Abstract
Circular RNAs (circRNAs) are rapidly coming to the fore as major regulators of gene expression and cellular functions. They elicit their influence via a plethora of diverse molecular mechanisms. It is not surprising that aberrant circRNA expression is common in cancers and they have been implicated in multiple aspects of cancer pathophysiology such as apoptosis, invasion, migration, and proliferation. We summarize the emerging role of circRNAs as biomarkers and therapeutic targets in cancer.
Keywords: circRNAs, cancer, biomarkers, diagnostics, therapeutics
Introduction
Our understanding of the human transcriptome has increased significantly by the discovery and understanding of the role of regulatory non-coding RNAs in physiology and diseases such as cancer (Vo et al., 2019). Among the non-coding regulatory transcripts, circRNAs have attracted intense research scrutiny in recent years (Chen, 2020). CircRNAs are single-stranded covalently closed continuous loop structures lacking free ends and a polyadenylate tail (Li X. et al., 2018b; Kristensen et al., 2019; Chen, 2020). Close to one-fifth of active genes in the human genome can potentially give rise to circRNAs (Salzman et al., 2012; Li X. et al., 2018b; Kristensen et al., 2019; Chen, 2020). CircRNAs are composed of exonic and/or intronic sequences and are primarily generated by back-splicing, a non-canonical alternative RNA splicing event mediated by the spliceosome and regulated by a combination of cis-elements and trans-factors (Chen and Yang, 2015; Li X. et al., 2018b; Kristensen et al., 2019; Chen, 2020). Due to the absence of free ends circRNAs are not susceptible to destruction by RNA degradation machinery and are more stable than linear RNAs (Lasda et al., 2014; Wang et al., 2015; Zhang Y. et al., 2016). The majority but not all circRNAs are non-coding and exhibit their biological functions by sequestration of miRNAs/proteins. Some circRNAs regulate transcription, splicing and may also be translated to polypeptides. CircRNAs are involved in the regulation of cancer hallmarks such as self-sustenance in growth signals, proliferation, angiogenesis, resistance to apoptosis, unlimited replicative potential, and metastasis (Shi, 2017; Bach et al., 2019; Vo et al., 2019). Here we summarize and catalog the advances in the use of circRNAs as biomarkers for cancer diagnosis and as therapeutic targets.
Biogenesis of Circular RNAs
In eukaryotes, the generation of a mature mRNA is a result of interaction between transcription, splicing, capping, polyadenylation, export, and degradation (Black, 2003; Moore and Proudfoot, 2009; Nilsen and Graveley, 2010). CircRNAs are formed by a specialized non-conventional alternative splicing referred to as back-splicing (Zhang X. O. et al., 2016). In contrast to the classical canonical splicing, during back-splicing, a downstream 5' splice-site is joined to an upstream 3' splice-site across a single or multiple exons leading to the formation of circRNA species (Figure 1A) (You et al., 2015; Li X. et al., 2018b; Kristensen et al., 2019; Chen, 2020).
Figure 1.
Biogenesis of circular RNAs. (A) During transcription, linear and back-splicing of exons rival each other. Back-splicing is facilitated by long flanking introns, cis-elements i.e., intronic complementary elements (ICS), and trans-factors i.e., RNA-binding proteins (RBPs). To facilitate back-splicing, a downstream splice-donor (SD) site is brought in close vicinity with an upstream splice-acceptor (SA) site via base-pairing interactions between ICS or dimerization of RBPs. An upstream branch point (BP) nucleophilically attacks a downstream SD site, which thereafter nucleophilically attacks an upstream SA site, resulting in the creation of exonic circRNAs or exon-intron circRNAs. (B) Alternative splicing events like exon-skipping often generate skipped exon bearing lariat precursors, which can participate in the genesis of exonic circRNAs. (C) Intronic lariat precursors generated by linear canonical splicing can escape lariat debranching and serve as a source for intronic circRNAs.
Two models have been proposed to explain the coupling of back-splicing to canonical splicing for circRNAs biogenesis (i) “exon-skipping” or “lariat-intermediate” model and (ii) “direct back-splicing” model (Lasda et al., 2014). In the “lariat intermediate” model, canonical splicing occurs first and generates an intronless linear RNA, and an intron lariat bearing skipped exons which eventually undergoes back-splicing (Zaphiropoulos, 1996; Kelly et al., 2015) (Figures 1B,C). In the “direct back-splicing” model back-splicing occurs first leading to the formation of a circRNA followed by the creation of a linear RNA (Li Y. et al., 2017) (Figure 1A). Based on their origin, circRNAs fall into three major classes, exonic, intronic, and exon-intron circRNAs. Except for splice-sites, no particular sequences are necessary for circularization, however, a median exonic length is required for back-splicing involving either single or multiple exons (Ashwal-fluss et al., 2014; Zhang et al., 2014).
CircRNA synthesis by back-splicing occurs both co-transcriptionally and post-transcriptionally and is favored by a high rate of transcription elongation (Ashwal-fluss et al., 2014; Zhang Y. et al., 2016; Vo et al., 2019). The ligation of a downstream 5′ splice-site with an upstream 3′ splice-site during back-splicing is not favored sterically leading to lower efficiency of back-splicing as opposed to conventional linear splicing (Jeck et al., 2013; Zhang Y. et al., 2016). Interestingly, alternative back-splicing events can also occur and generate multiple circRNA isoforms (Gao et al., 2016; Zhang X. O. et al., 2016). Just like linear RNAs, circRNAs too are subjected to widespread reversible modification, in particular N6-methyladenosine (m6A) modification, which may influence their cellular fate (Zhou et al., 2017).
Role of Cis-Elements and Trans-Factors in Circular RNA Formation
CircRNA formation by back-splicing is facilitated by cis-elements such as intronic complementary sequences (ICS), flanking circRNA forming exons and trans-factors like RNA-binding proteins (RBPs) (Figure 1A) (Jeck et al., 2013; Ashwal-fluss et al., 2014; Liang and Wilusz, 2014; Zhang et al., 2014). ICS facilitate RNA pairing, by bringing distal splice-sites close to each other, which promotes circularization (Jeck et al., 2013). In humans, both complementary inverted-repeat Alu elements located in introns, as well as non-repetitive complementary sequences in introns, promote RNA pairing and subsequent back-splicing (Jeck et al., 2013; Liang and Wilusz, 2014; Zhang et al., 2014; Starke et al., 2015). Trans-factors contribute to circRNA biogenesis by modulating back-splicing: (i) by directly bridging distal splice-sites (ii) by binding to ICS. Some examples of trans-factors are RBPs such as Quaking (QKI), Heterogeneous-nuclear ribonucleoprotein L (HNRNPL), and RNA-binding motif protein 20 (RBM20) (Figure 1A) (Conn et al., 2015; Errichelli et al., 2017). RBPs which bind to ICS and regulate circRNA biogenesis bear double-stranded RNA-binding domains (dsRBDs) and can stabilize or destabilize the base-pairing between ICSs to promote or prevent back-splicing. The dsRBDs which promote back-splicing are nuclear factor 90 (NF90) and nuclear factor (NF110) (Patiño et al., 2015; Li et al., 2017) whereas dsRBDs which prevent back-splicing include DExH-Box Helicase 9 (DHX9) and adenosine deaminase 1 acting on RNA (ADAR1) (Ivanov et al., 2015; Aktaş et al., 2017).
Mechanism of Action of Circular RNAs
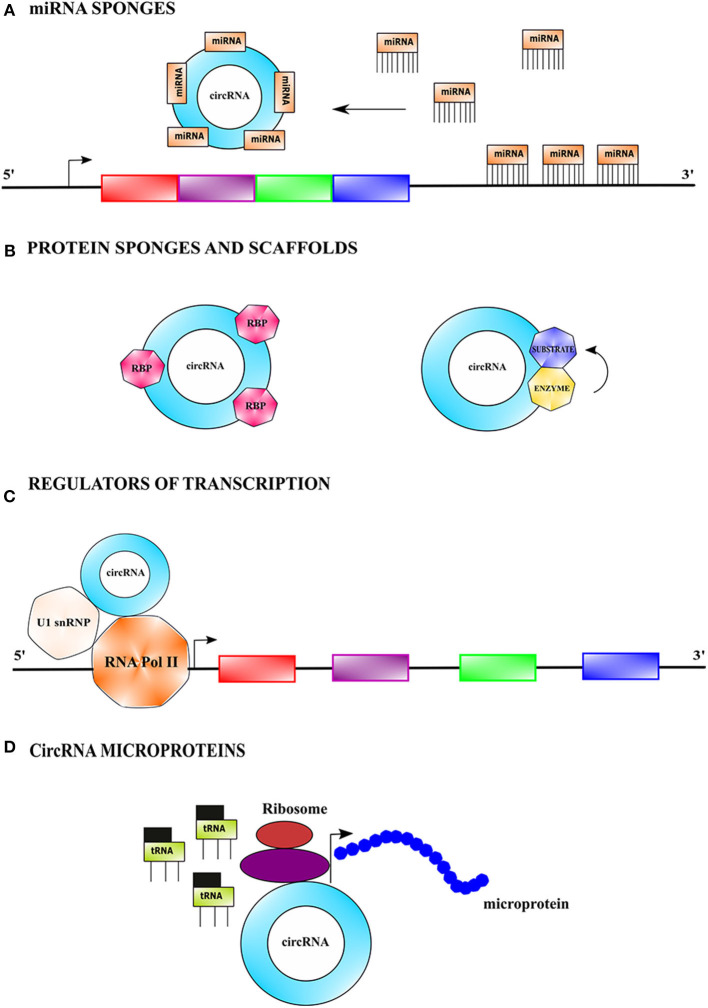
Circular RNAs as miRNA Sponges
CircRNAs competitively bind and sponge miRNAs leading to the stabilization of their target transcripts. They can have single or multiple binding sites for single or several miRNAs (Figure 2A). For instance, the expression of miR-7 target genes is regulated by CDR1as, which harbors >70 conserved binding sites for miR-7 (Hansen et al., 2013). Some circRNAs acting as miRNA sponges have oncogenic and tumor-suppressive properties (Kristensen et al., 2019). For example, circCCDC66 binds two miRNAs, miR-33b and miR-93, and promotes tumorigenesis in colorectal cancer by upregulation of c-MYC (Hsiao et al., 2017).
Figure 2.
Mechanistic action of circular RNAs. (A) CircRNAs can serve as miRNA sponges by competitively binding to miRNA(s) via base-pairing interactions, causing stabilization of target transcript(s) of the sequestered miRNA(s), and thus making it more available for translation. (B) CircRNAs can sponges protein by binding to them and thus indirectly regulating their functions. CircRNAs can also function as protein scaffolds by facilitating colocalization of an enzyme and its substrate to influence the kinetics of catalysis. (C) CircRNAs can modulate transcription by binding to RNA polymerase II complex bearing the U1 small nuclear ribonucleoprotein among other proteins and augment the function of certain proteins of the complex. (D) CircRNAs bearing internal ribosome entry site (IRES) elements and initiation codons can initiate translation in a cap-independent fashion and generate short polypeptides referred to as microproteins.
Circular RNAs as Protein Sponges
Interestingly, circRNAs can also bind to proteins and prevent their activity (Figure 2B) (Ashwal-fluss et al., 2014). For example, circPABPN1 binds to the Hu-antigen R (HuR) and prevents its binding to the cognate linear mRNA resulting in its reduced translation (Abdelmohsen et al., 2017). Certain circRNAs bind to multiple proteins and hold them together as a scaffold to facilitate their interaction. For example, circAMOTL1 simultaneously binds to both AKT1 and PDK1 in cardiac tissue serving as a scaffold to facilitate the phosphorylation of AKT1 (protein kinase B) by PDK1 (3-phosphoinositide-dependent protein kinase 1) (Zeng et al., 2017).
Circular RNAs as Regulators of Transcription and Splicing
Nuclear circRNAs also modulate transcription and splicing (Figure 2C). For example, the intronic ci-ankrd52 assembles at the transcription sites of its cognate gene and positively regulates RNA polymerase II driven transcription (Yang Y. et al., 2017). CircRNAs EIF3J and PAIP2 interact and form complexes with U1snRNP, which in turn interact with RNA polymerase II at the promoters of the parental genes, leading to transcriptional enhancement (Li Z. et al., 2015). Some circRNAs also regulate alternative splicing e.g., circ-UBR5 modulates RNA splicing by binding to splicing regulators such as QKI, NOVA1, and U1snRNA (Qin et al., 2018; Chen, 2020).
Functions of Circular RNAs Encoded Microproteins
Most circRNAs are noncoding but a few circRNAs have short open reading frames (ORFs) which are translated into short peptides referred to as microproteins (Figure 2D). Generally, microproteins are <100 amino acids in length and possess distinct functions as compared to the protein coded by their cognate linear mRNA (Hanada et al., 2009; Andrews and Rothnagel, 2014). CircRNAs undergo cap-independent translation facilitated by internal ribosomal entry sites (IRESs) and m6A modification in the 5'untranslated region (5'UTR) (Abe et al., 2015). Examples of microproteins encoding circRNAs include circFBXW7, circZNF609, circMbl, circPINTexon2, and circSHPRH (Motegi et al., 2006; Akhoondi et al., 2007; Yang Y. et al., 2018; Zhang M. et al., 2018; Zhang et al., 2019).
Circular RNAs as Cancer Biomarkers
CircRNAs have several attributes that make them potential biomarkers for cancer diagnosis and prognosis. They are more stable than linear RNAs due to lack of free 5′ and 3′ ends (Memczak et al., 2013; Li Z. et al., 2015; Zhang Z. et al., 2018; Vo et al., 2019), and often display tissue and developmental stage-specific expression pattern, and can be quantitatively detected by reverse transcription followed by real-time quantitative polymerase chain reaction (RT-qPCR) (Panda and Gorospe, 2018). Moreover, altered expression of circRNAs has been frequently observed in cancer tissues and/or in plasma, and saliva from cancer patients (Memczak et al., 2013, 2015; Bahn et al., 2015; Li Z. et al., 2015; Panda and Gorospe, 2018; Zhang Z. et al., 2018; Vo et al., 2019). Li et al. first reported the presence of circRNAs in exosomes in serum of cancer patients and several cancer cell types and coined the term exo-circRNAs (Li Y. et al., 2015). The presence of exo-circRNAs in a variety of human bodily fluids that may be assessed easily without biopsy, makes exo-circRNAs a good choice for cancer diagnosis (Bai et al., 2019; Geng X. et al., 2020). Numerous circRNAs are dysregulated in cancer but few have the potential to serve as biomarkers for cancer and are summarized and cataloged in Table 1.
Table 1.
Biological functions and roles of circular RNAs in cancer diagnostics and therapeutics.
