Abstract
Leptospirosis is a public health challenge in Sabah State of Malaysian Borneo. Rapid urbanization, rural-to-urban migration, and undocumented immigration in Sabah have increased the pressure on the urban garbage disposal system. Rodents and other small animals thrive under these conditions. We hypothesized that urban sanitation workers would be at risk of developing leptospirosis. In total, 303 urban sanitation workers with a mean age of 42.6 years were enrolled in this study. The serum samples collected from these workers were subjected to the microscopic agglutination test (MAT), PCR and nucleotide sequencing of the amplicons to confirm the presence of Leptospira. The phylogenetic analysis using the neighbor joining method was performed to assess whether they were pathogenic. In this study 43.8% (133/303) of the samples were MAT-seropositive and among them, 29 (21.8%) were positive by PCR. Nucleotide sequencing of the amplicons confirmed the presence of Leptospira. Phylogenetic analysis showed that our strains belonged to the pathogenic group of Leptospira. A high proportion of urban sanitation workers were seropositive for leptospirosis, and a considerable number were PCR positive for Leptospira, thereby indicating asymptomatic infections. Further research is needed to confirm whether this is a transient phenomenon or antibiotic therapy is required.
Subject terms: Microbiology, Bacteria, Clinical microbiology, Pathogens, Epidemiology
Introduction
Leptospirosis is the most common emerging zoonotic disease globally and it is expected to become more prominent worldwide because of climate change and the growing urban population1. Although leptospirosis is an underreported disease, 1.03 million people are infected worldwide and it causes 58,900 deaths every year2. Most of these deaths occur in tropical countries because the incidence of leptospirosis in the tropics is approximately 10 times higher than that in temperate regions3. Malaysia is a tropical country that has undergone rapid economic growth during recent decades and large numbers of people have moved into urban areas. At present, 76% of the total population lives in urban areas. Leptospirosis is an endemic notifiable disease in Malaysia, and it has been detected at alarming levels in different states throughout the country4.
Sabah is one of the 13 states of Malaysia and one of the two states located on the island of Borneo. According to data provided by the Sabah State Health Department, the incidence of leptospirosis is 4.07–26.68 per 100,000 people in the state. In the year 2000, a widely reported outbreak of leptospirosis occurred in Sabah that affected 304 athletes from 26 countries after they participated in the Eco-challenge race5. In 1999, another outbreak affected 46 patients after swimming in a creek in Beaufort district, Sabah6. Analyses have shown that the numbers of leptospirosis cases that occurred during outbreaks in Sabah only comprised a modest proportion of the total disease incidence, and most of these were related to multiple isolated cases reported from every district in Sabah. The high prevalence of leptospirosis is related to occupational activities, such as close contact with animals or exposure to water, mud, soil, vegetation, and waste contaminated with urine from animals with a high risk of infection7,8.
Among the infected animals, Leptospira spp. are most often associated with rodents, including the species that are frequently found in urban areas9. The prevalence of leptospirosis in urban habitats occupied by wildlife is higher than that found in natural environments, and this trend appears to be particularly significant for rodents10. In cities, the rodent populations are often larger and denser than those found in natural environments, which can lead to higher rates of contact with people and affect the risk of human diseases9. Leptospirosis is recognized as a hazard for certain occupations with an increased risk of exposure to infected animals, such as agricultural workers, sewage workers, military personnel, veterinarians and animal handlers5,11. However, the occupational risk of leptospirosis in Sabah remains unknown.
Urban sanitation workers are among the occupations that may have high risk of leptospirosis in Malaysia. Kota Kinabalu is the capital of Sabah and it has a population of 553,900. Rapid urbanization, migration of the rural population, an influx of undocumented immigrants, increased garbage disposal, and tropical weather have facilitated exponential increases in the rodent population in Kota Kinabalu, thereby exposing urban sanitation workers to Leptospira12. Pathogenic Leptospira species can cause a wide range of diseases in humans, where the complications can range from severe, such as Weil’s disease and pulmonary hemorrhagic syndrome, to mild flu-like symptoms13. Thus, we hypothesized that urban sanitation workers in Kota Kinabalu might be repeatedly exposed to Leptospira and have a high rate of seropositivity for Leptospira, and that some might be experiencing asymptomatic leptospirosis. However, no previous studies have investigated this important public health issue in Sabah. Therefore, the present study aimed to determine the consequences of repeated exposure to Leptospira among sanitation workers in Kota Kinabalu.
