Abstract
Long-chain fatty acids are widely used in food and chemical industries, and the enzymatic preparation of fatty acids is considered an environmentally friendly process. In the present study, long-chain fatty acids were prepared by the enzymatic hydrolysis of rapeseed oil with a genetically engineered lipase. Because thermophilic lipase has strong stability at higher temperatures, it was more suitable for the industrial production of long-chain fatty acids. Therefore, the thermophilic lipase BTL2 from Geobacillus thermocatenulatus was efficiently expressed in E. coli BL21(DE3) cells with an enzyme activity of 39.50 U/mg followed by gene codon optimisation. Experimental results showed that the recombinant lipase BTL2 exhibited excellent resistance to certain organic solvents (n-hexane, benzene, ethanol, and butanol). The metal cation Ca2+ and the non-ionic surfactant Triton-100X enhanced enzyme activity by 7.36% and 56.21% respectively. Moreover, the acid value of the liberated long-chain fatty acids by hydrolysing rapeseed oil was approximately 161.64 mg KOH/g at 50 °C in 24 h, the hydrolytic conversion rate was 91.45%, and the productivity was approximately 6.735 mg KOH/g h. These results suggested that the recombinant lipase BTL2 has excellent hydrolytic performance for rapeseed oil and showed great potential for the enzymatic preparation of long-chain fatty acids.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02517-6) contains supplementary material, which is available to authorized users.
Keywords: Thermophilic lipase, Escherichia coli, Enzymatic hydrolysis, Rapeseed oil
Introduction
Lipase (EC 3.1.1.3) is any enzyme that catalyses the hydrolysis of triglycerides. Its diverse array of uses can be seen in many fields ranging from food and medicine to detergents and biofuels (Sarmah et al. 2018). In recent years, thermophilic lipases have gained increasing attention because of its stability at elevated temperatures (He et al. 2016). Lipase 2 from Geobacillus thermocatenulatus (BTL2) belongs to the lipase family 1.5, which is a typical thermophilic lipase with excellent catalytic properties such as strong thermal stability, tolerance to organic solvents, and high substrate selectivity. Due to the interesting performance of lipase BTL2, in recent years, studies have mainly focused on heterologous expression (Yamada et al. 2016), the relationship between structure and function (Kajiwara et al. 2020), and improvement of enzyme activity (Godoy et al. 2019). Therefore, there is an urgent need to construct an engineered bacterial platform that can efficiently express lipase for in-depth applicable exploration. Moreover, the application of liquid lipase is mainly focused on eukaryotic expression systems such as Pichia pastoris (Wang et al. 2019). However, there are relatively few studies on prokaryotic expression systems such as Escherichia coli, which have outstanding protein expression ability with simple culture conditions and short growth cycle. Its unique intracellular expression can also enrich the protein content, which has unique advantages in the application of liquid lipase.
Fatty acids are some of the most widely used raw materials in oleochemicals, and they are mainly derived from natural oils and petroleum. In contrast, natural oils and fats derived from plants and animals are mixtures of glycerides rich in fatty acids that are renewable resources (Cui et al. 2019). Natural oils such as rapeseed oil, soybean oil, cottonseed oil, rice bran oil, and palm oil are inexpensive and readily available. Long-chain fatty acids (LCFAs) are organic fatty acids composed of 14 to 24 carbon atoms. They are widely found in animal fats, vegetable oils, and marine oils, and they are used in the pharmaceutical, cosmetic, fragrance, and detergent industries (Zhu et al. 2013). LCFAs can also be classified into two categories: saturated long-chain fatty acids and unsaturated long-chain fatty acids (ULCFAs), where ULCFAs are mainly composed of ω-3, ω-6 and ω-9 polyunsaturated LCFAs. The ω-3 and ω-9 polyunsaturated ULCFAs are essential nutrients that have important metabolic functions (Al-Jawadi et al. 2018).
LCFAs, especially ULCFAs, are widely used in the food and chemical industries, and are important for human health. Natural oils such as rapeseed oil (RSO) are rich in UCLFAs, which can be produced via enzymatic hydrolysis, and this method is environmentally friendly. Also, the preparation of LCFAs via the hydrolysis of RSO with lipase has recently become very popular with researchers due to the mild reaction conditions, simplicity, low formation of by-products, and unfavourable oxidation of unsaturated fatty acids (Li et al. 2018). However, lipase-mediated hydrolysis reactions are relatively rare at higher temperatures; therefore, using a thermophilic lipase would introduce a unique advantage to this application. Thermophilic lipase could also provide the stability that is more appropriate for the industrial production of LCFAs at higher temperatures. Therefore, in this study, we developed an enzymatic method to produce LCFAs using thermophilic lipase: The lipase gene BTL2 from G. thermocatenulatus was cloned and expressed in E. coli, and the recombinant lipase BTL2 was further characterised by measuring enzyme activity; The enzymatic hydrolysis of RSO into LCFAs using crude lipase BTL2 according to biochemical properties.
Materials and methods
Strains, plasmids and reagents
The gene BTL2 was optimised and synthesised by Synbio Technologies (Suzhou, China). Specifically, a series of chemically synthesised oligonucleotides were spliced into the desired gene sequence by PCR and then cloned the gene fragment into the pUC18 vector. E. coli DH5α and BL21(DE3) cells (Transgen, Beijing, China) were used for gene cloning and gene expression. All E. coli strains were cultured in LB medium. The pMD18-T vector was used as the cloning vector, and the pET28b ( +) vector was used as the expression vector. DNA polymerase, T4 DNA ligase, and restriction enzymes were purchased from Takara (Dalian, China). The enzymatic substrate of ρ-nitrophenylpalmitate (ρNPP) (99%) was obtained from Aladdin (Shanghai, China). All other chemicals were of analytical grade.
Optimization and synthesis of lipase gene BTL2
The gene BTL2 was obtained from GeneBank (accession no. X95309), and the CDS coding sequences (1254 bp) were optimised by Synbio Technologies (Suzhou, China). To optimise the gene for E. coli, some nucleotides of gene BTL2 were changed, the GC content was decreased to 53%, and the codon adaptation index (CAI) was increased to 0.86. The variation curve of the GC content was optimised, and stable hairpin structures were minimised.
