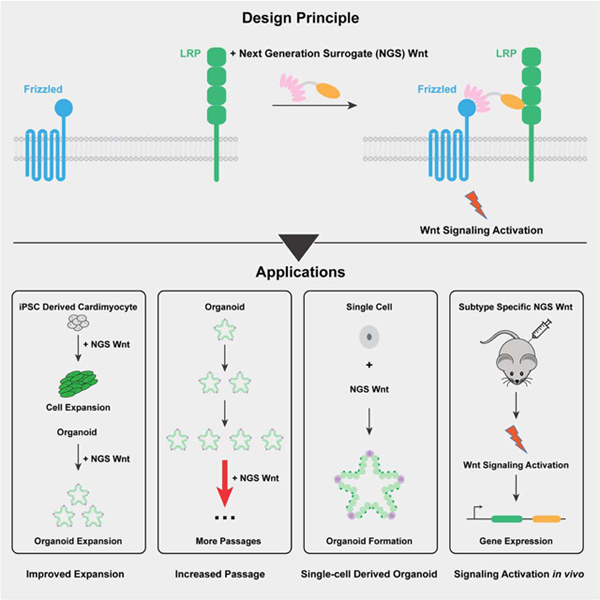
Summary
Modulation of Wnt signaling has untapped potential in regenerative medicine due to its essential functions in stem cell homeostasis. However, Wnt lipidation and Wnt-Frizzled (Fzd) cross-reactivity have hindered translational Wnt applications. Here, we designed and engineered water-soluble, Fzd-subtype specific ‘next generation surrogate’ (NGS) Wnts that hetero-dimerize Fzd and Lrp6. NGS Wnt supports long-term expansion of multiple different types of organoids, including kidney, colon, hepatocyte, ovarian and breast. NGS Wnts are superior to Wnt3a conditioned media in organoid expansion and single cell organoid outgrowth. Administration of Fzd-subtype specific NGS Wnt in vivo reveals that adult intestinal crypt proliferation can be promoted by agonism of Fzd5 and/or Fzd8 receptors while a broad spectrum of Fzd receptors can induce liver zonation. Thus, NGS Wnts offer a unified organoid expansion protocol and a laboratory “tool-kit” for dissecting the functions of Fzd subtypes in stem cell biology.
Keywords: Wnt, canonical Wnt signaling, surrogate Wnt, organoids, DARPin, Frizzled, stem cell, protein engineering, regenerative medicine
eTOC statement
Wnt is an essential cell growth factor used in organoid research. Garcia and colleagues developed next generation surrogate (NGS) Wnt that supports organoid expansion, single cell derived organoid outgrowth and long-term organoid maintenance. NGS Wnts also showed systemic Wnt activation when administrated in vivo.
Graphical Abstract
Introduction
The Wnt signaling pathway is essential for embryonic development, stem cell differentiation and regeneration of injured tissues (Barker and Clevers, 2006; Clevers and Nusse, 2012; Logan and Nusse, 2004; MacDonald et al., 2009; Niehrs, 2012). Wnts hetero-dimerize Frizzled (Fzd) family receptors and their co-receptors Lrp5/6 (Wehrli et al., 2000), triggering downstream signaling pathways including the canonical β-catenin cascade, and resulting in expression of genes regulating cell development (Clevers and Nusse, 2012; Logan and Nusse, 2004; Niehrs, 2012).
Due to its critical roles in development, Wnt has been widely used as an essential cell growth factor in organoids, a rapidly developing model system for tissue development (Clevers, 2016; Drost and Clevers, 2018; Sato et al., 2009). Currently, Wnt3a or the small molecule glycogen synthase kinase-3 (GSK3) inhibitor CHIR is commonly used as Wnt source. Recombinant Wnt3a is water insoluble and could not stably serve as a medium supplement (Janda and Garcia, 2015; Janda et al., 2012), thus, serum-stabilized Wnt3a conditioned media (CM) is a common substitute. However, the components of Wnt3a CM are not fully defined, introducing uncontrollable experimental variables. For example, the presence of serum in Wnt3a CM was reported to prevent long-term maintenance of human pancreatic organoids (Seino et al., 2018). On the other hand, CHIR targeted GSK3 regulates diverse cellular pathways other than Wnt signaling pathway (Jope and Johnson, 2004), leading to undesired off-target effects.
Another unsolved problem for Wnt signaling is the functional dissection of individual Fzd receptors. Despite the extensive literature characterizing the overall Wnt-Fzd signaling (Clevers, 2006; Nusse and Clevers, 2017), the linkage between specific Fzd receptors and their downstream responses remains elusive. Individual Wnt and Fzd have been shown to exhibit a high degree of sub-type cross-reactivity, rendering it technically challenging to isolate the effects of individual Fzd subtypes using natural Wnt molecules (Dijksterhuis et al., 2015; Voloshanenko et al., 2017). Moreover, recombinant Wnts are not water soluble and are thus difficult for in vitro mechanistic studies (Janda and Garcia, 2015; Janda et al., 2012; Willert et al., 2003). Considering the critical roles of Wnt in organoid culture and cell development, it is important to engineer water-soluble and Fzd-specific surrogate Wnt agonist.
Here, we report the development of Fzd-subtype specific NGS Wnts. Previously we reported bi-specific molecule scFv-DKK1c that dimerizes Fzd and Lrp and activates canonical Wnt signaling (Janda et al., 2017). These proof-of-concept molecules were only weakly potent and lacked Fzd-selectivity. Recently, two ‘tetrameric’ antibody-based Wnt agonists were reported confirming the ability of a Fzd/Lrp6 clustering agent to activate canonical Wnt signaling (Chen et al., 2020; Tao et al., 2019). However, in contrast to claims that tetrameric ligands are required for Wnt signal initiation, our previous bi-specific 1:1 surrogate Wnt scFv-DKK1c also displayed activity (Janda et al., 2017). This discrepancy requires clarification in order to make progress in generating therapeutic Wnt agonists.
In the current report, we have incorporated recently designed Fzd-subtype specific binders into a simple bi-specific format that results in extremely potent and selective agonism. Furthermore, their ease of production opens up access to the wider community as Wnt3a CM replacements. NGS Wnt potently supports different types of organoid growth, including colon, pancreas, ovarian, breast and hepatocyte. Notably, compared with Wnt3a CM, NGS Wnt supports better organoid proliferation, long-term organoid maintenance, and better single cell derived organoid outgrowth. Overall, NGS Wnt has the potential to be universally adopted as Wnt source for organoid expansion and maintenance. We also show that NGS Wnts are systemically active in vivo, and exhibit tissue selectivity in accord with the Fzd-subtype specificity of the NGS Wnt, allowing functional investigation of Fzd-specific agonism in regenerative medicine.
Results
Design of Fzd-subtype specific NGS Wnt
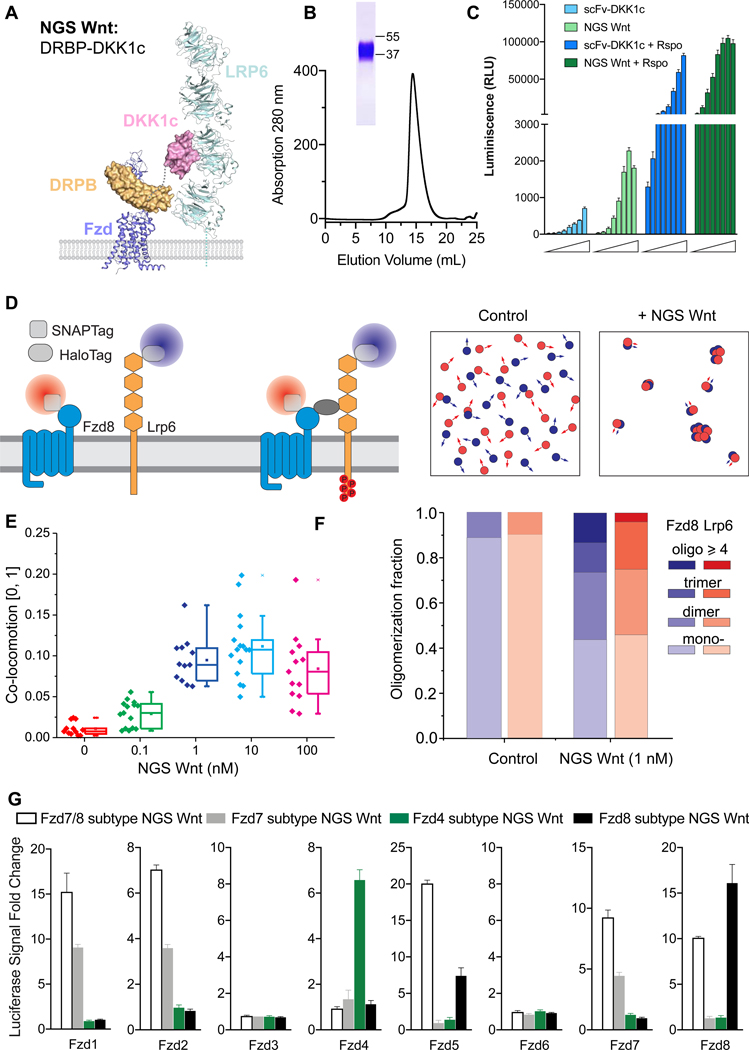
To design Fzd-subtype specific surrogate Wnts, we hypothesized that a bi-specific protein binding to Fzd receptor cystine rich domain (CRDs) and Lrp5/6 receptors simultaneously should mimic endogenous Wnt function, as previously shown for our “first generation surrogate Wnt scFv-DKK1c” (Janda et al., 2017). We chose Designed Repeat Protein Binder Fz7/8 (DRPB_Fz7/8) (Dang et al., 2019) as the Fzd binding module, due to its high affinity and broad binding spectrum for Fzd receptors, and fused with high affinity Lrp5/6 receptor binder DKK1c (Cheng et al., 2011) (Figure 1A). We purified this protein to homogeneity and denoted it as Fzd7/8 subtype NGS Wnt (activating Fzd 1, 2, 5, 7 and 8) (Figure 1B). We compared Fzd7/8 subtype NGS Wnt with the previously reported scFv-DKK1c using Wnt reporter assays in HEK 293STF cells (Janda et al., 2017). NGS Wnt induced stronger Wnt signaling at 5 nM concentration than 250 nM scFv-DKK1c (Figure 1C). In addition, NGS Wnt showed synergy with R-spondin (Rspo), a Wnt potentiator, and also exhibited higher Wnt response than scFv-DKK1c in the presence of Rspo (Figure 1C) (de Lau et al., 2011).