Methods
Study area, population, and ethics statement
A cross-sectional analysis was conducted in March 2017 among urban sanitary workers from Kota Kinabalu, Sabah, Malaysia. Only people who had worked outdoor environments for more than 6 months were enrolled in this study. Ethical clearance (JKEtika: 4/16[1]) was obtained from the Ethics Committee of the Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, Kota Kinabalu, Sabah, Malaysia (UMS). All methods were performed in accordance with the relevant guidelines and regulations. Written informed consent was obtained from all participants. We collected 10 mL of venous blood from each participant and separated the serum. The serum samples were transported by cold chain to the laboratory at UMS and Kota Kinabalu Public Health Laboratory. All of the serum samples were stored at – 80 °C until they were analyzed.
Determination of seroprevalence using microscopic agglutination test (MAT)
The MAT was conducted according to standard methods at Kota Kinabalu Public Health Laboratory to determine the antibody titers against Leptospira serovars. It is the state reference laboratory for MAT to diagnose leptospirosis. Serum samples were serially diluted in microtiter plates, and to each well were added 20 live Leptospira serovars recommended by the Institute of Medical Research Malaysia (L. meyeri serovar Melaka, L. kemamanensis serovar Terengganu, L. sarikeinsis serovar Sarawak, L. interrogans serovar Copenhageni, L. borgpetersenii serovar Hardjobovis, L. borgpetersenii serovar Lai, L. interrogans serovar Australis, L. interrogans serovar Autumnalis, L. interrogans serovar Bataviae, L. interrogans serovar Canicola, L. weilii serovar Celledoni, L. krischneri serovar Grippotyphosa, L. borgpetersenii serovar Hardjoprajitno, L. interrogans serovar Icterohaemorrhagiae, L. borgpetersenii serovar Javanica, , L. interrogans serovar Pyrogenes, L. borgpetersenii serovar Terrasovi, L. interrogans serovar Djasiman, L. biflexa serovar Patoc and, L. interrogans serovar Pomona). The Institute of Medical Research is the national reference laboratory, the recommended serovars are based on the background exposure and seroprevalence of leptospirosis in the country. The plates were then incubated for 2 h at 30 °C and examined by dark-field microscopy. A mixture was considered positive when it exhibited 50% agglutination and 50% of the cells were free compared with the control culture. MAT titers were reported as the reciprocal of the number of dilutions that still agglutinated 50% of the live bacterial antigen. A titer of ≥ 1:50 was taken as reactive to that serovar.
Determination of Leptospira by PCR
DNA was extracted from serum samples using a DNeasy Blood Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. To determine whether the serum samples from the urban sanitary workers contained Leptospira, nested PCR assays were performed using GoTaqGreen Master Mix (Promega Corporation, Madison, WI, USA) and primers generated from the flagellin b gene of Leptospira14. The first PCR primers were LflaB-F1 (5′-CTCACCGTTCTCTAAAGTTCAAC-3′) and L-flaB-R1 (5′- TGAATTCGGTTTCATATTTGCC-3′). The second PCR primers were L-flaB-F2 (5′-TGTGCACAAGACGATGAAAGC-3′) and L-flaB-R2 (5′-AACATTGCCGTACCACTCTG-3′).
Nucleotide sequencing was performed for the amplicons using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Gaithersburg, MD, USA). The purified amplicons were sequenced using an ABI3130 Genetic Analyzer (Applied Biosystems). The second primers were used for sequencing. All of the procedures were conducted according to the manufacturer’s instructions. PCR positive samples were taken as asymptomatic leptospirosis positive samples.
Sensitivity, specificity, positive and negative predictive values of MAT for asymptomatic leptospirosis
The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of MAT titer (1:50) to identify asymptomatic cases as determined by PCR were calculated by using MedCalc statistical software (https://www.medcalc.org/calc/diagnostic_test.php).
Phylogenetic analysis
Phylogenetic analysis was performed to determine the relatedness among our strains and pathogenic and intermediate pathogenic Leptospira strains. We also selected 30 newly reported species of Leptospira for phylogenetic analysis15. The nucleotide sequences of the flagellin b genes from pathogenic and intermediate pathogenic and newly reported Leptospira were retrieved from GenBank. Multiple sequence alignment was performed using CLUSTALW ver. 216. Phylogenetic analyses were conducted with the neighbor-joining method using the software MEGA ver. 7. The branching patterns were evaluated statistically by bootstrap analyses with 1,000 replicates.
