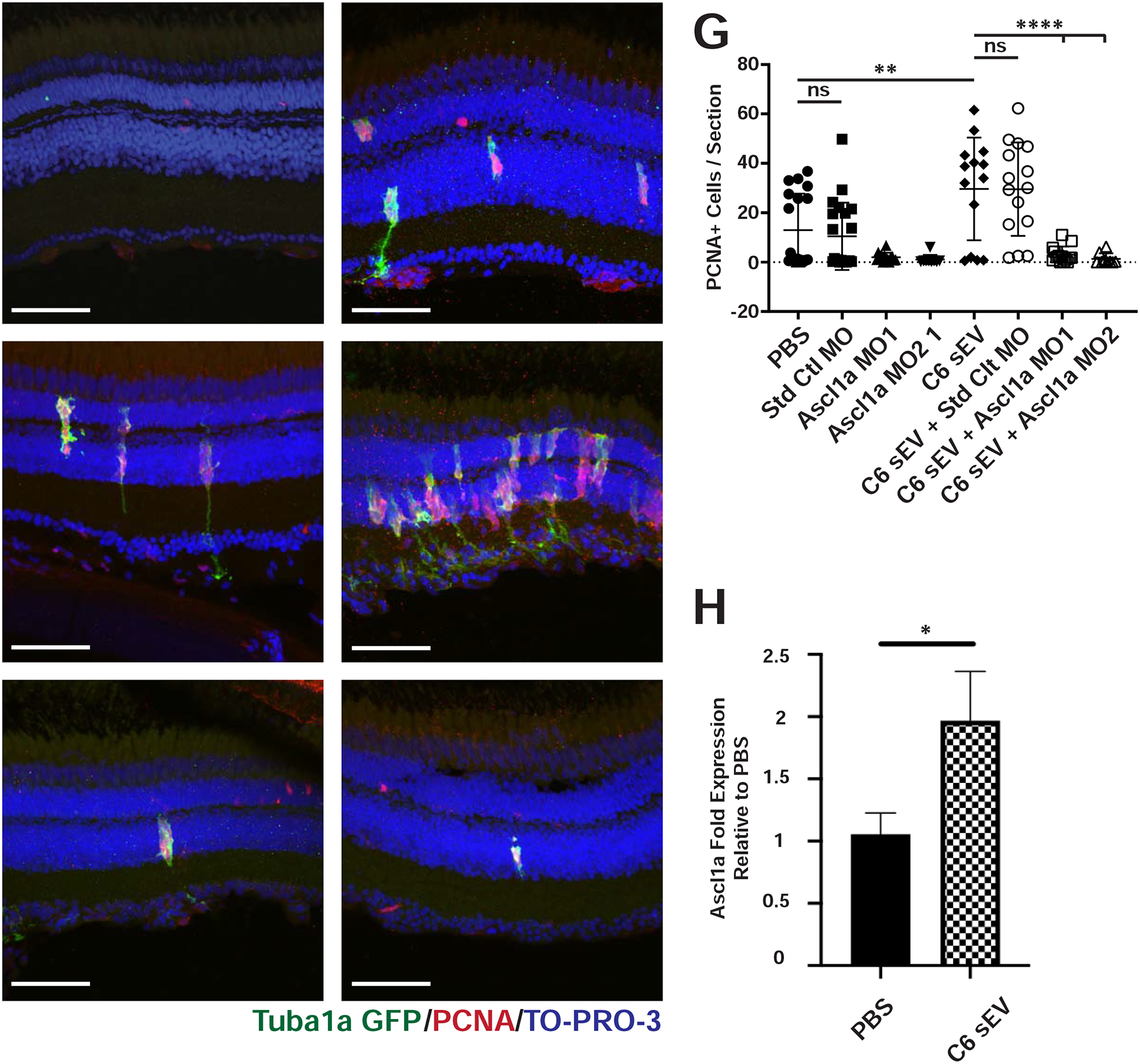
(A-B) PBS or C6 sEVs were injected into Tg(1016tuba1a:gfp) transgenic zebrafish eyes. Retinas were collected 72 hours after injection and immunostained with antibodies against PCNA to label proliferating cells and GFP to label dedifferentiated MG, respectively. Nuclei were stained with TO-PRO-3. C-F) Representative images of retinas from Tg(1016tuba1a:gfp) transgenic fish immunostained as in A and B. C) Control morpholinos were injected and electroporated into Tg(1016tuba1a:gfp) transgenic zebrafish retinas. D) Control morpholinos were injected and electroporated into Tg(1016tuba1a:gfp) transgenic zebrafish retinas in the presence of C6 sEVs. E) Morpholinos against ascl1a were injected and electroporated into Tg(1016tuba1a:gfp) transgenic zebrafish retinas. F) Morpholinos against ascl1a were injected and electroporated into Tg(1016tuba1a:gfp) transgenic zebrafish retinas in the presence of C6 sEVs. G) Quantification of PCNA+ cells across INL and ONL. Significance was calculated using one-way ANOVA with Holm-Sidak multiple comparison test, where **p-value=0.0083 ****p-value<0.0001. H) Zebrafish eyes were injected with PBS or C6 EVs. Retinas were isolated after 72 hours and pooled into groups of 3 for RNA purification. Data represent the mean +/− SEM with an N=5. Ascl1a expression was significantly higher among C6 EV injected conditions compared to PBS, where *p=0.036 using a two-tailed t-test. Scale bar, 50 μm.
2.3. Nanoparticle Tracking Analysis (NTA)
Particle sizes and numbers were analyzed using the Zetaview® Nanoparticle Tracking Video Microscope PMX-120 (Particle Matrix) and associated software. After optimization, settings were held constant across all replicate samples. Samples were diluted and particle counts and sizes were generated following the manufacturer’s protocols. The concentration of vesicles ranged from 108 to 1011 particles/mL. The average diameter of vesicles counted was ~100nm, corresponding to the size of small EVs.
2.4. EV Injections
EVs were injected into the vitreous of adult zebrafish eyes using an adapted protocol as previously described (Thummel et al., 2008). Briefly, a sapphire blade scalpel was used to make an incision in the cornea near the pupil after anesthetizing fish with 4% tricaine. A Hamilton syringe was inserted into the incision site and used to inject 0.5μl of solution into the vitreous. Fish were immediately placed into a recovery tank after injections.
2.5. Immunohistochemistry
Injected adult zebrafish eyes were removed and fixed overnight at 4.0°C in 4% PFA and 1X PBS at 72 hours post injection. Following fixation, eyes were washed in 1X PBS with 5% sucrose and cryo-protected in 30% sucrose in PBS for 4 hrs. at room temperature (RT), followed by a 2:1 mixture of OCT (Thermo Scientific) to 30% sucrose for 4 hrs. at RT and 1 hr. in straight OCT at RT before embedding in OCT. Eyes were cut into 15–20 micron sections using a Leica CM 1950, placed on Histobond slides (VWR), and dried on a slide warmer before immunostaining. Prior to immunohistochemistry (IHC), sections were subjected to antigen retrieval by incubation in 5mM sodium citrate, 0.05 % TWEEN 20, pH 6.0 for 10–20 mins. at 95°C. Slides were then rinsed with PBS and blocked in 3% donkey serum and 0.1% Triton X-100 for 2 hrs. at RT before primary antibodies were added. Slides were incubated with primary antibodies for 4 hrs. at RT or overnight at 4.0°C in 1% donkey serum and 0.05% TWEEN. The following primary antibodies were used at 1:500 dilution: mouse anti-PCNA (Sigma); rabbit anti-PCNA (Abcam); mouse anti-GFP (Invitrogen) and rabbit anti-GFP (Torrey Pines Biolabs). Following three 10 min. PBS washes, the following secondary antibodies were added for 4 hrs. at RT or overnight at 4.0°C in 1% donkey serum, 0.05% TWEEN at 1:500 dilution: donkey anti-mouse AF488, donkey anti-rabbit AF488, donkey anti-mouse Cy-3, donkey anti-rabbit Cy-3 (Jackson Immuno-Research). TO-PRO-3 was added with secondary antibodies at 1:1000 dilution (Thermo-Fisher Scientific). Following secondary antibody incubation, slides were washed three times with PBS for 10 mins. each before being air-dried and treated with Vectashield (Vector Labs) before being cover slipped.
2.6. Transwell Assays
C6 cells were cultured to ~80% confluency in T75 flasks (Corning). Cells were washed once in 1xPBS and then incubated with fresh media containing 4μL DiI per 7mL media at 37°C for 24 hrs. (DiIC18(3), Invitrogen). Cells were then trypsinized and seeded into Transwell chambers (0.4μm pore well, Costar) at 0.1×106 cells per well, and then incubated for 24 hrs. to allow adherence. Concurrently, human Müller cells were collected after trypsinization (Gibco) and stained using PKH67 (PKH67 GFP, Sigma). Müller cells were plated on coverslips, submerged in media within a 12 well dish (Corning), and incubated for 24 hours to allow adherence. The following day, both donor and recipient cells were washed three times in 1xPBS, and then co-cultured in fresh media for 48 hours. Following incubation, recipient cells were washed twice in 1x PBS and then fixed in 4% PFA for 20 minutes at RT (Sigma). Coverslips were then mounted on slides using Vectashield with DAPI (Vector).
2.7. Morpholino Injections
0.5 μl of 3’-lissamine labeled morpholinos (MO) from Gene Tools at a stock concentration of 1.5mM were injected into the intravitreal space of adult zebrafish eyes. The fish were allowed to recover for 1 hour after which 0.5μl of C6 sEVs or PBS were injected into the intravitreal space of adult zebrafish eyes followed by electroporation with two pulses of 75V for 50ms using a Biorad Gene Pulser Xcell (Ramachandran et al., 2010; Ramachandran et al., 2012; Thummel et al., 2008). The following morpholinos were used:
Control MO: 5’-CCTCTTACCTCAGTTACAATTTATA-3’
ASCL1a MO1: 5’-ATCTTGGCGGTGATGTCCATTTCGC-3’
ASCL1a MO2: 5’-AAGGAGTGAGTCAAAGCACTAAAGT-3’
2.8. Imaging and Scoring of Retinal Sections
Antibody stained retinal sections were imaged using a META Zeiss LSM 510 Meta confocal microscope under a 40X objective. Images were processed using ImageJ 2.0. For scoring of all retina sections, PCNA+ cells were counted across inner and outer nuclear layers in double blind experiments utilizing undergraduate researchers. Subsequent experiments utilized co-staining with antibodies against glutamine synthase or Tg(1016tuba1a:gfp)+ or Tg(GFAP:GFP)mi2001 lines which allowed determination of the number of PCNA+ cells that co-localized with these markers, primarily in the inner nuclear layer in addition to scoring of all PCNA+ cells across the inner and outer nuclear layers. Cells were counted from 2–4 non-consecutive sections and averaged for each eye, as indicated in respective figure legends. A Zeiss Axio Observer Z1 was used with a 40X objective.