Gene cloning, expression, and purification
Using a pUC18 vector carrying the synthetic gene BTL2 as a template, the primers BTL2-F/R were used to amplify the target gene fragment, and correct gene open reading frames (ORFs) were ligated into the pET28b ( +) vector, generating plasmids BTL2-pET28b ( +). Recombinant plasmids were verified by sequencing using the primers MCS-F/R and double restriction digests (Noc I and Xho I). Then, the correct plasmids were propagated and transformed into BL21(DE3) competent cells. The transformants were cultured in fresh LB medium for approximately 45 min; then, a kanamycin-resistant plate was uniformly coated with cells and was incubated overnight. Subsequently, single colonies were picked and cultured for 1 h. Positive clones were selected by PCR using the primers BTL2-F/R (Table 1) and were cultured 12–14 h as seed solution used to inoculate 200 mL fresh LB media (3% inoculum at 37 °C with shaking at 180 rpm) in a 500-mL Erlenmeyer flask. When the OD600 reached approximately 1.0 after 18–20 h, 1 mM of isopropyl-â-D-thiogalactoside (IPTG) was added to induce protein expression at 37 °C with shaking at 150 rpm for 4 h. The induced cells were resuspended in 20 mL of 50 mM Tris–HCl buffer at pH 8.0 and lysed by ultrasonic cell disruption. The crude enzyme extracts were obtained by centrifugation at 4 °C and analysed by SDS-PAGE; then, they were concentrated twice by centrifugal filter devices (Merck Millipore Ltd., Darmstadt, Germany) for protein purification. Additionally, separation and purification of lipase BTL2 by the protein purification system (NGC, Bio-Rad, US) based on the C-terminus of His-Tag by a nickel column using binding buffer (PBS, pH7.5, 10% glycerol), washing buffer (PBS, pH7.5 with 0, 10 mM and 30 mM imidazole) and elution buffer (PBS, pH7.5 with 300 mM imidazole).
Table 1.
Primers used in this study
| Primers | Sequences (5′–3′)a |
|---|---|
| BTL2-F | GCCATGGATGGCAAGTCCGCGCGCAA (Nco I) |
| BTL2-R | GGCTCGAGTTAATGGTGATGGTGATGATGCGGACG (Xho I) |
| MCS-F | CGAAATTAATACGACTCACTATAGG |
| MCS-R | CCCAAGGGGTTATGCTAGTTATTGC |
aRestriction enzymes site were marked in italic
Assay of lipase activity
Lipase activity was determined using ρ-nitrophenylpalmitate (ρNPP) as the substrate. Cleavage of ρNPP was measured at pH 8.0 and 60 °C using 50 mM Tris–HCl buffer. One unit was defined as the amount of recombinant lipase BTL2 that caused the release of 1 μmol of ρ-nitrophenol (ρNP) per min under the test conditions. The concentration of ρ-nitrophenol (CρNP) was determined by measuring the OD410 (Y = 0.0165X + 0.0053, R2 = 0.9999, Y: OD410; X: CρNP), and the protein concentration (Cp) of purified lipase BTL2 was determined by analysis of OD562 (Y = 0.0046X + 0.0031, R2 = 0.9980, Y: OD562; X: Cp) according to the bicinchoninic acid (BCA) protein assay kit (Sangon, Shanghai, China). An inactivated lipase solution was used as the blank control, and all experiments were performed in duplicate. The temperature was controlled using a water bath.
Effects of pH on the recombinant lipase
To investigate the effects of pH on lipase activity, the optimum pH and pH stability of recombinant lipase were determined by testing the enzyme activity at different pH levels with phosphate buffers (pH 6.0–7.0), Tris–HCl buffers (pH 7.5–8.0) and Gly–NaOH buffers (8.5–10.5) at 60 °C. For pH stability, the recombinant lipase BTL2 was incubated under the same conditions for 1 h followed by the enzyme activity assay.
Effects of temperature on the recombinant lipase
To evaluate the effects of temperature on lipase activity, the optimal temperature of the recombinant lipase was determined by testing the enzyme activity at different temperatures from 30 to 80 °C at pH 8.0. For thermal stability, the recombinant lipase was determined by incubating the lipase at the test temperature for 1 h followed by the enzyme activity assay.
Hydrolysis kinetics of the recombinant lipase
To determine the hydrolysis kinetics of ρNPP, the lipase activity assay was used, and the product ρNP was measured using various concentrations of ρNPP. Sample concentrations were analysed at substrate concentrations of 0.4, 0.8, 1, 1.2, 1.6, 2, and 4 μmol.
Effects of organic solvents, metal ions, and surfactants on lipase activity
Various metal ions, organic solvents, and surfactants were added to the solution containing the recombinant lipase and incubated for 1 h at pH 8.0 and 60 °C. The concentration of each metal ion sample (Mn2+, Mg2+, Zn2+, Ca2+, Co2+, Cu2+, Fe2+, Fe3+, Ni2+, and EDTA) was 20 mM. The volume/volume percentage of each organic solvent (methanol, ethanol, isopropanol, n-butanol, isopentanol, benzene, methylbenzene, styrene, n-hexane, n-heptane, acetone, and furfural) was 1%. The volume/weight percentage of each surfactant (Triton-100X, Tween 20, Tween 80, sodium dodecyl sulphate (SDS), gum arabic, polyethylene glycol) was 1%. The relative activity of the control samples without treatment was set at 100%.
Hydrolysis of rapeseed oil by the recombinant lipase
The hydrolytic reaction mixture comprised 5 g of RSO, 5 g of deionised water, 2500 U of crude lipase BTL2, 20 mM Ca2+, and 1% Triton-100X. The solution was mixed in a 150-mL single-necked flask equipped with a condensation tube, and hydrolysed at 50 °C for 36 h using a constant temperature magnetic stirrer. At the end of the reaction, the upper layer of hydrolysate was collected by centrifugation and dried, and then the acid value was measured using a 99% ethanol solution of KOH. Moreover, to compare the catalytic performance of recombinant lipase BTL2 with other commercial lipases, the LipozymeTL100L (Thermomyces lanuginose lipase, TLL) and Palatase 20000L (Rhizomucor miehei lipase, RML) from Novozymes were used to perform the same catalytic hydrolysis of RSO. Inactivated lipase solution was used as a blank control, and all experiments were performed in duplicate. The RSO conversation ratio was as follows (Eq. 1):
| 1 |
AV is the acid value of the sample after hydrolysis (mg KOH/g); AV′ is the acid value of the RSO (mg KOH/g); AV′′ is the acid value of the control sample (mg KOH/g); SV is the saponification value of RSO (mg KOH/g).