Results
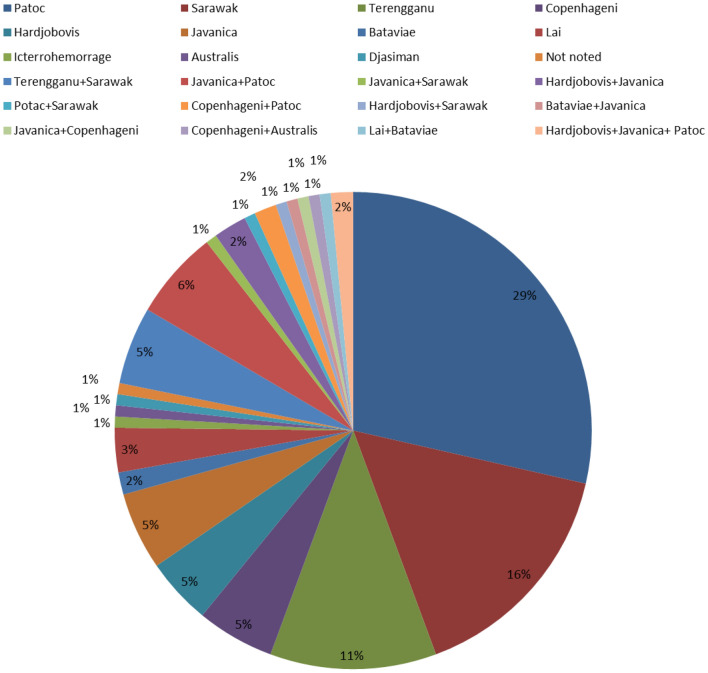
In total, 303 sanitation workers who fulfilled all of the criteria were enrolled in this study with 222 males (73.3%), 78 females (25.7%), and three participants with an unrecorded gender (1%). The male:female ratio was 2.8:1. The mean age was 42.6 years (range 23–60 years). With a cutoff value of ≥ 1:50, 43.8% (133/303) of the samples were seropositive for Leptospira. Among the 20 recommended Leptospira serovars used for MAT, only 11 were reactive with our serum samples. Among the 133 seropositive samples, 104 (78.2%) were reactive with one serovar, 27 (20.3%) were reactive with two serovars, and only two (1.5%) were reactive with three serovars. Antibodies to serovars Patoc, Sarawak, and Terengganu were detected in 56% of the serum samples that reacted to single serovars, and antibodies to serovars Terengganu + Sarawak and Javanica + Patoc were detected in 11% of the serum samples that reacted to two serovars (Fig. 1).
Figure 1.
Distribution of reactive serovars in microscopic agglutination test (MAT) seropositive (> 1:50 as the cutoff value) samples represented as a pie chart. Colors representing different Leptospira serovars are indicated above the pie chart.
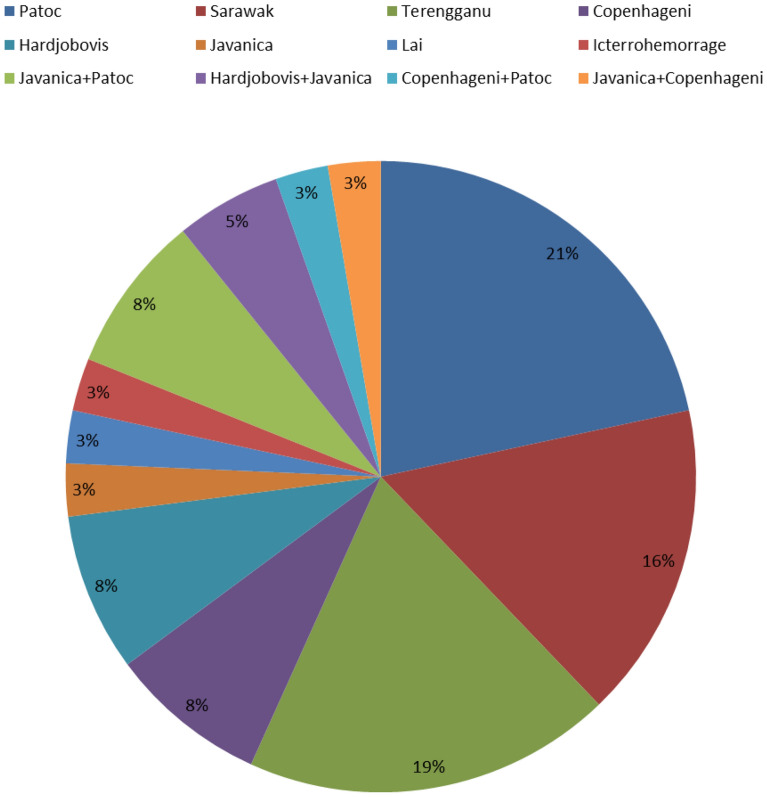
A 732-base pair amplicon generated by nested PCR indicated the presence of Leptospira. Among the 133 MAT-seropositive samples, 29 (21.8%) were positive by Leptospira-specific PCR. It may be mentioned here that PCR cannot distinguish between viable and nonviable organisms. Among the PCR-positive samples, Patoc, Sarawak, and Terengganu serovars accounted for 56% of the serum samples that reacted to single serovars in MAT, and Javanica + Patoc and Hardjobovis + Javanica accounted for 13% of the serum samples that reacted to two serovars in MAT (Fig. 2).