| circRNA | Associated Cancer (s) | Regulation (Up ↑, Down↓) | Biological Function | Target (s) | Type of model | Type of cell line (s) | Biomarker/therapeutics | References |
|---|---|---|---|---|---|---|---|---|
| circ_0009910 | AML | ↑ | Sponges miR-20a-5p and inhibits apoptosis. Overexpressing miR-20a-5p counteracts chemoresistance in vitro and in vivo through targeting KIF26B by modulating the activities of the MAPK/ERK and cAMP/PKA signaling pathways | miR-20a-5p | Human, Mouse; in vitro/in vivo | Human cell lines – Mono-Mac-6, KG-1, AML2, AML5 | +/− | Ping et al., 2019a |
| GC | ↑ | Associated with distant metastasis and differentiation; downregulation reduces GC cell proliferation, migration and invasion. Acts as oncogene that acts by inducing the EMT. Its knockdown suppresses the migration invasion and EMT of GC cells in vitro. | – | Human; in vitro | Human cell lines – BGC823, SGC7901, AGS, MGC803, MKN45, GES1 | +/+ | Liu M. et al., 2018 | |
| CML | ↑ | Promotes cell growth and imatinib resistance, reduces apoptosis and autophagic activation. It accelerates imatinib-resistance in cells by modulating ULK1-induced autophagy via targeting miR-34a-5p, providing a potential target in imatinib resistance of CML. | miR-34a-5p | Human; in vitro | Human cell lines – K562, K562/R | +/+ | Cao et al., 2020 | |
| OSC | ↑ | Sponges miR-449a which targets IL6R; prohibits cell-cycle arrest, promotes proliferation and inhibits apoptosis. | miR-449a | Human; in vitro | Human cell lines – MG63, Saos-2, U2OS, hFOB | +/+ | Deng et al., 2018 | |
| circ-vimentin | AML | ↑ | Associated with poor overall survival (OS), leukemia-free survival and vimentin expression. | – | Human; in vivo | – | +/− | Yi and Lin, 2018 |
| circ_0075001 | AML | ↑ | Lowers expression of components of Toll-like receptor signaling pathway | – | Human; in vitro | Human cell lines – NB-4, KASUMI-1, OCI-AML5, OCI-AML3, ME-1, MV4-11, K562 | +/− | Hirsch et al., 2017 |
| circ_0004277 | AML | ↓ | – | – | Human; in vivo | – | +/− | Li W. et al., 2017 |
| circ_100053 | CML | ↑ | Associated with clinical stage, BCR/ABL mutant status and imatinib resistance | – | Human; in vivo | – | +/− | Ping et al., 2019b |
| circ–RPL15 | CLL | ↑ | Inhibits miR-146b-3p mediated suppression of the RAS/RAF1/MEK/ERK pathway | miR-146b-3p | Human; in vitro | Human cell lines – MEC-1 and JVM-3 | +/+ | Wu Z. et al., 2019 |
| circ-CBFB | CLL | ↑ | Activates Wnt/β-catenin signaling pathway | miR-607 | Human; in vitro | Human cell line - MEC-1 | +/+ | Xia L. et al., 2018 |
| circ_0007841 | MM | ↑ | Correlated with chromosomal aberrations such as gain 1q21, t (4:14), mutations in ATR and IRF4 genes; miR-199a-3p affects the multi-chemoresistance of OS via targeting AK4; overexpression correlates with osteolytic bone destruction in MM; overexpressed in BTZ-resistant MM cell lines | miR-199a-3p | Human; in vitro | Human cell lines – THP-1, KM3, U266, RPMI-8226, KM3/BTZ, 146 U266/BTZ, RPMI-8226/BTZ | +/− | Gao et al., 2019 |
| circ_0000190 | MM | ↓ | Correlates with prognosis survival rates of MM patients, inhibits MM progression via modulating miR-767-5p/MAPK4 pathway | miR-767-5p | Human, Mouse; in vitro/in vivo | Human cell lines – MM.1S, NCI-H929 | +/+ | Feng et al., 2019 |
| GC | ↓ | Associated with tumor diameter, lymphatic metastasis, distal metastasis, TNM stage and CA19-9 level | – | Human; in vivo | – | +/– | Chen et al., 2017 | |
| OSC | ↓ | Correlated with bigger tumor size, advanced staging (IIB/III) and distant metastasis | miR-767-5p | Human, Mouse; in vitro/in vivo | Human cell lines –hFOB1.19, SAOS-2, MG63, U2OS, SJSA1, and HOS | +/– | Li et al., 2020 | |
| circ-SMARCA5 | MM | ↓ | Higher expression is correlated with lower β2-MG level and less advanced ISS stage; native resistance to drugs is common due to multiple chromosomal abnormities in the pathogenesis of MM | miR-767-5p | Human; in vitro | Human cell lines – NCI-H929, RPMI8226, U226, OPM2, JJN3 | +/+ | Liu H. et al., 2019 |
| GC | ↓ | Correlates with differentiation, lymph node metastasis, vascular invasion | – | Human; in vitro | Human cell lines – GES-1, MGC803, MKN45, AGS, MKN74, BGC-823, SGC-7901 | +/– | Cai et al., 2019 | |
| HCC | ↓ | Promotes apoptosis and expression of tumor suppressor TIMP3, inhibits proliferation, invasion and metastasis; reverses tumor growth along with decreased expression of MMP9 and MMP7 | miR-17-3p miR-181b-5p | Human; in vitro | Human cell lines – Huh7, HCCLM9, HepG2 | +/+ | Li Z. et al., 2019 | |
| circRNA_101237 | MM | ↑ | Increases significantly in bortezomib-resistant cell lines; overexpression is associated with a poor response to chemotherapy in MM patients | – | Human; in vitro | Human cell lines – THP-1, MM.1S, H929, MM.1S/BTZ, H929/BTZ | +/– | Liu and Wang, 2020 |
| HCC | ↑ | Associated with tumor size, lymph node metastasis, distant metastasis and TNM stage; cisplatin resistance associated with loss of Runt- associated transcription factor 3 and upregulation of cyclophilin B | – | Human; in vitro | Human cell lines – HCCLM3, Hep3B, MHCC97-H, cisplatin-resistant Huh7/DDP cells | +/– | Zhou et al., 2020 | |
| circ-APC | BCL | ↓(DLBCL) | Inhibits Wnt/β-catenin signaling | miR-888 | Human, Mouse; in vitro/in vivo | Human cell lines – SUDHL-3, U2932, TMD8, OCI-Ly3, L428, GM12878 | +/+ | Hu et al., 2019 |
| CDR1as | CRC | ↑ | Enhances EGFR/RAF1/MAPK pathway, induces cell growth, resistance to apoptosis and cell cycle arrest | miR-7 | Human; in vitro | Human cell lines –HCT-116, DLD-1, NCM460, CCD841CoN | +/+ | Weng et al., 2017 |
| HCC | ↑ | Promotes HCC progression by activating PI3K/AKT/mTOR pathway | miR-7 | Human; in vivo | – | +/+ | Xu et al., 2017 | |
| GC | ↑ | Activates PTEN/PI3K/AKT pathway | miR-7 | Human, Mouse; in vitro/in vivo | Human cell lines – MGC-803, HGC-27, GES-1 | +/+ | Pan et al., 2018 | |
| circCCDC66 | CRC | ↑ | Promote CRC growth and metastasis by stabilizing MYC mRNA | miRNA-33b, miR-93 | Human; in vitro/in vivo | Human cell lines – HCT-116, HT-29 | +/– | Hsiao et al., 2017 |
| circ_0004585 | CRC | ↑ | Associated with increased patient's tumor size | – | Human; in vivo | – | +/– | Tian et al., 2019 |
| circ_0007142 | CRC | ↑ | Regulates proliferation and invasion of CRC; upregulation is associated with lymphatic metastasis | miR-103a-2-5p | Human; in vitro | Human cell lines – HCT-116, HT-29, LoVo, HCO | +/– | Zhu et al., 2019 |
| circHUEW1 | CRC | ↑ | Associated with lympho-vascular invasion, lymph node metastasis, distant metastasis, and TNM stage; affects IGF2/β-catenin signaling pathway | miR-486 | Human; in vivo, in vitro | Human cell lines – HCT116, SW480 | +/+ | Chen H. Y. et al., 2020 |
| circ_0001178 | CRC | ↑ | Metastatic clinical features, advanced TNM stage and adverse prognosis; induces EMT through increasing ZEB1 expression | miR-382, miR-587 and miR-616 | Human, Mouse; in vitro/in vivo | Human cell lines – NCM460 cells, CRC LoVo, SW620 | +/+ | Ren et al., 2020 |
| circ_0005075 | CRC | ↑ | Downregulation modulated Wnt/β-catenin pathways and reduced cell proliferation and metastasis; Knockdown suppresses EMT progression by decreasing the levels of Vimentin and N-cadherin | – | Human; in vivo, in vitro | Human cell lines – HCO, SW480, SW620, HT29, HCT116, SW1116, LOVO | +/+ | Jin et al., 2019 |
| HCC | ↑ | Contributes to HCC proliferation, invasion, and metastasis | miR-23b-5p, miR-93-3p, miR581,miR-23a-5p | Human; in vivo | – | +/– | Shang et al., 2016 | |
| circHIPK3 | CRC | ↑ | Promotes CRC progression,increases expression of downstream oncogenic target genes, FAK, IGF1R, EGFR, and YY1 that activate PI3K/AKT and MEK/ERK signaling pathways to promote cancer progression and drug resistance | miR-7 | Human, Mouse; in vitro/in vivo | Human cell lines – FHC, HCT116, HT29, SW480, SW620, DLD1 | +/+ | Zeng et al., 2018 |
| OSC | ↓ | Correlates with Enneking stage and lung metastasis. | – | Human; in vitro | Human cell lines – SaoS2, HOS, KH-OS, MG63, 143B, U2OS | +/– | Kun-peng et al., 2018b | |
| BCa | ↓ | Reduces aggressiveness and metastasis by targeting the miR-558/heparanase axis | miR-558 | Human, Mouse; in vitro/in vivo | Human cell lines –T24T, UMUC3, SV-HUC-1, HUVEC, | +/+ | Li Y. et al., 2017 | |
| circ_0001649 | CRC | ↓ | – | – | Human; in vitro | Human cell line - H116 | +/– | Ji et al., 2018 |
| HCC | ↓ | Associated with tumor size, occurrence of tumor embolus; correlates with metastasis, | – | Human; in vitro | Human cell lines – HCC–LM3, MHCC-97L | +/– | Qin et al., 2016 | |
| LC | ↓(NSCLC) | associated with shorter OS, positive lymph node, and differentiation grade | miR-331-3p miR-338-5p | Human, Mouse; in vitro/in vivo | Human cell lines – A549, H358, H1299, H1581, 16HBE | +/– | Liu H. et al., 2018 | |
| circITGA7 | CRC | ↓(CRC) | Inactivates Ras signaling pathway; associated with tumor size, lymph metastasis, distant metastasis, and TNM stage | miR-370-3p | Human, Mouse; in vitro/in vivo | Human cell lines – SW480, RKO, Caco-2, SW620, LoVo, HCT116, DLD1, FHC | +/– | Li X. et al., 2018a |
| circ_0000711 | CRC | ↓ | – | – | Human; in vitro | Human cell lines – NCM460, HCT116, COLO205, HT29 | +/– | Li J. et al., 2018b |
| circ_0014717 | CRC | ↓ | Overexpression promotes G0/G1 phase arrest, reduces growth, invasion and distal metastasis | – | Human, Mouse; in vitro/in vivo | Human cell lines – HCT116, HT29, SW480, FHC | +/– | Wang F. et al., 2018 |
| circFBXW7 | BC | ↓(TNBC) | Inhibits tumor progression; up-regulates FBXW7 and degrades c-Myc, negatively correlated with metastasis | miR-197-3p | Human, Mouse; in vitro/in vivo | Human cell lines – MCF-10A, MCF-7, T47D, BT474, SKBR-3, MDA-MB-453, MDA-MB-468, MDAMB-231, BT549, HCC38, 4T1, MA-891 | +/+ | Ye F. et al., 2019 |
| circSEPT9 | BC | ↑(TNBC) | Activates LIF/Stat3 signaling pathway, correlates with lymph node metastasis | miR-637 | Human, Mouse; in vitro/in vivo | Human cell lines – MDA-MB-231, BT-549, MDA-MB-468, MDA-MB-453, SUM-159, MCF-10A | +/– | Zheng et al., 2020 |
| circ_0001785 | BC | ↑(BC) | Associated with histological grade, TNM stage and distant metastasis | – | Human; in vivo | – | +/– | Yin et al., 2018 |
| circCDYL | BC | ↑ | Regulates miR-1275-ATG7/ULK1 axis; downregulates the expression of autophagy associated genes AKT and ULK1; enhances malignant progression | miR-1275 | Human, Mouse; in vitro/in vivo | Human cell lines – MDA-MB231, MCF-7 | +/+ | Liang et al., 2020 |
| MM | ↑ | Promotes MM growth by targeting YAP; inhibits apoptosis | miR-1180 | Human, Mouse; in vitro/in vivo | Human cell lines – MM1.S, NCI-H929 | +/+ | Chen F. et al., 2020 | |
| circKIF4A | BC | ↑(TNBC) | Induces TNBC cell proliferation and migration regulating the expression of KIF4A; miR-375 can sensitize resistant cells to tamoxifen and partly reverse EMT | miR-375 | Human, Mouse; in vitro/in vivo | Human cell lines – MCF10A, MCF-7, T47D, BT474, KBR3, MDA-MB-453, MDA-MB-468, MDA-MB-231, BT549, HCC38 | +/– | Tang et al., 2019 |
| circPLK1 | BC | ↑(TNBC) | Promotes TNBC cell proliferation and metastasis by regulating PLK1 | miR-296-5p | Human, Mouse; in vitro/in vivo | Human cell lines – MCF10A, MDAMB-468, MDA-MB-453, MDA-MB-231, HCC38, BT549 | +/– | Kong Y. et al., 2019 |
| circHMCU | BC | ↑(BC) | Enhanced proliferation and metastasis; can modify EMT pathway, promotes mesenchymal phenotypes and inhibits epithelial phenotypes; stable and resistant to ActD treatment | let-7 family | Human, Mouse; in vitro/in vivo | Human cell lines –MDA-MB-231, MDA-MB-468, MCF7 | +/– | Song et al., 2020 |
| circ_0068033 | BC | ↓(BC) | Overexpression induces apoptosis | miR-659 | Human, Mouse; in vitro/in vivo | Human cell lines – MCF10A, MCF-7, T47D, MDA-MB-468 | +/– | Yuan et al., 2020 |
| circ_0005075 | HCC | ↑ | Contributes proliferation, invasion, and metastasis | miR-23b-5p, miR-93-3p, miR581 and miR-23a-5p | Human; in vivo | – | +/– | Shang et al., 2016 |
| circ_100338 | HCC | ↑ | Increased cell metastasis progression; regulates the MTOR signaling pathway | miR-141-3p | Human; in vitro | Human cell lines –Hep3B, BEL7402, MHCC97H, HCCLM6 | +/+ | Huang X. Y. et al., 2017 |
| circRHOT1 | HCC | ↑ | Promotes HCC progression, recruits TIP60,enhances invasion, inhibits apoptosis, and promotes metastasis | – | Human, Mouse; in vitro/in vivo | Human cell lines –Hep3B, Huh7 | +/+ | Wang L. et al., 2019 |
| circ_0091579 | HCC | ↑ | Associated with poor OS | – | Human; in vivo | +/– | Zhang C. et al., 2018 | |
| circ-HOMER1 | HCC | ↑ | Increases the expression of CXCL6; associated with larger tumor size, higher TNM stage, and worse prognosis | miR-1322 | Human; in vitro | Human cell lines – Sk-Hep-1, SMMC-7721, HCCLM3, Huh-7, HepG2 cells, L02 | +/– | Zhao M. et al., 2020 |
| circ_0016788 | HCC | ↑ | Downregulates miR-486/CDK4 expression; associated with poor OS | miR-486 | Human; in vivo | - | +/+ | Cheng et al., 2020 |
| circ_0078602 | HCC | ↓ | Associated with a poor prognosis | - | Human; in vivo | – | +/– | Kou et al., 2019 |
| circC3P1 | HCC | ↓ | Associated with TNM stage, tumor size and vascular invasion, overexpression decreased metastatic nodules | miR-4641 | Human, Mouse; in vitro/in vivo | Human cell lines –BEL7402, Hep3B, HuH7, MHCC97-L, HL-7702 | +/- | Zhong et al., 2018 |
| circ-ITCH | HCC | ↓ | Correlated with poor OS | - | Human; in vivo | – | +/– | Guo et al., 2017 |
| BCa | ↓ | Inhibits cell proliferation, migration and invasion through circ-ITCH/miR-17,miR-224/p21,PTEN signaling axis | miR-17,miR-224 | Human, Mouse; in vitro/in vivo | Human cell lines – EJ, T24, 253 J, RT4, TCC-SUP, UMUC, J82, 5637, SV-HUC | +/+ | Yang C. et al., 2018 | |
| CRC | ↓ | Overexpression reduces cell proliferation by downregulating c-Myc and cyclinD1 | miR-7, miR-20a | Human; in vitro | Human cell lines – HCT116, SW480 | +/+ | Huang et al., 2015 | |
| circMTO1 | HCC | ↓ | Promotes expression of a tumor suppressor p21 resulting in reduced tumor cell proliferation, metastasis and invasion | miR-9 | Human, Mouse; in vitro/in vivo | Human cell lines – HepG2, SMMC-7721, QGY-7701, SK-Hep1 | +/+ | Han et al., 2017 |
| circ_0013520 | GBM | ↑ | Correlated with tumor size, TNM and worse OS | – | Human; in vitro | Human cell lines – SHG-44, U251, HEB | +/– | Zhou and Fan, 2020 |
| circ_0004379 | GBM | ↑ | Correlated with tumor size, TNM and worse OS | – | Human; in vitro | Human cell lines – SHG-44, U251, HEB | +/– | Zhou and Fan, 2020 |
| circ-CDC45 | GBM | ↑ | Associated with larger tumor size, higher grade, and worse survival | miR-516b, miR-527 | Human; in vitro | Human cell lines – U87MG, U118, U251, LN229 | +/- | Liu J. et al., 2019 |