2.9. qRT/PCR
Whole retinas were dissected and immediately placed into TRIzol (Life Technologies) 72 hours post injection. qRT/PCR for Ascl1a was performed on total RNA isolated from dissected retinas in TRIzol according to the manufacturer’s protocol. cDNA templates were reverse transcribed with the Accuscript High Fidelity 1st Strand cDNA Synthesis Kit (Agilent). qPCR was performed with primers for Ascl1a (5’-TGAGCGTTCGTAAAAGGAAACT-3’ and 5’-CGTGGTTTGCCGGTTTGTAT-3’) and 18S rRNA (5’-TTACAGGGCCTCGAAAGAGA-3’ and 5’-AAACGGCTACCACATCCAAG-3’) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). Relative expression values were calculated using the ΔΔ Ct method and 18S rRNA for normalization. Statistical analysis was performed on log transformed expression values using two-tailed t-tests.
2.10. Western Blots
Protein lysates were collected using 1x RIPA lysis buffer (Millipore) and concentrations were determined using the Pierce BCA assay kit (Thermo Scientific). Proteins (1μg) were separated on 12% MINI-PROTEAN TGX pre-cast gels (Bio-Rad) and transferred onto PVDF membranes using the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked in 5% milk in TBS-T for 1 hour and then incubated with primary antibodies in 5% milk in TBS-T overnight at 4°C. Primary antibodies were used at the following concentrations: anti-TSG101 (Invitrogen) at 1:1000, anti-CD81 (Santa Cruz Biotechnology) at 1:1000, and anti-Histone H3 (Abcam) at 1:5000. The next day, blots were washed with 1X TBS-T 3 times, then incubated with secondary antibodies in 5% milk in TBS-T at room temperature for 45 minutes. The following secondary antibodies were used at a concentration of 1:10,000: anti-Mouse ECL (GE Healthcare) and anti-Rabbit IgG (Cell Signaling). Blots were washed with 1X TBS-T three times and then treated with SuperSignal™ West Fenmto Maximum Sensitivity Substrate (Thermo Scientific) to visualize bands.
2.11. Proteomics
Sample preparation for shotgun proteomic analysis of cellular and exosomal proteins was performed using S-traps (https://www.protifi.com/s-trap/) according to the manufacturer’s instructions. The resulting peptides were analyzed by high resolution LC-MS/MS. Briefly, peptides were autosampled onto a 200 × 0.1 mm (Jupiter 3 micron, 300A), self-packed analytical column coupled directly to a Q-exactive plus mass spectrometer (ThermoFisher) using a nanoelectrospray source and resolved using an aqueous to organic gradient. Both the intact masses (MS) and fragmentation patterns (MS/MS) of the peptides were collected in a data dependent manner utilizing dynamic exclusion to maximize depth of proteome coverage. The resulting peptide MS/MS spectral data were searched against the rat protein database to which common contaminants and reversed versions of each protein were appended using Sequest (https://link.springer.com/article/10.1016/1044-0305%2894%2980016-2). The resulting identifications were filtered and collated together at the protein level using Scaffold (http://www.proteomesoftware.com/).
2.12. RNAseq
Total RNA from cells and sEVs was isolated using TRIzol (Life Technologies). For sEVs, TRIzol was incubated with 100μl or less of concentrated sEVs for an extended 15 min. incubation period prior to chloroform extraction. RNA pellets were resuspended in 60μl of RNase-free water and then re-purified using miRNeasy (Qiagen). RNAseq libraries were prepared using 200 ng of RNA and the NEBNext® Small RNA Library Prep Set for Illumina® (NEB, Cat: E7330S). Size selection targeting 100–200 nucleotides was performed on small RNA libraries using the Pippin Prep instrument and 3% agarose dye free gel (Sage Science #CDF 3010). Libraries were sequenced using the NovaSeq 6000 with 150 bp paired end reads targeting 50M reads per samples. Reads were trimmed post sequencing to 50 bp SE. RTA (version 2.4.11; Illumina) was used for base calling. Cutadapt (https://github.com/marcelm/cutadapt) was used to trim adapters. TIGER (https://github.com/shengqh/TIGER), was used to perform read mapping, miRNA quantification and differential analysis. Specifically, Bowtie was used to map reads to the rat miRNAs in miRBase and the rat genome. DESeq2 was used to detect differential expression between exosome and cells. Genes with fold change greater than 2 and adjusted p-value less than 0.01 were considered differentially expressed.
2.13. Functional Enrichment Analysis
Gene ontology analyses were performed on proteomics data using WebGestalt (Liao et al., 2019). Only proteins enriched in sEVs with a value greater than 2-fold were used. Proteins were compared using Over-Representation Analysis (ORA) set to the genome protein-coding reference set. All groups were significant with an FDR < 0.05. For RNAseq data, predicted targets for the top 10 miRNAs enriched in sEVs were determined using MicroRNA Target Prediction Database (mirdb.org). Only the top 5 predicted targets with a target score > 80 were used.
2.14. Statistical Analysis
Student t-tests or One-way ANOVA analyses were used to calculate significance depending on how many conditions were simultaneously performed using GraphPad Prism 7 software. Multiple comparison tests with one-way ANOVA are specified in each figure legend. The threshold for significance (alpha) was 0.05. All data are represented by a mean value +/− standard error.
2.15. Antibodies Used
| Name | Company | Catalog Number | Concentration |
|---|---|---|---|
| Rabbit anti-TSG101 | Invitrogen | PA5–31260 | 1:1000 |
| Mouse anti-CD81 | Santa Cruz Biotechnology | Sc-166029 | 1:1000 |
| Rabbit anti-Histone H3 | Abcam | Ab1791 | 1:5000 |
| Anti-mouse ECL | GE Healthcare | NA931 | 1:10000 |
| Anti-Rabbit IgG | Cell Signaling | 7074 | 1:10000 |
| Mouse anti-PCNA | Sigma | P8825 | 1:500 |
| Rabbit anti-PCNA | Abcam | Ab18197 | 1:500 |
| Mouse anti-GFP | Invitrogen | MAS-15256 | 1:500 |
| Rabbit anti-GFP | Torrey Pines Biolabs | TP401 | 1:500 |
| Donkey anti-mouse AF488 | Jackson ImmunoResearch | 715-545-151 | 1:500 |
| Donkey anti-rabbit AF488 | Jackson ImmunoResearch | 711-545-152 | 1:500 |
| Donkey anti-mouse Cy3 | Jackson ImmunoResearch | 715-165-160 | 1:500 |
| Donkey anti-rabbit Cy3 | Jackson ImmunoResearch | 711-165-152 | 1:500 |
| TO-PRO-3 | Thermo Fisher Scientific | T3605 | 1:500 |
3. Results
3.1. In vivo screen of EVs capable of inducing MG-derived proliferation in undamaged zebrafish retinas
We performed a large-scale in vivo screen of EV preparations for their ability to stimulate MG proliferation in the retina of zebrafish, as marked by the expression of Proliferating Cell Nuclear Antigen (PCNA). As a proof of concept, we tested whether EVs could be taken up by MG using Transwell assays which showed that primary cultured human MG (Capozzi et al., 2014) were capable of direct uptake of fluorescently labeled EVs (Fig. S1). Thus, we set out to isolate EVs from a variety of sources and test whether intravitreal injection into zebrafish retinas could induce a regenerative response. EVs were purified by ultracentrifugation from tissue culture media and dissected tissue samples and then 0.5μl were intravitreally injected into the eyes of 5–15 six month-old AB zebrafish and compared to PBS vehicle control injections for each clutch of zebrafish tested. Fish were sacrificed 72 hours after injection, eyes dissected and fixed, cryo-protected, sectioned, and antibody stained for PCNA (Rajaram et al., 2014). For each eye, 2–4 nonconsecutive sections per retina (from ~60 sections) were scored in double blind counting assays for the average number of PCNA+ cells across the inner and outer nuclear layers (INL and ONL), excluding the circumferential germinal zone or ciliary marginal zone, which is known to proliferate through adulthood (Fischer et al., 2013; Stenkamp, 2007). To ensure that any increases in PCNA+ cells were not simply due to nonspecific injury from the injections themselves, we performed TUNEL staining to detect apoptotic cells at 48 and 72 hours after injection with PBS or with EVs from C6 glioma conditioned media (Fig. S2). No significant differences in TUNEL staining were detected between PBS and EV injected retinas. Injection of PBS alone induced a slight increase in PCNA+ cells compared to uninjected control retinas which showed little to no PCNA+ cells except for rare single PCNA+ cells in the ONL that were only observed in 5–10% of sections and likely correspond to rod precursors (Fig. S3).. All statistical analyses were performed by comparing the number of PCNA+ cells in EV injected retinas to PBS control injections.
In total, we screened 59 independent EV preparations from a variety of cultured stem cells, primary neuronal cultures, iPS cells undergoing a variety of differentiation conditions, cancer cell lines, and from wild type zebrafish retinas or zebrafish retinas undergoing regeneration after being subjected to constant intense light damage (Vihtelic and Hyde, 2000) (Fig. 1, Table S1). Injection of cell free media or large EVs (greater than 1000nm) did not result in increased levels of PCNA+ counts compared to PBS control injections (Fig. S4). Injection of microvesicles (150–500nm vesicles) resulted in a modest but significant increase in PCNA+ cells (p<0.05). The most significant induction of PCNA+ cells (p<0.001) was observed after injection of small EVs (sEVs; 40–100nm) (Fig. S4). For all sEV preparations, Nanosight tracking analysis revealed that particle size distribution from the different preparations was similar (40–100nm) and that all sEV preparations had relatively high numbers of particles (Table S1). Although the particle counts between different sEV preparations sometimes differed by an order of magnitude, there was no correlation between particle counts and the induction of PCNA+ cells at the numbers tested. However, serial dilution of C6 sEVs led to a corresponding decrease in PCNA+ cell numbers and heat or protease treatment also abolished activity (Fig. S5)
Fig 1. In vivo screen to identify EV sources capable of inducing increased numbers of PCNA+ cells.