Analytical methods
The OD600 and OD410 were analysed by an Eon Microplate Reader (Gene Company Limited, Chai Wan, Hong Kong). The purified lipase BTL2 was analysed by liquid chromatography-mass spectrometry (LC–MS), and this test was performed by FitGene Biotechnology Co., Ltd (Guangzhou, China). The mass spectrum data of enzymatically hydrolysed polypeptides were obtained, which were combined with a protein database (https://www.uniprot.org/taxonomy/33938) to search and analyse. Liquid chromatography (Dionex Ultimate 3000 RSLCnano) and mass spectrometer (Q Exactive) equipped with Acclaim™ PepMap™ 100 C18 (Thermo Scientific, Shanghai, China). Mobile phase A was 0.1% formic acid, mobile phase B was 0.1% formic acid, and 80% acetonitrile. The analysis process lasted 60 min and maintained a flow rate of 300 nL/min. The acid and saponification values were determined by acid–base titration. All analysis procedures were performed in duplicate.
Results and discussion
Plasmid construction, gene expression, and protein purification
The gene encoding the BTL2 has previously been cloned in E. coli and expressed (Velez et al. 2009), but its expression requires higher temperature induction at 42 °C and E. coli is known to have codon usage bias (CUB) (Rosano and Ceccarelli 2014). Therefore, it was necessary to optimise the codons in the gene sequence and use a strong promoter T7 to achieve efficient expression of lipase BTL2. To achieve high-efficiency expression of the lipase BTL2 in E. coli, the coding sequence (CDS) of the gene BTL2 was codon-optimised and synthesised. However, according to the crystal structure of lipase BTL2 (PDB: 2W22), as well as analysis of signal peptides using SignalP3.0 (Bendtsen et al. 2004), the N-terminal sequence of mature lipase started from the 29th amino acid, and the first to twenty-ninth amino acids are regarded as signal peptides. Because E. coli cannot recognise and cleave the signal peptide of lipase BTL2 (Paetzel et al. 1998), the amplified gene fragment (1170 bp) did not contain the gene sequence encoding the first 29 amino acids.
The recombinant plasmid, BTL2-pET28b ( +) (Fig. 1a), containing the gene BTL2, was transformed into E. coli BL21(DE3) competent cells by heat shock. The screened positive clones were induced to express lipase BTL2 by IPTG, and the SDS-PAGE analysis showed the presence of a protein band of approximately 43 kDa for purified protein, which was consistent with the theoretical molecular weight (Fig. 1b). Moreover, E. coli was better able to express the enzyme protein because of the codon optimisation of lipase gene BTL2, and the yield of purified lipase BTL2 was 2.2 mg/mL.
Fig. 1.
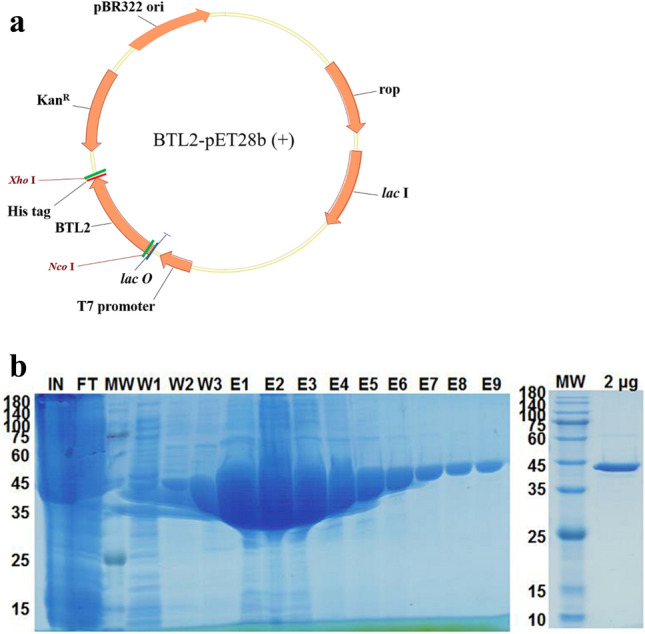
BTL2-pET28b ( +) recombinant plasmid construction (a), and protein purification profile of recombinant lipase BTL2 by SDS-PAGE (b). MW molecular weight marker, IN Input samples, FT flow-through samples, W washes samples (W1–W3), E eluted fractions samples (E1–E9). 2 μg was the loading amount of purified recombinant BTL2
Qualitative and quantitative analysis of BTL2 by LC–MS
LC–MS is a sampling system that uses both liquid chromatography to separate complex chemical components and mass spectrometry as a detector for quantitative and qualitative analysis, which can accurately and quickly identify proteins (Wu et al. 2018). According to the Mascot (https://www.matrixscience.com/) search results, it could be concluded that the recombinant protein had the highest similarity and also the highest score compared with the lipase BTL2 dated from G. thermocatenulatus, confirming that the recombinant protein identified was thermophilic lipase BTL2. Quantitative analysis also showed that the molecular weight of the recombinant lipase was 46.334 kDa and the isoelectric point was 6.68 (Table 2). Besides, the abundance index of purified lipase BTL2 was 40.23, which was much larger than that of other proteins, which indicated that the recombinant lipase BTL2 had a high protein abundance in the purification solution (approximately 90.04%), and showed that the result of protein purification was favourable.
Table 2.