Figure 2.
Distribution of reactive serovars detected by microscopic agglutination test (MAT) among samples that tested positive for Leptospira according to PCR represented as a pie chart. Colors representing different Leptospira serovars are indicated above the pie chart.
Compared to PCR the sensitivity, specificity, PPV, and NPV of MAT for the detection of asymptomatic leptospirosis were 100% [95% confidence intervals (CI) 88.1–100%], 62.0% (95% CI 56.0–67.8%), 21.8% (95% CI 19.3–24.5%), and 100%, respectively.
Nucleotide sequencing was successful for all of the PCR-positive samples and they were confirmed as Leptospira by aligning sequences using the Basic Local Alignment Search Tool (BLAST). All of the MAT-negative samples were also negative for Leptospira specific PCR.
Phylogenetic analysis showed that our strains belonged to the pathogenic group of Leptospira (Fig. 3). In particular, 21 belonged to a single cluster that mainly contained Leptospira interrogans strains. A single strain did not cluster with these strains. Seven of the strains belonged to the intermediate pathogenic group of Leptospira and they formed a single cluster with a Leptospira fainei strain detected in Japan.
Figure 3.
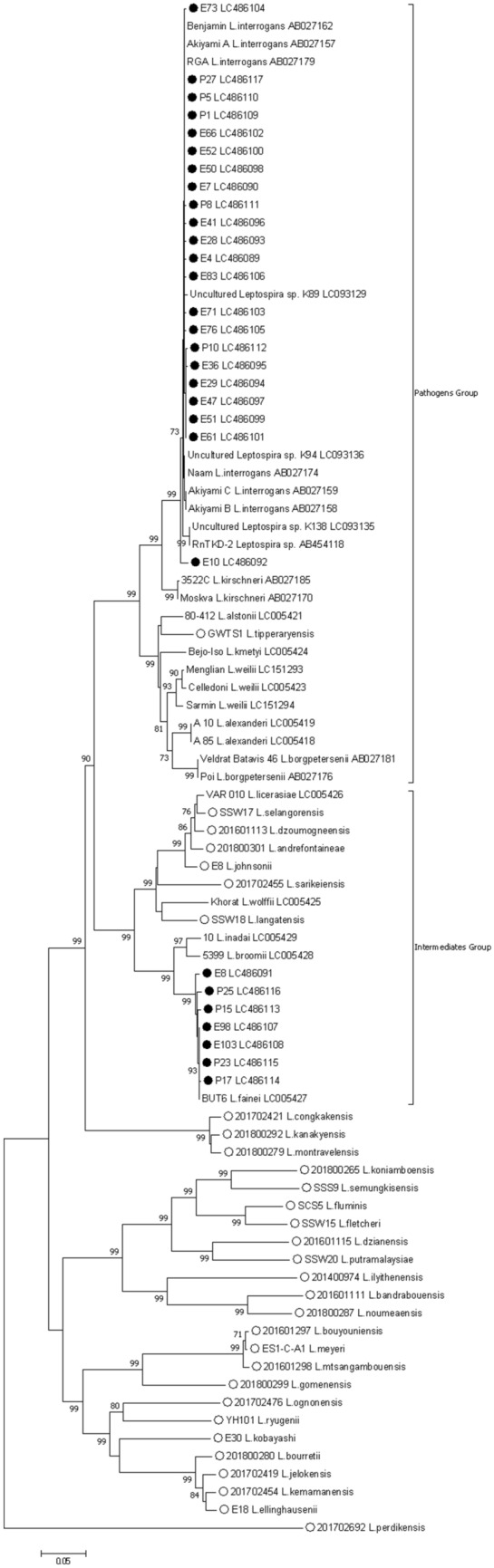
Phylogenetic tree constructed using the nucleotide sequences of the flagellin b gene in Leptospira. Nucleotide sequences of pathogenic, intermediate pathogenic, and recently reported new (open circle) strains were extracted from GenBank; our strains (closed circle) clustered with either pathogenic and intermediate pathogenic strains of Leptospira. Please note that generally the pathogenic potential of the newly reported Leptospira is unknown. Strains names are followed by GenBank accession numbers. The strains analyzed in this study are denoted by filled circles. The numbers adjacent to the nodes represent the bootstrap values, and values < 70% were omitted from this tree. The scale bar at the bottom indicates the genetic distance expressed as nucleotide substitutions per site.