Figure 1. Design concept of NGS Wnt.
(A) Design schematic of NGS Wnt. Fzd subtype (purple) specific binder DRPB (orange) is fused with Lrp5/6 (cyan) binder DKK1c (pink) using a flexible Gly-Ser linker (grey dash line). Figure is adapted from PDBs (3S8V, 3S94, 6BD4 and 6NDZ).
(B) Size exclusion chromatography of Fzd7/8 subtype NGS Wnt. The band on SDS-PAGE gel represents Fzd7/8 subtype NGS Wnt.
(C) NGS Wnt (156 pM to 20 nM) induces higher β-catenin signaling change than scFv-DKK1c (2 nM to 250 nM) with or without Rspo (25 nM). Data represent mean and S.E., N = 3 technical replicates from representative experiment.
(D) Schematic for characterizing NGS Wnt induced heterodimer-/oligomerization ofFzd8 and Lrp6 by single molecule imaging in live cells.
(E) Co-locomotion Lrp6 and Fzd8 measured within 20 min after addition of NGS Wnt at indicated concentrations. N > 12 for each condition. Error bar represents mean and S.E.
(F) Quantification of Lrp6 and Fzd8 receptor oligomer fractions. Control refers tountreated samples. More than 2800 individual complex intensities were examined for each receptor.
(G) Fzd subtype NGS Wnts specifically induce β-catenin signaling in through targeted Fzd receptors. All data represents mean and S.E., N = 3 technical replicates from representative experiment.
See also Figures S1, S2 and Video S1 and S2.
To further measure Fzd7/8 subtype NGS Wnt induced Fzd-Lrp5/6 receptor clustering on live cell membranes, we probed the dynamics of Fzd8 and Lrp6 using dual-color single-molecule fluorescence imaging (Figure 1D) (Janda et al., 2017). Upon addition of 0.1 nM and 1 nM NGS Wnt we detected co-locomotion (i.e. synchronous diffusion) of Fzd8 and Lrp6 (Figures 1E and S1). In contrast, co-locomotion was only detected at 100 nM using scFv-DKK1c (Janda et al., 2017). Further quantification of Fzd8/Lrp6 stoichiometry on live cell membrane using 1 nM NGS Wnt showed a principal cluster of 1:1 heterodimer, with higher-order species of dimers and trimers also observed (Figure 1F). This result strongly suggests that tetrameric ligands are not the minimal multimerization unit necessary to activate Wnt signaling (Chen et al., 2020; Tao et al., 2019), which is consistent with our previous scFv-DKK1c studies (Janda et al., 2017). However, multimeric ligands could certainly potentiate signaling through enhanced clustering. An additional consideration to understand the apparent discrepancy between our 1:1 NGS Wnt and the tetrameric ligands, is that the structural epitope on Fzd-CRD, where the surrogate ligand binds, is a very important determinant of signaling. Unfavorable binding epitopes in the context of a 1:1 ligand could likely be overcome by presentation of a 2:2 ligand. In the case of NGS Wnt, the DRBP binding epitope seems to be highly favorable (Dang et al., 2019), far superior to the original scFv-DKK1c in signaling potency, despite both being linked to DKK1c and similar affinity of scFv and DRPB to Fzd-CRDs (Dang et al., 2019; Janda et al., 2017).
Next, we sought to design NGS Wnts that are specific to each of the three major Fzd subtypes. Using DRPBs specific to Fzd subtypes (Dang et al., 2019), we developed three additional NGS Wnts, namely, Fzd4 subtype NGS Wnt (activating Fzd4 receptor), Fzd7 subtype NGS Wnt (activating Fzd1, 2 and 7 receptors) and Fzd8 subtype NGS Wnt (activating Fzd5 and 8 receptors). To test their selectivity to individual Fzd, we generated Fzd specific cell lines by expressing individual Fzd receptor in a “pan-Fzd receptor knockout” HEK 293T cell line (Voloshanenko et al., 2017). We subsequently treated these cell lines with Fzd subtype NGS Wnts. As expected, Fzd subtype NGS Wnts induced β-catenin signaling in a Fzd receptor subtype dependent way. For example, in cells expressing Fzd4 receptor, only Fzd4 subtype NGS Wnt induced β-catenin signaling while none of the other NGS Wnts showed any signaling. In Fzd2 subtype specific cells, only Fzd7 subtype NGS Wnt and Fzd7/8 subtype NGS Wnt demonstrated activation of Wnt signaling (Figures 1G and S2).
NGS Wnt expands human induced pluripotent stem cell (hiPSC)-derived cardiomyocyte (CM)
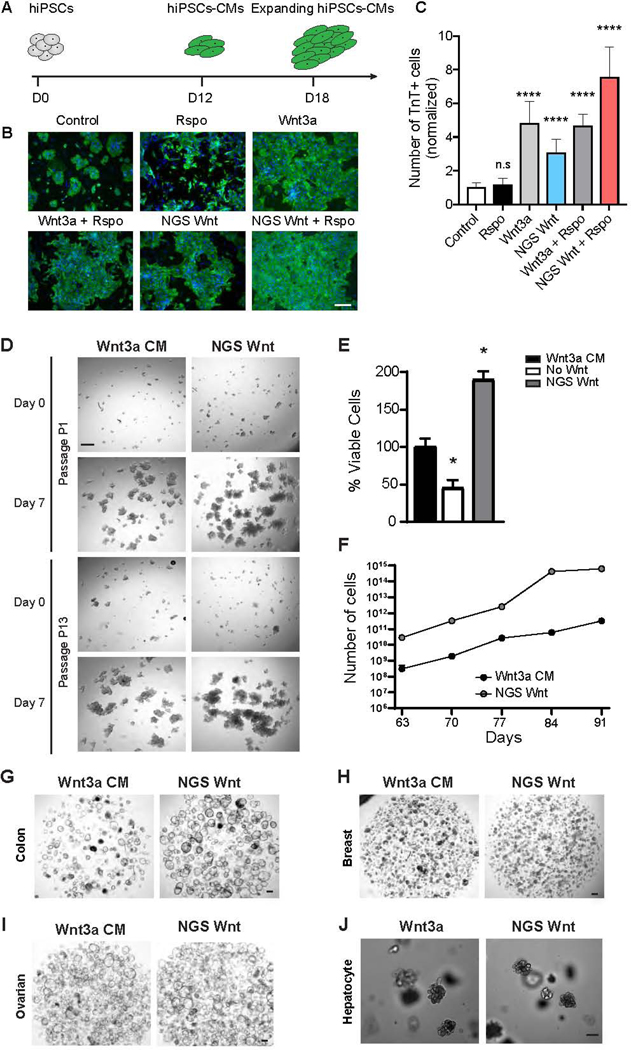
Wnt signaling plays pivotal roles in CM expansion during development to ensure growth of myocardial mass (Buikema et al., 2020; Buikema et al., 2013; Qyang et al., 2007).We applied broad spectrum Fzd7/8 subtype NGS Wnt to hiPSC-CMs and monitored their expansion (Figure 2A). Fzd7/8 subtype NGS Wnt alone resulted in a 3.1-fold increase (normalized to media control) in hiPSC-CM expansion, shown by troponin T positive cell number (Figures 2B and 2C). Moreover, treatment with NGS Wnt and Rspo together exhibited synergistic effects on hiPSC-CM expansion, whereas Rspo treatment alone showed no increase in hiPSC-CM number (Figure 2C). These data demonstrate that NGS Wnt recapitulates natural Wnt in expansion of hiPSC-CMs.
Figure 2. NGS Wnt promotes hiPSC-CM expansion and supports proliferation of multiple organoids.
(A) Experimental diagram of hiPSC-CM expansion. At day 12 (D12), hiPSC-derived CMs were treated with Wnt activators and expansion was monitored at day 18 (D18).