Quantitative analysis of BTL2 by LC–MS
| Lipase | Mass of protein | Isoelectric point | emPAIa |
|---|---|---|---|
| BTL2 | 46,334 | 6.68 | 40.23 |
aThe protein abundance index, indicating the abundance of the protein in the sample, the larger the value, and the higher the abundance. The total emPAI of lipase BTL2 was 44.68
Biochemical analysis of recombinant lipase BTL2
pH effects on lipase activity
The pH of a solution is an important property that affects the individual amino acid residues of a protein, and its enzyme activity (Lessard and Hill 2000). The recombinant lipase BTL2 expressed in E. coli had catalytic activity between pH 6.0 and 10.5 (Fig. 2). At pH 8.0, the catalytic activity of lipase BTL2 was the highest (~ 38.44 U/mg). Moreover, alkali adaptability of this lipase was also observed (Fig. 2). After incubation for 1 h, the recombinant lipase exhibited higher enzyme activity at pH values from 7.5 to 8.5, than at other pH values. At pH 8.0, this lipase showed the highest activity of ~ 35.53 U/mg. This lipase resembles other bacterial lipases, which typically exhibit high pH stability and adaptability. For example, lipase lipSM54 from Stenotrophomonas maltophilia GS11 was found to have an enzyme activity in the pH range of 7.0 to 8.0, and its optimal activity was observed at pH 8.0 (Li et al. 2016).
Fig. 2.
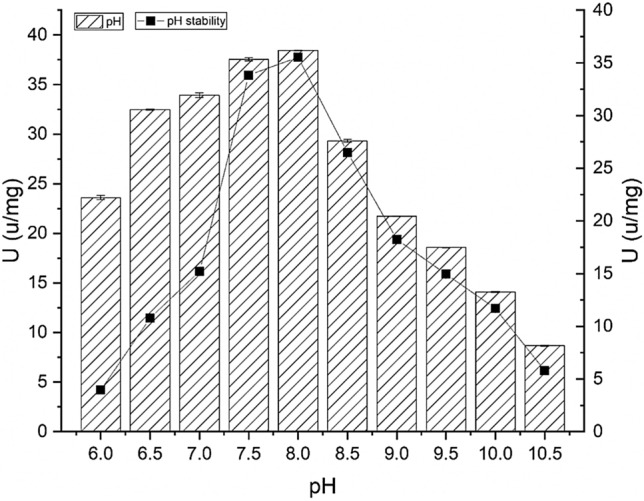
Effects of pH on recombinant lipase BTL2. The optimal pH was measured at 60 °C with varying buffer pH; lipase BTL2 was incubated in various pH buffers for 1 h before enzyme activity analysis to verify pH stability
The superior adaptability of lipase BTL2 to pH is closely related to its primary structure. The stability of covalent bonds on the primary structure controls the enzyme conformation in an acid–base environment and maintains enzyme activity. It is generally accepted that an inappropriate reaction pH may change the ionisation state of key functional amino acid residues (Fernandes et al. 2004). Amino acid residues located in the lid and catalytic centre of lipase contribute more to the stability of lipase BTL2 and could be especially affected by pH. Furthermore, the ionic bonds that help to maintain the rigidity of three-dimensional enzyme structures can change in various environments, which would cause enzyme denaturation or the loss of enzyme activity at inappropriate pH conditions (Ibarra-Molero et al. 1999).
Temperature effects on lipase activity
G. thermocatenulatus is adapted to a unique thermal environment; therefore, its endogenous lipase is very stable at high temperatures (Rua et al. 1997). Based on our analysis, the optimal temperature for the lipase expressed in E. coli was 60 °C (39.50 U/mg), and this lipase was catalytically active in the temperature range of 30 °C to 80 °C (Fig. 3). At temperatures lower than the optimum, the reaction rate constant decreased (Gomes et al. 2003), and the lower temperatures hindered the activation of lipase because the lid structure of the active centre was only partially open (Brzozowski et al. 2000; Murakami et al. 2005). Additionally, at temperatures lower than optimum, lipase activity increased with increasing temperature. The increase in temperature improved the spatial structure of lipase, and consequently its catalytic performance. However, at temperatures higher than optimum, lipase activity decreased sharply because of the denaturation of lipase. High temperatures cause the loss of ionic bonds, hydrogen bonds, and other forces that maintain the structure. The α-helical content of the enzyme decreases (Goncalves et al. 2014), but the β-sheet content increases (Cordeiro et al. 2004). These changes likely resulted in a conformational change in the lipase during inactivation, possibly leading to denaturation.
Fig. 3.
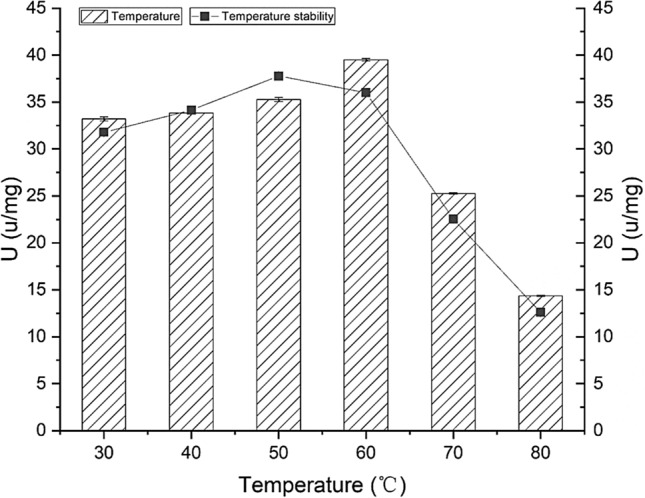
Effects of temperature on recombinant lipase BTL2. The optimal temperature was measured at pH 8.0 with varying temperatures; lipase BTL2 was incubated at various temperatures for 1 h before lipase activity analysis to verify temperature stability
Based on the thermostability analysis, after 1 h of incubation at 80 °C, 70 °C, and 60 °C, the recombinant lipase activity was approximately 12.56 U/mg, 22.53 U/mg, and 36.00 U/mg, respectively (Fig. 3). These values were approximately 33.25%, 59.63%, and 95.29% of the maximum activity at 50 °C (37.78 U/mg), respectively. These results showed that the recombinant lipase BTL2 was similar to other bacterial lipases in thermostability. However, the activity decreased when the lipase was incubated at high temperatures (> 60 °C) for a long time, which could be due to the heat-induced denaturation of lipase.