Discussion
In the present study, we determined that about 40% of the urban sanitation workers who were frequently exposed to the environment were seropositive for Leptospira and one-third of this population were PCR positive for Leptospira, but none complained of illness, thereby indicating that they had asymptomatic Leptospira infections. We did not assess fever or other symptoms of leptospirosis because the samples were collected during the workday, and thus we considered all participants to be healthy. Compared with urban sanitation workers in Kelantan (25.5%) and Selangor (34.8%) states, the seroprevalence of Leptospira was considerably higher in the present study17,18. The detection of DNA from pathogenic Leptospira species by PCR is one of the confirmatory tests employed for detecting leptospirosis19. In the present study, BLAST and phylogenetic analyses confirmed that pathogenic Leptospira isolates were detected by PCR. In addition to a positive PCR result, a fourfold increase in the antibody titer between acute and convalescent serum samples based on MAT or a high (≥ 1:400) MAT titer are confirmatory tests for leptospirosis19. Although these MAT criteria are recommended for the diagnosis of symptomatic leptospirosis, but there are no similar diagnostic criteria for asymptomatic infections. Clearly, PCR may be the most suitable test of choice for the diagnosis of asymptomatic cases. We found that the MAT titer was not related to the positivity by PCR in the diagnosis of asymptomatic leptospirosis. The MAT titer was > 1:50 for all of the PCR-positive samples, but not all samples with a MAT titer ≥ 1:50 were PCR-positive. However, all samples with MAT titer of < 1:50 were PCR negative for Leptospira, thereby indicating that MAT can be used to determine the absence of asymptomatic leptospirosis, but PCR is required to confirm the diagnosis of asymptomatic leptospirosis in cases where the MAT titer is > 1:50. Statistical analyses also support these findings. The MAT titer reflects the presence of both immunoglobulin G (IgG) and M (IgM) antibodies against Leptospira, and thus the MAT-positive cases may have exhibited high levels of IgG but not IgM, thereby indicating past exposure. These subjects may have been repeatedly exposed to Leptospira, so they might have exhibited higher levels of IgG in their serum.
The risk of infection after contact with a Leptospira-contaminated environment depends on the ability of bacteria to survive, persist and infect new hosts20. The factors that favor the environmental recycling and transmission of Leptospira species are not well understood20–22. However, the capacity of Leptospira for biofilm formation and cell aggregation might allow the maintenance of a sufficient concentration of bacteria to achieve an infection23–26.