(B) Representative immunofluorescence images for cardiac troponin T (TnT) (green) and DAPI (blue) in hiPSC-CMs treated with indicated Wnt activators or control. Scale bar represents 100 μm.
(C) Quantification of the TnT positive cell number with the indicated Wnt activators orcontrol. **** indicates P<0.0001 by unpaired t-test. Data were from 3 independent experiments and 2 replicates. Results are expressed as mean and S.D..
(D) Representative brightfield images of human cystic fibrosis colon organoid cultures in either Wnt3a CM or NGS Wnt. N=5. Scale bar represents 100 μm.
(E) Quantification of viable cells (as percentage) of human cystic fibrosis colon organoids after first passage (7 days) cultured in Wnt3a CM, no Wnt source or NGS Wnt. * indicates P<0.05.
(F) Growth curves of human cystic fibrosis colon organoids were analyzed from passage P9 to P13. Cell number counts are expressed as mean and S.D. of three independent assays. * indicates P<0.05.
(G-J) Representative brightfield images of human colon, ovarian, breast and mouse hepatocyte organoids expanded with Wnt3a CM/Wnt3a or NGS Wnt at 0.5 nM. N=10 for colon and N=3 for other organoid types. Scale bar represents 100 μm.
See also Figures S3 and S4
NGS Wnt supports organoid growth
Wnt is an essential factor for the growth and maintenance of organoids. To test the feasibility of NGS Wnt in organoid expansion, we substituted Wnt3a CM with Fzd7/8 subtype NGS Wnt, which signals through a broader spectrum of Fzd receptors, in mouse colon organoid media. Indeed, Fzd7/8 subtype NGS Wnt supports colon organoid growth even at 0.1 nM to 0.5 nM concentration while previous scFv-DKK1c supports colon organoids at 180 nM (Figures S1G and S1H). We further reasoned Fc conjugation could improve NGS Wnt stability at 37 °C culturing condition, and also increase the potency through avidity. We compared NGS Wnt and NGS Wnt-Fc to Wnt3a, Wnt3aCM and scFv-DKK1c using Wnt reporter assays (Figures S3A-S3C). Fc tagged NGS Wnt showed slightly better Wnt response (Emax) (Figure S3B) but demonstrated significantly improved EC50 (Figure S3C). Therefore, Fc-tagged Fzd7/8 subtype NGS Wnt is used for all subsequent organoid experiments and simply denoted as NGS Wnt. To further compare NGS Wnt to recombinant Wnt3a, we noted improved efficacy of NGS Wnt in cultured P26N human colon organoids (Figures S3D, S3E and S3F), highlighting the superiority of NGS Wnt.
We further tested NGS Wnt in human colon organoids from both cystic fibrosis (Dekkers et al., 2013) (Figure 2D) patients and healthy donors (Figures 2G and S3G), and compared its efficacy with currently used Wnt3a CM. Both NGS Wnt and Wnt3a CM supported long-term expansion of cystic fibrosis and healthy colon organoids after multiple passages. Remarkably, NGS Wnt showed improved better expansion rates and in vitro growth than Wnt3a CM at both the first passage (Figures 2E and S3H) and late passages from P9 to P13 (Figures 2F and S3I). NGS Wnt also retained potency after long-term storage (Figures S4A-S4D). Moreover, colon organoids expanded with traditional Wnt3a CM showed signs of differentiation (small, dark and compact organoids). In contrast, NGS Wnt-supported colon organoids are more proliferative (cystic organoids with thin walls) (Figure 2G).
Subsequently, we compared NGS Wnt with Wnt3a CM/Wnt3a in multiple types of organoid models. We applied NGS Wnt to ovarian, breast and hepatocyte organoids (Hu et al., 2018; Kopper et al., 2019; Peng et al., 2018; Sachs et al., 2018). NGS Wnt is sufficient to expand all these organoid models at 0.5 nM for multiple passages (Figures 2H, 2I and 2J). To confirm NGS that Wnt can be used universally, we also successfully cultured human colon organoids from different donors and at different laboratories (Figure S4E). In addition, NGS Wnt supported colon organoids retained proper cell polarity and can be redirected to a differentiated state by withdrawal of growth factors (Figures S4F-I).
Next, we examined whether NGS Wnt could prolong organoid life span. Using previously established culture protocol, human kidney tubular epithelial organoids (tubuloids) can only can be rapidly expanded for up to approximately 10 passages after which they significantly slowdown in growth rate (Schutgens et al., 2019). Addition of NGS Wnt to the culture medium did not significantly change tubuloid expansion up to about passage 10. Remarkably, tubuloids grown with NGS Wnt could still be passaged beyond passage 10 at similar frequency and ratios (Figures 3A and S4J). Furthermore, histological characterization did not reveal any phenotypic difference between tubuloids cultured with or without NGS Wnt (Figure S4K). In summary, these data suggest that NGS Wnt supports a wide variety of organoid models and outperforms previously described Wnt3a CM.
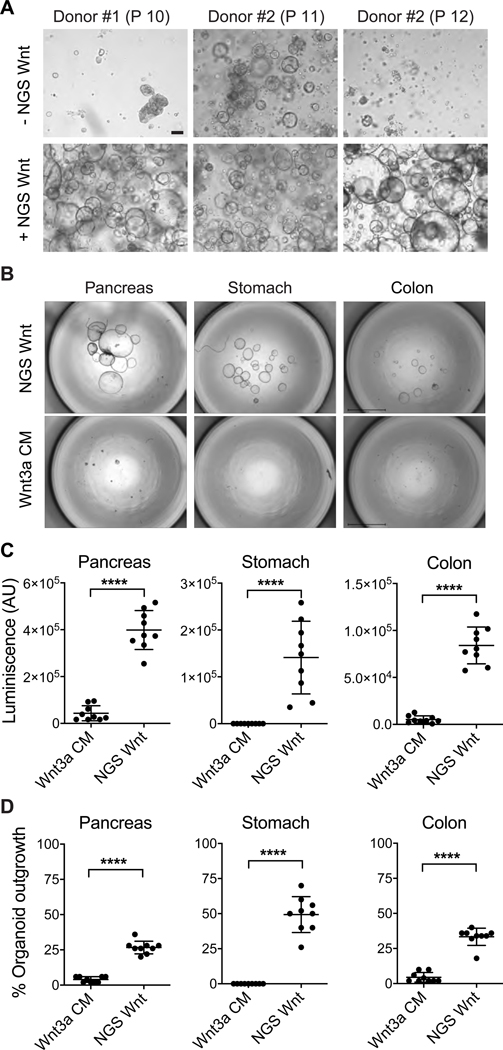
Figure 3. NGS Wnt increases human tubuloid life span and improves organoid outgrowth efficiency from single cells.
(A) Representative brightfield images of human tubuloids without/with NGS Wnt at indicated passage number. Scale bar represents 100 μm.
(B) Representative images of human pancreas, stomach and colon organoids at 2 weeks after seeding 50 single cells, cultured with either Wnt3a CM or NGS Wnt. N=1 for individual organoid type. Scale bars represent 2 mm.
(C-D) Quantification of organoid outgrowth by CellTiter-Glo assay (C) and organoid counting (D). Mean and S.E. for 9 technical replicates are displayed. **** indicates P<0.0001.
See also Figure S4.
NGS Wnt supports organoid outgrowth from single cells
The success of NGS Wnt in multiple organoids prompted us to test whether it is capable of supporting single cell derived organoid outgrowth. CRISPR-Cas9-mediated genome engineering of healthy organoids to recapitulate tumor progression requires single cell derived organoid outgrowth (Cong et al., 2013; Drost et al., 2017; Drost et al., 2015; Matano et al., 2015). However, the success rate of single cell organoid expansion is extremely low using previously reported Wnt3a CM. We therefore chose pancreas, stomach and colon organoids, and compared NGS Wnt with Wnt3a CM (Bartfeld et al., 2015; Boj et al., 2015; Sato et al., 2011). Notably, Wnt3a CM failed to support stomach organoid outgrowth from single cell while NGS Wnt demonstrated tremendous improvement (Figures 3B-D). We also detected significantly improved organoid expansion by NGS Wnt in colon and pancreas organoids (Figures 3B-D). Overall, compared to Wnt3a CM, NGS Wnt augmented single cell derived organoid outgrowth in all three chosen organoid types and potentially other untested organoid models. Such improvements in organoid expansion protocol could be extremely useful for high-throughput experiments, such as establishing a patient organoid Biobank (van de Wetering et al., 2015) and CRISPR-Cas9-mediated genomic editing (Drost et al., 2015).
NGS Wnt deconvolutes Fzd receptor pleiotropy during intestine and liver homeostasis
To extend upon our studies examining NGS Wnts in vitro and ex vivo, we then tested if our NGS Wnts could activate Wnt signaling in vivo and whether the NGS Wnts could be employed to dissect Fzd subtype functions during organ homeostasis. The NGS Wnts were thus evaluated in mice to survey effects on adult intestinal stem cell expansion and liver zonation, both representing archetypal Wnt-responsive processes (Clevers, 2006; Nusse and Clevers, 2017). We generated adenoviruses for in vivo delivery of NGS Wnts as described (Dang et al., 2019; Janda et al., 2017; Kuhnert et al., 2004; Yan et al., 2017).