Study of the reaction kinetics of recombinant lipase
At fixed concentrations of substrate (138.1 µg/mL) and purified lipase BTL2 (44 µg), the concentration of released ρ-nitrophenol increased with the reaction time, and the lipase-catalysed hydrolysis of ρNPP followed a typical Michaelis–Menten equation. According to the equation, the Michaelis–Menten constant (Km) of recombinant lipase BTL2 was 1.5831 μmol/L, the maximum reaction rate (Vmax) was 1.2003 μmol/min/mg, and the catalytic constant (Kcat) was 0.9269/s.
Catalytic performance stability is important for the possible applications of lipase because the accumulation of the hydrolysate at the reaction interface would affect the lid structure of the active centre, which in turn leads to deactivation (Zhang et al. 2015). The linear kinetics of lipase BTL2 demonstrated that the catalytic capacity of the recombinant lipase was stable and that it maintained a normal catalytic ability; this could contribute to the enzyme’s resistance to denaturation by inhibiting the accumulation of the hydrolysate at the reaction interface.
Effects of metal ions, organic solvents and surfactants on lipase activity
The effects of metal ions on lipase are shown in Table 3. It was found that Ca2+ enhanced the recombinant lipase activity of lipase BTL2 compared to the control non-ion treatment by 7.36%. Ca2+ plays an important role in the structure and function of lipases (El Khattabi et al. 2003). According to previous studies (Won et al. 2005), various lipases are activated by Ca2+. Moreover, the structure site of bacterial lipase associated with Ca2+ and the mechanism of activation of Ca2+ have also been elucidated (Amada et al. 2001). The activation of lipase by Ca2+ may be related to the structure of lipase (Malhotra et al. 2000; Hide et al. 1992). Approximately 94.34% and 95.65% of its activity were retained for 1 h in the presence of Mg2+ and Mn2+, respectively. Meanwhile, after incubation with Zn2+, Co2+, Cu2+, Fe2+, Fe3+ or EDTA for 1 h, the activity of lipase BTL2 decreased dramatically; the relative activity only remained in the presence of Fe2+ (1.87%), Fe3+ (5.04%), and Co2+ (10.50%). These results suggest that metal ions may affect the enzyme activity to varying degrees; these ions may interact with amino acid residues in lipase (Chor Leow et al. 2007), consequently causing the deactivation of lipase.
Table 3.
Effects of organic solvents, metal ions, and surfactants on lipase activity
| Effects of lipase activity | Relative activity (%) | |
|---|---|---|
| Influencing factors | Enzyme activity (U/mg) | |
| Control samples | 31.67 ± 0.26 | 100% |
| Metal ions | ||
| Mn2+ | 30.29 ± 0.25 | 95.65 |
| Mg2+ | 29.88 ± 0.06 | 94.34 |
| Zn2+ | 4.18 ± 0.20 | 13.18 |
| Ca2+ | 34.00 ± 0.06 | 107.36 |
| Co2+ | 3.32 ± 0.04 | 10.50 |
| Cu2+ | 3.99 ± 0.31 | 12.61 |
| Fe2+ | 0.59 ± 0.07 | 1.87 |
| Fe3+ | 1.60 ± 0.06 | 5.04 |
| Ni2+ | 12.08 ± 0.08 | 38.15 |
| EDTA | 6.00 ± 0.34 | 18.94 |
| Organic solvents | ||
| Methanol | 22.88 ± 0.18 | 72.24 |
| Ethanol | 33.98 ± 0.18 | 107.29 |
| Isopropanol | 15.00 ± 0.01 | 47.36 |
| n-Butanol | 32.60 ± 0.07 | 102.93 |
| Isopentanol | 17.33 ± 0.22 | 54.71 |
| Benzene | 33.98 ± 0.18 | 72.24 |
| Methylbenzene, | 25.74 ± 0.01 | 81.29 |
| styrene | 25.11 ± 0.01 | 79.28 |
| n-Hexane | 37.34 ± 0.14 | 117.90 |
| n-Heptane | 24.21 ± 0.47 | 76.44 |
| Acetone | 26.06 ± 0.02 | 82.29 |
| Furfural | 21.40 ± 0.22 | 67.57 |
| Surfactants | ||
| Triton-100X | 49.47 ± 0.56 | 156.21 |
| Tween 20 | 1.73 ± 0.01 | 5.48 |
| Tween 80 | 5.37 | 16.95 |
| SDS | 3.09 ± 0.43 | 9.77 |
| Gum arabic | 33.72 ± 0.17 | 106.48 |
| Polyethylene glycol | 40.30 ± 0.12 | 127.26 |
The effects of organic solvents on lipase activity were investigated, as shown in Table 3. After 1-h incubation with 1% organic solvent, the enzyme activity of lipase BTL2 changed significantly and showed different degrees of decline. The lipase activity decreased to 72.24%, 81.29%, 82.29%, 76.44%, and 79.28% after treatment with methanol, methylbenzene, acetone, n-heptane, and styrene, respectively, when compared to the control. However, lipase BTL2 showed good tolerance to n-hexane, benzene, ethanol and butanol, and the lipase activity remained at 117.90%, 107.36%, 107.29%, and 102.93%, respectively. The tolerance of lipase BTL2 to organic solvents indicates its potential for catalytic synthesis in the organic phase. Furthermore, the lipase activity decreased sharply and only retained enzyme activities of 67.60%, 54.71%, and 47.36% when treated with furfural, isopentanol and isopropanol, respectively. This was likely because the organic solvent was able to destroy the secondary structure of lipase, causing the denaturation of lipase BTL2 (Yenenler et al. 2018). Therefore, during lipase-catalysed organic synthesis, it is necessary to select a suitable reaction solvent or to improve the tolerance of lipase by immobilisation (Hu et al. 2018; Adlercreutz 2013).
Further investigation of the effects of surfactants on lipase activity is shown in Table 3. It was found that the surfactants gum arabic and polyethylene glycol slightly activated lipase BTL2 and increased its enzyme activity by 6.48% and 27.26%, respectively. Meanwhile, non-ionic surfactant Triton-100X greatly activated lipase BTL2 and enhanced its enzyme activity by 56.21%. Tween 20, Tween 80, and SDS also significantly inhibited the activity of lipase BTL2; the lipase BTL2 retained only 5.48%, 16.95% and 9.77% of its enzyme activity, respectively. Surfactants are known to affect the lid structure of the lipase catalytic centre, and different surfactants may promote or inhibit the opening of the structure, thereby leading to diverse enzyme activity (Singh et al. 2007; Delorme et al. 2011). These findings were consistent with previous studies (Quyen et al. 2003); therefore, concerning future enzymatic applications, the appropriate concentration of Triton-100X could be added to enhance lipase activity.