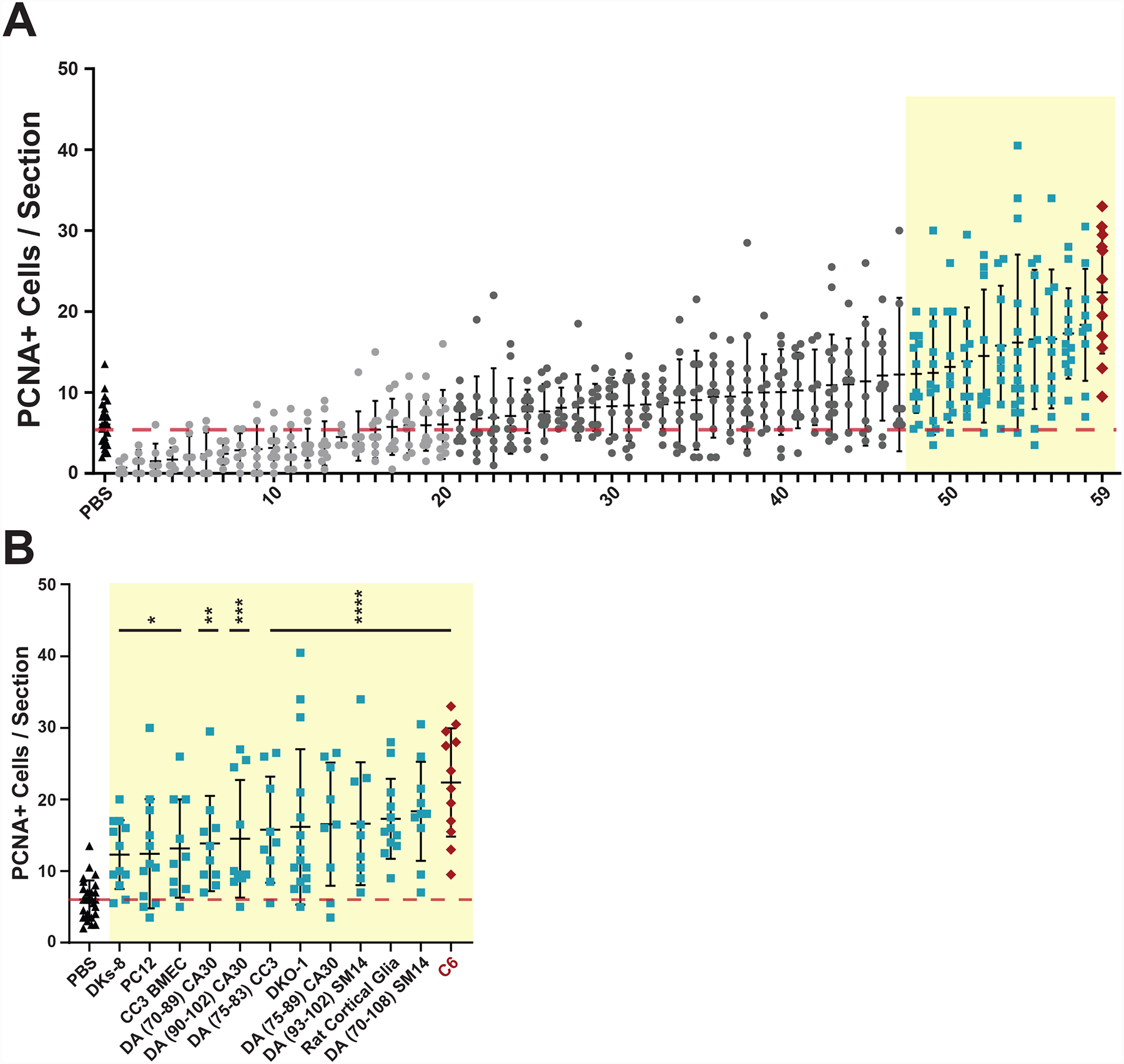
EVs were isolated from conditioned media or dissociated tissues and intravitreally injected into undamaged wild type AB zebrafish eyes. After 72 hours, retinas were dissected, sectioned, and immunostained with antibodies against Proliferating Cell Nuclear Antigen (PCNA). A) 59 independent EV preparations were tested and PCNA+ cells were counted across the inner nuclear layer (INL) and outer nuclear layer (ONL) and compared to control PBS injections. Each data point represents PCNA+ cells from a single retina and consists of average counts from 2–4 nonconsecutive sections from the same eye. Light gray EV samples (1–20) led to PCNA counts less than or equal to PBS control background levels (red dotted line). Dark gray EV samples (21–47) induced non-significant PCNA counts slightly greater than background. Light blue EV samples (48–58) induced significant (p-values <0.05) PCNA counts greater than control PBS injections. C6 EVs (red diamonds) induced the most significant PCNA+ counts compared to PBS controls. One-way ANOVA with Dunnett multiple comparison tests (to PBS injection) were used to determine significance. The identify of each EV preparation can be found in Table S1. B Enlargement of EV preparations from (A) that produced significant increases in PCNA counts compared to PBS controls with the indicated source of EVs shown along the X axis. *p-value <0.05, **p-value = 0.0034, ***p-value = 0.0008, ****p-value <0.0001.
Twelve sEV preparations induced statistically significant increases in PCNA+ counts across the INL and ONL of injected wild type retinas (Fig. 1B). Subsequent co-staining of sections with antibodies against glutamine synthase (GS), a marker of MG (Rajaram et al., 2014), showed that only a subset of EV preparations induced proliferation of PCNA+ cells in the INL that co-localize with MG (Fig. S6). Of the 7 lines secreting EVs that induced the most significant increase in PCNA+ cells (p<0.001) 0.001)(Fig. 1B), 4 were derived from human iPS cells differentiating over time into the dopaminergic (DA) lineage (Neely et al., 2017). These EVs most commonly induced proliferation of cells in the ONL (Fig. S6). EVs that predominantly induced proliferation of cells in the INL include those derived from DKO-1 mutant KRAS colorectal cancer cells (Shirasawa et al., 1993), primary cultures of rat hippocampal glia (astrocytes), and C6 rat glioma cells (Grobben et al., 2002) (Fig. S6). For this paper, we focused on induction of proliferating cells in the INL that co-localize with markers consistent with MG-derived proliferation.
3.2. C6 EVs induce proliferation in MG-derived cells
EVs from C6 glioma cells were the most consistent at inducing statistically significantly increased levels of PCNA+ cells in the INL and the majority of those cells co-localized with GS (Fig. S6) and with GFP using Tg(1016tuba1a:gfp) zebrafish in which GFP expression marks dedifferentiated MG and proliferating progenitor cells (Fausett and Goldman, 2006) (Fig. 2A, B). To further test whether a canonical regenerative response was initiated, we co-injected C6 EVs into the Tg(1016tuba1a:gfp) line in the presence or absence of antisense morpholinos targeting ascl1a. Ascl1a is a transcription factor that is required for MG-derived retinal regeneration in zebrafish (Fausett et al., 2008; Ramachandran et al., 2011; Rao et al., 2017). Compared to control MO injection, co-injection of two independent ascl1a MOs resulted in a complete suppression of sEV-induced proliferation (Fig. 2C–G).
Decreased levels of proliferation after injection of morpholinos targeting ascl1a suggest that C6 EVs can induce expression of Ascl1a. To test this, we isolated RNA after intravitreal injection of C6 EVs and performed qRT/PCR with primers against ascl1a. As shown in Fig. 2H, we observed a significant increase in ascl1a levels after C6 EV injection when compared to control PBS injections.
3.3. C6 exosomes induce MG-derived proliferation
As EV purification protocols are being refined and optimized, additional purification steps and new standards are being adopted regarding the use of protein markers for specific subclasses of EVs (Jeppesen et al., 2019; Thery et al., 2018). To more precisely define the identity of the C6 EVs that are responsible for increased numbers of PCNA+ cells after intravitreal injection, we purified sEVs using iodixanol density gradient fractionation (Li et al., 2018) (Fig. 3A). This allowed for separation of the sEV preparations used in the initial screen into 12 fractions corresponding to dense, non-vesicular protein-rich fractions (marked by histone H3), small intermediate density vesicular fractions that include classical exosomes (marked by TSG101 and CD81), and larger, lipid-rich, vesicles (Fig. 3B).
Fig 3. Gradient Purified C6 sEVs induce proliferation.