We previously established that overexpression of scFv-DKK1c alone does not significantly perturb homeostasis of intestine or liver, but their combination with Rspo dramatically enhances intestinal proliferation and liver pericentral gene expression (Janda et al., 2017; Yan et al., 2017). Therefore, mice were infected with adenoviruses expressing different Fzd subtype NGS Wnts or mouse IgG2a Fc (Ad-Fc) as negative control with or without Ad-Rspo2. As expected (Yan et al., 2017), adenoviruses expressing NGS Wnts alone did not induce homeostatic intestinal crypt proliferation (Figure 4A). In contrast, when combined with Ad-Rspo2, adenoviruses expressing Fzd8 subtype NGS Wnt, but not Fzd4 or Fzd7 subtype NGS Wnt, induced crypt hyperplasia (Figure 4B), villus hyperproliferation (Figure 4C), marked expansion of mitotic Ki67+ epithelium (Figures 4A and 4E) and Wnt target CD44 (Figures 4D and 4F) staining from the crypt extending towards the peri-villus tip region. Notably, increasing doses of Ad-Fzd8 subtype NGS Wnt beyond 104 pfu and Ad-Fzd7/8 subtype NGS Wnt beyond 106 pfu produced decreasing synergy with Ad-Rspo2, potentially paralleling the in vitro TOP-Flash data (Figure S2). Overall, these data inferred a selective intestinal susceptibility to Fzd5/8 agonism versus other Fzd subtypes.
Figure 4. NGS Wnts reveal functionally relevant Fzd receptor profiles in adult intestinal crypt and liver homeostasis.
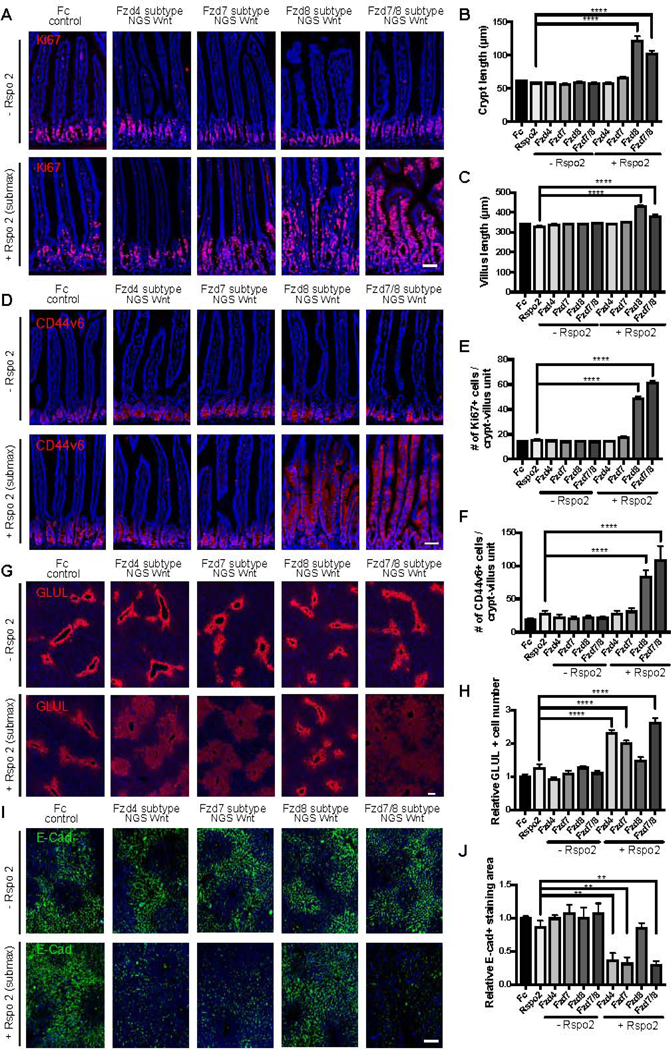
(A-F) Analysis of representative jejunum cross-sectional immunofluorescence images for (A) Ki67 (red) and (D) CD44v6 (red) following adenovirus administration. DAPI staining is represented in blue. Crypt length (B), villus length (C), Ki67+ cells/crypt-villus unit (E), CD44v6+ cells/crypt-villus unit (F) were measured in each condition shown in (A) and (D). Scale bars represent 50 μm.
(G-J) Analysis of representative liver cross-sectional immunofluorescence images for GLUL (G) and E-Cadherin (E-Cad) (I) following adenovirus administration. (H) The number of GLUL positive hepatocytes around the central vein was measured in each condition shown in (G). (J) The relative E-Cadherin+ cells within a fixed area was measured in each condition shown in (I). Scale bars represent 100 μm. Data are expressed as mean ± S.E. N=4 animals per group and experiments were repeated twice. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparison test(**P< 0.01; ****P< 0.0001).
In the liver, Wnts are expressed in the central vein and activate pericentral gene expression such as Glutamine synthetase (GLUL) and Axin2 in adjacent hepatocytes (Wang et al., 2015). Conversely, Wnt pathway activation represses periportal genes such as Arginase 1 (Arg1) (Benhamouche et al., 2006) and E-Cadherin (Cdh1) (Rocha et al., 2015). Thus,the Wnt/β-catenin signaling pathway is thus a major regulator of metabolic liver zonation (Benhamouche et al., 2006). We found that single treatments with Ad-NGS Wnts alone (106 pfu) or Ad-Rspo2 alone did not significantly perturb liver GLUL expression (Figure 4G and 4H). However, combinational infection of Ad-Fzd4, Ad-Fzd7 or Ad-Fzd7/8 subtype NGS Wnts with Ad-Rspo2 induced a synergistic expansion of the GLUL-expressing pericentral region compared to control mice while inhibiting staining for the Wnt-repressed periportal marker E-Cadherin (Figures 4G-J). This demonstrates clear reciprocal changes of augmented pericentral and diminished periportal markers induced by these NGSs Wnts, while the Fzd subtype specificity of the active NGS Wnts strongly parallels our prior defined activity spectrum of DRPB-based Fzd subtype antagonists (Dang et al., 2019). Decreasing synergy with Ad-Rspo2 was observed with Ad-NGS Wnt doses higher than 106 pfu. In marked contrast, Ad-Fzd8 subtype NGS Wnt + Ad-Rspo2 did not affect liver GLUL expression over Ad-Fzd8 subtype NGS Wnt doses from 104-109 pfu (Figure 4G) despite this treatment markedly augmenting intestinal proliferation (Figure 4A-F). In summary, NGS Wnts deconvoluted Fzd subtype receptor pleiotropy in intestine and liver homeostasis where these two Wnt-responsive organs exhibited markedly distinct Fzd receptor agonist response profiles.
Discussion
We report here the development of NGS Wnt, which is water-soluble and highly potent agonist and can serve as an easily produced resource to the community as a Wnt replacement in a broad array of experimental situations. NGS Wnt has a broad application to support the growth and maintenance of various organoids and single cell derived organoid outgrowth. More importantly, the defined composition of NGS Wnt would also minimize experimental variations and improve result consistency. Overall, NGS Wnt significantly improves current protocols in organoid research, and potentially could replace Wnt3a CM to unify organoid protocols.
We also developed Fzd-subtype specific NGS Wnts to investigate individual Fzd subtype receptor’s function in vivo. Adenovirally-expressed Fzd8-, but not Fzd4- or Fzd7-, subtype NGS Wnt synergized with Ad-Rspo2 to induce intestinal Ki67+ crypt proliferation and expression of the Wnt target gene CD44 in the intestine. On the contrary, Fzd4- and Fzd7-subtype NGS Wnt but not the Fzd8-subtype NGS Wnt synergized with Ad-Rspo2 to upregulate pericentral zonation while repressing periportal zonation. Both of these intestinal and liver phenotypes from Fzd subtype-selective NGS Wnts fully phenocopy the synergistic effect between Rspo2 and the first generation surrogate Wnt agonist scFv-DKK1c that targets a broad range of multiple Fzds (Janda et al., 2017; Yan et al., 2017). Thus, NGS Wnts identify distinct Fzd subtype profiles that are functionally relevant and sufficient to mediate archetypal Wnt responses in different organs such as the intestine and liver.
There are marked parallels between the gain-of-function phenotypes of the Fzd-subtype specific NGS Wnts described here, and our prior loss-of-function analysis of Fzd-subtype antagonists (Dang et al., 2019). Notably, the proliferative intestine gain-of-function phenotype of the Fzd8-subtype NGS Wnt but not Fzd4- or Fzd7-subtype NGS Wnts strongly correlates with our recent loss-of-function data showing that the antagonists DRPB_Fz8, but not DRPB_Fz4 or Fz7, produce intestinal crypt and villus loss in mice (Dang et al., 2019). This concurrence strongly argues for an essential role of Fzd5 and/or Fzd8 during intestinal crypt homeostasis. On the other hand, we previously described that Fzd4-, Fzd7- and Fzd8 subtype antagonists each individually reduced liver GLUL expression, suggesting that a broad spectrum of Fzd receptors combinatorically maintain hepatic zonation (Dang et al., 2019). Here, the ability of both Fzd4- and Fzd7-subtype NGS Wnts to upregulate GLUL and pericentral zonation correlates with our previous description of reciprocal GLUL suppression by the inhibitory DRPB_Fz4 or Fz7 antagonists (Dang et al., 2019). The inability of the Fzd8-subtype NGS Wnt to induce pericentral GLUL expression despite extensive adenovirus dose titration does contrast with GLUL repression by the cognate Fzd8 subtype antagonist, conceivably resulting from saturating amounts of Wnts binding to Fzd5 and/or Fzd8 already present nearby central veins in vivo.