| circNFIX | GBM | ↑ | Predicts poor prognosis; promotes cell propagation and migration; knockdown enhances TMZ sensitivity in resistant cells; regulates NOTCH pathway | miR-132, miR-34a-5p | Human, Mouse; in vitro/in vivo | Human cell lines – HA1800, SF-539, SHG-44, U87 | +/+ | Xu et al., 2018; Ding et al., 2020 |
| circ_0013958 | LC | ↑(LAC) | Associated with the TNM stage and lymphatic metastasis | miR-134 | Human; in vitro | Human cell lines – A549, H1299, BEAS-2B | +/– | Zhu X. et al., 2017 |
| circFARSA | LC | ↑(NSCLC) | Promotes cell migration and invasion; upregulates FASN | miR-330-5p, miR-326 | Human; in vitro/in vivo | Human cell line – A549 | +/– | Hang et al., 2018 |
| CRC | ↑ | Promotes proliferation, migration, and invasion;regulates miR-330-5p/LASP1 axis | miR-330-5p | Human, Mouse; in vitro/in vivo | Human cell lines – FHC, LS174T, RKO, HT29, HCT116, SW480 | +/+ | Lu C. et al., 2020 | |
| circ_0014130 | LC | ↑(NSCLC) | Associated with tumor volume, distant metastasis;upregulates Bcl2 | miR-136-5p | Human, Mouse; in vitro/in vivo | Human cell lines – PC-9, A549 | +/– | Geng Y. et al., 2020 |
| circ_0000792 | LC | ↑(LAD) | Overexpression is correlated with T stage, distant metastasis | – | Human; in vivo | – | +/– | Li, 2018 |
| circ_100876 | LC | ↑(NSCLC) | Related to carcinogenesis of NSCLC and it might serve as a potential prognostic biomarker and therapeutic target | – | Human; in vivo | – | +/+ | Yao J. T. et al., 2017 |
| circFADS2 | LC | ↑ | Associated with advanced TNM stage, lymph node metastasis, poor differentiation, and shorter OS; induces progression, invasion and proliferation | miR-498 | Human; in vitro | Human cell line – HepG2 | +/+ | Zhao F. et al., 2018 |
| CRC | ↑ | Associated with distant metastasis | - | Human; in vivo | – | +/– | Xiao et al., 2020 | |
| circPVT1 | LC | ↑(NSCLC) | Associated with distant metastasis; promotes cell proliferation, migration and invasion, and inhibits apoptosis through upregulated E2F2 and E2F2-related protein expression | miR-125b | Human, Mouse; in vitro/in vivo | Human cell lines – A549, H292, SPC-A1, H1299, H1650, H1975, SK-MES-1, HBE | +/+ | Li X. et al., 2018c |
| OSC | ↑ | Associated to chemoresistance and lung metastasis; knockdown decreases ABCB1 expression, promotes chemoresistance; knockdown partly reverses the doxorubicin and cisplatin resistance | – | Human; in vitro | Human cell lines – SaoS2, KHOS, U2OS, MG63 | +/+ | Kun-peng et al., 2018b | |
| ALL | ↑ | Promotes cell proliferation and inhibits apoptosis | let-7,miR-125 | Human; in vivo, in vitro | Human ALL cell lines | +/+ | Hu et al., 2018 | |
| circ_0067934 | LC | ↑(NSCLC) | Tumor-promoting circRNA; induces cell proliferation, metastasis and invasion | – | Human; in vitro | Human cell lines – A549, H1299, SK-MES-1, PC-9, BEAS-2B | +/- | Wang and Li, 2018 |
| ESCC | ↑ | Promotes proliferation and migration | – | Human; in vitro | Human cell lines – TE-13, ECA-109 | +/+ | Zong et al., 2018b | |
| LSCC | ↑ | Promote cell proliferation and metastasis | miR-1324 | Human; in vitro | Human cell lines – TU212, TU686, 16HBE | +/– | Chu, 2020 | |
| HCC | ↑ | Enhances migration, invasion and proliferation of cells; regulates Wnt/β-catenin signaling pathway | miR-1324 | Human; in vitro | Human cell lines – BEL7402, Hep3B, HuH7, MHCC97-L, HL-7702 | +/+ | Zhu et al., 2018 | |
| circPRKCI | LC | ↑ | Upregulation increased proliferation and tumorigenesis | miR-545, miR-589 | Human, Mouse; in vitro/in vivo | Human cell lines – A549, NCI-H1975, NCI-H1703, NCI-H226, NCI-H46, PC9, NCI-H1299, SPC-A1, HCC827, HBE | +/– | Qiu et al., 2018 |
| circ_0000064 | LC | ↑ | Promotes cell proliferation and inhibits cell apoptosis, enhances expression of bcl-2; overexpression is correlated with TNM stage, lymph node metastasis, | – | Human; in vitro | Human cell lines – A549, H1229 | +/+ | Luo et al., 2017 |
| circ_0016760 | LC | ↑(NSCLC) | miR-1287 directly targets GAGE1, higher expression associated with shorter OS, correlated with lymph node metastasis | miR-1287 | Human, Mouse; in vitro/in vivo | Human cell lines – A549, H358, H1299, H1975 | +/– | Li Y. et al., 2018 |
| circ_102231 | LC | ↑(LAC) | Promotes lung cancer cells proliferation, migration and invasion | – | Cell line model (in vitro exp) | Human cell lines – BEAS-2B, A549 | +/+ | Zong et al., 2018a |
| circRNA_103809 | LC | ↑ | Regulates miR-4302/ZNF121/MYC loop; promotes tumor growth, cell proliferation and invasion, associated with tumor stage and lymph node metastasis | miR-4302 | Human, Mouse; in vitro/in vivo | Human cell lines – A549, H125, 95D, NCI-H292, H1975, HBE | +/+ | Liu W. et al., 2018 |
| CRC | ↓ | Promote apoptosis through FOXO4 activity | miR-532-3p | Human; in vitro | Human cell lines – SW620, HCT116, COCA-2, HT29, FHC | +/+ | Bian et al., 2018 | |
| circ_0005962 | LC | ↑(LAC) | – | – | Human; in vivo | – | +/– | Liu X. X. et al., 2019 |
| circ_0086414 | LC | ↓(LAC) | Associated with EGFR mutations | – | Human; in vivo | – | +/– | Liu X. X. et al., 2019 |
| circ-PRMT5 | LC | ↑(NSCLC) | Correlated with larger tumor, LNM, poor OS and progression free survival; upregulates EZH2 | – | Human, Mouse; in vitro/in vivo | Human cell lines – HBE, A549, 95-D, HCC827, H1299, SK-MES-1 | +/– | Wang Y. et al., 2019 |
| GC | ↑ | Promotes GC cell growth, clone formation, migration and invasion and inhibits apoptosis | miR-145, miR-1304 | Human; in vitro | Human cell lines – AGS, MKN-28, MKN45, BGC823, MGC803, SGC7901, GES-1 | +/+ | Du et al., 2019 | |
| circ-RAD23B | LC | ↑(NSCLC) | Regulates miR-593e3p/CCND2 axis; increases cell invasion via miR-653e5p/TIAM1 pathway | miR-593e3p, miR-653e5p | Human; in vitro | Human cell lines – 16HBE, H1299, H1581, H358, A549 | +/+ | Han et al., 2019 |
| circ_0102533 | LC | ↑(NSCLC) | Associated with tumor type, TNM stages, lymph nodes metastasis and distant metastasis or recurrence | – | Human; in vitro | Human cell lines – A549, H1299, H1792, SK-MES-1, SPC-A1 | +/– | Zhou X. et al., 2018 |
| circ_0079530 | LC | ↑ | Enhances cell proliferation and invasion | – | Human; in vitro | Human cell lines – A549, H1299, H460, Calu1, BEAS-2B | +/– | Li J. et al., 2018a |
| circFGFR3 | LC | ↑(NSCLC) | Increases cell invasion and proliferation, regulates Gal-1, pAKT, and p-ERK1/2 | miR-22-3p | Human; in vitro | Human cell lines – 95C, 95D, A549, H460 | +/– | Qiu B. Q. et al., 2019 |
| circ_000984 | LC | ↑(NSCLC) | Promotes cell proliferation and metastasis; regulates Wnt/β-catenin signaling | – | Human; in vitro | Human cell lines – H1975, SPC-A1, H1299, HCC827, PC 9, A549, BEAS-2B | +/– | Li X. et al., 2019 |
| circ_0001946 | LC | ↑(LAC) | Regulates SIRT1 that activates Wnt/β-catenin signaling pathway | miR-135a-5p | Human, Mouse; in vitro/in vivo | Human cell lines – H1299, A549, Calu3, SPC-A1, BEAS-2B | +/– | Yao et al., 2019 |
| GBM | ↓ | Reduces the migration, invasion, and proliferation of GBM cells | miR-671-5p | Human, Mouse; in vitro/in vivo | Human cell lines – U87, U251, HM | +/+ | Li, 2019a | |
| circ_0037515 | LC | ↓(NSCLC) | – | – | Human; in vivo | – | +/– | Zhao D. et al., 2020 |
| circ_0037516 | LC | ↓(NSCLC) | – | – | Human; in vivo | – | +/- | Zhao D. et al., 2020 |
| circ_0033155 | LC | ↓(NSCLC) | Reduces cell proliferation, colony formation and migration, correlated with lymphatic metastasis | – | Human; in vitro | Human cell lines – HCC827, H1975 | +/+ | Gu et al., 2018 |
| circ_100395 | LC | ↓ | Promotes LC malignancy regulating miR-1228/TCF21 axis | miR-1228 | Human; in vitro | Human cell lines – A549, H460, Beas-2B | +/+ | Chen D. et al., 2018 |
| circ-FOXO3 | LC | ↓(NSCLC) | Promotes NSCLC development; releases FOXO3; miR-155 and FOXO transcription factors affect chemoresistance | miR-155 | Human; in vitro | Human cell lines – A549, SPC-A1, NCI-H1299, NCI-H1650, SK-MES-1 | +/+ | Zhang Y. et al., 2018 |
| circ_0056616 | LC | ↑(LAD) | Upregulation is correlated with TNM stage and lymph node metastasis | – | Human; in vitro | Human cell lines – PC9, PC14, HEK293T | +/– | He Y. et al., 2020 |
| circ_0010882 | GC | ↑ | Contributes to the proliferation of GC cells, migration, invasion, and apoptosis through modulating PI3K/Akt/mTOR pathway | – | Human; in vitro | Human cell lines – HGC-27, MKN-45, SGC-7901, BGC-823, GES-1 | +/– | Peng et al., 2020 |
| circ-DCAF6 | GC | ↑ | Enhances GC progression | miR-1231, miR-1256 | Human; in vitro | Human cell lines – AGS, BGC823, MGC803, GES1 | +/– | Wu L. et al., 2019 |
| circ_0000419 | GC | ↓ | Associated with tumor stage, lymphatic and distal metastasis, venous and perineural invasion | hsa-miR-141-5p, hsa-miR-589-3p | Human; in vitro | Human cell lines – BGC-823, HGC-27, MGC-803, SGC-7901, GES-1 | +/– | Tao et al., 2020 |
| circ_0006156 | GC | ↓ | Associated with lymph node metastasis, nerve invasion and degree of tumor differentiation | – | Human; in vivo | – | +/– | He F. et al., 2020 |
| circ_0001821 | GC | ↓ | Negatively associated with tumor depth and lymph node metastasis | – | Human; in vitro | Human cell lines – SGC-7901, HGC-27, BGC-823, AGS, MKN-1 | +/- | Kong S. et al., 2019 |
| circCCDC9 | GC | ↓ | Upregulation sponges miR-6792-3p that targets CAV1, a tumor suppressor gene | miR-6792-3p | Human, Mouse; in vitro/in vivo | Human cell lines – GES-1, AGS, BGC-823, HGC-27, MGC-803, MKN-28, MKN-45, SGC-7901 | +/+ | Luo Z. et al., 2020 |
| circRHOBTB3 | GC | ↓ | Prevents the growth of cells, promotes expression of p21 | miR-654-3p | Human, Mouse; in vitro/in vivo | Human cell lines – AGS, HGC27, MKN45 | +/+ | Deng et al., 2020 |
| circ_100269 | GC | ↓ | Resists GC development | miR-630 | Human; in vitro | Human cell lines – AGS, MKN28, MKN45, BGC823, MGC803, SGC7901, GES1 | +/+ | Zhang Y. et al., 2017 |
| circ_0000745 | GC | ↓ | Associated with tumor differentiation | – | Human; in vivo | – | +/– | Huang M. et al., 2017 |
| circPSMC3 | GC | ↓ | Contributes to GC progression by regulating PTEN/miRNA-296-5p axis; PTEN regulates chemoresistance | miRNA-296-5p | Human, Mouse; in vitro/in vivo | Human cell lines – BGC823, MGC803, SGC7901, AGS, MKN45 | +/- | Rong et al., 2019 |
| circ-KIAA1244 | GC | ↓ | Associated with TNM stage, lymphatic metastasis | – | Human; in vivo | – | +/– | Tang et al., 2018 |
| circYAP1 | GC | ↓ | Decreases GC cell growth and invasion; regulates miR-367-5p/p27 Kip1 axis | miR-367-5p | Human, Mouse; in vitro/in vivo | Human cell lines – GES-1, HGC-27 | +/– | Liu H. et al., 2018 |
| circ_0006848 | GC | ↓ | Correlates with tumor differentiation and tumor size | – | – | – | +/– | Lu et al., 2019a |
| circ_0000520 | GC | ↓ | Associated with TNM stage | – | Human; in vitro | Human cell lines – MKN-45, BGC-823, MGC- 80 803, AGS | +/– | Sun et al., 2017 |
| circ_0001895 | GC | ↓ | Associated with cell differentiation, Borrmann type, and tissue CEA expression | – | Human; in vitro | Human cell lines – GES-1, AGS, BGC-823, HGC27, MGC-803, SGC-7901 | +/– | Shao et al., 2017 |
| circ_0005556 | GC | ↓ | Downregulation correlated with differentiation, TNM stage and lymphatic metastasis | – | Human; in vivo | – | +/– | Yang L. et al., 2019 |
| circ_0067582 | GC | ↓ | – | – | Human; in vivo | – | +/– | Yu et al., 2020 |
| circ_0000467 | GC | ↑ | Promotes proliferation, migration, and invasion of GC cells,inhibits tumor apoptosis | – | Human; in vitro | Human cell lines – HGC-27, MGC-803, AGS, NUGC-3, GES-1 | +/– | Lu et al., 2019b |
| circ_102958 | GC | ↑ | Overexpression is correlated with TNM stage | – | Human; in vivo | – | +/– | Wei et al., 2019 |
| circFUT8 | BCa | ↓ | Inhibits migration and invasion of Bca cells through silencing KLF10-mediated Slug signaling, inhibitory effect on lymphatic metastasis | miR-570-3p | Human, Mouse; in vitro/in vivo | Human cell lines – SV-HUC-1, T24, UM-UC-3 | +/– | He Y. et al., 2020 |
| circ_0071662 | BCa | ↓ | Inhibits cell proliferation and invasion; upregulates HPGD and NF2 | miR-146b-3p | Human; in vivo | – | +/+ | Abulizi et al., 2019 |
| circ_0018289 | CC | ↑ | Associated with poor disease free survival | – | Human; in vivo | – | +/– | He Q. et al., 2020 |
| circ_0001038 | CC | ↑ | Promotes cell metastasis; suppresses inhibition of oncogenic targets like CNNM3 and MACC1 | miR-337-3p | Human; in vivo | – | +/– | Wang Y. et al., 2020 |
| circEIF4G2 | CC | ↑ | Induce cell growth and migration | miR-218 | Human; in vitro | Human cell lines – HeLa, CasKi, C33A, SiHa cells | +/– | Mao et al., 2019 |
| circCLK3 | CC | ↑ | Promotes cell proliferation, EMT, migration and invasion | miR-320a | Human, Mouse; in vitro/in vivo | Human cell lines – SiHa, HeLa, CaSki, C-33A, MS751 | +/– | Hong et al., 2019 |
| circ_0000388 | CC | ↑ | Induces proliferation, migration, invasion,inhibit apoptosis; regulates miR-377-3p/TCF12 axis | miR-377-3p | Human; in vitro | Human cell lines – HeLa, SiHa | +/– | Meng et al., 2020 |
| circ_0101996 | CC | ↑ | – | – | Human; in vivo | – | +/– | Wang Y-M. et al., 2017 |
| circ_0101119 | CC | ↑ | – | – | Human; in vivo | – | +/– | Wang Y-M. et al., 2017 |
| circ_0104649 | CC | ↑ | – | – | Human; in vivo | – | – | Wang Y-M. et al., 2017 |
| circ_0104443 | CC | ↑ | – | – | Human; in vivo | – | – | Wang Y-M. et al., 2017 |
| circFoxO3a | CC | ↓ | Correlates with stromal invasion, positive lymph node metastasis and poor prognosis | – | Human; in vivo | – | +/– | Tang et al., 2020 |
| circ_0081001 | OSC | ↑ | Overexpression was associated with poor prognosis | – | Human; in vitro | Human cell lines – MG63,KHOS,U2OS | +/– | Kun-peng et al., 2018a |
| circ_0002052 | OSC | ↓ | Overexpression suppresses OS cell proliferation, migration and invasion while promoting apoptosis; regulates circ_0002052/miR-1205/APC2/Wnt/b-catenin pathway | miR-1205 | Human; in vitro | Human cell lines – hFOB 1.19, 293T | +/+ | Wu Z. et al., 2018 |
| circ-SLC7A5 | ESCC | ↑ | Overexpression correlated with TNM stage and poor OS | – | Human; in vitro | Human cell lines – K30, K70, K140, K180, K150, K450, T10, T12w | +/– | Wang Q. et al., 2020 |
| circ_0004771 | ESCC | ↑ | Increases the expression of CDC25 | miR-339-5p | Human; in vitro | Human cell lines – FHC, HCT-116, SW480 | +/– | Huang E. et al., 2020 |
| CRC | ↑ | – | – | Human; in vitro | Human cell lines – FHC, HCT-116, SW480 | +/– | Pan et al., 2019 | |
| circ_0092125 | OSCC | ↓ | Correlated with tumor size, TNM stage, and lymph node metastasis | - | Human; in vitro | Human cell lines – SCC15, SCC25, CAL27 | +/– | Gao et al., 2020 |
| circ_0001874 | OSCC | ↑ | Associated with TNM stage and tumor grade | – | Human; in vivo | – | +/– | Zhao S. Y. et al., 2018 |