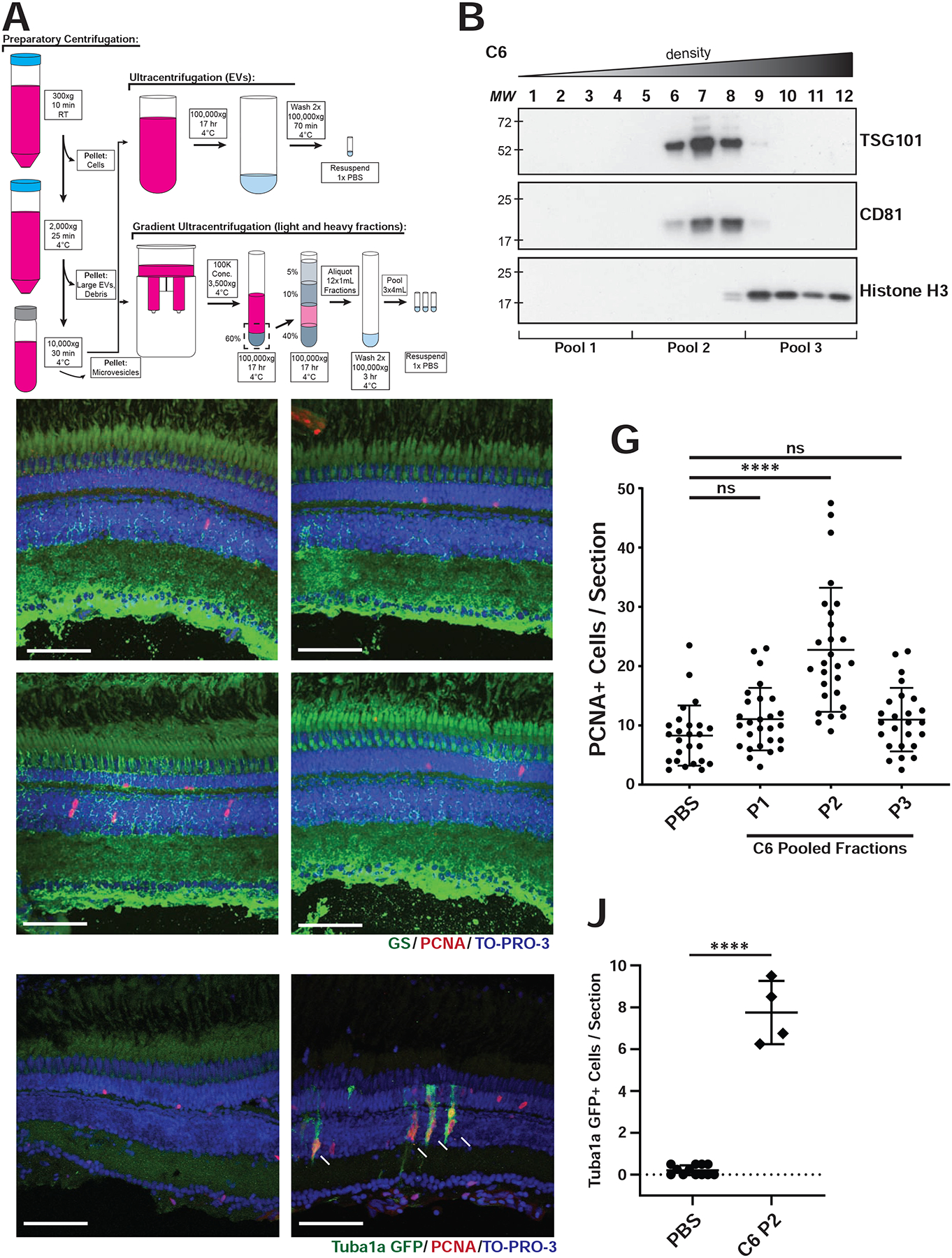
A) Differential ultracentrifugation steps during sEV purification. B) Representative western blots of C6 density gradient fractions using antibodies against CD81, TSG101, or histone H3. C-F) After iodixanol gradient ultracentrifugation, fractions were combined into three pools (four fractions per pool; P1, P2, P3). Intravitreal injections into wild type AB fish were performed with either PBS, P1, P2, or P3. Retinas were collected 72 hours after injection and representative images are shown after immunostaining with antibodies against PCNA and glutamine synthase (GS). G) Quantification of PCNA+ cells after injection as in C-F, averaged across 2–4 non-consecutive retinal sections per eye, across INL and ONL. Significance was calculated using one-way ANOVA with Tukey multiple comparison test, where ****p-value<0.0001. H, I) Representative images of retinas from Tg(1016tuba1a:gfp) fish injected with either PBS (H) or C6 P2 sEVs (I). Sections were stained with antibodies against GFP and PCNA. J) Quantitation of GFP+ cells from experiments in H, I. Significance was calculated using ANOVA, where ****p-value<0.0001. Scale bar, 50 μm.
For intravitreal injections of gradient purified particles, we combined fractions into pools based on vesicle marker profiles (Fig. 3B). Pool 1 (P1) contained fractions 1–4, composed of large vesicles, Pool 2 (P2) contained fractions 5–8, composed of sEVs, and Pool 3 (P3) contained fractions 9–12, composed primarily of nonvesicular lipoproteins, ribonucleoproteins, and protein aggregates. After injection into wild type AB fish and co-staining for both GS and PCNA, the highest proliferative activity was found to reside in P2, where significantly higher PCNA+ cells were observed in the INL compared to the other fractions and to the PBS control (Fig. 3C–G)). The PCNA+ cells were observed adjacent to and/or closely associated with GS-stained MG processes. Neither P1 nor P3 induced a proliferative response above control PBS injections. When we injected P2 sEVs into the Tg(1016tuba1a:gfp) line we detected a significant increase in the number of GFP+ cells (indicating dedifferentiated MG) that co-localized or were adjacent to PCNA+ cells, consistent with MG-derived progenitor cells (Fig. 3H–J)). We also injected P2 sEVs into Tg(gfap:gfp) fish which express GFP in MG (Bernardos and Raymond, 2006). Again, the resulting PCNA+ cells co-localized with and adjacent to GFP+ MG in the INL at 72 hr post injection (Fig. S7)), consistent with the idea that the MG are dedifferentiating into a progenitor state.
3.4. Proteomic analysis of C6 P2 exosomes
To begin to characterize what factors might be responsible for inducing proliferation after intravitreal injection of C6-derived sEVs, we performed proteomic analysis of P2 and identified enriched proteins compared to C6 cellular levels. From total spectral counts, 1849 unique proteins were identified with 33% enriched in C6 cells, 16% enriched in P2 sEVs, and ~50% shared between both (Fig. 4A–C; Table S2)). Most proteins known to be enriched in sEVs were found in P2 including traditional exosome markers β1-integrin, CD29, CD63, CD9, Syntenin-1, and Caveolin-1 (Thery et al., 2018). Some proteins were found at or below the limits of detection in the cellular proteome, but were readily identified in P2 sEVs, indicating either rapid secretion and/or degradation in cells. Fig. 4A includes all proteins detected in either the cellular or sEV proteomes (or both), including those where the cellular levels were at or below the limits of detection. Fig. 4B includes only those proteins where fold enrichment values could be calculated, i.e. cellular levels well above background. The volcano plot in Fig. 4C is derived from those proteins included in Fig. 4B and depicts individual proteins enriched in either C6 cells (red) or sEVs (blue) with corresponding p-values.
Fig 4. Proteomics analysis for C6 cells and EVs.
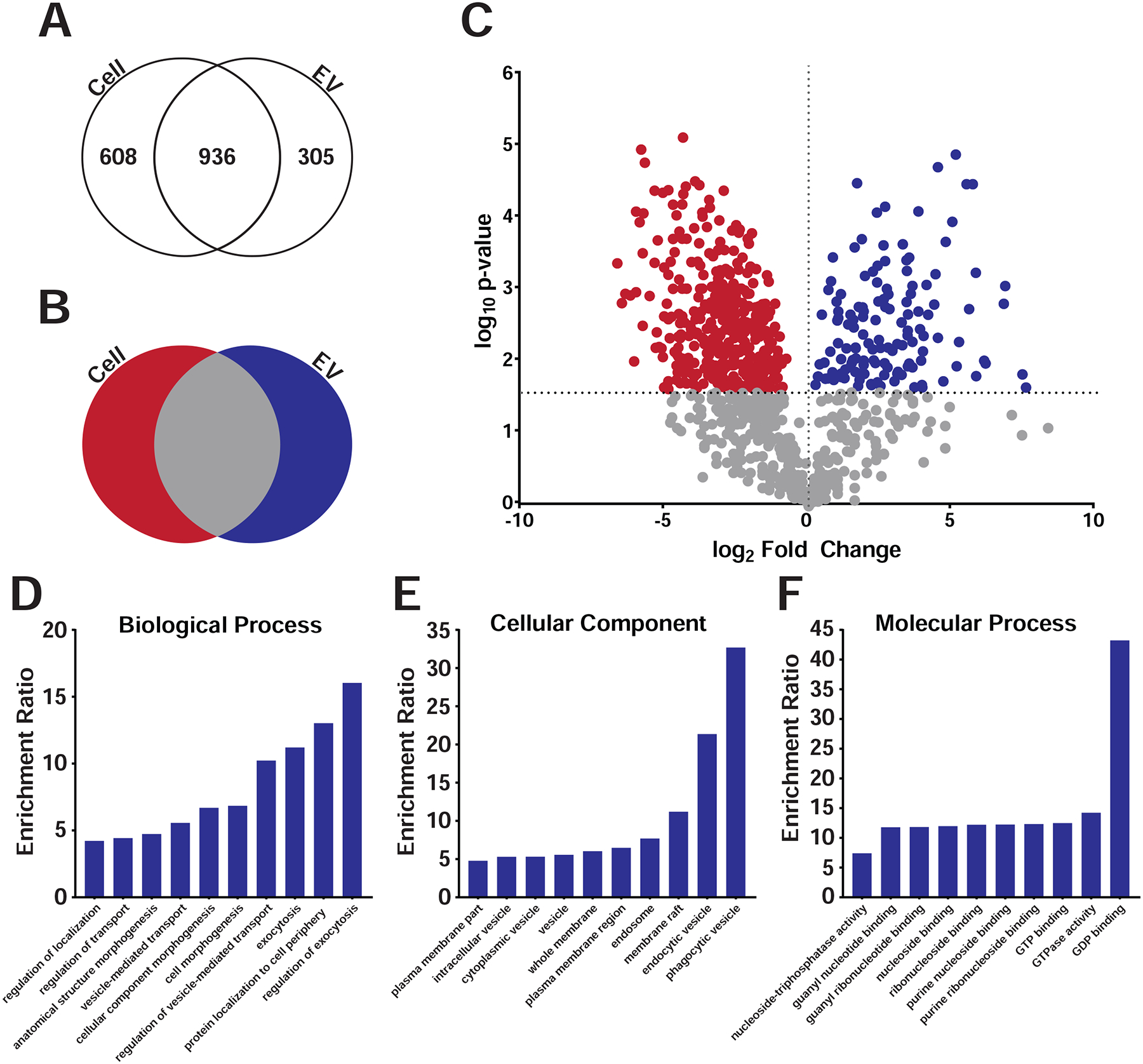
A) Gradient purified sEVs from C6 fractions were subjected to mass spectroscopy and protein levels compared between sEVs and parent C6 cellular levels. Venn diagram showing total detectable spectral counts found in cells, EVs, or both. B) Venn diagram showing enrichment of proteins in cells, EVs or both after excluding proteins with little to no detectable cellular levels. C) Volcano plot of proteomic analysis plotting p-value versus the fold change between cells (red) and EVs (blue) after excluding proteins with little to no detectable cellular levels. Red and blue dots indicate individual proteins enriched in either cells (red) or sEVs (blue) above a p-value threshold of p<0.02696 using the Benjamini-Hochberg test. D-F) Gene Ontology analyses of proteins in P2 sEVs showed enrichment in categories as shown.