Taken together, our results suggest that NGS Wnts represent a pharmacologic approach to attribute specific Fzd subtype receptors with biological functions in vivo. Combined with our previously engineered Fzd subtype specific Wnt antagonists (Dang et al., 2019), these molecules may serve as probes to bi-directionally regulate Wnt signaling through selective targeting of Fzd subtype receptors. Such a “tool-kit” provides an orthogonal approach to the in vivo interrogation of Fzd receptor function and should deepen the current understanding of Wnt signaling in physiology and disease.
Limitation of study
While our experiments show that NGS Wnt has robust activity in supporting organoid cultures, and Fzd-subtype specific systemic activity, our in vivo results fall short of demonstrating therapeutic effects in organ repair, evaluation of possible toxicities, and effects on intestinal stem cells. Our earlier studies show that exogenous administration of surrogate Wnts as monotherapy, in a background of endogenous Wnt, do not affect Lgr5+ intestinal stem cells. Rather, surrogate Wnts predominantly act on intestinal transit-amplifying cells and only when given as combination treatment with R-spondins (Yan et al., 2017). These areas remain under investigation for these NGS Wnts. Furthermore, while the NGS Wnt design is a bi-specific molecule that, by TIRF imaging, forms a majority of 1:1 Fzd8/Lrp6 complexes on cells, it is possible that activated Wnt/Fzd/Lrp complexes form higher order clusters through a variety of mechanisms not investigated here. Thus, recent tetrameric antibody-based Wnt agonists that show highly potent Wnt activity, may do so through enhanced clustering of 1:1 complexes, amplifying the signal. Finally, the full extent to which NGS Wnt recapitulates the exact signaling and genetic programs of endogenous Wnts is not definitively shown in our work, and will require careful gene expression analysis.
STAR METHOD TEXT
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, K. Christopher Garcia (kcgarcia@stanford.edu).
Materials Availability
Plasmids generated in this study will be deposited to Addgene. Academic labs will have access for non-profit research with Material Transfer Agreement. Fc tagged NGS Wnt recombinant protein for organoid research will be commercially available through U-Protein Express BV.
Data and Code Availability
This study did not generate/analyze datasets/code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tissues
All experimentation using human tissues described herein in Figures 2, 3, S3 and S4A-B was approved by the ethical committee at University Medical Center Utrecht (UMCU; TcBio #14–008, #12–093 and 14–472). Informed consent for tissue collection, generation, storage, and use of the organoids was obtained from the patients at Wilhelmina Children’s Hospital (WKZ) and University Medical Center Utrecht (UMCU).
The biopsy samples for human colon organoid research described in Figure S4E were acquiredin the context of clinical trials (Hatano et al., 2013b; Hunt et al., 2013; Somsouk et al., 2014) and observational studies for a large number of laboratory-based collaborators (Hatano et al., 2013a; Josefsson et al., 2013; Leeansyah et al., 2013; Yukl et al., 2014) by Drs. Deeks and Somsouk (UCSF) that have been working together to recruit and perform more than 1,000 research colonoscopies and sigmoidoscopies on over 500 HIV-infected and uninfected individuals.
Tissues for human tubuloids data described here in Figures 3A were derived under approval of the medical ethical committee of the Erasmus Medical Center (Rotterdam, the Netherlands). The parents of all patients participating in the biobank study signed informed consent forms approved by the responsible authority. All organoid culture details are described in the METHOD DETAILS section.
Vertebrate animals
All animal experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee (IACUC) at Stanford University and University of California, San Francisco.
Cell lines
Expi293F and HEK293F cells were grown in FreeStyle™ 293 Expression Medium and Expi293™ Expression Medium at 37°C, respectively, with humidified atmosphere of 8% CO2 on a shaker at 130 rpm. HEK293STF, HEK293 FzdKO and L cell, were maintained at 37 °C in a 5% CO2 environment using Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS.
METHOD DETAILS
NGS Wnt expression
NGS Wnts with a C-terminal 6 X His-tag were cloned into the lentiviral vectors RKS or pHR vector as reported (Bandaranayake et al., 2011). The detailed amino acid sequence for each NGS Wnt is shown below. RKS vector is courtesy of David J. Rawlings laboratory and pHR vector is courtesy of Ronald Vale laboratory. Lentivirus were packaged as reported (Bandaranayake et al., 2011). HEK293F cells were stably transduced with lentivirus and protein expression were induced by adding 10 mM sodium butyrate at a cell density of 2 × 106/ mL. All NGS Wnts were purified by Ni2+-Nitrolotriacetic acid (NTA) affinity column chromatography and eluted using 20 mM HEPES, pH 7.2, 300 mM NaCl and 300 mM imidazole. Proteins were further purified using size exclusion chromatography using a Superdex S200 column (GE Healthcare) in HBS (20 mM HEPES, pH 7.2, 300 mM NaCl) buffer. The proteins were concentrated using MilliporeSigma™ Amicon™ Ultra-15 Centrifugal Filter Units, snap frozen in liquid nitrogen with addition of 10% glycerol and stored at −80 °C for future usage.
Amino acid sequence of Fzd7/8 subtype NGS Wnt:
SELGTRLIRAALDGNKDRVKDLIENGADVNASLMSGATPLHAAAMNGHKEVVKLLISKGADVNAQSVAGSTPLDAAAFSGHKEVVKLLISKGADVNAVNAAGLTPLHAAADNGHKEV VKLLISKGADVNAKADHGMTPLHFAAQRGHKEVVKLLISKGADLNTSAKDGATPLDMARESGNEEVVKLLEKQLEGSGSGGSGSGKMYHTKGQEGSVCLRSSDCASGLCCARHF WSKICKPVLKEGQVCTKHRRKGSHGLEIFQRCYCGEGLSCRIQKDHHQASNSSRLHTCQRH
Amino acid sequence of Fzd7 subtype NGS Wnt:
SELGTRLIRAALDGNKDRVKDLIENGADVNASLMSGATPLHAAAMNGHKEVVKLLISKGADVNAQSVAGSTPLDAAAFSGHKEVVKLLISKGADVNAVNAAGLTPLHDAADDGHNEV VKLLISKGADVNAKADHGMTPLHFAAQRGHKEVVKLLISKGADLNTSAKDGATPLDMARESGNEEVVKLLEKQLEGSGSGGSGSGKMYHTKGQEGSVCLRSSDCASGLCCARHF WSKICKPVLKEGQVCTKHRRKGSHGLEIFQRCYCGEGLSCRIQKDHHQASNSSRLHTCQRH
Amino acid sequence of Fzd8 subtype NGS Wnt:
SELGKRLIMAALDGNKDRVKDLIENGADVNASLVSGATPLHAAAMNGHKEVVKLLISKGADVNAQSAAGSTPLAAAAINGHKEVVKLLISKGADVNAVTAAGMTPLHAAAANGHKEV VKLLISKGADVNAKADRGMTPLHFAAWRGHKEVVKLLISKGADLNTSAKDGATPLDMARESGNEEVVKLLEKQLEGSGSGGSGSGKMYHTKGQEGSVCLRSSDCASGLCCARHF WSKICKPVLKEGQVCTKHRRKGSHGLEIFQRCYCGEGLSCRIQKDHHQASNSSRLHTCQRH
Amino acid sequence of Fzd4 subtype NGS Wnt:
KMYHTKGQEGSVCLRSSDCASGLCCARHFWSKICKPVLKEGQVCTKHRRKGSHGLEIFQRCYCGEGLSCRIQKDHHQASNSSRLHTCQRHGSGSGGSGSGSELGKRLIRAALD GNKDRVKDLIENGADVNASLMSGTTPLYAAAMNGHKEVVKLLISKGADVNAQSVAGSTPLVAAANFGHNEVVKLLISKGADVNAVTAFGVTPLHAAAADGHKEVVKLLISKGADVNA KAGRGMTPLHIAAFRGHKEVVKLLISKGADLNTSAKDGATPLDMARESGNEEVVKLLEKQLE
Subsequently, for initial organoid growth experiments, only the Fzd7/8 subtype NGS Wnt is used given the broader spectrum of activities on Fzd receptors (Fzd 1, 2, 5, 7 and 8) compared to other Fzd subtype NGS Wnts. NGS Wnt used for subsequent organoid experiments were expressed with Fc tag (except Figure S1G). NGS Wnt is transiently expressed in Expi293F™ cells following manufacturer’s protocol (Thermo Fisher Scientific ExpiFectamine™ 293 Transfection Kit). Briefly, 300 mL of Expi293F™ cells were grown to 3 × 106 cells/mL. DNA was transiently transfected using Gibco™ ExpiFectamine™ 293 Reagent. Enhancers were added 18 hrs post transfection and cells were incubated for another 72 hrs before harvesting. Protein was purified as described above.