| circ_0001971 | OSCC | ↑ | Upregulation is associated with TNM stage | – | Human; in vivo | – | +/– | Zhao S. Y. et al., 2018 |
| circ-CCND1 | LSCC | ↑ | Improve the stability of CCND1 mRNA; increases LSCC growth | HuR; miR-646 | Human, Mouse; in vitro/in vivo | Human cell lines –AMC-HN-8, Hep-2, LSC-1, TU212, TU177, TU686, SCC10A, NP-69 | +/– | Zang et al., 2020 |
| circFLNA | LSCC | ↑ | Induces migration of LSCC cells by targeting miR486-3p/FLNA axis; high level of FLNA implicates poor survival and drug resistance | miR486-3p | Human; in vitro | Human cell lines –Tu212, SCC-2, SCC40 | +/+ | Wang J. X. et al., 2019 |
| circMATR3 | HSCC | ↑ | Upregulation of oncogene USP28 that contributes to MYC stability | miR-188-5p, miR-448 | Human; in vitro | Human cell line - FaDu | +/– | Wang Z. et al., 2020 |
| circMORC3 | HSCC | ↓ | Associated with T stages and tumor sizes | – | – | – | +/– | Zheng and Chen, 2020 |
| circMYBL2 | AML | ↑ | Increases the translational efficiency of FLT3 kinase; knockdown impairs the cytoactivity of FLT3-ITD AML cells, including quizartinib-resistant cells | – | Human, Mouse; in vitro/in vivo | Human cell lines – MV4-11, MOLM-13,THP-1, HL60, NB4 and ML-2,U937 | +/+ | Sun et al., 2019 |
| Circ-DLEU2 | AML | ↑ | Induces cell proliferation and reduces apoptosis | miR-496 | Human, Mouse; in vitro/in vivo | Human cell lines – MOLM-13, HL-60, MV-4-11 | +/+ | Wu D. M. et al., 2018 |
| f-circPR | AML | ↑ | Promoted cell proliferation | – | – | – | +/+ | Guarnerio et al., 2016 |
| f-circM9 | AML | ↑ | Favors leukemia progression | – | – | – | +/+ | Guarnerio et al., 2016 |
| circ_001569 | CRC | ↑ | Promotes cell proliferation and invasion | miR-145 | Human; in vitro | Human cell lines – SW480, HCT116, SW620, LOVO | +/+ | Xie et al., 2016 |
| circ_0007534 | CRC | ↑ | Increase in the Bcl2/Bax ratio in CRC cells and inhibits apoptosis | – | Human; in vitro | Human cell lines – SW620, HCT116, LoVo, SW480, HT29 | +/+ | Zhang R. et al., 2018 |
| circ_0000069 | CRC | ↑ | Knockdown induces G0/G1 arrest and inhibits cancer progression | – | Human; in vitro | Human cell lines – HT29, LoVo, HCT-116, SW480 | +/+ | Guo et al., 2016 |
| circ_0020397 | CRC | ↑ | Upregulates TERT and thereby induces cell proliferation | miR-138 | Human; in vitro | Human cell lines – LoVo, HCT116, SW480, SW620 | +/+ | Zhang X. et al., 2017 |
| circBANP | CRC | ↑ | Promotes CRC cell proliferation; induces p-Akt protein expression | – | Human; in vitro | Human cell lines – HT29, HCT116 | +/+ | Zhu M. et al., 2017 |
| circ5615 | CRC | ↑ | promotes CRC progression through miR-149-5p/TNKS axis | miR-149-5p | Human, Mouse; in vitro/in vivo | Human cell lines – HCT 116, LoVo, HT-29, SW480, NCM460 | +/+ | Ma et al., 2020 |
| circPTK2 | CRC | ↑ | Promotes EMT of CRC cells via expression of mesenchymal marker vimentin | – | Human, Mouse; in vitro/in vivo | Human cell lines – HCT15, SW620, SW480, LOVO | +/+ | Yang H. et al., 2020 |
| LC | ↓(NSCLC) | Overexpression augments T1F1γ expression, reduces TGF β induced EMT | miR-429/miR-200b3p | Human, Mouse; in vitro/in vivo | Human cell lines – BEAS-2B, A549, H1299, H1650, SPC-A1, Calu3,H226, H520, SK-MES-1 | +/+ | Wang L. et al., 2018 | |
| circ_0060745 | CRC | ↑ | Promotes CSE1L-mediated CRC cell proliferation and metastasis | miR-4736 | Human; in vitro | Human cell lines – NCM460, HT29, LOVO, PKO, SW480 | +/+ | Wang and Wang, 2020 |
| circ_0008285 | CRC | ↓ | Inhibits CRC cell proliferation and migration; regulates PI3K/AKT pathway | miR-382-5p | Human; in vitro | Human cell lines – SW480, RKO, HCT8, SW620, HCT116, DLD1,FHC | +/+ | Wang et al., 2020 |
| circ-0001313 | CRC | ↑ | Inhibits apoptosis regulating PI3K/Akt signaling pathway | miRNA-510-5p | Human; in vitro | Human cell lines – SW620, HCT116, SW480, HT-29, LoVo, NCM460 | +/+ | Tu et al., 2020 |
| circDDX17 | CRC | ↓ | inhibits cell proliferation, migration, invasion, and promotes apoptosis | hsa-miR-21-5p | Human; in vitro | Human cell lines – SW480, SW620, HT29, LoVo, HCT116, RKO | +/+ | Li X-N et al., 2018 |
| circ-ABCB10 | BC | ↑ | Knockdown suppresses proliferation and induces apoptosis | miR-1271 | Human; in vitro | Human cell lines – MCF-7, MDA- MB-231, MDA-MB-468, MDA-MB-453 | +/+ | Liang et al., 2017 |
| circEHMT1 | BC | ↓ | Inhibits metastasis by regulating circEHMT1/miR-1233-3p/KLF4 axis | miR-1233-3p | Human, Mouse; in vitro/in vivo | Human cell lines – ZR-75-1, MCF-7, MB-468, T47D, SK-BR3, MDA-MB-231, BT-549, HMEpC | +/+ | Lu M. et al., 2020 |
| circ_0011946 | BC | ↑ | Promotes migration and invasion | miR26a/b | Human; in vitro | Human cell lines – HS-578T, T47D, MCF-7, BT549, MDA-MB-231, SKBR-3 | +/+ | Zhou J. et al., 2018 |
| circGFRA1 | BC | ↑(TNBC) | Promotes cell proliferation and inhibits apoptosis; regulates GFRA1 expression | miR-34a | Human, Mouse; in vitro/in vivo | Human cell lines – MCF10A, SKBR3, T47D, BT474, MCF-7, BT-483, BT-20, BT549, MDA-MB-468, MDA-MB-231 | +/+ | He et al., 2017 |
| circ_0001982 | BC | ↑ | Promotes tumorigenesis | miR-143 | Human; in vitro | Human cell lines – MDA-MB-231, MCF-7, MDAMB-468, MDA-MB-435s | +/+ | Tang et al., 2017 |
| circTADA2A | BC | ↓ | Possesses tumor-suppressor capability, restores the expression of SOCS3, suppressed cell proliferation, migration, invasion, clonogenicity | miR-203a-3p | Human, Mouse; in vitro/in vivo | Human cell lines – MCF-7 MDA-MB-231 | +/+ | Xu et al., 2019 |
| circ-10720 | HCC | ↑ | Promotes migration, invasion and EMT by stabilizing vimentin | miR-1246, miR-578, miR-490-5p | Human, Mouse; in vitro/in vivo | Human cell lines – PLC-PRF-5, SMMC-7721, HEK-293T | +/+ | Meng et al., 2018 |
| circPTGR1 | HCC | ↑ | Knockdown promotes expression of epithelial markers and reduces the levels of mesenchymal markers | miR-449a | Human, Mouse; in vitro/in vivo | Human cell lines – HepG2, L-O2, SMCC7721, HEP3B, HUH7, MHCC97-L, MHCC 97H, HCC-LM3 | +/+ | Wang G. et al., 2019 |
| circTRIM33–12 | HCC | ↓ | Upregulates TET1 expression; suppresses tumor proliferation, migration, invasion | miR-191 | Human, Mouse; in vitro/in vivo | Human cell lines – MHCC97-L, HCC97-H, HCCLM3, SMMC-7721 | +/+ | Zhang P. F. et al., 2019 |
| circ-BIRC6 | HCC | ↑ | Knockdown reduces Bcl2 mRNA and protein levels | miR3918 | Human, Mouse; in vitro/in vivo | Human cell lines – SKHEP-1, Huh-7 MHCC97H | +/+ | Tang et al., 2015 |
| circ_0070269 | HCC | ↓ | Increases expression of NPTX1,that inhibits aggressive tumor behavior | miR182 | Human, Mouse; in vitro/in vivo | Human cell lines – Hep3B, SMMC-7721, HepG2, PLC, Huh-7,LO2 | +/+ | Zhang P. F. et al., 2019 |
| circADAMTS13 | HCC | ↓ | Acts as a tumor suppressant; inhibits HCC proliferation | miR-484 | Human; in vitro | Human cell lines – PLC/PRF/5, SK-Hep-1, Hep3B, HepG2 | +/+ | Qiu L. et al., 2019 |
| cZNF292 | GBM | ↑ | Promotes angiogenesis; regulates STAT3/5/β-catenin pathway | – | Human; in vitro | Human cell lines – U87MG and U251 | +/+ | Yang P. et al., 2019 |
| circ_0037251 | GBM | ↑ | Enhances GBM progression, upregulates mTOR; inhibits cell apoptosis and G1 phase arrest | miR-1229-3p | Human, Mouse; in vitro/in vivo | Human cell lines – U373, U251, HEK293T | +/+ | Cao et al., 2019 |
| circMAPK4 | GBM | ↑ | Suppress apoptosis through decreased phosphorylation of p38/MAPK | miR-125a-3p | Human, Mouse; in vitro/in vivo | Human cell lines – U138, U373, U87 | +/+ | He et al., 2020a |
| circ-U2AF1 | GBM | ↑ | Enhances cell proliferation, migration, and invasion, increases expression of NOVA2 | miR-7-5p | Human, Mouse; in vitro/in vivo | Human cell lines – U87MG, U251, U87, HEB | +/+ | Li, 2019a |
| circNT5E | GBM | ↑ | Upregulates NT5E, SOX4, PI3KCA; promotes cell proliferation, migration, and invasion | miR-422a | Human, Mouse; in vitro/in vivo | Human cell lines – U87, U251 | +/+ | Wang R. et al., 2018a |
| circ_0029426 | GBM | ↑ | Promotes cell proliferation, migration and invasion, and inhibits cell apoptosis | miR-197 | Human; in vitro | Human cell lines – U87, U251, LN229, U87MG, A172, NHA | +/+ | Zhang G. et al., 2019 |
| circ-TTBK2 | GBM | ↑ | Regulates circ-TTBK2/miR-217/HNF1β/Derlin-1 axis; promotes cell proliferation, migration, and invasion, while inhibiting apoptosis | miR-217 | Human, Mouse; in vitro/in vivo | Human cell lines – U87, U251, HEK293T | +/+ | Zheng et al., 2017 |
| circMMP9 | GBM | ↑ | Upregulates the expression of (CDK4) and aurora kinase A; promotes proliferation, migration and invasion abilities | miR-124 | Human, Mouse; in vitro/in vivo | – | +/+ | Wang R. et al., 2018b |
| circ_0020123 | LC | ↑(NSCLC) | Upregulates ZEB1 and EZH2 for tumor growth and EMT | miR-144 | Human, Mouse; in vitro/in vivo | Human cell lines – PC9, H1573, A549, SK-MES-1, H1299, Calu-3 | +/+ | Qu et al., 2018 |
| f-circEA-4a | LC | ↑(NSCLC) | Induces cell proliferation, metastasis and invasion | – | Human; in vitro | Human cell lines –A549, HT1299 | +/+ | Tan et al., 2018 |
| f-circEA-2a | LC | ↑(NSCLC) | Promotes cell migration and invasion | – | Human; in vitro | Human cell lines –A549, HT1299 | +/+ | Tan et al., 2018 |
| circ_104916 | GC | ↓ | Inhibits cell proliferation, migration and EMT | – | Human, Mouse; in vitro/in vivo | Human cell lines – AGS, MKN-28, NCI-N87, MKN-45,GES1 | +/+ | Li J. et al., 2017 |
| circPDSS1 | GC | ↑ | Enhances expression of NEK2 leading to cell migration and proliferation | miR-186-5p | Human; in vitro | Human cell lines – MGC-803, HGC-27, BGC-823, GES-1 | +/+ | Ouyang et al., 2019 |
| circ_0023642 | GC | ↑ | Promotes cell proliferation and metastasis; upregulates N-cadherin, Vimentin and Snail expression | – | Human; in vitro | Human cell lines – MGC-803, MNK-45,SGC-7901, HGC-27, GES1 | +/+ | Zhou L. H. et al., 2018 |
| circATAD1 | GC | ↑ | Increases cell progression, upregulates YY1 | miR-140-3p | Human; in vivo, in vitro | Human cell lines – GES1, SGC7901, BGC-823, AGS, MGS-803 | +/+ | Zhang L. et al., 2020 |
| circFN1 | GC | ↑ | Promotes viability and inhibits apoptosis; facilitates CDDP resistance in vitro | miR-182-5p | Human, Mouse; in vitro/in vivo | Human cell lines – SGC7901CDDP, BGC823C DDP, SGC7901, SGC823) | +/+ | Huang X. X. et al., 2020 |
| circCACTIN | GC | ↑ | Induces EMT and regulates Smad signaling, promote metastatic conversion, angiogenesis | miR-331-3p | Human, Mouse; in vitro/in vivo | Human cell lines – GES1, BGC-823, MGC-803, SGC-7901c | +/+ | Zhang L. et al., 2019 |
| circ-CEP85L | GC | ↓ | Inhibits tumor growth, proliferation and invasion of GC cells | miR-942-5p | Human, Mouse; in vitro/in vivo | Human cell lines – MGC-803, AGS, KATOIII, BGC-823, HGC-27, MKN-45 | +/+ | Lu J. et al., 2020 |
| circMYLK | BCa | ↑ | Augments proliferation, migration, the tube formation of HUVEC, and EMT; stabilizes VEGFA | miR-29a | Human, Mouse; in vitro/in vivo | Human cell lines – EJ, T24, 5673, BIU-87 | +/+ | Zhong et al., 2017 |
| circACVR2A | BCa | ↓ | Upregulates EYA4 expression; suppresses proliferation, migration and invasion and metastasis through miR-626/EYA4 axis | miR-626 | Human, Mouse; in vitro/in vivo | Human cell lines – T24, UM-UC-3, RT4, J82, 5637, HT-1376, TCCSUP, SV-HUC-1 | +/+ | Dong et al., 2019 |
| circ_0061140 | Ovarian cancer | ↑ | Regulates miR370/FOXM1 pathway; promotes cell proliferation, migration, and the EMT | miR-370 | Human, Mouse; in vitro/in vivo | Human cell lines – SKOV3, A2780, OV2008, IGROV1, ES-2 | +/+ | Chen Q. et al., 2018 |
| circUBAP2 | OSC | ↑ | Stable expression of Bcl-2; promotes OS growth and inhibits apoptosis | miR-143 | Human, Mouse; in vitro/in vivo | Human cell lines – hFOB 1.19, MG63, U2OS | +/+ | Zhang H. et al., 2017 |
| circ_001564 | OSC | ↑ | Inhibits cell cycle arrest in G0/G1 phase and apoptosis | miR-29c-3p | Human; in vitro | Human cell lines – U2OS, Saos-2, HOS, MG-63 | +/+ | Song and Li, 2018 |
| circNASP | OSC | ↑ | Augments FOXF1 expression that leads to proliferation and invasion of OS cells, positively correlates with the tumor size and metastasis | miR-1253 | Human; in vitro | Human cell lines – 143B and MG63 | +/+ | Huang et al., 2018 |
| circ_0000337 | ESCC | ↑ | Promotes cell proliferation, migration, and invasion | miR-670-5p | Human; in vitro | Human cell lines – KYSE-150, TE-1, HET-1A | +/+ | Song et al., 2019 |
| circUHRF1 | OSCC | ↑ | Modulates the transcription factor c-Myc; regulates circUHRF1/miR-526b-5p/c-Myc/TGF-β1/ESRP1 axis | miR-526b-5p | Human, Mouse; in vitro/in vivo | Human cell lines – SCC25, CAL27, SCC15, TSCCA | +/+ | Zhao W. et al., 2020 |
| circ_0082182 | CRC | ↑ | Correlated with lymph node metastasis | – | Human; in vitro | Human cell lines – HCT116, SW480, SW620, NCM460 | +/– | Ye D. et al., 2019 |
| circ_0000370 | CRC | ↑ | Correlated with lymph node metastasis | – | Human; in vitro | Human cell lines – HCT116, SW480, SW620, NCM460 | +/– | Ye D. et al., 2019 |
| circ_0035445 | CRC | ↓ | Correlated with TNM stage | – | Human; in vitro | Human cell lines – HCT116, SW480, SW620 and normal cell line-NCM460 | +/– | Ye D. et al., 2019 |
| circ5615 | CRC | ↑ | Promotes CRC progression through miR-149-5p/TNKS axis | miR-149-5p | Human, Mouse; in vitro/in vivo | Human cell lines – HCT 116, LoVo, HT-29, and SW480, NCM460 and HEK-293T | +/+ | Ma et al., 2020 |
| circ_0060745 | CRC | ↑ | Regulates miR-4736/SCE1L | miR-4736 | Human; in vitro | Human cell lines – human colon epithelial cell line (NCM460) and human CRC cell lines (HT29, LoVo, PKO, and SW480) | +/– | Wang and Wang, 2020 |
| circ_0026344 | CRC | ↓ | Correlated with metastasis | miR-21,miR-31 | Human, Mouse; in vitro/in vivo | Human cell lines – HCT116, SW480, HT29, SW620, NCM460, FHC, HEK293T | +/– | Yuan et al., 2018 |
| circ_0000567 | CRC | ↓ | Correlated with lymph metastasis, distal metastasis and TNM stage | – | Human; in vitro | Human cell lines – FHC, SW480, RKO, CACO2, SW620, HCT116 | +/– | Wang J. et al., 2018 |
| circ_0003906 | CRC | ↓ | Correlated with poor differentiation and lymphatic metastasis | – | Human; in vitro | Human cell lines: NCM460, SW480, SW620, HCT8, HCT116, HT29, LoVo | +/– | Zhuo et al., 2017 |
AML, Acute Myeloid Leukemia; CML, Chronic Myeloid Leukemia; CLL, Chronic Lymphocytic Leukemia; MM, Multiple Myeloma; BCL, B cell Lymphoma; CRC, Colorectal Cancer; BC, Breast Cancer; HCC, Hepatocellular Carcinoma; GBM, Glioblastoma; LC, Lung Cancer; NSCLC, Non-Small Cell Lung carcinoma; LAC, Lung Adenocarcinoma; GC, Gastric Cancer; BCa, Bladder Cancer; CC, Cervical Cancer; OSC, Osteosarcoma; ESCC, Esophageal Squamous Cell Cancer; OSCC, Oral Squamous Cell Carcinoma; LSCC, Laryngeal Squamous Cell Cancer; HSCC, Hypopharyngeal Squamous Cell Carcinoma.