Bioinformatic analyses of the most enriched proteins in C6 sEVs identified several expected protein classes, including proteins involved in endo- and exocytosis and regulators of such trafficking including the Rab family of GTPases (Fig. 4 and Table S3, Table S4A–C).). The protein with the highest spectral counts enriched in P2 sEVs was lactadherin, also known as MFG-E8 or SED1 (Ensslin and Shur, 2007; Stubbs et al., 1990; Taylor et al., 1997). MFG-E8 is a secreted protein that contains EGF and Factor VIII domains that was originally identified on milk fat globules and thought to mediate adhesion to integrin-expressing cells (Taylor et al., 1997). MFG-E8 binds to phosphatidylserine on the surface of membrane vesicles or apoptotic cells (Hanayama et al., 2002; Oshima et al., 2002), and has also been shown to accumulate on exosomes (Veron et al., 2005). It also plays a role in photoreceptor-RPE interactions (Nandrot et al., 2007).
3.5. RNAseq of C6 exosomes
Besides protein cargo, EVs carry a variety of RNAs, the best characterized being miRNA (Cha et al., 2015; Patton et al., 2015; Skog et al., 2008; Valadi et al., 2007). We purified small RNAs from gradient purified C6 sEVs and performed RNAseq to identify differentially enriched miRNAs between C6 cells and sEVs. Analysis of the data identified numerous miRNAs enriched in C6 exosomes (Fig. 5A; Table S5). Previously, miRNAs were identified in EVs from neural progenitor cells that were proposed to inhibit inflammatory signaling and prevent microglia activation (Bian et al., 2020). Interestingly, we detected little overlap in the two data sets which could be consistent with P2 sEVs activating a regenerative response as opposed to blocking an inflammatory response. The Reh lab identified two miRNAs (miR-25 and miR-124) whose overexpression can induce Ascl1 expression during conversion of mouse MG into neuronal/progenitor cell phenotypes (Wohl et al., 2019). miR-25 was not enriched in C6 sEVs compared to parental C6 cells whereas miR-124 showed enrichment in sEVs but was expressed at low to undetectable levels in C6 cells.
Fig 5. RNAseq analysis for C6 cells and sEVs.
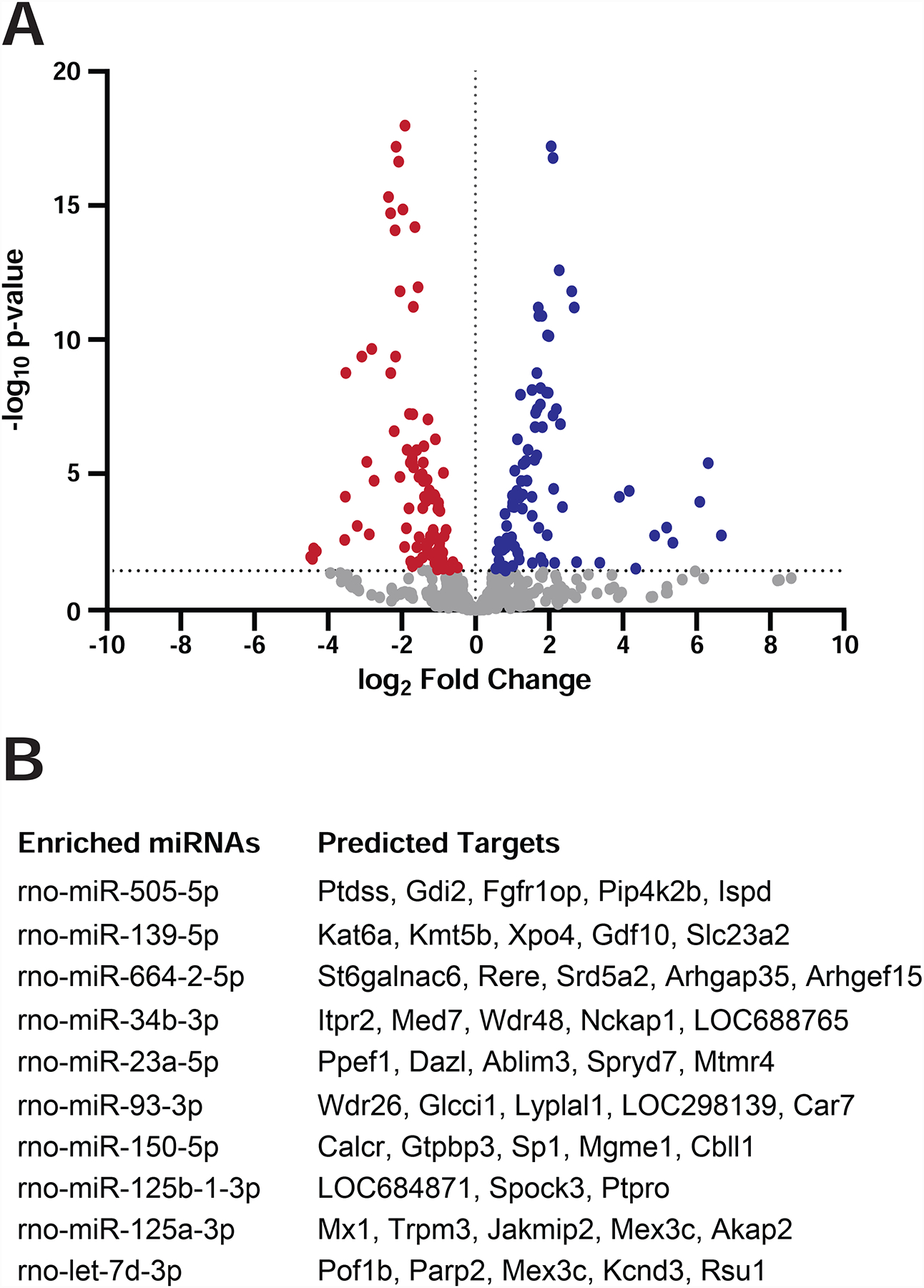
Small RNAs were isolated and purified from C6 cells and from gradient sEVs and subjected to RNA sequencing to identify differentially enriched miRNAs. A) The volcano plot shows p-values versus fold change levels between cells (red) and sEVs (blue). Dots represent individual miRNAs. B) Predicted mRNA targets for the most enriched miRNAs were determined using the MicroRNA Target Prediction Database.
We used the MicroRNA Target Prediction Database to identify mRNA targets for the most enriched miRNAs in P2 sEVs (Fig. 5B). Gene Ontology analyses of these predicted targets did not result in significant enrichment of any specific category or biological process. Full analysis of the differentially enriched EV miRNAs and identification and validation of their mRNA targets will require complementary mRNAseq experiments in cells exposed to C6 sEVs.
4. Discussion
We conducted an in vivo screen to identify EV sources capable of eliciting MG-derived proliferation after intravitreal injection into zebrafish eyes. While we initially targeted EVs prepared from a variety of stem cells and induced pluripotent stem cells that might be suitable for future therapeutic use, some of the best sources for inducing regenerative responses came from cancer cell lines. Among all the lines that produced EVs that increased levels of PCNA+ cells after injection, we observed distinct differences in terms of the localization of proliferating cells in the zebrafish retina. EVs from iPSCs differentiating into mature dopaminergic neurons tended to induce PCNA+ cells that were found mostly in the ONL. In contrast, EVs from C6 glioma cells induced PCNA+ cells in the INL which mostly co-localize with MG markers. It will be interesting to determine the exact origin and lineage of the different populations of PCNA+ cells induced by specific EV preparations to discover whether different sources of EVs induce distinctly different responses. Proteomic analysis of breast cancer derived EVs revealed that the cell of origin can often be inferred based on EV cargo content (Wen et al., 2019). This raises the possibility that the glial origin of C6 cells might result in membrane and cargo content that preferentially drives uptake by MG and whose identify could help in future EV targeting experiments, perhaps including MFG-E8.
While we focused mostly on EVs that induced increased numbers of PCNA+ cells derived from MG in the INL, some of the PCNA+ cells are likely to be rod precursors (Otteson et al., 2002; Raymond et al., 2006), microglia (Conedera et al., 2019; Mitchell et al., 2018; Mitchell et al., 2019), or other cells that might be preferentially sensitive to damage induced by the injected EVs. Induction of PCNA expression could be part of a regenerative response, but could also be due to damage by delivery of specific EV cargo or lipid content. TUNEL staining (Fig. S2) argues against extensive non-specific damage due to C6 sEV injections, but it remains possible that some EVs might induce damage and that some PCNA+ cells could be a response to such damage.
4.1. In vivo screening for EVs that induce retina regeneration
The rationale for the in vivo screen used here was driven by increasing interest in the roles of EVs in cell-cell communication and by findings that retinal cells can take up EVs and induce changes in gene expression (Katsman et al., 2012; Mead and Tomarev, 2017; Yu et al., 2016; Zhou et al., 2018). A challenge for the screen was to efficiently isolate EVs from numerous sources grown in the absence of serum to avoid contamination of mostly bovine EVs. The majority of EV preparations did not induce a significant increase in PCNA+ cells, some even led to slightly decreased levels of PCNA+ cells compared to PBS. This could indicate that some EVs can suppress proliferation, but the changes are so modest that the effects should be carefully interpreted since we observed variation from fish to fish and from preparation to preparation.