NGS Wnt used in the human colon, ovarian, breast, pancreas and stomach organoid experiments was expressed and purified by U-Protein Express. The NGS Wnt construct was codon optimized, cloned into pUPE expression vector (U-Protein Express BV) with a C-terminal Fc tag. NGS Wnt was transiently expressed in HEK293E (U-Protein Express BV) cells. Six days after transfection, the media were harvested by centrifugation and followed by MabSelect Sure LX Sepharose purification (GE Healthcare Catalog # 17547401). The bound recombinant protein was eluted using 20 mM citrate, 150 mM NaCl, pH 3.0, and 5 ml fractions were collected in 12 ml tubes containing 1 ml 1 M K2HPO4/KH2PO4 pH 8.0 buffer for neutralisation to pH 7. The NGS Wnt was subsequently purified by size exclusion chromatography using Superdex200 26/600 column (GE Healthcare) in 25 mM Tris, pH 8.2 and 500 mM NaCl. The fractions were pooled and sterile filtered before used in organoid growth experiments. The full amino acid sequence of Fzd7/8 subtype NGS Wnt (with Fc tag) used in organoid research is shown below.
Amino acid sequence of Fzd7/8 subtype NGS Wnt with Fc:
GSSELGTRLIRAALDGNKDRVKDLIENGADVNASLMSGATPLHAAAMNGHKEVVKLLISKGADVNAQSVAGSTPLDAAAFSGHKEVVKLLISKGADVNAVNAAGLTPLHAAADNGHK EVVKLLISKGADVNAKADHGMTPLHFAAQRGHKEVVKLLISKGADLNTSAKDGATPLDMARESGNEEVVKLLEKQLEGSGSGGSGSGKMYHTKGQEGSVCLRSSDCASGLCCAR HFWSKICKPVLKEGQVCTKHRRKGSHGLEIFQRCYCGEGLSCRIQKDHHQASNSSRLHTCQRHAAAENLYFQGSSEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISR TPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVK GFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKGAA
NGS Wnt luciferase reporter assays
To assess the potency of Fzd7/8 subtype NGS Wnt, 293STF cells were treated with NGS Wnt at various concentrations in the presence or absence of Rspo2. Luciferase reporter assays were performed as previous reported (Dang et al., 2019). To perform NGS Wnt reporter assays in a Fzd specific manner, Fzd-knockout human embryonic kidney (HEK293T) cell line, with Fzd1, 2, 4, 5, 7 and 8 knockout, was used. The Frizzled receptor knockout 293T cell line was generously provided by Michael Boutros laboratory at German Cancer Research Center (Voloshanenko et al., 2017). Fzd knockout cell line was stably transfected with Firefly reporter as previously reported in the Garcia laboratory (Janda et al., 2017) followed by puromycin (Thermo Fisher # A1113803) selection. Cells were maintained in DMEM media with 10% FBS and incubated at 37°C incubator with 5% CO2. These cells were plated to 96-well plates (Corning # 353072). Individual Fzd receptor in pRK5 vector was transiently expressed (Yu et al., 2012) in the cell line using FuGENE® HD Transfection Reagent (Promega) following manufacturer’s protocol. These cells were stimulated with different concentrations of NGS Wnts the following day. The cells were lysed 20 hrs after NGS Wnt stimulation following manufacturer’s protocol as detailed in Dual Luciferase Assay kit (Promega). Luminance signals were recorded using SpectraMax Paradigm or SpectraMax i3x. Data were plotted and analyzed by GraphPad Prism 8.
Single molecule fluorescence microscopyand receptor heterodimerization analysis
NGS Wnt induced heterodimerization/oligomerization of receptors was characterized by single molecule imaging in live cells. Fzd8 and Lrp6 fused to an N-terminal SNAP-tag and HaloTag, respectively, were transiently expressed in HeLa cells and labelled with photostable fluorophores DY649 and tetramethylrhodamine (TMR), respectively. Single molecule imaging of Fzd8 and Lrp6 were carried out at room temperature by total internal reflection fluorescence microscopy. Dimerization/oligomerization of receptors were quantified by co-locomotion analysis as described previously (Janda et al., 2017). IWP-2, an inhibitor of endogenous Wnt secretion, was used for maintaining the basal low level of receptor co-localization.
Expansion of hiPSC-CMs
Previously described wildtype human induced pluripotent stem cells (hiPSCs) from the Stanford Biobank were differentiated into cardiomyocytes (hiPSC-CMs) within 11 days. After dissociation, cells were re-plated into Matrigel-coated 96-well plates in equal densities and allowed to recover for 24 hours. At day 12, hiPSC-CMs were treated with Rspo 1 (10 nM), NGS Wnt (10 nM), NGS Wnt (10 nM) + Rspo 1 (10 nM), Wnt3a (100 ng/mL), Wnt3a (100 ng/mL) + Rspo 1 (10 nM) or media alone (Control) for a consecutive 6 days. At day 18, cells were fixed and stained for cardiac troponin T with nuclear dye DAPI. Troponin T and DAPI positive cell numbers were quantified with Image J software.
Human colon organoid culture assay
P26N, normal adult human colon organoids were cultured as previously described (Janda et al., 2017). Briefly, the colon organoids were cultured in the presence of 3 μM IWP-2 and different concentrations of recombinant mWnt3a or Fc conjugated NGS Wnt for 7 days. Luminescence signals were recorded using CellTiter-Glo® Luminescent Cell Viability Assay.
Mouse colon organoid formation and imaging
Mouse colon was isolated and flushed with cold Phosphate-Buffered Saline (PBS). Colon was cut longitudinally and into small pieces before being incubated in isolation buffer, PBS with 3 mM ethylenediaminetetraacetic acid (EDTA), for 60 min rotating at 4°C. Colon pieces were placed in 20 mL of cold isolation buffer and shaken for 3 min. Fragments were allowed to settle and the supernatant was collected. This process was repeated 3–4 times until supernatant was clear. All suspensions were pooled and centrifuged at 500 g for 10 min at 4°C and resuspended in cold PBS supplemented with 10% FBS. Next, crypts were counted and centrifuged at 700 g for 5 min at 4°C. Crypt pellet was then resuspended in Matrigel to a concentration of 200–500 crypts per 50 μL Matrigel drop. Matrigel was allowed to solidify in a CO2 incubator (5 % CO2, 37 °C) and 600 μL of complete organoid media was added to the well. Complete organoid media adapted from (Sato et al., 2011): DMEM/F12 with GlutaMAX, Penicillin/Streptomycin (100 μg/mL), HEPES (10mM), murine recombinant EGF (50 ng/mL), murine recombinant Noggin (100 ng/mL), N2 Supplement (1X), B27 Supplement minus Vitamin A (1X), N-acetylcysteine (1 mM), Rspo 2 (25 nM), and supplemented with NGS Wnt at varying concentrations (0 nM, 0.1 nM, 0.5 nM, 1 nM, 5 nM and 10 nM) to test organoid growth. Images were taken on days 4–11 using an inverted microscope Keyence BZ-X700 with CDD cooling camera Keyence and BZ-X analyzer software.
Western protocol for Wnt3a
Wnt3aCM was harvested from L-Cells stably transfected with a pcDNA-3.1 vector encoding mWnt3a. Conditioning was optimized by selection of clonal lines, and performed in the presence of DMEM-F12 medium supplemented with 10% FBS. Purified mWnt3a was obtained from Time Bioscience. All samples (20 μl/lane) were run under reducing conditions on a 12-comb 4–15% PAA gel (BioRad Mini-Protean TGX gel 456–1085). Upon transfer to a PVDF membrane (Millipore Immobilon IPVH00010), blocking of nonspecific binding (1 hr) and 3 subsequent antibody incubations (below) were performed at room temperature, in PBS buffer supplemented with 5% non-fat milk and 0,1% Tween20. HRP enzyme activity was visualized using ECL (Pierce Supersignal West Dura Extended #34075).
Rat anti h/m Wnt3a (R&D Systems) at 2 μg/ml for 4 hrs.
Rabbit anti-rat (Southern Biotech) at 0.5 μg/ml for 1 hr.
Poly-HRP anti-rabbit IgG (Immunologic) at a dilution of 1:4 for 1 hr.
Human colon, ovarian and breast organoid expansion
Human colon, ovarian and breast organoids were expanded as described in (Boj et al., 2017; Kopper et al., 2019; Sachs et al., 2018; van de Wetering et al., 2015). Briefly, organoids were split by mechanical disruption (colon and ovarian samples) or using TrypLE (breast) embedded in Matrigel, platted in different wells (24-well plates) and cultured in the corresponding organoid culture media containing different source of Wnt activation (Wnt3a CM or NGS Wnt). Final concentration for NGS Wnt was 0.5 nM while Wnt3aCM was used at 50% v/v for colon organoids and 25% v/v for breast and ovarian organoids.
Human kidney tubuloids expansion and histology
Human kidney tubuloids were cultured and passaged as previously described (Schutgens et al., 2019) in the absence and presence of 0.5 nM NGS Wnt. Tissues were fixed in 4% paraformaldehyde, dehydrated and embedded in paraffin. Sections were subjected to H&E staining protocol as previously described (Schutgens et al., 2019).