Hematological Malignancies
Acute Myeloid Leukemia (AML)
Circ_0009910 is overexpressed in the bone marrow of AML patients, which correlates with poor overall survival (OS) (Ping et al., 2019a). It sponges miR-20a-5p and knocking it down induces apoptosis in AML cells (Ping et al., 2019a). Circ-vimentin is upregulated in AML patients and its elevated expression is an independent poor prognostic factor for OS and leukemia-free survival (LFS) in AML patients (Yi and Lin, 2018). Hirsh et al. examined the expression of circ_0075001 in a cohort of NPM1 wild-type and mutated AML patients and found it to be positively correlated with expression of the cognate linear RNA but independent of the NPM1 mutational status (Hirsch et al., 2017). However, high circ_0075001 expression levels defined patient subgroups characterized by lower expression of components of the Toll-like receptor (TLR) signaling pathway which is associated with a more immature AML phenotype (Hirsch et al., 2017). Circ_0004277 was downregulated in AML patients expression, however, its expression is restored in AML patients subjected to chemotherapy indicating it as a potential diagnostic marker and treatment target in AML (Li W. et al., 2017).
Chronic Myeloid Leukemia (CML)
Circ_100053 was upregulated in peripheral blood mononuclear cells and serum of CML patients and was associated with clinical stage and BCR/ABL mutation status (Ping et al., 2019b). Elevated CircRNA_100053 levels predicted a poor outcome in CML patients and imatinib resistance (Ping et al., 2019b).
Chronic Lymphocytic Leukemia (CLL)
Circ-RPL15 was upregulated in CLL patients and correlated with poor OS and immunoglobulin heavy-chain variable region (IGHV) mutation used in the validation of CLL prognosis (Wu Z. et al., 2019). Circ-RPL15 sequesters miR-146b-3p and activates RAS/RAF1/MEK/ERK pathway to promote CLL development (Wu Z. et al., 2019). Circ-CBFB levels are also elevated in CLL patients and its expression can distinguish CLL patients from healthy controls (Xia L. et al., 2018). It sponges miR-607, which targets FZD3, an activator of Wnt/β-catenin signaling in CLL (Xia L. et al., 2018). Higher expression of circ-CBFB predicted reduced OS in CLL patients and may serve as a prognostic marker for CLL (Xia L. et al., 2018).
Multiple Myeloma (MM)
Elevated expression of circ_0007841 in MM correlated with chromosomal aberrations such as gain 1q21, t (4:14), mutations in ATR, and IRF4 genes, however, its function in MM needs further investigation (Gao et al., 2019). Feng et al. observed lower levels of circ_0000190 in MM tissues and peripheral blood, which correlated with the prognosis and OS of MM patients (Feng et al., 2019). Circ-SMARCA5 is downregulated in MM and its higher expression correlates with lower β2-microglobulin (MG) level and less advanced International Staging System stage (Liu H. et al., 2019). Circ-SMARCA5 downregulation correlates with reduced OS, progression-free survival (PFS), and treatment response in MM patients (Liu H. et al., 2019). CircRNA_101237 level was upregulated in MM patients and has high diagnostic accuracy for MM (Liu and Wang, 2020). Its expression was elevated in patients with 13q14 deletion, 1q21 amplification, p53 deletion, and t(4,14) and t(14,16) gene mutations, but was decreased in those with t(11,14) gene mutations; also the upregulation was associated with a poor response to chemotherapy (Liu and Wang, 2020).
B Cell Lymphoma (BCL)
Circ-APC (circ_0127621) was downregulated in diffuse large B-cell lymphoma (DLBCL) and its levels in plasma can distinguish patients from healthy controls (Hu et al., 2019). Moreover, DLBCL patients with lower circ-APC levels were more likely to exhibit an advanced Ann Arbor stage, shorter OS, resist chemotherapy and display a low International Prognostic Index (Hu et al., 2019).
Solid Tumors
Colorectal Cancer (CRC)
CDR1as was upregulated in CRC tissues and its overexpression correlated with poor survival (Weng et al., 2017). Its upregulation is an independent risk factor for OS and enhanced EGFR/RAF1/MAPK pathway by inhibiting miR-7 tumor-suppressor activity (Weng et al., 2017). Similarly, circCCDC66 was also elevated in CRC patients and its high expression correlated with poor prognosis (Hsiao et al., 2017). It sponges miRNA-33b and miR-93 to promote CRC growth and metastasis by stabilizing MYC mRNA (Hsiao et al., 2017). Increased expression of circ_0004585 in the CRC tissues was associated with increased tumor size (Tian et al., 2019). Circ_0007142 (Zhu et al., 2019). Circ_0007142 regulates invasion of CRC by sponging miR-103a-2-5p and its upregulation, associated with poor differentiation and lymphatic metastasis (Zhu et al., 2019). Upregulation of circ-HUEW1 in CRC tissues was associated with lymphovascular invasion, lymph node and distant metastasis, and Tumor, Node, and Metastasis (TNM) stage (Chen H. Y. et al., 2020). It sponges miR-486 and regulates the IGF2/β-catenin signaling pathway by targeting PLAGL2 (Chen H. Y. et al., 2020). CRC patients with higher circ_0001178 were more likely to have metastatic features, advanced TNM stage, and adverse prognosis (Ren et al., 2020). It sponges miR-382, miR-587, and miR-616, all of which target ZEB1 (Ren et al., 2020). Circ_0005075 was also highly expressed in CRC and is associated with depth of invasion and advanced TNM stage, and is a prognostic factor affecting both OS and disease-free survival (DFS) in CRC patients (Jin et al., 2019). CircHIPK3 is upregulated in CRC tissues and serves as an independent prognostic factor of poor OS and positively correlates with metastasis and advanced clinical stage (Zeng et al., 2018). Circhipk3 sponges miR-7 and promotes CRC progression by increasing the expression of its target genes FAK, IGF1R, EGFR, and YY1 (Zeng et al., 2018). Ye et al. discovered a 3-circRNA signature as a non-invasive biomarker for CRC diagnosis, they observed elevated expression of circ_0082182 and circ_0000370, and downregulated circ_0035445 levels in the plasma of CRC patients (Ye D. et al., 2019). The upregulation of circ_0082182 and circ_0000370 was strongly associated with lymph node metastasis, while the circ_0035445 downregulation was connected with the TNM stage (Ye D. et al., 2019). Also, circ_0082182 and circ_0035445 showed a difference between preoperative and postoperative stages, while circ_0000370 had no significant difference between these two stages (Ye D. et al., 2019). Circ_0007534 was upregulated in plasma of CRC patients, which correlated with the progression of clinical classifications, metastatic phenotype, poor differentiation, and poor prognosis in CRC patients (Zhang W. et al., 2018). Increased circ_0007534 expression was associated with poor prognosis in CRC patients (Zhang W. et al., 2018). Overexpression of circFADS2 was closely related to the size, differentiation, infiltration depth, lymphatic and distant metastasis, and TNM stage of CRC patients (Xiao et al., 2020). Patients with increased circFADS2 levels had a poor OS and had a better predictive value when combined with the TNM stage (Xiao et al., 2020). Circ5615 was upregulated in CRC was an independent prognosis factor for CRC, and it was associated with a higher T stage and poor OS in CRC patients (Ma et al., 2020). Patients with elevated expression of circ-FARSA had a poor OS and the circRNA promotes CRC progression by regulating the miR-330-5p/LASP1 axis (Lu C. et al., 2020). Overexpression of circ_0060745 in CRC tissues was significantly associated with shorter OS, advanced clinical stage, nodal classification, metastasis classification, and liver metastasis (Wang and Wang, 2020). It promoted CRC progression by sponging miR-4736 and regulating SCE1L expression (Wang and Wang, 2020). Downregulation of circ_0001649 in CRC tissues and patient serum was negatively associated with CRC differentiation (Ji et al., 2018). CircITGA7 was downregulated in CRC tissues and was negatively associated with tumor size, lymph metastasis, distant metastasis, and TNM stage (Li X. et al., 2018a). It sponges miR-370-3p and inactivates Ras signaling pathway by upregulating neurofibromin 1 (NF1) (Li X. et al., 2018a). Circ_0000711 was downregulated in CRC tissues and could act as a diagnostic marker for CRC (Li J. et al., 2018b). Downregulation of circ_0014717 in CRC tissues was associated with distant metastasis, TNM stage, and poor OS (Wang F. et al., 2018). Its overexpression promoted cell-cycle arrest by increasing p16 expression leading to reduced growth and invasion of CRC cells (Wang F. et al., 2018). Circ_0004771 was upregulated in serum exosomes of CRC patients and had good diagnostic potential (Pan et al., 2019). Circ_0026344 was downregulated in CRC samples of stage III/IV as compared to tissues with stage I/II (Yuan et al., 2018). Lower circ_0026344 expression correlated with metastasis and predicted poor prognosis in CRC patients (Yuan et al., 2018). Circ_0000567 expression was downregulated in CRC tissues and was associated with tumor size, lymph metastasis, distal metastasis, and TNM stage (Wang J. et al., 2018). Circ_0003906 was downregulated in CRC tumors and its lower expression was associated with a higher incidence of poor differentiation, lymphatic metastasis, and is an independent risk factor for survival of CRC patients (Zhuo et al., 2017). Analysis of serum from CRC patients revealed overexpression of exo-circ-PNN and it can serve as a potential non-invasive biomarker for CRC detection (Xie Y. et al., 2020).
Breast Cancer (BC)
CircFBXW7 was downregulated in triple-negative breast cancer (TNBC) and was correlated with poor clinical outcomes (Ye F. et al., 2019). Its expression was also negatively associated with tumor size, lymph node metastasis, and is an independent prognostic factor for TNBC (Ye F. et al., 2019). CircFBXW7 sponges miR-197-3p and also encodes for a microprotein FBXW7-185aa that upregulates the tumor-suppressor, FBXW7 (Ye F. et al., 2019). The upregulation of circSEPT9 in TNBC tissues was associated with advanced clinical stage and poor prognosis (Zheng et al., 2020). It decoys miR-637 and modulates, leukemia inhibitory factor (LIF) expression to activate TNBC progression (Zheng et al., 2020). Circ_0001785 was upregulated in BC plasma samples and correlated with histological grade, TNM stage, and distant metastasis (Yin et al., 2018). Autophagy-associated circCDYL was upregulated in tissues and serum from BC patients (Liang et al., 2020). Higher circCDYL levels were associated with estrogen receptor (ER) negative status, higher Ki67 index, larger tumor size, and more lymphatic metastasis (Liang et al., 2020). Moreover, BC patients with high serum circCDYL had a poorer OS compared to early BC and benign patients (Liang et al., 2020). CircKIF4A was overexpressed in TNBC tissues, which correlated with tumor size, lymph node metastasis, and TNM stage (Tang et al., 2019). It induced TNBC cell proliferation and migration by sponging miR-375 and regulating KIF4A expression (Tang et al., 2019). The higher expression levels of circKIF4A correlated with poor OS in TNBC patients (Tang et al., 2019). CircPLK1 was also upregulated in TNBC tissues and correlated with larger tumor size, lymph node positivity, advanced TNM stage and poor OS (Kong Y. et al., 2019). It promotes TNBC metastasis by sponging miR-296-5p and regulating PLK1 expression (Kong Y. et al., 2019). Song et al. observed a higher expression of cytoplasmic circHMCU in BC tissues (Song et al., 2020). CircHMCU sequestered members of let-7 family and enhanced proliferation and metastasis in BC (Song et al., 2020). Its upregulation was associated with histological grade, lymph node metastasis, TNM stage, and poor prognosis (Song et al., 2020). Downregulation of circ_0068033 in BC tissues was associated with tumor size and TNM stage (Yuan et al., 2020). It sequesters miR-659 and its overexpression induces apoptosis (Yuan et al., 2020). Overexpression of circGFRA1 in TNBC tissues was associated with tumor size, TNM staging, lymph node metastasis, and histological grade (He et al., 2017). Patients with upregulated circGFRA1 had shorter OS and DFS (He et al., 2017). CircGFRA1 increases proliferation and inhibit apoptosis by regulating the expression of its cognate gene GFRA1 by sponging miR-34a (He et al., 2017). CircTADA2A-E6 and circTADA2A-E5/E6 generated from the TADA2A gene were downregulated in BC (Xu et al., 2019). Lower expression of circTADA2A-E6 was associated with increased lymphatic metastasis and advanced clinical stage (Xu et al., 2019). BC patients with downregulated circTADA2A-E6 had a poor prognosis with shorter DFS and OS, whereas no association was identified between DFS or OS and circTADA2A-E5/E6 levels (Xu et al., 2019).
Hepatocellular Carcinoma (HCC)
The upregulation of circ_0005075 in HCC tissues was associated with tumor size and had good diagnostic potential (Shang et al., 2016). It decoys miR-23b-5p, miR-93-3p, miR581, and miR-23a-5p and contributes to HCC proliferation, invasion, and metastasis (Shang et al., 2016). Overexpression of circ_100338 was related to low OS and metastatic progression in HCC patients with HBV infection (Huang X. Y. et al., 2017). It decoys miR-141-3p and increases metastatic progression in HCC (Huang X. Y. et al., 2017). CircRHOT1 was upregulated in HCC, its expression was higher in stage III HCC tissues than in stage I/II (Wang L. et al., 2019). HCC patients with higher circRHOT1 expression had poor prognosis (Wang L. et al., 2019). Circ_0091579 was overexpressed in HCC tissues and its upregulation was associated with poor OS of HCC patients (Zhang C. et al., 2018). Interestingly, exposure of HCC samples to cisplatin upregulated circRNA_101237, and its expression correlated with tumor size, lymph node metastasis, distant metastasis, and TNM stage (Zhang C. et al., 2018). CircRNA_101237 was upregulated in HCC tissues and serum samples, and was correlated with tumor size, lymph node metastasis, distant metastasis, and TNM stage (Zhou et al., 2020). Elevated serum circRNA_101237 levels was an independent predictor of poor OS and prognosis in HCC patients (Zhou et al., 2020). Circ-HOMER1 was also upregulated in HCC tissues and associated with larger tumor size, higher TNM stage, and poor prognosis (Zhao M. et al., 2020). It decoys miR-1322 and upregulates CXCL6 (Zhao M. et al., 2020). Circ_0016788 was upregulated in HCC tissues and was associated with poor OS, higher performance status score, larger tumor size, increased Barcelona clinic liver cancer (BCLC) stage, abnormal aspartate aminotransferase, abnormal alpha-fetoprotein and abnormal carbohydrate antigen 199 levels (Cheng et al., 2020). CDR1as was upregulated in HCC samples and was one of the independent factors of hepatic microvascular invasion and had the potential predictive ability (Xu et al., 2017). CDR1as promotes HCC progression by activating PI3K/AKT/mTOR pathway by sponging miR-7 (Xu et al., 2017). Circ_0078602 was downregulated in HCC tissues and was associated with poor prognosis (Kou et al., 2019). CircC3P1 was also downregulated in HCC and was negatively correlated with TNM stage, tumor size, vascular invasion, and lower OS in HCC patients (Zhong et al., 2018). It sponges miR-4641 which targets PCK1 (Zhong et al., 2018). Circ_0001649 was downregulated in HCC and the decreased expression associated with tumor size and occurrence of tumor embolus (Qin et al., 2016). Guo et al. observed the downregulation of circ-ITCH in HCC tissues, which correlated with the poor OS, whereas upregulated circ-ITCH associated with favorable survival in HCC patients (Guo et al., 2017). CircMTO1 was downregulated in HCC tissues and its decreased expression was associated with poor prognosis of HCC patients (Han et al., 2017). Downregulation of circTRIM33-12 was observed in HCC tissues and associated with tumor proliferation, migration, invasion, and immune evasion, and it also served as an independent risk factor for OS and recurrence-free survival (RFS) of HCC patients after surgery (Zhang P. F. et al., 2019). CircTRIM33-12 reduces HCC metastasis and immune evasion by upregulating TET1 expression by sponging miR-191 (Zhang P. F. et al., 2019). CircADAMTS13 was downregulated in HCC tissues and this correlated with the absence of liver cirrhosis, larger tumor size, more severe BCLC stage, and poor patient prognosis (Qiu L. et al., 2019). CircADAMTS13 serves as a tumor-suppressor by sponging miR-484 (Qiu L. et al., 2019). Circ_0070269 was downregulated in HCC tissues and its low expression was correlated with advanced TNM stage, large tumor size, lymph node metastasis, poor OS, and metastasis-free survival of HCC patients (Xiaotong et al., 2019). Circ_0070269 inhibits HCC progression by regulating the miR-182/NPTX1 axis (Xiaotong et al., 2019). Lower expression levels of circSMARCA5 in tissues and plasma samples of HCC patients has good diagnostic potential (Li Z. et al., 2019). Downregulation of circSMARCA5 was associated with tumor differentiation, TNM stage, cancer invasion, and cancer diameter (Li Z. et al., 2019). Adipose-secreted exo-circ-deubiquitination (circ-DB) was upregulated in HCC patients with higher body fat ratios (Zhang H. et al., 2019). It promotes HCC growth and reduces DNA damage by suppression of miR-34a and the activation of USP7 (Zhang H. et al., 2019). Depletion of circ-DB suppressed HCC growth and metastasis in vivo (Zhang H. et al., 2019).