Textbooks and other publications often mention the presence of asymptomatic leptospirosis, but its true frequency is unknown. Asymptomatic leptospirosis was reported based on the detection of IgM in subjects from an area with a previous Leptospira outbreak in Nicaragua27 but Leptospira was not detected by culture or molecular methods. In addition, human asymptomatic leptospirosis was reported from a rural area of Thailand28, where only one of 37 human samples was positive for Leptospira by PCR, and the study site was selected because human cases occurred 1–2 months previously. Indeed, Leptospira can be isolated from blood or cerebrospinal fluid several months after an acute episode29. Therefore, these studies might have detected the aftereffects of an acute episode of leptospirosis rather than asymptomatic infections. In a study conducted in the Seychelles, positive Leptospira PCR tests confirmed asymptomatic cases in 9% of the healthy adult males with manual occupations29. In general, it is accepted that humans can excrete Leptospira for weeks to months after an infection30; however, a recent study in the Peruvian Amazon found that asymptomatic individuals without serological evidence of recent exposure were shedding either pathogenic or intermediately pathogenic Leptospira in their urine31. Another study in India used PCR to find that Leptospira was excreted in the urine by 30.2% of the asymptomatic individuals investigated32. It is not known whether the excretion of Leptospira in urine is a characteristic of asymptomatic leptospirosis, and we did not assess the excretion of Leptospira in our subjects. Thus, further investigations are required.
Leptospira serotyping is based on the use of specific monoclonal antibodies, and it allows the distinction of > 300 serovars based on the structural heterogeneity of the surface-exposed lipopolysaccharide1. Associations among serovars and animal reservoir hosts have been demonstrated with this method33. A recent article showed that a particular molecular type of Leptospira is found in a unique ecosystem1. Therefore, determining the types of Leptospira present in a given area could have great public health, diagnostic, and epidemiological implications. Over 200 Leptospira serovars have been identified in Malaysia, mostly in rodents, thereby making them the most important sources of human infections and environmental contamination34. In Malaysia, various serovars have been detected in different areas. In Sarawak, reactions to Patoc and Celledoni were the predominant serovars and only nine of the serovars tested were reactive in the samples obtained from planters35. In Kelantan, the predominant serovars were reactive to Patoc, Bataviae, and Javanica, and 12 of the tested serovars were reactive in town service workers36. The predominant serovars reactive with MAT in the present study were Patoc, Sarawak, and Terengganu, which accounted for more than half of the serum samples tested. Among the 20 serovars tested in our study, 11 serovars were that reacted in MAT, thereby indicating that these serovars might be the specific focus when conducting MAT, which may decrease the workload when performing MAT in the laboratory. Similarly, PCR also obtained positive results for the serum samples that were mainly reactive with the Patoc, Sarawak, and Terengganu serovars by MAT, and they accounted for more than half of the serum samples tested by PCR. Only eight of the 20 serovars tested by MAT were identified by PCR.
The use of personal protective equipment (PPE) and hygiene measures appears to be important for avoiding contact with bacteria, ensuring health protection, and preventing disease37. Sanitation workers typically use PPE during their work, but we did not determine the extent of PPE usage among the participants in this study. Overall, the prevalence of Leptospira increases as the anthropogenic influence across the landscape increases, accordingly, a significantly higher proportion of infected rodents are observed in urban locations9. The survival and persistence of Leptospira in the environment depend on its interactions with other microorganisms and reservoir hosts. Recently, an environmental DNA-based approach was developed to identify these factors38. Rapid economic development has occurred in Sabah, and it has greatly influenced the urbanization of the area. Thus, an environmental DNA-based approach might be useful for identifying the factors responsible for the environmental survival of Leptospira under changing conditions. It is not clear whether these asymptomatic infections are transient or whether appropriate antibiotic treatment is necessary. The long-term consequences of human asymptomatic infections with Leptospira need to be explored in details.
In conclusion the high seroprevalence of Leptospira among the urban sanitation workers in Kota Kinabalu indicates that a considerable risk is associated with this occupation. Furthermore, a large number of these workers were afflicted with asymptomatic leptospirosis. Because of the nature of their occupation, urban sanitation workers are repeatedly exposed to Leptospira, and thus more studies are needed to determine whether asymptomatic leptospirosis is a transient state or whether antibiotic treatment is required. We also determined that a MAT titer < 1:50 indicates the absence of asymptomatic leptospirosis, whereas a MAT titer ≥ 1:50 indicates possible asymptomatic leptospirosis, which should be confirmed by PCR, although a MAT titer of 1:50 can indicate past exposure as well. The seroprevalence of Leptospira varies among different areas depending on the Leptospira burden, rodent population and other factors, so we advise caution regarding the use of this cutoff value because variations might exist, and it should be independently verified.