Compared to the levels of proliferation typically observed using a variety of retina damage models in zebrafish (Lenkowski and Raymond, 2014), injection of EVs led to far fewer PCNA+ cells. The notion that a single injection of EVs into an otherwise undamaged zebrafish retina could replicate effects observed after more extensive damage was not unexpected. Even in fish, retina regeneration is a multi-step process; it may be that EVs, and possibly combinations of EVs from multiple sources, will need to be delivered over time to induce a complete regenerative response, especially for application to mammalian retina regeneration. Further, even though we focused on induction of PCNA+ cells in the INL that are derived from MG, it is highly likely that the injected EVs are being taken up by a variety of cells. Ideally, targeting molecules will be identified that direct uptake by MG. Nevertheless, the use of transgenic lines expressing markers of MG and dedifferentiated MG and the appearance of clusters of PCNA+ cells along MG processes after C6 sEV injection are all consistent with induction of MG-derived proliferative cells, as is the reduction in PCNA levels after Ascl1 knockdown.
4.2. sEVs carry cargo capable of inducing MG-derived proliferation
Intravitreal injection of C6-derived sEVs could induce retina regeneration in multiple ways. One mechanism could be that the sEVs bind to MG and induce a signal transduction cascade that initiates proliferation without actually being internalized. A second mechanism could be that sEVs are endocytosed and activate endosomal receptors such as Toll-like receptors (Fabbri et al., 2013). We identified 138 proteins enriched in C6 sEVs with one of the most abundant being MFG-E8 (Ensslin and Shur, 2007; Stubbs et al., 1990; Taylor et al., 1997). While MFG-E8 plays a role in photoreceptor-RPE interactions (Nandrot et al., 2007), that role would seem to be unrelated to activation of MG, unless reduced levels of MFG-E8 lead to photoreceptor death. Future work will be devoted to examination of additional candidates.
Proteomic analysis of C6 sEVs did not show significant enrichment of any of the best characterized proteins involved in retina regeneration (Wan and Goldman, 2016). This could simply be due to a lack of enrichment in sEVs compared to cellular levels. For example, STAT3 (Nelson et al., 2012), MAPK (Fischer et al., 2009; Wan et al., 2014) and PTB (Zhou et al., 2020) were readily detectable in C6 cells but with much lower levels in sEVs. β-catenin was detected in both cells and sEVs with a slight enrichment in sEVs. It is possible that the modest effects we observe on regeneration after intravitreal injection might be due to limiting amounts of key factors in sEVs. However, it could also be that unknown factors within the sEVs stimulate expression of key inducers of retina regeneration such as Ascl1 which was not identified in C6 sEVs but was found to increase after intravitreal injection.
Despite the caveats, our data raise the possibility that EVs could be used as a therapeutic agent to induce retina regeneration. Chen and colleagues demonstrated that delivery of multiple factors using AAV vectors in mice can generate rod photoreceptors in a MG-derived pathway (Yao et al., 2016; Yao et al., 2018). AAV2 vectors that are associated with exosomes are also capable of gene delivery in the murine retina (Wassmer et al., 2017). Beyond viral vectors, Reh and colleagues showed that genetic delivery of Ascl1 and a general histone deacetylase inhibitor could stimulate MG-derived regeneration in mice (Jorstad et al., 2017). Thus, retinal delivery of genes or other cargo shows great promise to promote endogenous retina regeneration. EVs provide an alternative method of delivery that bypasses concerns about viral vectors and could potentially overcome obstacles related to genetic delivery. As an initial attempt to determine whether our approach in zebrafish might extend to mice, we intravitreally injected a subset of the EVs from our large screen into mice and our findings from those experiments will be reported separately. Moving forward, it will be interesting to determine whether the efficiency of EV-mediated regeneration can be enhanced using EVs loaded with factors capable of inducing retinal regeneration and expressing surface proteins that target uptake by MG. Indeed, one possibility is that MFG-E8 somehow plays a role in targeting MG for uptake. Should MFG-E8 or other proteins be identified that can target EVs to MG, it could be especially attractive as a delivery vehicle because it has been found that proteins associated with, or on the surface of EVs, are more active when delivered to recipient cells than when delivered as recombinant or purified proteins (Higginbotham et al., 2011).
Supplementary Material
Figure S1. Transfer of EVs between donor and recipient cells.
A) Schematic of Transwell assay. C6 cells were DiI-labeled, washed, and seeded as donor cells on top of a 0.4uM polyester Transwell membrane. Primary human Müller glia (HMGs; (Capozzi et al., 2014; Madamanchi et al., 2014)) were PKH67-labeled, washed, and seeded as recipient cells in the bottom of the Transwell culture dish. After 24 hours, cells were washed and co-cultured in serum-free media for 48 hours. B) Representative confocal images of donor and recipient cells. When C6 donor cells were co-cultured with HMG recipient cells, co-localization (yellow) was visualized, indicating uptake of C6-derived EVs by HMCs. C) Enlarged images of cells depicted in B. Scale bar, 50 μm.
Figure S2. TUNEL Staining to Detect Apoptosis after EV injection.
AB wild type zebrafish were injected with either PBS or C6 sEVs. After recovery for either 48 hours or 72 hours, TUNEL staining was performed. Red-PNCA; Blue–TOPRO-3. Scale bar, 50 μm. ONL–outer nuclear layer; INL–inner nuclear layer; GCL–ganglion cell layer. (A) PBS injection (B) C6 sEV injections analyzed 48 hours post-injection. (C) C6 sEVs injection analyzed at 72 hours post-injection. (D) Scatterplot of TUNEL staining analysis. Each data point is from a separate eye and is an average of 2–4 non-consecutive sections, counting all TUNEL+ cells across all retinal layers. One-way ANOVA with Tukey multiple comparison tests were used for quantitation. Error bars are mean ±SEM; n=8.
Figure S3. Uninjected Retina
Z-stack of an uninjected retina from a Tg(GFAP:GFP)mi2001 transgenic zebrafish. Representative sections are shown with (A) showing a single PCNA+ cell in the ONL, most likely a rod precursor cell. Most sections had no detectable PCNA+ cells across all retinal layers (B). Green–GFAP:GFP; red–PCNA; blue–TOPRO-3. Scale bar, 50 μm. ONL–outer nuclear layer, INL–inner nuclear layer, GCL–ganglion cell layer.
Figure S4. Induction of proliferation by C6 EVs.
The various stages of EV purification from C6 cell media were tested for their ability to induce proliferation as determined by PCNA+ cell counts. The indicated fractions (see Fig. 4) or control PBS were injected into wild type zebrafish eyes. After 72 hours, retinas were dissected, sectioned, and immunostained with antibodies against PCNA. Each data point represents the average PCNA+ counts from 2–4 nonconsecutive retinal sections per eye, across the INL and ONL. Significance was calculated by one-way ANOVA, where *p-value=0.0257 and ***p-value=0.0001.
Figure S5. Dilution of EVs
C6 EVs were diluted to test whether the induction of PCNA+ cells was dose dependent.
Figure S6. Glutamine Synthase Co-Localization
Representative images of PBS control injected eyes (A), injection of EVs from IPS-derived cortical glutamatergic neurons at day 2–3 of differentiation (#16 from Fig. 2 and Table S1) (B), injection of EVs from iPS-derived cortical glutamatergic neurons at day 4–6 of differentiation (#30 from Fig. 2 and Table S1), (C) injection of EVs from CA30 mesencephalic dopaminergic neurons (#53 from Fig. 2 and Table S1) (D) and injection of EVs from C6 glioma cells (#59 from Fig. 2 and Table S1) (E) in AB wildtype zebrafish. Immunostaining was performed with antibodies against PCNA to label proliferating cells, co-stained with glutamine synthase (GS) which labels MG. Nuclei were stained with ToPRO-3. Scale bar, 50 μm.
Figure S7. Proliferating cells are derived from MG.
Tg(GFAP:GFP)mi2001 transgenic zebrafish retinas were injected with either PBS (A) or C6 P2 sEVs (B). After 72 hours, retinas were dissected, sectioned, and immunostained with antibodies against PCNA and GFP. Scale bar, 50 μm.
Table S1. Sources of EVs for in vivo screen.
The identity of the cell sources for the EVs tested in Fig. 2 are shown as well as Nanosight Tracking (NTA) data for each preparation. All preparations had vesicles from 40–100nm in diameter. PCNA+ cell counts for each preparation and statistics are as shown. One-way ANOVA with multiple comparisons (to PBS injection) was used to determine significance. *p-value <0.05, **p-value = 0.0034, ***p-value = 0.0008, ****p-value <0.0001.
Table S2. C6 P2 sEV Proteomics
P2 fractions after iodixanol gradient purification were subjected to mass spectrometry. Identified proteins and the number of spectral counts for each are as indicated. The far-right columns indicate the average number of spectral counts between cellular and sEV levels for each protein.
Table S3. Most Enriched sEV Proteins
Proteins that were the most enriched in sEVs from P2 pooled fractions are shown. Both the fold change and the average number of spectral counts are shown with the table ordered based on average spectral counts in sEVs.
Table S4A-C. Gene Ontology of P2 sEV Enriched Proteins
Gene ontology analysis was performed on proteomics data using WebGestalt (Liao et al., 2019). Only proteins enriched in P2 sEVs with a value greater than 2 were compared using Over-Representation Analysis (ORA) set to the genome protein-coding reference set. All groups were significant with an FDR < 0.05.
Table S5. RNA Sequencing
Total RNA from C6 cells and gradient purified sEVs was isolated, small RNAs purified, and RNA sequencing was performed using the Illumina platform. TIGER (https://github.com/shengqh/TIGER) was used to perform read mapping, miRNA quantification and differential analysis. Bowtie was used to map reads to the rat miRNAs in miRBase and the rat genome and DESeq2 was used to detect differential enrichment between exosome and cells. Raw data are shown from triplicate libraries with log2 fold changes and p values as shown. Adjusted p-values less than 0.01 were considered differentially expressed and were included in the volcano plot in Fig. 6.
Highlights.
Intravitreal injection of extracellular vesicles can induce proliferation
In vivo screen identified 12 sources of extracellular vesicles capable of inducing proliferation
Extracellular vesicles from C6 cells promote Müller glia-derived proliferation
Extracellular vesicles from C6 cells induce Ascl1a expression
Acknowledgements
We would like to thank the labs of Drs. Vivian Gama, Ethan Lippmann, Aaron Bowman, Diana Neely, Alissa Weaver, Ron Emeson, and John Penn from Vanderbilt and Dr. David Gamm from the University of Wisconsin, Madison for cell lines or culture media for EV isolation. The (Tg(1016tuba1a:GFP)) transgenic zebrafish line was provided by Dr. Daniel Goldman from the University of Michigan, and the Tg(GFAP:GFP)mi2001 line was provided by Dr. Pam Raymond from the University of Michigan. We would like to thank Dr. Bryan Mills in the Vanderbilt Cell Imaging Shared Resource for assistance with image analysis, and Qiang Guan for zebrafish maintenance and husbandry.