Enzymatic hydrolysis of rapeseed oil
To evaluate the potential of lipase BTL2 to produce LCFAs, the enzymatic hydrolysis of RSO was analysed considering the factors likely to affect the activity of the recombinant BTL2. The ability of BTL2 to hydrolyse RSO to produce LCFAs was investigated with a water to oil ratio of 1:1 (g/g), an enzyme activity of 2500 U, and the addition 1% of Ca2+ and Triton-100X at 50 °C for 36 h. This study found that the recombinant lipase BTL2 had an excellent biocatalytic ability to achieve mild hydrolysis of triglycerides to form LCFAs (Fig. 4).
Fig. 4.
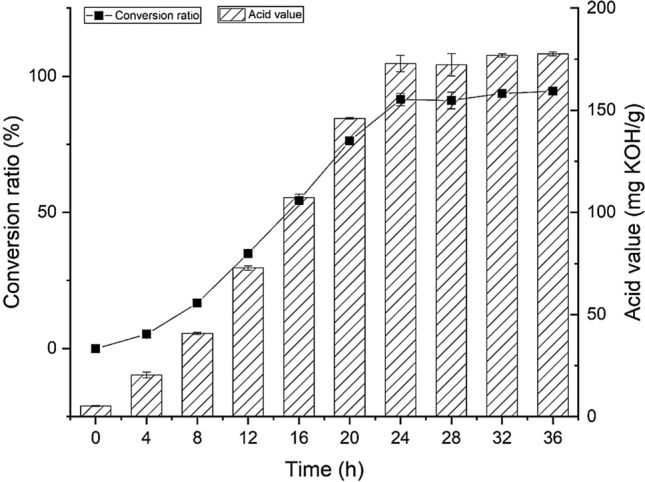
Reaction time curve of recombinant lipase BTL2 to release long-chain fatty acids. The hydrolysis reaction of rapeseed oil catalysed by recombinant lipase BTL2 was carried out under 50 °C in 32 h
The acid value (AV) and saponification value (SV) of RSO were 5.26 and 182 mg KOH/g, respectively. AV directly reflects the change in the content of fatty acids. As the reaction time increased, the AV also increased, indicating that the content of LCFAs also undergoing the same trend change. The AV increased slowly from 0 to 8 h, and from 8 to 24 h, the AV almost increased quickly. This indicates that the lipase gradually opens the lid structure and exposes the active catalytic centre, and the hydrolysis reaction proceeds rapidly at the oil–water interface. From 24 to 36 h during hydrolytic reaction, the AV trended to be stable, showing that the reaction rate gradually decreased with the extension of time, and the conversion rate slowly increased and tended to be steady. Moreover, the hydrolysis reaction of RSO catalysed by lipase BTL2 was reversible, and as the products accumulated, the reverse reaction gradually intensified. Therefore, the hydrolysis time of RSO was determined to be 24 h. The acid value of the final liberated fatty acid was 161.64 mg KOH/g, and the conversion rate was 91.45%.
Moreover, two commercial liquid lipases were selected to compare the catalytic performance of crude lipase BTL2 expressed in E. coli, and the catalytic performance was defined as the conversion rate of released long-chain fatty acid per unit enzyme activity and unit time. As shown in Table 4, the catalytic performance of lipase BTL2 was 7.04% higher than that of T. lanuginose lipase, indicating that these two thermophilic lipases were equivalent in terms of enzymatic hydrolysis of RSO; while the catalytic performance of lipase BTL2 was much better than that of R. miehei lipase, which was as high as 90.00%. Besides, a previous study investigated liquid lipases for hydrolysis of RSO at 37 °C, and the commercial lipases from C. rugosa and Rhizopus sp. resulted in the conversion rate of nearly 100% over 24 h (Wu et al. 1996). This showed that the recombinant lipase BTL2 from G. thermocatenulatus had excellent hydrolysis activity for rapeseed oil at higher temperatures, and it’s a promising biocatalyst for the preparation of LCFAs, especially ULCFAs. Furthermore, this recombinant lipase BTL2 could also be used in the field of environmental remediation to treat industrial or food waste oils to reduce pollution.
Table 4.
Comparison of the hydrolytic performance from various lipases
| Lipases | Quantity/kU | Conversion rate/%a | Hydrolytic propertyb |
|---|---|---|---|
| Recombinant lipase BTL2 | 2.5 | 91.45 ± 2.23 | 1.52 |
| T. lanuginose lipase | 2.8 | 95.55 ± 1.9 | 1.42 |
| R. miehei lipase | 5.0 | 96.13 ± 2.68 | 0.80 |
a5 g RSO was hydrolysed by various lipases at 50 °C in 24 h
bRSO hydrolysis conversion rate for unit enzyme activity and per hour
Conclusions
The thermophilic lipase BTL2 was successfully expressed in E. coli after gene codon optimisation with the enzyme activity 39.50 U/mg. Besides, this recombinant lipase BTL2 was active in a weakly basic environment at high temperatures and exhibited good resistance to organic solvents, especially n-hexane, benzene, ethanol and butanol, while Ca2+ and Triton-100X could active lipase BTL2. To our knowledge, this is the first report of enzymatic hydrolysis of RSO to produce LCFAs at 50 °C in 24 h with a yield of 91.45% using crude lipase BTL2, indicating that recombinant lipase BTL2 was a promising biocatalyst for the preparation of long-chain fatty acids.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Availability of data and material
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study. Code availability NCBI: X95309; PDB: 2W22.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
Conceptualization, Methodology, Software, Investigation, Writing—original draft: ZJ; Validation, Formal analysis, Visualization, Software: TM; Supervision, Data curation: LW and WZY; Resources, Writing—review & editing, Supervision, Data curation: LPM, XJL and WZM.