Glioblastoma (GBM)
Lyu et al. using circRNA microarrays identified several differentially expressed circRNAs in GBM (Zhou and Fan, 2020). Enhanced expression of circ_0013520 and circ_0004379 correlated with tumor size, TNM stage, and worse OS in GBM patients (Zhou and Fan, 2020). Circ-CDC45 was also elevated in GBM and associated with larger tumor size, higher grade, and poor OS in glioma (Liu J. et al., 2019). Circ-CDC45 serves as a sponge for miR-516b and miR-527 which functions as tumor-suppressor in GBM (Liu J. et al., 2019). Exosomal circNFIX was upregulated in the serum of temozolomide (TMZ) resistant patients and predicted poor prognosis (Ding et al., 2020). It sequesters miR-132 in GBM cells and its knockdown enhanced TMZ-sensitivity (Ding et al., 2020). Circ_0029426 was upregulated in GBM tissues and this was associated with tumor size and World Health Organization grading (Zhang G. et al., 2019). Circ_0029426 was an independent prognostic factor for GBM and correlated with the poor OS (Zhang G. et al., 2019). It promotes GBM progression by sequestering miR-197 (Zhang G. et al., 2019).
Lung Cancer (LC)
Circ_0013958 was upregulated in lung adenocarcinoma (LUAD) tissues and plasma of patients and was associated with TNM stage and lymphatic metastasis (Zhu X. et al., 2017). Circ_0013958 decoys miR-134 and upregulates CCND1 in LUAD (Zhu X. et al., 2017). CircFARSA was upregulated in tissues and plasma of non-small-cell lung carcinoma (NSCLC) patients (Hang et al., 2018). High expression of circFARSA correlated with cell migration and invasion (Hang et al., 2018). CircFARSA sequesters miR-330-5p and miR-326, leading to the upregulation of the oncogene fatty acid synthase (FASN) (Hang et al., 2018). Circ_0014130 was also overexpressed in NSCLC tissues and correlated with tumor volume, distant metastasis, and poor prognosis (Geng Y. et al., 2020). Li et al. observed the upregulation of circ_0000792 in LUAD tissues, which correlated with T stage, distant metastasis, and smoking status (Li, 2018). Overexpression of circ_100876 in NSCLC tissues was correlated with tumor stage, lymph node metastasis, and reduced OS in NSCLC patients (Yao J. T. et al., 2017). Circ_100876 acts by sequestering miR-136 which targets MMP13 (Yao J. T. et al., 2017). Microarray analysis revealed the upregulation of circFADS2 in LC tissues and this correlated with advanced TNM stage, lymph node metastasis, poor differentiation, and shorter OS of NSCLC patients (Zhao F. et al., 2018). CircFADS2 induces NSCLC progression by sponging miR-498 (Zhao F. et al., 2018). Overexpression of circPVT1 in NSCLC tissues and serum samples was associated with distant metastasis (Li X. et al., 2018c). Circ_0067934 was upregulated in NSCLC tissues and its overexpression was correlated with TNM stage, lymph node status, and distant metastasis (Wang and Li, 2018). Overexpression of circ_0067934 is associated with poorer OS and is an independent poor prognostic factor for NSCLC patients (Wang and Li, 2018). CircPRKCI was upregulated in LUAD tissues and associated with tumor size, TNM stage, poor prognosis, and shorter OS (Qiu et al., 2018). Higher circPRKCI increased proliferation and tumorigenesis of LUAD by sponging miR-545 and miR-589 and upregulating E2F7 (Qiu et al., 2018). Circ_0000064 was upregulated in LUAD tissues and its higher expression levels correlated with T stage, lymphatic metastasis and TNM stage (Luo et al., 2017). Increased circ_0000064 inhibited Caspase-3, Caspase-9, and Bax, and enhanced Bcl-2 expression in LUAD (Luo et al., 2017). Overexpression of circ_0016760 in LUAD tissues correlated with TNM stage, lymph node metastasis, smoking status, differentiation grade and shorter OS; promotes NSCLC development by sponging miR-1287 that targets G-antigen 1 (GAGE1) and is an independent predictor for the survival of NSCLC patients after surgery (Li J. et al., 2018a). Yao et al. reported the upregulation of circRNA_100876 in NSCLC tissues and it was associated with lymph node metastasis, tumor staging, and shorter OS (Yao J. T. et al., 2017). CircRNA_102231 overexpression in LUAD tissues correlated with advanced TNM stage (III-IV), lymph node metastasis, and poor OS (Zong et al., 2018a). CircRNA_103809 was significantly overexpressed in LUAD tissues and its higher expression correlated with a poor OS (Liu W. et al., 2018). CircRNA_103809 enhanced LUAD progression by regulating the miR-4302/ZNF121/MYC loop (Liu W. et al., 2018). Circ_0005962 was appreciably upregulated and circ_0086414 was downregulated in early-stage LUAD and this 2-circRNA signature is a promising diagnostic biomarker for early LUAD (Liu X. X. et al., 2019). Higher plasma levels of circ_0086414 were associated with EGFR mutations (Liu X. X. et al., 2019). Upregulation of circ-PRMT5 were observed in NSCLC tissues and they were associated with larger tumors, lymph node metastasis, later clinical TNM stage, poor OS and PFS in NSCLC patients and is an independent prognostic factor for NSCLC patients (Wang Y. et al., 2019). Circ-RAD23B overexpression in NSCLC tissues was associated with lymph node metastasis, lower differentiation grade, and poor OS (Han et al., 2019). Circ-RAD23B enhanced cell growth by regulating the miR-593-3p/CCND2 axis and increased cell invasion by regulating the miR-653e5p/TIAM1 pathway (Han et al., 2019). Circ_0102533 was elevated in NSCLC tissues and whole blood samples up and its regulation was significantly associated with tumor type, TNM stages, lymph nodes metastasis, and distant metastasis or recurrence (Zhou X. et al., 2018). Circ_0102533 was useful in the detection of stage I-II NSCLC patients and elevated circ_0102533 levels in whole blood was acceptable as a blood-based tumor marker for NSCLC screening (Zhou X. et al., 2018). Circ_0079530 functions as an oncogene in NSCLC by enhancing cell proliferation and invasion and its overexpression was associated with tumor size and lymph node metastasis (Li J. et al., 2018a). CircFGFR3 was significantly upregulated in LC tissues and its overexpression was closely associated with poor prognosis and reduced OS after surgery (Qiu B. Q. et al., 2019). CircFGFR3 increased NSCLC cell invasion and proliferation by regulating Gal-1, pAKT, and p-ERK1/2 by sponging miR-22-3p (Qiu B. Q. et al., 2019). Elevated circ_000984 levels in NSCLC tissues correlated with advanced TNM stage, lymph nodes metastasis, poor OS, and lower DFS in NSCLC patients (Li X. et al., 2019). Circ_000984 activated Wnt/β-catenin signaling and its overexpression is an independent prognostic indicator for NSCLC patients (Li X. et al., 2019). Overexpression of circ_0001946 in LUAD tissues was associated with a higher TNM stage, tumor size, and low OS (Yao et al., 2019). Circ_0001946 enhanced LUAD progression by sponging miR-135a-5p and stabilizing its target SIRT1, which activates Wnt/β-catenin signaling pathway (Yao et al., 2019). Circ_0037515 and circ_0037516 were significantly downregulated in NSCLC tissues and have the potential for diagnosis (Zhao D. et al., 2020). Reduced levels of circ_0033155 in NSCLC tissue was associated with lymphatic metastasis, and its overexpression reduced cell proliferation, colony formation and migration, and increased the level of PTEN in NSCLC (Gu et al., 2018). Downregulation of circ_100395 in LC tissues was associated with metastasis and poor prognosis (Chen D. et al., 2018). Overexpression of circ_100395 reduced malignancy by regulating the miR-1228/TCF21 axis (Chen D. et al., 2018). Downregulation of circ_0001649 in NSCLC tissues was associated with positive lymph node, smoking status, and differentiation grade (Liu T. et al., 2018). Patients with downregulated circ_0001649 had shorter OS and it could be a prognostic biomarker for NSCLC (Liu T. et al., 2018). CircRNA_0056616 was upregulated in tissues and plasma of LUAD patients and it was correlated with TNM stage and lymph node metastasis (He F. et al., 2020). Circ_0000190 and circ_000164 were overexpressed in plasma and tissues from LC patients and expression of circ_0000190 was associated with late-stage, extra-thoracic metastasis, poor survival, and prognosis (Luo Y. H. et al., 2020). These exosomal circRNAs are easily detectable in liquid biopsy and may serve as potential biomarkers for LC (Luo Y. H. et al., 2020).
Gastric Cancer (GC)
Deregulation of circRNAs has been reported in many gastric cancers and they can potentially serve as useful prognostic markers and therapeutic targets (Naeli et al., 2020). Elevated CDR1as levels in GC tissues was an independent risk factor and linked to the poor OS in GC patients (Pan et al., 2018). CDR1as enhances the development of GC by activating PTEN/PI3K/AKT pathway by sponging miR-7 (Pan et al., 2018). Overexpression of circ_0010882 in the plasma of GC patients was a prognostic factor for OS and correlated with the poor OS (Peng et al., 2020). Circ_0010882 contributes to GC cells proliferation, migration, invasion, and apoptosis by modulating PI3K/AKT/mTOR pathway (Peng et al., 2020). The upregulation of circ-DCAF6 was associated with depth of invasion, lymph node invasion, and TNM stage in GC patients and is an independent risk factor for OS (Wu L. et al., 2019). Circ-PRMT5 was upregulated in GC tissues and it was associated with tumor size, TNM stages, degree of differentiation, lymph node metastasis, and distant metastasis (Wu L. et al., 2019). GC patients with reduced circPRMT5 expression had better prognosis and OS than those with increased levels (Wu L. et al., 2019). Circ-PRMT5 promoted GC cell growth, migration, and invasion by sponging miR-145 and miR-1304 and upregulating MYC expression (Du et al., 2019). Circ_0009910 expression was significantly increased in GC tissues and correlated with clinical stage, distant metastasis, and differentiation (Liu M. et al., 2018). Patients with elevated circ_0009910 had a poor OS compared to patients with decreased expression (Liu M. et al., 2018). Circ_0000419 was downregulated in GC plasma and exosomes and this negatively correlated with tumor stage, lymphatic and distal metastasis, venous, and perineural invasion (Tao et al., 2020). Circ_0000419 is predicted to sponge miR-141-5p and miR-589-3p and its downregulation significantly correlate with Borrmann type and differentiation grade (Tao et al., 2020). Patients with downregulated circ_0000419 had a poor OS and DFS (Tao et al., 2020). Downregulation of circ_0006156 in GC tissues was associated with lymph node metastasis, nerve invasion, and degree of tumor differentiation, besides low expression of circ_0006156 correlated with progression-free survival, and OS of GC patients (He Y. et al., 2020). Circ_0001821 was significantly downregulated in GC tissues, and whole-blood specimens of GC patients (Kong S. et al., 2019). Downregulation of circ_0001821 was negatively associated with tumor depth and lymph node metastasis (Kong S. et al., 2019). The combined use of circulating circ_0001821 with the existing tumor markers yielded good diagnostic potential in GC (Kong S. et al., 2019). Downregulation of circCCDC9 in GC tissues was negatively associated with tumor size, lymph node invasion, advanced clinical stage, and OS (Luo Z. et al., 2020). CircCCDC9 sponges miR-6792-3p which targets CAV1 a tumor-suppressor gene in GC (Luo Z. et al., 2020). Downregulation of circRHOBTB3 in GC tissues was associated with poor differentiation and unfavorable prognosis in GC patients (Deng et al., 2020). CircRHOBTB3 has a tumor-suppressor activity and inhibits growth of GC cells by sponging miR-654-3p and promoting the expression of its target p21 (Deng et al., 2020). CircRNA_100269 was downregulated in GC tissues and its lower expression was associated with histological subtypes and node invasion (Zhang Y. et al., 2017). GC patients with low circRNA_100269 levels had poor OS than patients with higher levels (Zhang Y. et al., 2017). Downregulation of circRNA_100269 promoted GC development by releasing its inhibitory effect on oncogenic miR-630 (Zhang Y. et al., 2017). Circ_0000745 was downregulated in GC tissues and plasma samples of GC patients and was associated with tumor differentiation and TNM stage (Huang M. et al., 2017). The use of circ_0000745 in plasma combined with carcinoembryogenic antigen showed potential for use as a diagnostic marker for GC (Huang M. et al., 2017). Downregulation of circPSMC3 was observed in plasma and tissue samples from GC patients and is negatively correlated with TNM stage, lymphatic metastasis, and reduced OS in GC patients (Rong et al., 2019). CircPSMC3 contributed to GC progression by regulating PTEN by sponging miRNA-296-5p (Rong et al., 2019). Circ-KIAA1244 was downregulated in plasma and tissues from GC patients and was negatively associated with the TNM stage, lymphatic metastasis, and reduced OS (Tang et al., 2018). Downregulation of circ-KIAA1244 was an independent prognostic indicator of OS for GC patients (Tang et al., 2018). Downregulation of circ_0000190 was observed in tissues and plasma samples of GC patients and is correlated with tumor diameter, lymphatic metastasis, distal metastasis, TNM stage, and CA19-9 levels (Chen et al., 2017). Chen et al. observed downregulation of circSMARCA5 in GC tissues and it correlated with differentiation, lymph node metastasis, vascular invasion, poor OS and DFS in GC patients, moreover, low circSMARCA5 expression was an independent prognostic factor for survival of GC patients (Cai et al., 2019). CircYAP1 was downregulated in GC tissues and was correlated with poor prognosis and reduced OS in GC patients (Liu H. et al., 2018). CircYAP1 expression was higher in early-stage GC patients and such patients were more sensitive to chemotherapy (Liu H. et al., 2018). CircYAP1 decreased cell growth and invasion by sponging miR-367-5p to upregulate p27 Kip1 (Liu H. et al., 2018). Lower expression of circ_0006848 in GC tissues correlated with tumor differentiation and tumor size (Lu et al., 2019a). Levels of circ_0000520 were also decreased in tissues and plasma of GC patients and correlated negatively with the TNM stage in tissues and with CEA expression in plasma (Sun et al., 2017). Circ_0001895 was significantly downregulated in GC tissues and its lower expression was associated with cell differentiation, Borrmann type, and CEA expression (Shao et al., 2017). Circ_0005556 was downregulated in GC tissues and its low expression closely correlated with poor differentiation, TNM stage, and lymphatic metastasis (Yang L. et al., 2019). GC patients with decreased circ_0005556 levels had a shorter OS than those with higher levels (Yang L. et al., 2019). Circ_0067582 was downregulated in GC tissues and is correlated with increased tumor diameter and high CA19-9 (Yu et al., 2020). Circ_0067582 downregulation was associated with a better prognosis after surgery (Yu et al., 2020). Circ_0000467 was overexpressed in GC tissue and plasma and this was correlated with the TNM stage (Lu et al., 2019b). Diagnostic potential of circ_0000467 was found to be superior to other common plasma biomarkers such as CEA and carbohydrate antigens-724 (CA-724) (Lu et al., 2019b). Elevated circRNA_102958 levels were observed in GC tissues and it was significantly correlated with the TNM stage (Wei et al., 2019). Overexpression of circ-ATAD1 was observed in GC and associated with deeper invasion, positive lymph node metastasis, advanced TNM stages, and adverse prognosis (Zhang L. et al., 2020). It promotes GC tumorigenesis by regulating the miR-140-3p/YY1 signaling axis (Zhang L. et al., 2020). CircSHKBP1 was overexpressed in tumors and serum exosomes of GC patients, and it correlated with advanced pathological staging and poor OS (Xie M. et al., 2020). CircSHKBP1 promotes GC progression by sponging miR-582-3p to increase HuR levels and promoting VEGF stability, and also by binding HSP90 to prevent its interaction with STUB1 (Xie M. et al., 2020).
Bladder Cancer (BCa)
Downregulation of circFUT8 in BCa tissues was correlated with poor prognosis, high histological grade, lymph node metastasis, and poor survival rate (He Q. et al., 2020). Circ_0071662 was downregulated in BCa tissues and this correlated with poor prognosis, lymph node invasion and distal metastasis, and poor OS (Abulizi et al., 2019). Overexpression of circ_0071662 inhibited cell proliferation and invasion by sponging miR-146b-3p and upregulating its targets, hydroxy prostaglandin dehydrogenase (HPGD) and neurofibromin 2 (NF2) (Abulizi et al., 2019). Circ-ITCH was downregulated in BCa and this was associated with the histological grade of BCa patients (Yang C. et al., 2018). BCa patients with decreased circ-ITCH expression had poor OS than those with higher levels (Yang C. et al., 2018). Upregulation of circ-ITCH inhibited cell proliferation, migration, and invasion through circ-ITCH/miR-17, miR-224/p21, PTEN signaling axis (Yang C. et al., 2018). CircACVR2A was downregulated in BCa tissues and cell lines and its downregulation was correlated with advanced pathological stage, high grade, lymphatic metastasis, and poor OS (Dong et al., 2019). CircACVR2A reduces proliferation, migration, and invasion of BCa cells by sponging miR-626 to regulate EYA4 expression (Dong et al., 2019).