Funding
This work was supported by the National Institutes of Health [grant number: UO1EY027265].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest: None
References
- Ahmad I, Del Debbio CB, Das AV, Parameswaran S, 2011. Müller glia: a promising target for therapeutic regeneration. Investigative ophthalmology & visual science 52, 5758–5764. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond P.a., 2007. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA, 2006. GFAP transgenic zebrafish. Gene Expr Patterns 6, 1007–1013. [DOI] [PubMed] [Google Scholar]
- Bian B, Zhao C, He X, Gong Y, Ren C, Ge L, Zeng Y, Li Q, Chen M, Weng C, He J, Fang Y, Xu H, Yin ZQ, 2020. Exosomes derived from neural progenitor cells preserve photoreceptors during retinal degeneration by inactivating microglia. J Extracell Vesicles 9, 1748931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ, 2006. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol 172, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A, 2006. Müller cells in the healthy and diseased retina. Progress in retinal and eye research 25, 397–424. [DOI] [PubMed] [Google Scholar]
- Capozzi ME, McCollum GW, Penn JS, 2014. The role of cytochrome P450 epoxygenases in retinal angiogenesis. Invest Ophthalmol Vis Sci 55, 4253–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehajic-Kapetanovic J, Eleftheriou C, Allen AE, Milosavljevic N, Pienaar A, Bedford R, Davis KE, Bishop PN, Lucas RJ, 2015. Restoration of Vision with Ectopic Expression of Human Rod Opsin. Curr Biol 25, 2111–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Beckler MD, Weaver AM, Vickers K, Prasad N, Levy S, Zhang B, Coffey RJ, Patton JG, 2015. KRAS-dependent sorting of miRNA to exosomes. eLife 4, e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Thery C, 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Conedera FM, Pousa AMQ, Mercader N, Tschopp M, Enzmann V, 2019. Retinal microglia signaling affects Muller cell behavior in the zebrafish following laser injury induction. Glia 67, 1150–1166. [DOI] [PubMed] [Google Scholar]
- Ensslin MA, Shur BD, 2007. The EGF repeat and discoidin domain protein, SED1/MFG-E8, is required for mammary gland branching morphogenesis. Proc Natl Acad Sci U S A 104, 2715–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Paone A, Calore F, Galli R, Croce CM, 2013. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol 10, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber DB, Katsman D, 2016. Embryonic Stem Cell-Derived Microvesicles: Could They be Used for Retinal Regeneration? Adv Exp Med Biol 854, 563–569. [DOI] [PubMed] [Google Scholar]
- Fausett BV, Goldman D, 2006. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. Journal of Neuroscience 26, 6303–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Gumerson JD, Goldman D, 2008. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. Journal of Neuroscience 28, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Bosse JL, El-Hodiri HM, 2013. The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Experimental eye research 116, 199–204. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Scott MA, Ritchey ER, Sherwood P, 2009. Mitogen-activated protein kinase-signaling regulates the ability of Muller glia to proliferate and protect retinal neurons against excitotoxicity. Glia 57, 1538–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobben B, De Deyn PP, Slegers H, 2002. Rat C6 glioma as experimental model system for the study of glioblastoma growth and invasion. Cell Tissue Res 310, 257–270. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M, Sanchez-Madrid F, 2013. Transfer of extracellular vesicles during immune cell-cell interactions. Immunological reviews 251, 125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S, 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187. [DOI] [PubMed] [Google Scholar]
- Higginbotham James N., Demory Beckler M, Gephart Jonathan D., Franklin Jeffrey L., Bogatcheva G, Kremers G-J, Piston David W., Ayers Gregory D., McConnell Russell E., Tyska Matthew J., Coffey Robert J., 2011. Amphiregulin Exosomes Increase Cancer Cell Invasion. Current Biology 21, 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ, 2019. Reassessment of Exosome Composition. Cell 177, 428–445 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, Reh TA, 2017. Stimulation of functional neuronal regeneration from Muller glia in adult mice. Nature 548, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, 2016. The biology and function of exosomes in cancer. J Clin Invest 126, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl MO, Reh TA, 2010. Regenerative medicine for retinal diseases: activating endogenous repair mechanisms. Trends Mol Med 16, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsman D, Stackpole EJ, Domin DR, Farber DB, 2012. Embryonic stem cell-derived microvesicles induce gene expression changes in Muller cells of the retina. PloS one 7, e50417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C, 2016. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 113, E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenkowski JR, Raymond PA, 2014. Muller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res 40, 94–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wong DK, Hong KY, Raffai RL, 2018. Cushioned-Density Gradient Ultracentrifugation (C-DGUC): A Refined and High Performance Method for the Isolation, Characterization, and Use of Exosomes. Methods Mol Biol 1740, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B, 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47, W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SLN, Breakefield XO, Weaver AM, 2017. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends in Cell Biology 27, 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren RE, Pearson R.a., MacNeil a., Douglas RH, Salt TE, Akimoto M, Swaroop a., Sowden JC, Ali RR, 2006. Retinal repair by transplantation of photoreceptor precursors. Nature 444, 203–207. [DOI] [PubMed] [Google Scholar]
- Madamanchi A, Capozzi M, Geng L, Li Z, Friedman RD, Dickeson SK, Penn JS, Zutter MM, 2014. Mitigation of oxygen-induced retinopathy in alpha2beta1 integrin-deficient mice. Invest Ophthalmol Vis Sci 55, 4338–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B, 2018. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough IJ, Vincent JP, 2016. Exosomes in developmental signalling. Development 143, 2482–2493. [DOI] [PubMed] [Google Scholar]
- Mead B, Tomarev S, 2017. Bone Marrow-Derived Mesenchymal Stem Cells-Derived Exosomes Promote Survival of Retinal Ganglion Cells Through miRNA-Dependent Mechanisms. Stem Cells Transl Med 6, 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DM, Lovel AG, Stenkamp DL, 2018. Dynamic changes in microglial and macrophage characteristics during degeneration and regeneration of the zebrafish retina. J Neuroinflammation 15, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DM, Sun C, Hunter SS, New DD, Stenkamp DL, 2019. Regeneration associated transcriptional signature of retinal microglia and macrophages. Sci Rep 9, 4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, Vader P, 2019. Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp Mol Med 51, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC, 2007. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc Natl Acad Sci U S A 104, 12005–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely MD, Davison CA, Aschner M, Bowman AB, 2017. From the Cover: Manganese and Rotenone-Induced Oxidative Stress Signatures Differ in iPSC-Derived Human Dopamine Neurons. Toxicol Sci 159, 366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Gorsuch R.a., Bailey TJ, Ackerman KM, Kassen SC, Hyde DR, 2012. Stat3 defines three populations of Müller glia and is required for initiating maximal müller glia proliferation in the regenerating zebrafish retina. The Journal of comparative neurology 520, 4294–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Aoki N, Kato T, Kitajima K, Matsuda T, 2002. Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur J Biochem 269, 1209–1218. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Cirenza PF, Hitchcock PF, 2002. Persistent neurogenesis in the teleost retina: evidence for regulation by the growth-hormone/insulin-like growth factor-I axis. Mechanisms of Development 117, 137–149. [DOI] [PubMed] [Google Scholar]
- Patton JG, Franklin JL, Weaver AM, Vickers K, Zhang B, Coffey RJ, Ansel KM, Blelloch R, Goga A, Huang B, L’Etoille N, Raffai RL, Lai CP, Krichevsky AM, Mateescu B, Greiner VJ, Hunter C, Voinnet O, McManus MT, 2015. Biogenesis, delivery, and function of extracellular RNA. J Extracell Vesicles 4, 27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.a., Barber a.C., Rizzi M, Hippert C, Xue T, West EL, Duran Y, Smith a.J., Chuang JZ, Azam S.a., Luhmann UFO, Benucci a., Sung CH, Bainbridge JW, Carandini M, Yau K-W, Sowden JC, Ali RR, 2012. Restoration of vision after transplantation of photoreceptors. Nature 485, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Baulier E, Ke Y, Young A, Ahmedli NB, Schwartz SD, Farber DB, 2018. Human embryonic stem cells extracellular vesicles and their effects on immortalized human retinal Muller cells. PloS one 13, e0194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram K, Harding RL, Hyde DR, Patton JG, 2014. miR-203 regulates progenitor cell proliferation during adult zebrafish retina regeneration. Dev Biol 392, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D, 2010. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nature cell biology 12, 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao X-F, Goldman D, 2012. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nature cell biology 14, 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D, 2011. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A 108, 15858–15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MB, Didiano D, Patton JG, 2017. Neurotransmitter-Regulated Regeneration in the Zebrafish Retina. Stem cell reports 8, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stahl PD, 2019. Extracellular vesicles: a new communication paradigm? Nat Rev Mol Cell Biol 20, 509–510. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ, 2006. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Developmental Biology 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PD, Morelli AE, 2014. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 14, 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roska B, Sahel JA, 2018. Restoring vision. Nature 557, 359–367. [DOI] [PubMed] [Google Scholar]
- Shifrin DA Jr., Demory Beckler M, Coffey RJ, Tyska MJ, 2013. Extracellular vesicles: communication, coercion, and conditioning. Mol Biol Cell 24, 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Furuse M, Yokoyama N, Sasazuki T, 1993. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 260, 85–88. [DOI] [PubMed] [Google Scholar]