Acknowledgements
The authors acknowledge the patience and cooperation of the urban sanitation workers of Kota Kinabalu, as well as the generous support and cooperation of the authorities. We thank Rashida Binti Mohammad, Kota Kinbalu Public Health Laboratory for microscopic agglutination test. We thank Azalina Farina for excellent technical assistance. The study was supported in part by a grant from Malaysia One Health University Network (MyOHUN) (Grant number: GL00164-AK-2016), Skim Dana Nic Grant (Grant number: SDN0067-2019) and by Launching Grant for Centre of Excellence from Universiti Malaysia Sabah (Grant Number: AM 18006).
Author contributions
K.A. conceptualized the study. D.M. and N.A.Y. curated data. D.M., N.A.Y., K.A.L., R.O., M.R.H., L.S. and K.A. analyzed data. M.S.J., K.A.L., R.O., M.R.H., L.S. and K.A. acquired funds. D.M., N.A.Y., M.S.J., A.B.A., K.A.L., R.O., M.R.H. and L.S. performed investigations. D.M., N.A.Y. and A.B.A. performed laboratory procedures. M.S.J., K.A.L., R.O., M.R.H., L.S. and K.A. performed project administrations. M.S.J. and K.A. supervised. K.A. wrote the original daraft. D.M., N.A.Y., M.S.J., A.B.A., K.A.L., R.O., M.R.H., L.S. and K.A. reviewed, edited and wrote the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mohammad Saffree Jeffree and Daisuke Mori.
References
- 1.Guglielmini J, et al. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019;13:e0007374. doi: 10.1371/journal.pntd.0007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa F, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl. Trop. Dis. 2015;9:e0003898. doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartskeerl RA, Collares-Pereira M, Ellis WA. Emergence, control and re-emerging Leptospirosis: dynamics of infection in the changing world. Clin Microbiol. Infect. 2011;17:494–501. doi: 10.1111/j.1469-0691.2011.03474.x. [DOI] [PubMed] [Google Scholar]
- 4.Garba B, Bahaman AR, Khairani-Bejo S, Zakaria Z, Mutalib AR. Retrospective study of leptospirosis in Malaysia. EcoHealth. 2017;14:389–398. doi: 10.1007/s10393-017-1234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider MC, et al. Leptospirosis: a silent epidemic disease. Int. J. Environ. Res. Public Health. 2013;10:7229–7234. doi: 10.3390/ijerph10127229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koay TK, Nirmal S, Noitie L, Tan E. An epidemiological investigation of an outbreak of leptospirosis associated with swimming, Beaufort, Sabah. Med. J. Malaysia. 2004;59:455–459. [PubMed] [Google Scholar]
- 7.Wynwood SJ, et al. Leptospirosis from water sources. Pathog. Glob. Health. 2014;108:334–338. doi: 10.1179/2047773214Y.0000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Jalii IM, Bahaman AR. A review of human leptospirosis in Malaysia. Trop. Biomed. 2004;21:113–119. [PubMed] [Google Scholar]
- 9.Blasdell KR, Morand S, Perera D, Firth C. Association of rodent-borne Leptospira spp. with urban environments in Malaysian Borneo. PLoS Negl. Trop. Dis. 2019;13:e0007141. doi: 10.1371/journal.pntd.0007141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen-Ranberg EU, Pipper C, Jensen PM. Global patterns of Leptospira prevalence in vertebrate reservoir hosts. J. Wildlife Dis. 2016;52:468–477. doi: 10.7589/2014-10-245. [DOI] [PubMed] [Google Scholar]
- 11.Prevention, C. f. D. C. a. Leptospirosis: Risk of Exposure, https://www.cdc.gov/leptospirosis/exposure/index.html (2015).