Cervical Cancer (CC)
Circ_0018289 was upregulated in CC tissues and this correlated with tumor size and lymph node metastasis and poor DFS in CC patients (He et al., 2020b). Overexpression of circ_0001038 in CC tissues was associated with lymph node invasion, myometrial invasion, and unfavorable outcome (Wang Y. et al., 2020). It promotes metastasis by sequestering miR-337-3p and upregulating it targets, Cyclin A, CBS Domain Divalent Metal Cation Transport Mediator 3 (CNNM3), and Metastasis Associated In Colon Cancer 1 (MACC1) (Wang Y. et al., 2020). CircEIF4G2 was upregulated in CC tissues and this correlated with tumor size and lymph node metastasis (Mao et al., 2019). Elevated expression of circEIF4G2 was correlated with worse prognosis in CC patients and induced cell growth and migration by sponging miR-218 and increasing the expression of its target HOXA1 (Mao et al., 2019). Increased expression of circCLK3 in CC tissues was associated with poor tumor differentiation, advanced International Federation of Gynecology and Obstetrics (FIGO) stages and depth of stromal invasion, and indicated poor OS and DFS (Hong et al., 2019). It decoys miR-320a to remove its suppressive effects on FoxM1 and promotes cell proliferation, EMT, migration, and invasion (Hong et al., 2019). Higher circ_0000388 levels in CC patients were significantly associated with FIGO stage, lymph node metastasis, and depth of invasion (Meng et al., 2020). Circ_0000388 increased the proliferation, migration, and invasion, and reduced apoptosis of CC through regulating the miR-377-3p/ TCF12 axis (Meng et al., 2020). Wang et al. observed that 4 circRNAs namely, circ_0101996, circ_0104649, circ_0104443, and circ_0101119 were significantly upregulated in peripheral whole blood from CC patients (Wang Y-M. et al., 2017). Combined detection of circ_0101996 and circ_0101119 could easily distinguish CC patients from healthy controls (Wang Y-M. et al., 2017). CircFoxO3a was significantly downregulated in the serum of CC patients and correlated with deep stromal invasion, positive lymph node metastasis, and poor prognosis (Tang et al., 2020). CircFoxO3a downregulation is a poor prognostic factor for both OS and recurrence-free survival, independent of positive lymph node metastasis in CC patients (Tang et al., 2020).
Osteosarcoma (OSC)
Circ_0081001 was overexpressed in OSC tissues and serums samples and was associated with poor prognosis, and may serve as an independent prognostic factor and biomarker for OSC diagnosis and prognosis (Kun-peng et al., 2018a). Circ_0002052 was also upregulated in OSC tissues and associated with advanced stage, tumor size, metastasis, and poor survival rate in OSC patients (Jing et al., 2020). Circ-0002052 promotes OSC development by activating Wnt/β-catenin signaling by sponging miR-382 (Jing et al., 2020). CircPVT1 was significantly upregulated in OSC tissues and serum samples (Kun-peng et al., 2018b). Moreover, levels of circPVT1 were higher in patients with lung metastasis or chemoresistance (Kun-peng et al., 2018b). Increased expression of circPVT1 correlated with advance Enneking stage, chemoresistance, and lung metastasis, and was found to be a better diagnostic marker than alkaline phosphatase (ALP) for OSC (Kun-peng et al., 2018a). CircHIPK3 was downregulated in OSC tissues and plasma samples (Xiao-Long et al., 2018). Lower circHIPK3 levels correlated with Enneking stage, lung metastasis, lower OS, and poor prognosis in OSC patients (Xiao-Long et al., 2018). Circ_0000190 was found in the extracellular nanovesicles and transmitted from healthy cells to OSC cells to impede cancer development (Li et al., 2020). Reduced expression of circ_0000190 correlated with bigger tumor size, advanced staging (IIB/III), and distant metastasis and is a potential biomarker for OSC (Li et al., 2020).
Head and Neck Squamous Cell Cancer (HNSCC)
Esophageal Squamous Cell Cancer (ESCC)
Overexpression of circ-SLC7A5 in ESCC plasma samples was correlated with TNM stage and poor OS (Wang Q. et al., 2020). Elevated circ-0004771 levels were associated with heavier tumor burden and poor prognosis (Huang E. et al., 2020). Circ_0067934 was upregulated in ESCC tissues and its increased expression correlated with poor differentiation, I-II T stage, and I-II TNM stage (Zong et al., 2018b).
Oral Squamous Cell Carcinoma (OSCC)
Lower expression of circ_0092125 in OSCC correlated with tumor size, TNM stage, and lymph node metastasis in OSCC patients (Gao et al., 2020). Downregulation of circ_0092125 was associated with shorter OS and was an independent risk factor for OSCC prognosis (Gao et al., 2020). Zhao et al. compared circRNAs levels in the saliva between OSCC patients and healthy donors, and observed upregulation of circ_0001874 and circ_0001971 in the saliva of OSCC patients and this correlated with tumor stage and TNM (Zhao S. Y. et al., 2018).
Laryngeal Squamous Cell Carcinoma (LSCC)
Circ_0067934 was upregulated in LSCC tissues and its overexpression was associated with larger tumor size, stronger lymph node metastasis, distant metastasis, and poor prognosis with lower OS rate (Chu, 2020). Upregulated circ-CCND1 levels in LSCC correlated with tumor size, poor differentiation, advanced TNM stage, and poor prognosis (Zang et al., 2020). It binds to HuR and miR-646 to enhance the stability of CCND1 mRNA (Zang et al., 2020). CircFLNA upregulation in LSCC was associated with lymph node metastasis (Wang J. X. et al., 2019). CircFLNA increased the migration of LSCC cells by targeting the miR486-3p/FLNA axis (Wang J. X. et al., 2019).
Hypopharyngeal Squamous Cell Carcinoma (HSCC)
CircMATR3 was upregulated in HSCC tissues and was associated with advanced clinical stage, poor lymph node metastasis, and poor survival of HSCC patients (Wang Z. et al., 2020). CircMATR3 binds to miR-188-5p and miR-448, both having a common target, USP28 (Wang Z. et al., 2020). CircMORC3 downregulation in HSCC tissues and plasma samples was associated with tumor stage and tumor size (Zheng and Chen, 2020).
Circular RNAs in Cancer Therapeutics
Recent advances in RNA-based therapeutics coupled with aberrant expression of circRNAs in cancers makes them attractive therapeutic tools (Liu et al., 2017; Yang Z. et al., 2017; Lei et al., 2019). For example, circRNAs with multiple binding sites for oncogenic proteins or miRNAs can be introduced exogenously to restore the normal regulatory network to control proliferation and apoptosis in cancer (Tay et al., 2015). To facilitate this, multiple strategies to manipulate circRNA levels are currently under investigation and have good prospect for being developed into circRNA-based therapeutic strategies in near future.
The easiest approach to inhibit circRNA expression is RNA interference, using small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs) or by employing chemically modified antisense oligonucleotides (ASOs) complementary to the back-splice junction, latter is preferred for in vivo applications (Cortés-López and Miura, 2016; Santer et al., 2019). Furthermore, complete knockdown of circRNA, CDR1as, by CRISPR/Cas9 genome-editing has been achieved and CDR1as loss-of-function mutant mice were generated (Piwecka et al., 2017). Another possibility is use of the CRISPR/Cas13 RNA knockdown system, wherein circRNA silencing is attained by targeting the CRISPR/Cas13 guide RNA to the back-splice junction of the circRNA (Santer et al., 2019). CircRNA overexpression is usually achieved by retroviral, lentiviral, adenoviral, or adeno-associated virus (AAV) vector constructs bearing circRNA sequence flanked by introns containing intronic complementary sequences (ICS) and splicing signals (Wang K. et al., 2017; Bai et al., 2018; Xia P. et al., 2018). Additionally, antisense oligonucleotides (ASOs) can also be used to enhance circRNA expression, by targeting splice-sites or splice-enhancers to increase the efficiency of back-splicing (Zhang et al., 2014). Apart from this, non-viral systems for circRNA overexpression have also been explored, most notable being in vitro synthesis of circRNAs followed by their in vivo delivery. Exogenous circRNA production first involves the synthesis of linear RNA by in vitro transcription, followed by circularization by employing self-splicing introns or by T4 RNA ligase (Santer et al., 2019). CircRNAs with therapeutic potential are discussed below and summarized in Table 1.
Hematological Malignancies
Acute Myeloid Leukemia (AML)
CircMYBL2 is significantly upregulated in AML patients with FLT3-ITD mutations and it increases the translational efficiency of FLT3 transcript, by facilitating binding of polypyrimidine tract binding protein 1 (PTBP1) to FLT3 transcript (Sun et al., 2019). Downregulation of circMYBL2 reduced levels of FLT3 kinase and inhibited proliferation and promoted differentiation of FLT3-ITD AML (Sun et al., 2019). Overexpression of circ-DLEU2 promoted AML by sponging miR-496 and increasing levels of its target, PRKACB (Wu D. M. et al., 2018). Tumor growth due to overexpression of circ-DLEU2 in vivo was reversed by its knockdown (Wu D. M. et al., 2018). Guarnerio et al. demonstrated that the well-established oncogenic chromosomal translocations such as PML/RARα and MLL/AF9 give rise to fusion circRNAs (f-circRNA), f-circPR, and f-circM9, respectively (Guarnerio et al., 2016). Expression of these f-circRNAs in mouse embryonic fibroblasts promoted cell proliferation and transformed foci-forming capability (Guarnerio et al., 2016). Consistent with its role in promoting cell proliferation, knockdown of f-circM9 increased apoptosis in AML cells (Guarnerio et al., 2016). Presence of f-circM9 conferred protection to leukemic cells in a mice model upon treatment by arsenic trioxide and cytarabine (Guarnerio et al., 2016).
Acute Lymphoid Leukemia (ALL)
Higher expression of circ-PVT1 contributes to ALL progression by sponging let-7 and miR-125 (Hu et al., 2018). Knockdown of circ-PVT1 inhibits cell proliferation and induces apoptosis by reducing expression of c-Myc and Bcl-2, which are targets of let-7 and miR-125 respectively (Hu et al., 2018).
Chronic Myeloid Leukemia (CML)
Circ_0009910 was upregulated in CML and promotes imatinib resistance by sequestering miR-34a-5p which targets ULK1 (Cao et al., 2020). Knockdown of circ_0009910 reduced cell growth and imatinib resistance, along with increased apoptosis and autophagic activation (Cao et al., 2020).
Multiple Myeloma (MM)
Circ-CDYL facilitated MM growth by sponging miR-1180 and increasing the expression of its target YAP (Chen F. et al., 2020). Downregulation of circ-CDYL induces apoptosis by downregulating YAP (Chen F. et al., 2020).
B-Cell Lymphoma (BCL)
Circ-APC was significantly downregulated in DLBCL (Hu et al., 2019). In cytoplasm circ-APC sponges miR-888 leading to an increase in levels of its target APC, whereas in nucleus it binds to APC promoter and recruits the DNA demethylase TET1 to transcriptionally upregulate APC (Hu et al., 2019). Ectopically expressed circ-APC acts as a tumor-suppressor and acts by inhibiting Wnt/β-catenin signaling in DLBCL (Hu et al., 2019).
Solid Tumors
Colorectal Cancer (CRC)
CDR1as which acts as miR-7 RNA sponge is overexpressed in CRC and confers an aggressive oncogenic phenotype (Weng et al., 2017). CDR1as downregulation resulted in inhibition of CRC progression (Weng et al., 2017). EGFR expression is regulated by circHIPK3 which is upregulated in CRC tissues (Zeng et al., 2018). Similar to CDR1as, circHIPK3 also functions as a sponge for miR-7. The knockdown of circHIPK3 inhibited cell proliferation, migration, invasion, and metastasis (Zeng et al., 2018). Circ_001569 was significantly upregulated in CRC tissues and promoted cell proliferation and invasion (Xie et al., 2016). Mechanistically, circ_001569 performs a tumor-promoting function by sponging miR-145 and upregulating its targets E2F5, BAG4, and FMNL2 (Xie et al., 2016). Downregulation of circ_001569 resulted in reduced cell invasion and migration (Xie et al., 2016). Circ_0007534 upregulation was associated with a metastatic phenotype and evasion of apoptosis in CRC (Zhang R. et al., 2018). Silencing of circ_0007534 reduced Bcl2/Bax ratio in CRC cells and induced apoptosis (Zhang R. et al., 2018). Levels of circ_0000069 were also elevated in CRC tissues and its knockdown induced cell-cycle arrest and inhibited cancer progression (Guo et al., 2016). Circ_ITCH acts as a sponge for miR-7 and miR-20a and is significantly downregulated in CRC tissues (Huang et al., 2015). Overexpression of circ_ITCH reduced cell proliferation in CRC by downregulating c-Myc and cyclinD1 (Huang et al., 2015). CircRNA_103809 is also downregulated in CRC patients and its silencing promotes cell proliferation and migration via miR-532-3p/FOXO4 axis (Bian et al., 2018). Interestingly, telomerase reverse transcriptase (TERT) is one of the targets of the tumor-suppressor miR-138 which is sponged by circ_0020397(Zhang X. et al., 2017). Circ_0020397 is upregulated in CRC tissues and its downregulation resulted in lower TERT levels and reduced cell proliferation (Zhang X. et al., 2017). CircBANP was significantly upregulated in CRC tissues and cell lines and its silencing suppressed CRC cell proliferation and reduced p-Akt protein expression (Zhu M. et al., 2017). Circ5615 is upregulated in CRC tissues and functions by sequestering miR-149-5p which targets tankyrase (TNKS), an activator of Wnt/β-catenin stabilization (Ma et al., 2020). Downregulation of circ5615 inhibited proliferation and promoted cell-cycle arrest (Ma et al., 2020). CircFARSA is upregulated in CRC tissues and sequesters miR-330-5p, leading to the upregulation of LASP1 (LIM and SH3 protein 1) (Lu C. et al., 2020). The silencing of circFARSA inhibited proliferation, migration, and invasion of CRC cells (Lu C. et al., 2020). CircPTK2 is elevated in CRC tissues and functions by promoting EMT of CRC cells by binding to vimentin protein at Serine 38, 55, and 82 residues (Yang H. et al., 2020). CircPTK2 knockdown reduced tumorigenicity and metastatic potential of CRC cells (Yang H. et al., 2020). Circ_0060745 promotes CRC metastasis by sequestering miR-4736 and stabilizing its target CSE1L (chromosome segregation 1-like) (Wang and Wang, 2020). The knockdown of circ_0060745 suppressed CRC cell migration and invasion (Wang and Wang, 2020). Circ_0008285 is downregulated in CRC tissues and cell lines (Wang and Wang, 2020). It acts by suppressing PI3K/AKT signaling via miR-382-5p/PTEN axis, leading to inhibition of cell proliferation and migration in CRC (Wang and Wang, 2020). Circ_0001313 is highly expressed in CRC tissues and modulates tumorigenesis by sponging miR-510-5p to elevate AKT2 expression (Tu et al., 2020). Depletion of circ-0001313 decreased proliferation and induced apoptosis in CRC cells (Tu et al., 2020). CircDDX17 is significantly downregulated in CRC tissues and its silencing promoted CRC cell proliferation, migration, invasion, and inhibited apoptosis (Li X-N et al., 2018).
Breast Cancer (BC)
Circ-ABCB10 sponges miR-1271 in BC, its depletion suppresses proliferation and induces apoptosis in BC cells (Liang et al., 2017). CircEHMT1 was downregulated in BC tissues and promotes metastasis by upregulating MMP2 through circEHMT1/miR-1233-3p/KLF4 axis (Lu M. et al., 2020). Overexpression of circEHMT1 inhibited migration and invasion of BC cells by reducing MMP2 expression (Lu M. et al., 2020). Circ_0011946 functions by regulating the expression of replication factor C subunit 3 (RFC3), and silencing it suppressed migration and invasion of BC cells (Zhou J. et al., 2018). CircGFRA1 was upregulated in TNBC cells and functions by regulating the expression of its cognate GFRA1 (GDNF Family Receptor Alpha 1) transcript by sequestering miR-34a, and its knockdown induces apoptosis (He et al., 2017). Circ_0001982 was overexpressed in BC tissues and promotes BC tumorigenesis by sponging miR-143 (Tang et al., 2017). Silencing of circ_0001982 suppressed cell proliferation, invasion, and induced apoptosis in BC cells (Tang et al., 2017). Interestingly, circTADA2A-E6 and circTADA2A-E5/E6, originating from the TADA2A gene, were significantly downregulated in TNBC patients (Xu et al., 2019). CircTADA2A-E6 displays tumor-suppressor properties and functions as a miR-203a-3p sponge and restores the expression of its target SOCS3 (Xu et al., 2019). The knockdown of circTADA2A-E6 promotes proliferation, clonogenicity, migration, and invasion in BC cells (Xu et al., 2019). CircFBXW7 is downregulated in TNBC cell lines, it codes for a microprotein with tumor-suppressive functions in TNBC (Ye F. et al., 2019). Overexpression of circFBXW7 suppressed cell proliferation, migration, and reversed tumor growth in TNBC cells (Ye F. et al., 2019). CircCDYL promoted autophagy by the miR-1275-ATG7/ULK1 axis to enhance the malignant progression of BC cells, its knockdown slows down tumorigenesis by modulating autophagy (Liang et al., 2020).