- Silva AK, Luciani N, Gazeau F, Aubertin K, Bonneau S, Chauvierre C, Letourneur D, Wilhelm C, 2015. Combining magnetic nanoparticles with cell derived microvesicles for drug loading and targeting. Nanomedicine 11, 645–655. [DOI] [PubMed] [Google Scholar]
- Simons M, Raposo G, 2009. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol 21, 575–581. [DOI] [PubMed] [Google Scholar]
- Skog J, Würdinger T, Rijn S.v., Meijer DH, Gainche L, W.T. C Jr, Carter BS, Krichevsky AM, Breakefield XO, 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology 10, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL, 2007. Neurogenesis in the fish retina. International Review of Cytology 259, 173–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JH, Tian Y, Funderburgh J, Pellegrini G, Zhang K, Goldberg JL, Ali RR, Young M, Xie Y, Temple S, 2018. Regenerating Eye Tissues to Preserve and Restore Vision. Cell stem cell 22, 834–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JD, Lekutis C, Singer KL, Bui A, Yuzuki D, Srinivasan U, Parry G, 1990. cDNA cloning of a mouse mammary epithelial cell surface protein reveals the existence of epidermal growth factor-like domains linked to factor VIII-like sequences. Proc Natl Acad Sci U S A 87, 8417–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MR, Couto JR, Scallan CD, Ceriani RL, Peterson JA, 1997. Lactadherin (formerly BA46), a membrane-associated glycoprotein expressed in human milk and breast carcinomas, promotes Arg-Gly-Asp (RGD)-dependent cell adhesion. DNA Cell Biol 16, 861–869. [DOI] [PubMed] [Google Scholar]
- Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr., Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr., Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, 2018. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR, 2008. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Developmental Neurobiology 68, 392–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, Thery C, 2016. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO, 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- van Niel G, D’Angelo G, Raposo G, 2018. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Veron P, Segura E, Sugano G, Amigorena S, Thery C, 2005. Accumulation of MFG-E8/lactadherin on exosomes from immature dendritic cells. Blood Cells Mol Dis 35, 81–88. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR, 2000. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol 44, 289–307. [DOI] [PubMed] [Google Scholar]
- Wan J, Goldman D, 2016. Retina regeneration in zebrafish. Current Opinion in Genetics and Development 40, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhao XF, Vojtek A, Goldman D, 2014. Retinal injury, growth factors, and cytokines converge on β-catenin and pStat3 signaling to stimulate retina regeneration. Cell Reports 9, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer SJ, Carvalho LS, Gyorgy B, Vandenberghe LH, Maguire CA, 2017. Exosome-associated AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection. Sci Rep 7, 45329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S, Moller A, 2019. Breast Cancer-Derived Exosomes Reflect the Cell-of-Origin Phenotype. Proteomics 19, e1800180. [DOI] [PubMed] [Google Scholar]
- Wiklander OPB, Brennan MA, Lotvall J, Breakefield XO, El Andaloussi S, 2019. Advances in therapeutic applications of extracellular vesicles. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms E, Cabanas C, Mager I, Wood MJA, Vader P, 2018. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol 9, 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohl SG, Hooper MJ, Reh TA, 2019. MicroRNAs miR-25, let-7 and miR-124 regulate the neurogenic potential of Muller glia in mice. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I, Dror S, Kenific CM, Lyden D, 2019. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell 49, 347–360. [DOI] [PubMed] [Google Scholar]
- Yao K, Qiu S, Tian L, Snider WD, Flannery JG, Schaffer DV, Chen B, 2016. Wnt Regulates Proliferation and Neurogenic Potential of Müller Glial Cells via a Lin28/let-7 miRNA-Dependent Pathway in Adult Mammalian Retinas. Cell Reports 17, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, Qiu S, Wang YV, Park SJH, Mohns EJ, Mehta B, Liu X, Chang B, Zenisek D, Crair MC, Demb JB, Chen B, 2018. Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 560, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Shao H, Su C, Jiang Y, Chen X, Bai L, Zhang Y, Li Q, Zhang X, Li X, 2016. Exosomes derived from MSCs ameliorate retinal laser injury partially by inhibition of MCP-1. Sci Rep 6, 34562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z, Xiao Q, Wang B, Wu W, Sun Y, Zhou Y, Tang C, Liu F, Wang L, Feng C, Liu M, Li S, Zhang Y, Xu H, Yao H, Shi L, Yang H, 2020. Glia-to-Neuron Conversion by CRISPR-CasRx Alleviates Symptoms of Neurological Disease in Mice. Cell 181, 590–603 e516. [DOI] [PubMed] [Google Scholar]
- Zhou J, Benito-Martin A, Mighty J, Chang L, Ghoroghi S, Wu H, Wong M, Guariglia S, Baranov P, Young M, Gharbaran R, Emerson M, Mark MT, Molina H, Canto-Soler MV, Selgas HP, Redenti S, 2018. Retinal progenitor cells release extracellular vesicles containing developmental transcription factors, microRNA and membrane proteins. Sci Rep 8, 2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Transfer of EVs between donor and recipient cells.
A) Schematic of Transwell assay. C6 cells were DiI-labeled, washed, and seeded as donor cells on top of a 0.4uM polyester Transwell membrane. Primary human Müller glia (HMGs; (Capozzi et al., 2014; Madamanchi et al., 2014)) were PKH67-labeled, washed, and seeded as recipient cells in the bottom of the Transwell culture dish. After 24 hours, cells were washed and co-cultured in serum-free media for 48 hours. B) Representative confocal images of donor and recipient cells. When C6 donor cells were co-cultured with HMG recipient cells, co-localization (yellow) was visualized, indicating uptake of C6-derived EVs by HMCs. C) Enlarged images of cells depicted in B. Scale bar, 50 μm.
Figure S2. TUNEL Staining to Detect Apoptosis after EV injection.
AB wild type zebrafish were injected with either PBS or C6 sEVs. After recovery for either 48 hours or 72 hours, TUNEL staining was performed. Red-PNCA; Blue–TOPRO-3. Scale bar, 50 μm. ONL–outer nuclear layer; INL–inner nuclear layer; GCL–ganglion cell layer. (A) PBS injection (B) C6 sEV injections analyzed 48 hours post-injection. (C) C6 sEVs injection analyzed at 72 hours post-injection. (D) Scatterplot of TUNEL staining analysis. Each data point is from a separate eye and is an average of 2–4 non-consecutive sections, counting all TUNEL+ cells across all retinal layers. One-way ANOVA with Tukey multiple comparison tests were used for quantitation. Error bars are mean ±SEM; n=8.
Figure S3. Uninjected Retina
Z-stack of an uninjected retina from a Tg(GFAP:GFP)mi2001 transgenic zebrafish. Representative sections are shown with (A) showing a single PCNA+ cell in the ONL, most likely a rod precursor cell. Most sections had no detectable PCNA+ cells across all retinal layers (B). Green–GFAP:GFP; red–PCNA; blue–TOPRO-3. Scale bar, 50 μm. ONL–outer nuclear layer, INL–inner nuclear layer, GCL–ganglion cell layer.
Figure S4. Induction of proliferation by C6 EVs.
The various stages of EV purification from C6 cell media were tested for their ability to induce proliferation as determined by PCNA+ cell counts. The indicated fractions (see Fig. 4) or control PBS were injected into wild type zebrafish eyes. After 72 hours, retinas were dissected, sectioned, and immunostained with antibodies against PCNA. Each data point represents the average PCNA+ counts from 2–4 nonconsecutive retinal sections per eye, across the INL and ONL. Significance was calculated by one-way ANOVA, where *p-value=0.0257 and ***p-value=0.0001.
Figure S5. Dilution of EVs
C6 EVs were diluted to test whether the induction of PCNA+ cells was dose dependent.
Figure S6. Glutamine Synthase Co-Localization
Representative images of PBS control injected eyes (A), injection of EVs from IPS-derived cortical glutamatergic neurons at day 2–3 of differentiation (#16 from Fig. 2 and Table S1) (B), injection of EVs from iPS-derived cortical glutamatergic neurons at day 4–6 of differentiation (#30 from Fig. 2 and Table S1), (C) injection of EVs from CA30 mesencephalic dopaminergic neurons (#53 from Fig. 2 and Table S1) (D) and injection of EVs from C6 glioma cells (#59 from Fig. 2 and Table S1) (E) in AB wildtype zebrafish. Immunostaining was performed with antibodies against PCNA to label proliferating cells, co-stained with glutamine synthase (GS) which labels MG. Nuclei were stained with ToPRO-3. Scale bar, 50 μm.
Figure S7. Proliferating cells are derived from MG.
Tg(GFAP:GFP)mi2001 transgenic zebrafish retinas were injected with either PBS (A) or C6 P2 sEVs (B). After 72 hours, retinas were dissected, sectioned, and immunostained with antibodies against PCNA and GFP. Scale bar, 50 μm.
Table S1. Sources of EVs for in vivo screen.
The identity of the cell sources for the EVs tested in Fig. 2 are shown as well as Nanosight Tracking (NTA) data for each preparation. All preparations had vesicles from 40–100nm in diameter. PCNA+ cell counts for each preparation and statistics are as shown. One-way ANOVA with multiple comparisons (to PBS injection) was used to determine significance. *p-value <0.05, **p-value = 0.0034, ***p-value = 0.0008, ****p-value <0.0001.
Table S2. C6 P2 sEV Proteomics
P2 fractions after iodixanol gradient purification were subjected to mass spectrometry. Identified proteins and the number of spectral counts for each are as indicated. The far-right columns indicate the average number of spectral counts between cellular and sEV levels for each protein.
Table S3. Most Enriched sEV Proteins
Proteins that were the most enriched in sEVs from P2 pooled fractions are shown. Both the fold change and the average number of spectral counts are shown with the table ordered based on average spectral counts in sEVs.
Table S4A-C. Gene Ontology of P2 sEV Enriched Proteins
Gene ontology analysis was performed on proteomics data using WebGestalt (Liao et al., 2019). Only proteins enriched in P2 sEVs with a value greater than 2 were compared using Over-Representation Analysis (ORA) set to the genome protein-coding reference set. All groups were significant with an FDR < 0.05.
Table S5. RNA Sequencing
Total RNA from C6 cells and gradient purified sEVs was isolated, small RNAs purified, and RNA sequencing was performed using the Illumina platform. TIGER (https://github.com/shengqh/TIGER) was used to perform read mapping, miRNA quantification and differential analysis. Bowtie was used to map reads to the rat miRNAs in miRBase and the rat genome and DESeq2 was used to detect differential enrichment between exosome and cells. Raw data are shown from triplicate libraries with log2 fold changes and p values as shown. Adjusted p-values less than 0.01 were considered differentially expressed and were included in the volcano plot in Fig. 6.