- 12.Atil A, et al. Occupational determinants of leptospirosis among urban service workers. Int. J. Environ. Res. Public Health. 2020 doi: 10.3390/ijerph17020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haake DA, Levett PN. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015;387:65–97. doi: 10.1007/978-3-662-45059-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koizumi N, et al. Molecular and serological investigation of Leptospira and leptospirosis in dogs in Japan. J. Med. Microbiol. 2013;62:630–636. doi: 10.1099/jmm.0.050039-0. [DOI] [PubMed] [Google Scholar]
- 15.Vincent AT, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019;13:e0007270. doi: 10.1371/journal.pntd.0007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 17.Zainuddin MA, et al. Seroprevalence of leptospirosis among town service workers in Kelantan, Malaysia. Southeast Asian J. Trop. Med. Public Health. 1917;48:1222–1229. [Google Scholar]
- 18.Samsudin S, et al. Seroprevalence of leptospiral antibodies among healthy municipal service workers in Selangor. Adv. Public Health. 2015;208145:6. [Google Scholar]
- 19.Waggoner JJ, et al. Reverse-transcriptase PCR detection of Leptospira: absence of agreement with single-specimen microscopic agglutination testing. PLoS ONE. 2015;10:e0132988. doi: 10.1371/journal.pone.0132988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barragan V, Olivas S, Keim P, Pearson T. Critical knowledge gaps in our understanding of environmental cycling and transmission of Leptospira spp. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.01190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganoza CA, et al. Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med. 2006;3:e308. doi: 10.1371/journal.pmed.0030308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau CL, et al. Leptospirosis in American Samoa 2010: epidemiology, environmental drivers, and the management of emergence. Am. J. Trop. Med. Hyg. 2012;86:309–319. doi: 10.4269/ajtmh.2012.11-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ristow P, et al. Biofilm formation by saprophytic and pathogenic leptospires. Microbiology. 2008;154:1309–1317. doi: 10.1099/mic.0.2007/014746-0. [DOI] [PubMed] [Google Scholar]
- 24.Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi T, et al. Characterizing interactions of Leptospira interrogans with proximal renal tubule epithelial cells. BMC Microbiol. 2018;18:64. doi: 10.1186/s12866-018-1206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trueba G, Zapata S, Madrid K, Cullen P, Haake D. Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. Int. Microbiol. 2004;7:35–40. [PubMed] [Google Scholar]
- 27.Ashford DA, et al. Asymptomatic infection and risk factors for leptospirosis in Nicaragua. Am. J. Trop. Med. Hyg. 2000;63:249–254. doi: 10.4269/ajtmh.2000.63.249. [DOI] [PubMed] [Google Scholar]
- 28.Kurilung A, et al. Molecular detection and isolation of pathogenic Leptospira from asymptomatic humans, domestic animals and water sources in Nan province, a rural area of Thailand. Res. Vet. Sci. 2017;115:146–154. doi: 10.1016/j.rvsc.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Bovet P, Yersin C, Merien F, Davis CE, Perolat P. Factors associated with clinical leptospirosis: a population-based case-control study in the Seychelles (Indian Ocean) Int. J. Epidemiol. 1999;28:583–590. doi: 10.1093/ije/28.3.583. [DOI] [PubMed] [Google Scholar]
- 30.Bal AE, et al. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J. Clin. Microbiol. 1994;32:1894–1898. doi: 10.1128/JCM.32.8.1894-1898.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganoza CA, et al. Asymptomatic renal colonization of humans in the peruvian Amazon by Leptospira. PLoS. Trop. Dis. 2010;4:e612. doi: 10.1371/journal.pntd.0000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivasankari K, Shanmughapriya S, Natarajaseenivasan K. Leptospiral renal colonization status in asymptomatic rural population of Tiruchirapalli district, Tamilnadu, India. Pathog. Glob. Health. 2016;110:209–215. doi: 10.1080/20477724.2016.1222054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levett PN. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neela VK, et al. An outbreak of leptospirosis among reserve military recruits, Hulu Perdik, Malaysia. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:523–528. doi: 10.1007/s10096-018-03450-6. [DOI] [PubMed] [Google Scholar]
- 35.Ridzuan JM, Aziah BD, Zahiruddin WM. Study on seroprevalence and leptospiral antibody distribution among high-risk planters in Malaysia. Osong Public Health Res. Perspect. 2016;7:168–171. doi: 10.1016/j.phrp.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafei MN, et al. Seroprevalence of leptospirosis among town service workers on northeastern state of Malaysia. Int. J. Collab. Res. Intern. Med. Public Health. 2012;4:395–403. [Google Scholar]
- 37.Fernandes M, Vieira ML, Carreira T, Teodosio R. Sanitation workers from Portugal: Is there evidence of Leptospira spp? J. Infect. Public Health. 2019 doi: 10.1016/j.jiph.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Sato Y, et al. Environmental DNA metabarcoding to detect pathogenic Leptospira and associated organisms in leptospirosis-endemic areas of Japan. Sci. Rep. 2019;9:6575. doi: 10.1038/s41598-019-42978-1. [DOI] [PMC free article] [PubMed] [Google Scholar]