Funding
This work was supported by the Guangdong Special Support Program (No. 2017TX04Z109), the National Natural Science Foundation of China (No. 51606201), Science and Technology Planning Project of Guangdong Province (No. 2016A010104008), the Natural Science Foundation of Guangdong Province (No. 2017A010104010), the National Key research and development program of China (No. 2019YFB1504003), the National Natural Science Foundation of China (No. 51903236) and the Projects of International Cooperation and Exchanges NSFC (No. 51861145103).
Contributor Information
Pengmei Lv, Email: lvpm@ms.giec.ac.cn.
Jingliang Xu, Email: xujl@zzu.edu.cn.
References
- Adlercreutz P. Immobilisation and application of lipases in organic media. Chem Soc Rev. 2013;42(15):6406–6436. doi: 10.1039/c3cs35446f. [DOI] [PubMed] [Google Scholar]
- Al-Jawadi A, Moussa H, Ramalingam L, Dharmawardhane S, Gollahon L, Gunaratne P, Rahman RL, Moustaid-Moussa N. Protective properties of n-3 fatty acids and implications in obesity-associated breast cancer. J Nutr Biochem. 2018;53:1–8. doi: 10.1016/j.jnutbio.2017.09.018. [DOI] [PubMed] [Google Scholar]
- Amada K, Kwon HJ, Haruki M, Morikawa M, Kanaya S. Ca2+-induced folding of a family 1.3 lipase with repetitive Ca2+ binding motifs at the C-terminus. Febs Lett. 2001;509(1):17–21. doi: 10.1016/s0014-5793(01)03108-8. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Savage H, Verma CS, Turkenburg JP, Lawson DM, Svendsen A, Patkar S. Structural origins of the interfacial activation in Thermomyces (Humicola) lanuginosa lipase. Biochemistry-Us. 2000;39(49):15071–15082. doi: 10.1021/bi0013905. [DOI] [PubMed] [Google Scholar]
- Chor Leow T, Noor Zaliha Raja Abd Rahman R, Basri M, Salleh AB (2007) A thermoalkaliphilic lipase of Geobacillus sp T1. Extremophiles 11 (3):527–535. 10.1007/s00792-007-0069-y [DOI] [PubMed]
- Cordeiro Y, Kraineva J, Ravindra R, Lima L, Gomes MPB, Foguel D, Winter R, Silva JL. Hydration and packing effects on prion folding and beta-sheet conversion—High pressure spectroscopy and pressure perturbation calorimetry studies. J Biol Chem. 2004;279(31):32354–32359. doi: 10.1074/jbc.M404295200. [DOI] [PubMed] [Google Scholar]
- Cui Y, Thomas-Hall SR, Schenk PM. Phaeodactylum tricornutum microalgae as a rich source of omega-3 oil: progress in lipid induction techniques towards industry adoption. Food Chem. 2019 doi: 10.1016/j.foodchem.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Delorme V, Dhouib R, Canaan S, Fotiadu F, Carriere F, Cavalier JF. Effects of surfactants on lipase structure, activity, and inhibition. Pharm Res-Dordr. 2011;28(8):1831–1842. doi: 10.1007/s11095-010-0362-9. [DOI] [PubMed] [Google Scholar]
- El Khattabi M, Van Gelder P, Bitter W, Tommassen J. Role of the calcium ion and the disulfide bond in the Burkholderia glumae lipase. J Mol Catal B-Enzym. 2003;22(5–6):329–338. doi: 10.1016/s1381-1177(03)00047-x. [DOI] [Google Scholar]
- Fernandes MLM, Krieger N, Baron AM, Zamora PP, Ramos LP, Mitchell DA. Hydrolysis and synthesis reactions catalysed by Thermomyces lanuginosa lipase in the AOT/Isooctane reversed micellar system. J Mol Catal B-Enzym. 2004;30(1):43–49. doi: 10.1016/j.molcatb.2004.03.004. [DOI] [Google Scholar]
- Godoy CA, Klett J, Di Geronimo B, Hermoso JA, Guisan JM, Carrasco-Lopez C (2019) Disulfide engineered lipase to enhance the catalytic activity: a structure-based approach on BTL2. Int J Mol Sci 20 (21). 10.3390/ijms20215245 [DOI] [PMC free article] [PubMed]
- Gomes I, Gomes J, Steiner W. Highly thermostable amylase and pullulanase of the extreme thermophilic eubacterium Rhodothermus marinus: production and partial characterization. Bioresour Technol. 2003;90(2):207–214. doi: 10.1016/s0960-8524(03)00110-x. [DOI] [PubMed] [Google Scholar]
- Goncalves KM, Barbosa LRS, Lima LMTR, Cortines JR, Kalume DE, Leal ICR, Mariz e Miranda LS, de Souza ROM, Cordeiro Y (2014) Conformational dissection of Thermomyces lanuginosus lipase in solution. Biophys Chem 185:88–97. 10.1016/j.bpc.2013.12.001 [DOI] [PubMed]
- He HM, Han HB, Shi H, Tian YY, Sun FX, Song Y, Li QS, Zhu GS. Construction of thermophilic lipase-embedded metal organic frameworks via biomimetic mineralization: a biocatalyst for ester hydrolysis and kinetic resolution. Acs Appl Mater Interfaces. 2016;8(37):24517–24524. doi: 10.1021/acsami.6b05538. [DOI] [PubMed] [Google Scholar]
- Hide WA, Chan L, Li WH. Structure and evolution of the lipase superfamily. J Lipid Res. 1992;33(2):167–178. [PubMed] [Google Scholar]
- Hu YL, Dai LM, Liu DH, Du W, Wang YJ. Progress & prospect of metal-organic frameworks (MOFs) for enzyme immobilization (enzyme/MOFs) Renew Sustain Energy Rev. 2018;91:793–801. doi: 10.1016/j.rser.2018.04.103. [DOI] [Google Scholar]
- Ibarra-Molero B, Loladze VV, Makhatadze GI, Sanchez-Ruiz JM (1999) Thermal versus guanidine-induced unfolding of ubiquitin. An analysis in terms of the contributions from charge-charge interactions to protein stability. Biochemistry-Us 38(25):8138–8149. 10.1021/bi9905819 [DOI] [PubMed]
- Kajiwara S, Yamada R, Matsumoto T, Ogino H. N-linked glycosylation of thermostable lipase from Bacillus thermocatenulatus to improve organic solvent stability. Enzyme Microb Tech. 2020 doi: 10.1016/j.enzmictec.2019.109416. [DOI] [PubMed] [Google Scholar]