Mouse hepatocyte organoids
Primary mouse hepatocytes were isolated and cultured according to the protocol described previously (Peng et al., 2018). Briefly, dissociated hepatocytes were plated in 3D Matrigel and cultured in media containing 0.5 nM NGS Wnt or 1.25 nM recombinant Wnt3a protein (R&D system), 100 ng/mL Rspo 3 (Peprotech), 25 ng/mL EGF (Peprotech), 50 ng/mL HGF (Peprotech), 100 ng/mL TNFa (Peprotech), 10 mM Y27632 (Peprotech) and 1 mM A83–01 (Tocris). Medium was replaced every 2 – 3 days. Hepatocyte organoids were passaged every 2 weeks with dispase (Stem Cell Technologies), followed by incubation with TrypLE, and re-plated in Matrigel.
Human organoid single cell outgrowth experiment
Human pancreas, stomach and colon organoids were cultured in the previously described standard organoid culture media (Bartfeld et al., 2015; Boj et al., 2015; Sato et al., 2011). Organoids were collected, pelleted, resuspended in TrypLE Express 1X (Gibco #12604013) and incubated for 5 minutes at 37°C in order to dissociate the culture into a single cell suspension. DAPI was added to the suspension and living cells were FACS sorted (BD FACSARIA II) into collection tubes. 50 cells were seeded per well into 96 well plates and standard media were added containing 10 mM ROCK inhibitor Y-27632 (Abmole, M1817) and either Wnt3a CM (50%) (Homemade) or 0.5 nM NGS Wnt. Media was refreshed every 4 days. Organoids were incubated for 2 weeks at 37°C and 5% CO2 after which organoid growth was assessed by means of organoid counting and cell viability quantification using CellTiter-Glo® Luminescent Cell Viability Assay (Promega # G7572). 50 μL of CellTiter-Glo® solution was added directly to the organoids in 100 μL medium. Luminescence was measured using Spark® multimode microplate reader (TECAN).
Human tissue processing and colon organoid formation
To isolate crypts from biopsies, reported in Figure S4E, biopsy material was washed with ice-cold Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) and stripped off the underlying muscle layers by curved forceps to remove villi and debris. After that, the biopsies were cut into small pieces and washed for 8–10 times with wash buffer: DMEM/F12 containing 10% FBS, 1x Penicillin/Streptomycin and 0.5 mM DTT. Next, the biopsies were incubated together with collagenase-B for 15 min. 5 mM of EDTA (5 mM) was added to the media and the crypts were filtered using a 70 μm strainer. The crypt suspension was centrifuged for 5 min at 80 g, 4°C, and the pellet was resuspended in 20 mL of wash buffer, followed by centrifugation for 5 min at 200 g, 4°C. The pellet was washed again with DMEM/F12 supplemented with 10% FBS and centrifuged for 5 min at 200 g, 4°C. The pellet was resuspended at this point with ice cold Matrigel in a 2:1 dilution and 50 μL drops of cells mixed with Matrigel were placed in a pre-warmed 24 well plate. After polymerization of the gel in 37°C, 5% CO2 incubator, the basement membrane matrix was overlayed with 500 μL of complete media supplemented with 10μM Y-27632 (ROCK inhibitor). The organoids were cultured for 7–14 days with media change every 2–3 days. The organoid cultures were split every 10 days. The complete culture medium contains the key ingredients to stimulate the Wnt signaling pathway and other growth factors or inhibitors, including 0.5 nM NGS Wnt, 1 μg/mL Rspo 1, 50 ng/mL Noggin, 100 ng/mL EGF, 500 nM A-83–01 (TGF-β inhibitor) and 10 μM SB202190 (p38 inhibitor). For differentiation of the organoids into the major intestinal cell types, the media was changed from expansion media to a differentiating media (removing NGS Wnt, Nicotinamide and SB202190). The human colon organoids, reported in Figure S4E, were fixed with 4% paraformaldehyde. The organoids were blocked by a blocking solution (1% Fish skin gelatine, 5% FCS, 1% sera, 20mM glycine and 0.1% saponin in PBS). Primary antibody (E-cadherin antibody (1.B.54): sc-71009, Santa Cruz Biotechnology) and secondary antibody (goat anti mouse Alexa 488, Invitrogen) were used for E-cadherin staining. Phalloidin was used for Actin staining.
Western blotting of non-differentiated organoids and differentiated organoids
For western blot analysis, organoids were washed in ice-cold DPBS for at least 30 min with 3 to 5 buffer changes on ice. Supernatant was removed and organoids were lysed in Ripa Lysis and Extraction buffer (Thermo Fisher Scientific) containing protease and phosphatase inhibitors (Sigma-Aldrich Inc). Protein concentration was determined by DC Assay (Bio-Rad). For each sample, 30 μg of protein was subjected to gel electrophoresis and transferred to nitrocellulose membrane (Thermo Fisher Scientific). Membrane was blocked for 45 min with 5% non-fat dry milk (Bio-Rad) in TBS containing 0.2% Tween-20 and incubated over night with primary antibody (1:1000) diluted in blocking buffer. The following primary antibodies were used; mouse anti-human Mucin2 (Santa Cruz Biotechnology), mouse anti-human Villin (Santa Cruz Biotechnology), rabbit anti-human ChgA (Novus Biologicals) and mouse anti-human GAPDH (Abcam). Membranes were washed and then incubated for 1h with HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (Jackson Immuno Research) and visualized by using ECL (Roche) or SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
RNA extraction and real-time PCR analysis
RNA was extracted using RNA Stat-60 RNA Extraction Reagent (Amsbio) following the manufacturers protocol. An amount of RNA corresponding to 500 ng was reverse-transcribed with AMV reverse transcriptase (Promega) with random primers. cDNA was diluted 1:10 in the PCR reactions. The gene expressions levels were measured using SybrGreen Mastermix (Thermo Fisher Scientific) following the manufactures instruction. Results were expressed as 2-ΔΔCT. For normalization of expression levels, 18S RNA was used as a housekeeping gene. Primer sequences used are reported in Table S1.
In vivo experiments
Adult C57BL/6J mice (Jackson Laboratory, ME, USA) between 9–10 weeks old were injected intravenously with adenoviruses expressing mouse IgG2a Fc (Fc) or different Fzd subtype-specific NGS Wnt (1 × 104 pfu or 1 × 106 pfu per mouse for intestine and 1 × 106 pfu per mouse for liver) with or without Ad-Rspo2 (2.5 × 108 pfu per mouse). A submaximal dose of Ad-Rspo2 (2.5 × 108 pfu per mouse) was intentionally selected to maximize detection of synergy. Intestinal tissues and livers were collected and fixed in 10% formalin 7 days after adenovirus injection (n=4 mice per group). Extensive adenovirus titration for in vivo i.v. injection was carefully determined by thoroughly testing a wide range of doses (104 to 109 pfu) since progressive loss of phenotypes could be observed with higher amounts. Intestine and liver required different amount of adenovirus to induce phenotypes where 104 pfu (Fzd8 subtype NGS Wnt) or 106 pfu (Fzd7/8 subtype NGS Wnt) were needed to demonstrate Ki67+ crypt proliferation whereas 106 pfu was necessary for GLUL+ pericentral zonation. Paraffin-embedded sections were incubated in citrate antigen retrieval solution and blocked with 10% normal donkey serum. For intestine immunofluorescence, sections were stained with anti-Ki67 (Biolegend, 1:250) or anti-CD44v6 (Thermo Fisher Scientific, 1:300) antibody. For the liver, sections were stained with anti-Glutamine Synthetase (Millipore, 1:50) or anti-E-Cadherin (BD Biosciences, 1:300) antibody. Images were captured on a fluorescence microscope (KEYENCE BZ-X710) or a Zeiss Axio-Imager Z1 with ApoTome and analyzed using Image J. All animal experiments were repeated twice and were conducted in accordance with procedures approved by the IACUC at Stanford University.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics was performed using GraphPad Prism 8 as indicated in the figure legends. Mean and S.D. or S.E. are reported in the figure legends.
Supplementary Material
Movie S1 Real time single molecule imaging of Fzd8 (blue) and Lrp6 (red) without treatment. (Related to Figure 1)
Scale bar represents 2 μm.
Movie S2 Real time single molecule imaging of Fzd8 (blue) and Lrp6 (red) after treatment with 1 nM Fzd7/8 subtype NGS Wnt for 20 min. (Related to Figure 1)
Scale bar represents 2 μm.
Table S1 Primer sequences in qRT-PCR, related to STAR Methods.
Highlights.
NGS Wnts activate Wnt signaling through targeted Frizzled subtype receptors
NGS Wnt supports expansion of multiple types of organoids
NGS Wnt supports single cell derived organoid outgrowth
NGS Wnts deconvolute Frizzled subtype receptor dependency in tissue homeostasis
Acknowledgments:
The work here is supported by the Ludwig Foundation (K.C.G. and C.J.K.); Mathers Fund (K.C.G.); HHMI (K.C.G. and R.N.); NIH grant NIH grants U01DK085527, U19AI116484, R01NS100904 and U01CA217851 (C.J.K.); NIH grant 1R01DK115728 (C.J.K. and K.C.G.); P.D. is a Mark Foundation Momentum Fellow supported by The Mark Foundation for Cancer Research.