- Lessard LP, Hill CG. Effect of pH on the production of lipolyzed butter oil by a calf pregastric esterase immobilized in a hollow-fiber reactor: II Multiresponse kinetics. Biotechnol Bioeng. 2000;70(3):332–341. doi: 10.1002/1097-0290(20001105)70:3<332::Aid-bit10>3.0.Co;2-g. [DOI] [PubMed] [Google Scholar]
- Li M, Yang LR, Xu G, Wu JP. Cloning and characterization of a novel lipase from Stenotrophomonas maltophilia GS11: the first member of a new bacterial lipase family XVI. J Biotechnol. 2016;228:30–36. doi: 10.1016/j.jbiotec.2016.04.034. [DOI] [PubMed] [Google Scholar]
- Li GH, Chen JZ, Ma X, Zhang Z, Liu N, Wang Y. Enzymatic preparation and facile purification of medium-chain, and medium- and long-chain fatty acid diacylglycerols. LWT-Food Sci Technol. 2018;92:227–233. doi: 10.1016/j.lwt.2018.02.032. [DOI] [Google Scholar]
- Malhotra R, Noorwez SM, Satyanarayana T. Production and partial characterization of thermostable and calcium-independent alpha-amylase of an extreme thermophile Bacillus thermooleovorans NP54. Lett Appl Microbiol. 2000;31(5):378–384. doi: 10.1046/j.1472-765x.2000.00830.x. [DOI] [PubMed] [Google Scholar]
- Murakami MT, Arni RK, Vieira DS, Degreve L, Ruller R, Ward RJ. Correlation of temperature induced conformation change with optimum catalytic activity in the recombinant G/11 xylanase A from Bacillus subtilis strain 168 (1A1) Febs Lett. 2005;579(28):6505–6510. doi: 10.1016/j.febslet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Paetzel M, Dalbey RE, Strynadka NCJ. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396(6707):186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- Quyen DT, Schmidt-Dannert C, Schmid RD. High-level expression of a lipase from Bacillus thermocatenulatus BTL2 in Pichia pastoris and some properties of the recombinant lipase. Protein Express Purif. 2003;28(1):102–110. doi: 10.1016/s1046-5928(02)00679-4. [DOI] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:17. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua ML, SchmidtDannert C, Wahl S, Sprauer A, Schmid RD. Thermoalkalophilic lipase of Bacillus thermocatenulatus large-scale production, purification and properties: aggregation behaviour and its effect on activity. J Biotechnol. 1997;56(2):89–102. doi: 10.1016/S0168-1656(97)00079-5. [DOI] [PubMed] [Google Scholar]
- Sarmah N, Revathi D, Sheelu G, Rani KY, Sridhar S, Mehtab V, Sumana C. Recent advances on sources and industrial applications of lipases. Biotechnol Progr. 2018;34(1):5–28. doi: 10.1002/btpr.2581. [DOI] [PubMed] [Google Scholar]
- Singh A, Van Hamme JD, Ward OP. Surfactants in microbiology and biotechnology: Part 2 Application aspects. Biotechnol Adv. 2007;25(1):99–121. doi: 10.1016/j.biotechadv.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Velez AM, Iemma MR, Silva AJ, Horta ACL, Giordano RC, Giordano RL. Influence of operational conditions in the expression of Bacillus thermocatenulatus lipase BTL2 in Escherichia coli. New Biotechnol. 2009;25:S219–S220. doi: 10.1016/j.nbt.2009.06.180. [DOI] [Google Scholar]
- Wang JR, Wu ZZ, Zhang TY, Wang YH, Yang B (2019) High-level expression of Thermomyces dupontii thermophilic lipase in Pichia pastoris via combined strategies. 3 Biotech 9(2). 10.1007/s13205-019-1597-8 [DOI] [PMC free article] [PubMed]
- Won K, Kim S, Kima KJ, Park HW, Moon SJ. Optimization of lipase entrapment in Ca-alginate gel heads. Process Biochem. 2005;40(6):2149–2154. doi: 10.1016/j.procbio.2004.08.014. [DOI] [Google Scholar]
- Wu XY, Jaaskelainen S, Linko YY. An investigation of crude lipases for hydrolysis, esterification, and transesterification. Enzyme Microb Tech. 1996;19(3):226–231. doi: 10.1016/0141-0229(95)00239-1. [DOI] [Google Scholar]
- Wu YP, Guo WB, Zhao JB, Ding LS, Chen XH. Isolation and identification of a novel LCI like antibacterial protein from Bacillus sp MD-5 reveals its potential application in controlling Staphylococcus aureus in food industry. Food Control. 2018;89:142–149. doi: 10.1016/j.foodcont.2018.01.026. [DOI] [Google Scholar]
- Yamada R, Kimoto Y, Ogino H. Combinatorial library strategy for strong overexpression of the lipase from Geobacillus thermocatenulatus on the cell surface of yeast Pichia pastoris. Biochem Eng J. 2016;113:7–11. doi: 10.1016/j.bej.2016.05.005. [DOI] [Google Scholar]
- Yenenler A, Venturini A, Burduroglu HC, Sezerman OU. Investigating the structural properties of the active conformation BTL2 of a lipase from Geobacillus thermocatenulatus in toluene using molecular dynamic simulations and engineering BTL2 via in-silico mutation. J Mol Model. 2018;24(9):13. doi: 10.1007/s00894-018-3753-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ge J, Liu Z. Enhanced activity of immobilized or chemically modified enzymes. Acs Catal. 2015;5(8):4503–4513. doi: 10.1021/acscatal.5b00996. [DOI] [Google Scholar]
- Zhu X, Ye A, Verrier T, Singh H. Free fatty acid profiles of emulsified lipids during in vitro digestion with pancreatic lipase. Food Chem. 2013;139(1–4):398–404. doi: 10.1016/j.foodchem.2012.12.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study. Code availability NCBI: X95309; PDB: 2W22.