Footnotes
Declaration of Interests: Y.M., L.T.D., D.B. and K.C.G. are inventors on patent applications submitted by Leland Stanford Junior University that covers the use of NGS Wnt.H.C. is inventor on several patents related to organoid technology. W.H. works at U-Protein Express BV, a contract research organization that produces recombinant proteins and antibodies as commercial activity.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Bandaranayake AD, Correnti C, Ryu BY, Brault M, Strong RK, and Rawlings DJ (2011). Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Research 39, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, and Clevers H. (2006). Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov 5, 997–1014. [DOI] [PubMed] [Google Scholar]
- Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, and Clevers H. (2015). In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 148, 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, Vasseur-Cognet M, Kuo CJ, Kahn A, Perret C, et al. (2006). Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Developmental cell 10, 759–770. [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. (2015). Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 160, 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Vonk AM, Statia M, Su JY, Vries RRG, Beekman JM, and Clevers H. (2017). Forskolin-induced Swelling in Intestinal Organoids: An In Vitro Assay for Assessing Drug Response in Cystic Fibrosis Patients. J Vis Exp, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema JW, Lee S, Goodyer WR, Maas RG, Chirikian O, Li G, Miao Y, Paige SL, Lee D, Wu H, et al. (2020). Wnt Activation and Reduced Cell-Cell Contact Synergistically Induce Massive Expansion of Functional Human iPSC-Derived Cardiomyocytes. Cell Stem Cell 27, 50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema JW, Mady AS, Mittal NV, Atmanli A, Caron L, Doevendans PA, Sluijter JPG, and Domian IJ (2013). Wnt/beta-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development 140, 4165–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lu C, Ouyang B, Zhang H, Huang Z, Bhatia D, Lee S-J, Shah D, Sura A, Yeh W-C, et al. (2020). Development of Potent, Selective Surrogate WNT Molecules and Their Application in Defining Frizzled Requirements. Cell Chemical Biology. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Biechele T, Wei Z, Morrone S, Moon RT, Wang L, and Xu W. (2011). Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nature structural & molecular biology 18, 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2006). Wnt/β-catenin signaling in development and disease. Cell 127, 469–480. [DOI] [PubMed] [Google Scholar]
- Clevers H. (2016). Modeling Development and Disease with Organoids. Cell 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- Clevers H, and Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang LT, Miao Y, Ha A, Yuki K, Park K, Janda CY, Jude KM, Mohan K, Ha N, Vallon M, et al. (2019). Receptor subtype discrimination using extensive shape complementary designed interfaces. Nature Structural & Molecular Biology 26, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VSW, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297. [DOI] [PubMed] [Google Scholar]
- Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NWM, Bijvelds MJC, Scholte BJ, et al. (2013). A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature Medicine 19, 939–945. [DOI] [PubMed] [Google Scholar]
- Dijksterhuis JP, Baljinnyam B, Stanger K, Sercan HO, Ji Y, Andres O, Rubin JS, Hannoush RN, and Schulte G. (2015). Systematic Mapping of WNT-FZD Protein Interactions Reveals Functional Selectivity by Distinct WNT-FZD Pairs. J Biol Chem 290, 6789–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, and Clevers H. (2018). Organoids in cancer research. Nat Rev Cancer 18, 407–418. [DOI] [PubMed] [Google Scholar]
- Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, et al. (2017). Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. (2015). Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47. [DOI] [PubMed] [Google Scholar]
- Hatano H, Somsouk M, Sinclair E, Harvill K, Gilman L, Cohen M, Hoh R, Hunt PW, Martin JN, Wong JK, et al. (2013a). Comparison of HIV DNA and RNA in gut-associated lymphoid tissue of HIV-infected controllers and noncontrollers. Aids 27, 2255–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Yukl SA, Ferre AL, Graf EH, Somsouk M, Sinclair E, Abdel-Mohsen M, Liegler T, Harvill K, Hoh R, et al. (2013b). Prospective Antiretroviral Treatment of Asymptomatic, HIV-1 Infected Controllers. PLoS Pathog 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HL, Gehart H, Artegiani B, Lopez-Iglesias C, Dekkers F, Basak O, van Es J, Lopes S, Begthel H, Korving J, et al. (2018). Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 175, 1591–1606. [DOI] [PubMed] [Google Scholar]
- Hunt PW, Shulman NS, Hayes TL, Dahl V, Somsouk M, Funderburg NT, McLaughlin B, Landay AL, Adeyemi O, Gilman LE, et al. (2013). The immunologic effects of maraviroc intensification in treated HIV-infected individuals with incomplete CD4+ T-cell recovery: a randomized trial. Blood 121, 4635–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Dang LT, You CJ, Chang JL, de Lau W, Zhong ZDA, Yan KS, Marecic O, Siepe D, Li XN, et al. (2017). Surrogate Wnt agonists that phenocopy canonical Wnt and beta-catenin signalling. Nature 545, 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, and Garcia KC (2015). Wnt acylation and its functional implication in Wnt signalling regulation. Biochemical Society Transactions 43, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, and Garcia KC (2012). Structural Basis of Wnt Recognition by Frizzled. Science 337, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jope RS, and Johnson GVW (2004). The glamour and gloom of glycogen synthase kinase-3. Trends BiochemSci 29, 95–102. [DOI] [PubMed] [Google Scholar]
- Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, et al. (2013). The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 110, E4987-E4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopper O, de Witte CJ, Lohmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H, et al. (2019). An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nature Medicine 25, 838–849. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang H-T, Chu P, Lee M, Yuan J, Nusse R, and Kuo CJ (2004). Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proceedings of the National Academy of Sciences 101, 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, Somsouk M, Deeks SG, Martin JN, Moll M, et al. (2013). Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood 121, 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, and Nusse R. (2004). The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20, 781–810. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, and He X. (2009). Wnt/β-catenin signaling: components, mechanisms, and diseases. Developmental cell 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, and Sato T. (2015). Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nature Medicine 21, 256–262. [DOI] [PubMed] [Google Scholar]
- Niehrs C. (2012). The complex world of WNT receptor signalling. Nature reviews Molecular cell biology 13, 767–779. [DOI] [PubMed] [Google Scholar]
- Nusse R, and Clevers H. (2017). Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999. [DOI] [PubMed] [Google Scholar]
- Peng WC, Logan CY, Fish M, Anbarchian T, Aguisanda F, Alvarez-Varela A, Wu P, Jin YH, Zhu JJ, Li B, et al. (2018). Inflammatory Cytokine TNF alpha Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture. Cell 175, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. (2007). The renewal and differentiation of Isl1(+) cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell 1, 165–179. [DOI] [PubMed] [Google Scholar]
- Rocha AS, Vidal V, Mertz M, Kendall TJ, Charlet A, Okamoto H, and Schedl A. (2015). The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Reports 13, 1757–1764. [DOI] [PubMed] [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, et al. (2018). A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 172, 373–386. [DOI] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M, et al. (2019). Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nature Biotechnology 37, 303–313. [DOI] [PubMed] [Google Scholar]
- Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, Kawasaki K, et al. (2018). Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 22, 454–467. [DOI] [PubMed] [Google Scholar]
- Somsouk M, Dunham RM, Cohen M, Albright R, Abdel-Mohsen M, Liegler T, Lifson J, Piatak M, Gorelick R, Huang Y, et al. (2014). The Immunologic Effects of Mesalamine in Treated HIV-Infected Individuals with Incomplete CD4+T Cell Recovery: A Randomized Crossover Trial. Plos One 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Mis M, Blazer L, Ustav M, Steinhart Z, Chidiac R, Kubarakos E, O’Brien S, Wang XM, Jarvik N, et al. (2019). Tailored tetravalent antibodies potently and specifically activate Wnt/Frizzled pathways in cells, organoids and mice. eLife 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. (2015). Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 161, 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshanenko O, Gmach P, Winter J, Kranz D, and Boutros M. (2017). Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families. Faseb J 31, 4832–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao LD, Fish M, Logan CY, and Nusse R. (2015). Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 524, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, and DiNardo S. (2000). arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407, 527–530. [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, and Nusse R. (2003). Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452. [DOI] [PubMed] [Google Scholar]
- Yan KS, Janda CY, Chang JL, Zheng GXY, Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, et al. (2017). Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature 545, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HM, Ye X, Guo N, and Nathans J. (2012). Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Development 139, 4383–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl SA, Sinclair E, Somsouk M, Hunt PW, Epling L, Killian M, Girling V, Li PL, Havlir DV, Deeks SG, et al. (2014). A comparison of methods for measuring rectal HIV levels suggests that HIV DNA resides in cells other than CD4 R T cells, including myeloid cells. Aids 28, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 Real time single molecule imaging of Fzd8 (blue) and Lrp6 (red) without treatment. (Related to Figure 1)
Scale bar represents 2 μm.
Movie S2 Real time single molecule imaging of Fzd8 (blue) and Lrp6 (red) after treatment with 1 nM Fzd7/8 subtype NGS Wnt for 20 min. (Related to Figure 1)
Scale bar represents 2 μm.
Table S1 Primer sequences in qRT-PCR, related to STAR Methods.
Data Availability Statement
This study did not generate/analyze datasets/code.