Hepatocellular Carcinoma (HCC)
CDR1as is overexpressed in HCC resulting in enhanced proliferation and invasion (Yu et al., 2016). Knockdown of CDR1as resulted in increased availability of miR-7 and downregulation of its target genes CCNE1 and PIK3CD, leading to inhibition of cell proliferation and invasion (Yu et al., 2016). Levels of circMTO1 were decreased in HCC, its overexpression in HCC cells sponges oncogenic miR-9 to promote the expression of tumor-suppressor p21 resulting in reduced tumor cell proliferation, metastasis, and invasion (Han et al., 2017). Expression of circ-10720 promotes EMT by inducing transcription factor, Twist1 (Meng et al., 2018). It promotes migration, invasion, and EMT by stabilizing mesenchymal marker vimentin by sponging miR-1246, miR-578, and miR-490-5p (Navarro, 2019). Depletion of circ-10720 inhibited Twist1-induced metastasis (Meng et al., 2018; Navarro, 2019). Elevated circRNA-100338 induced mTOR signaling via the circRNA-100338/miR-141-3p/RHEB axis (Huang X. Y. et al., 2020). The depletion of circ_100338 reduced the activity of mTOR signaling pathway and suppressed HCC tumorigenesis and progression (Huang X. Y. et al., 2020). Circ_0067934 functions by modulating the miR-1324/FZD5/Wnt/β-catenin axis to enhance migration, invasion, and proliferation of HCC cells (Zhu et al., 2018). Silencing of circ_0067934 suppressed proliferation, migration, and invasion of HCC cells (Zhu et al., 2018). CircSMARCA5 is downregulated in HCC tissues and inhibits proliferation, invasion, and metastasis of HCC cells by promoting the expression of the tumor-suppressor TIMP3 by sequestering miR-17-3p and miR-181b-5p (Li Z. et al., 2019). Overexpression of circSMARCA5 inhibits the proliferation and migration of HCC cells (Li Z. et al., 2019). CircPTGR1 promoted HCC progression via the miR-449a/MET pathway and its knockdown reduced HCC progression (Chen et al., 2015; Wang G. et al., 2019). CircRHOT1 facilitated HCC progression by recruiting TIP60, a histone acetyltransferase to the nuclear orphan receptor NR2F6 promoter to enhance its expression (Wang L. et al., 2019). CRISPR/Cas9-based depletion of circRHOT1 suppressed proliferation, migration, and invasion, and promoted apoptosis in HCC cells (Wang L. et al., 2019). CircTRIM33-12 modulates TET1-induced DNA demethylation by sponging miR-191 (Zhang P. F. et al., 2019). Overexpression of circTRIM33-12 inhibited proliferation and invasion of HCC cells (Zhang P. F. et al., 2019). Circ-BIRC6 facilitates HCC progression by acting as a miR-3918 sponge and thus targeting the miR-3918/Bcl2 axis (Tang et al., 2015). Its knockdown resulted in decreased HCC cell proliferation, migration, and invasion, and enhanced apoptosis (Tang et al., 2015). Circ_0070269 levels are downregulated in HCC tissues and it facilitates HCC progression by regulating the miR-182/NPTX1 axis (Zhang P. F. et al., 2019). Its overexpression suppresses the proliferation, and invasion of HCC cells (Zhang P. F. et al., 2019). CircADAMTS13 was downregulated in HCC tissues (Qiu L. et al., 2019). It sequesters oncogenic miR-484, and overexpression of circADAMTS13 resulted in a significant reduction in HCC cell proliferation (Qiu L. et al., 2019).
Glioblastoma (GBM)
CircNFIX acts as a sponge for miR-34a-5p which targets the Notch signaling pathway in GBM cells (Xu et al., 2018). The knockdown of circNFIX inhibited cell proliferation and migration of GBM cells by downregulating NOTCH1 (Xu et al., 2018). CircRNA cZNF292 is an oncogenic circRNA that promotes angiogenesis in GBM (Yang P. et al., 2019). Downregulation of cZNF292 reduced proliferation in GBM cells and suppressed human glioma tube formation by modulating Wnt/β-catenin signaling pathway (Yang P. et al., 2019). Circ_0037251 enhances GBM progression by sponging miR-1229-3p and upregulating mTOR (Cao et al., 2019). Knockdown of circ_0037251 inhibited the expression of mTOR leading to increased apoptosis and promoting cell-cycle arrest (Cao et al., 2019). CircMAPK4 functions as an oncogene to enhance GBM cell survival by sponging miR-125a-3p and regulating the p38/MAPK pathway, its downregulation induces apoptosis of GBM cells (He et al., 2020a). Circ-U2AF1 enhanced glioma cell proliferation, migration, and invasion by sponging miR-7-5p and increasing the expression of NOVA2 (Li, 2019b). The knockdown of circ-U2AF1 decreased the migration and invasion abilities of glioma cells by downregulating NOVA2 (Li, 2019a). Circ_0001946 was downregulated in GBM cells and functions by sponging miR-671-5p (Li, 2019a). Overexpression of circ_0001946 reduced the migration, invasion, and proliferation of GBM cells by inhibiting the pro-tumorigenic effects of miR-671-5p (Li, 2019a). CircNT5E promotes GBM tumorigenesis by sponging miR-422a and its CRISPR/Cas9-mediated deletion suppressed proliferation, migration, and invasion of GBM cells (Wang R. et al., 2018a). Circ_0029426 facilitates tumorigenesis by sequestering miR-197, its silencing suppressed proliferation, migration, and invasion, and promoted apoptosis of GBM cells (Zhang G. et al., 2019). Circ-TTBK2 promotes GBM malignancy by modulating the miR-217/HNF1β/Derlin-1 pathway, and its knockdown blocked GBM progression (Zheng et al., 2017). CircMMP9 elicits its oncogenic function by sequestering miR-124 and upregulating the expression of its targets, cyclin-dependent kinase 4 (CDK4), and aurora kinase A (AURKA) (Wang R. et al., 2018b). Silencing of circMMP9 inhibited proliferation, migration, and invasion of GBM cells (Wang R. et al., 2018b).
Lung Cancer (LC)
CircRNA_103809 functions as a miR-4302 sponge leading to the ZNF121-mediated increase in MYC expression (Liu W. et al., 2018). Downregulation of circRNA_103809 resulted in delayed tumor growth and inhibited cell proliferation and invasion in LC cells (Liu W. et al., 2018). Circ_0020123 sequesters miR-144 and causes upregulation of ZEB1 and EZH2 which are critical for EMT and its knockdown suppresses NSCLC growth and metastasis (Qu et al., 2018). CircFADS2 sponges tumor-suppressor miR-498, its silencing reduced invasion and proliferation in LC cells (Zhao F. et al., 2018). Circ_0000064 levels were elevated in LC tissues and its ablation attenuates cell proliferation and promotes cell apoptosis in LC cells (Luo et al., 2017). CircRNA_102231 is overexpressed in LUAD tissues and its inhibition resulted in reduced cell proliferation, and invasion (Zong et al., 2018b). Circ_0033155 is downregulated in NSCLC tissues, and its overexpression resulted in reduced cell proliferation, migration, and colony formation in NSCLC (Gu et al., 2018). CircRNA_100876 acts as a miR-136 decoy, which targets MMP13 (Yao J. T. et al., 2017). Its silencing suppressed MMP13 expression and increased extracellular matrix formation (Yao J. T. et al., 2017). CircPTK2 was downregulated in NSCLC cells during TGF-β induced EMT (Wang L. et al., 2018). CircPTK2 functions as the miR-429/miR-200b3p sponge and reduced the expression of tumor-suppressor T1F1γ, consistent with this its overexpression in NSCLC cells augments T1F1γ expression and reduces TGF-β induced EMT (Wang L. et al., 2018). CircPVT1 facilitates the increased expression of E2F2 by sponging miR-125b, and its downregulation increased apoptosis via E2F2 signaling pathway (Li X. et al., 2018c). Tan et al. identified the oncogenic, f-circEA-4a in plasma of NSCLC patients with EML4-ALK fusion (Tan et al., 2018). Its silencing reduced cell proliferation, metastasis, and invasion (Tan et al., 2018). The same group identified another oncogenic fusion-circRNA, f-circEA-2a produced from EML4-ALK fusion bearing an “AA” motif at the junction site. Its overexpression was reported to promote cell migration and invasion in NSCLC cells (Tan et al., 2018). Lower expression levels of circ-FOXO3 were observed in NSCLC tissues and its overexpression reduced NSCLC development by sponging miR-155 and releasing repression of FOXO3 (Zhang Y. et al., 2018).
Gastric Cancer (GC)
CDR1as modulates PTEN/PI3K/AKT signaling pathway and confers an aggressive oncogenic phenotype to GC cells (Pan et al., 2018). Downregulation of CDR1as induced cell death and restricts GC progression (Li X. et al., 2019). Circ_100269 is downregulated in GC tissues and its overexpression sponges oncogenic miR-630 suppressing GC growth (Zhang Y. et al., 2017). Circ_104916 was downregulated in GC tissues and cell lines, its overexpression suppressed cell proliferation, migration, and EMT (Li J. et al., 2017). CircPDSS1 sponges tumor-suppressing miR-186-5p and upregulate the oncogene NEK2 in GC tissues, and its depletion inhibited cell proliferation (Ouyang et al., 2019). Circ_0023642 is upregulated in GC and regulates the EMT signaling pathway, and its depletion results in tumor inhibition, reduced cell proliferation, and metastasis due to the downregulation of N-cadherin, Vimentin and Snail (Zhou L. H. et al., 2018). Circ-ATAD1 promotes GC progression by modulating the miR-140-3p/YY1/PCIF1 signaling axis (Zhang L. et al., 2020). Consistent with its oncogenic function, the depletion of circATAD1 reduced cell viability and colony formation of GC cells (Zhang L. et al., 2020). Interestingly, circFN1 was highly expressed in cisplatin-resistant GC tissues and promoted cisplatin-resistance by enhancing cell viability and suppressing apoptosis, by sequestering miR-182-5p (Huang X. X. et al., 2020). The knockdown of circFN1 promotes cisplatin-sensitivity and apoptosis in GC cells (Huang X. X. et al., 2020). CircCACTIN promotes GC progression by sponging miR-331-3p and increasing expression of TGFBR1 (Transforming growth factor-β receptor type 1) (Zhang L. et al., 2019). Knockdown of circCACTIN suppressed proliferation, migration, invasion, and EMT of GC cells (Zhang L. et al., 2019). Circ-CEP85L is downregulated in GC tissues, it acts as miR-942-5p sponge leading to the upregulation of NFKBIA (NFKB Inhibitor Alpha) (Lu J. et al., 2020). Consistent with this overexpression of circ-CEP85L inhibited proliferation and invasion of GC cells (Lu J. et al., 2020).
Bladder Cancer (BCa)
CircRNA-MYLK augments proliferation, migration, tube formation of human umbilical vein epithelial cells (HUVEC) and EMT by sponging miR-29a, and stabilizing its target VEGFA in BCa cells (Zhong et al., 2017). The depletion of circRNA-MYLK decreased proliferation, motility, and induced apoptosis in BCa (Zhong et al., 2017). CircACVR2A is downregulated in BCa tissues, it sponges miR-626 to upregulate the expression of the tumor-suppressor EYA4 (Dong et al., 2019). Consistent with this overexpression of circACVR2A suppressed proliferation, migration, and invasion of BCa cells and metastasis (Dong et al., 2019). In contrast to CRC, circHIPK3 is downregulated in BCa and serves as a sponge for miR-558 (Li Y. et al., 2017). It prevents angiogenesis by inhibition of heparanase (HPSE), a positive regulator of VEGF expression (Li Y. et al., 2017). Overexpression of circHIPK3 can be used to reduce aggressiveness and metastasis in BCa cells by targeting the miR-558/heparanase axis (Li Y. et al., 2017). CircITCH was downregulated in BCa samples, overexpression of circITCH upregulates p21 and PTEN expression by sponging oncogenic miRNAs, miR-17/miR-224, leading to inhibition of BCa cell proliferation, migration, and invasion (Yang C. et al., 2018).
Ovarian Cancer
Circ_0061140 is upregulated in ovarian cancer cell lines and regulates the miR-370/FOXM1 pathway by sequestering miR-370 (Chen Q. et al., 2018). Knockdown of circ_0061140 suppressed proliferation and migration in GC cells (Chen Q. et al., 2018). CDR1as expression is upregulated in OC tissues and it correlated with poor prognosis for TNM stages, lymph node metastasis, and reduced OS (Luo Y. H. et al., 2020). CDR1as sponges miR-641 causing up-regulation of ZEB1 and MDM2 expression to promote OC (Luo Y. H. et al., 2020). A large number of circRNAs are misexpressed in primary and metastatic sites of epithelial ovarian carcinoma and their expression exhibits an inverse trend as compared to their linear counterparts in many cancer-related pathways and signaling pathways like NFkB, PI3k/AKT, and TGF-β (Ahmed et al., 2016). Accumulating evidence suggest that circRNA are associated with the initiation and progression of OC (Shabaninejad et al., 2019).
Osteosarcoma (OSC)
CircUBAP2 acts miR-143 sponge and upregulates its target Bcl-2 in OSC (Zhang H. et al., 2017). Depletion of circUBAP2 suppressed proliferation and induced apoptosis in OSC cells (Zhang H. et al., 2017). Circ_0009910 sequesters miR-449a which targets IL6R (interleukin 6 receptor), and its knockdown induced cell-cycle arrest, inhibited proliferation and induced apoptosis is OSC cells (Deng et al., 2018). CircPVT1 was upregulated in the OSC tissues and chemoresistant cell lines, its silencing reversed chemoresistance by decreasing the expression of ABCB1 (ATP Binding Cassette Subfamily B Member 1) (Kun-peng et al., 2018b). Circ_001564 promotes tumorigenicity by sequestering miR-29c-3p, its depletion suppressed the proliferative activity, induced cell-cycle arrest, and promoted apoptosis (Song and Li, 2018). Circ_0002052 was downregulated in OSC tissues and suppresses Wnt/β-catenin signaling pathway by promoting APC2 expression via sponging miR-1205 (Wu Z. et al., 2018). Overexpression of circ_0002052 suppresses migration and invasion in OSC cells (Wu Z. et al., 2018). CircNASP functions by sponging miR-1253 leading to the upregulation of FOXF1 (Huang et al., 2018). Ablation of circNASP by siRNAs inhibits the proliferation, cell-cycle progression, and invasion in OSC cells (Huang et al., 2018).
Head and Neck Squamous Cell Carcinoma (HNSCC)
Esophageal Squamous Cell Carcinoma (ESCC)
Circ_0067934 was upregulated in ESCC tumor tissues and cell lines, also its silencing inhibited proliferation and migration of ESCC cells (Xia et al., 2016). Circ_0000337 was upregulated in ESCC tissues and sequesters miR-670-5p, its depletion inhibits cell proliferation, migration, and invasion (Song et al., 2019).
Oral Squamous Cell Carcinoma (OSCC)
CircUHRF1 functions as a miR-526b-5p sponge and positively regulates c-Myc, which induces TGF-β1 and ESRP1 (Epithelial Splicing Regulatory Protein 1) expression (Zhao W. et al., 2020). The knockdown of circUHRF1 reduces migration, invasion, and EMT of OSCC cells (Zhao W. et al., 2020).
Conclusion and Future Perspectives
CircRNAs which were considered mere splicing artifacts until a few years ago are poised to occupy a center stage in the world of regulatory RNAs. CircRNAs regulate the cellular transcriptome by diverse mechanisms and contribute to a range of cellular functions. They are involved in regulating all the major hallmarks of cancer and can serve as promising biomarkers for cancer diagnosis and prognosis. Unfortunately, so far none of these circRNAs have reached the clinics, and evaluation of a combination of circRNAs as a signature for diagnosis and correlation with clinical features is the likely way forward. CircRNAs also have immense potential for use as therapeutic targets. Novel and effective therapies can be designed by either modulating the endogenous expression circRNAs or by exogenous delivery of artificially engineered circRNAs. At present, the use of circRNAs as therapeutic agents is restricted to the bench and warrants further investigation for clinical use. The circRNA which may be suitable for therapeutic targeting, may also be different for distinct cancer types. However, the aberrant CDR1as expression is common to several cancer types, and targeting it for treating many different cancer types has shown promising results in vitro and in vivo. Development of RNA-based therapeutics for targeting CDR1as for clinical use has the potential to emerge as a single-target therapy for multiple cancers and is worth further investigation.
Author Contributions
PK and VS conceived and designed the manuscript. AR, SB, VS, and PK wrote the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by extramural grants from the Government of India (DST-SERB CRG/2018/000492 to PK) and (DST-SERB ECR/2017/001953 and DBT-RLS 102/IFD/SAN/3499/2016-17 to VS) and intramural funds from the Centre for Human Disease Research (to PK and VS) and OPERA award (to VS) from BITS Pilani.
Glossary
5′UTR, 5′Untranslated region; ABCB1, ATP Binding Cassette Subfamily B Member 1; ADAR1, Adenosine deaminase 1 acting on RNA; ALP, Alkaline phosphatase; AML, Acute myeloid leukemia; APC, Adenomatous polyposis coli; ASOs, Antisense oligonucleotides; AURKA, Aurora kinase A; BAG4, BAG cochaperone 4; BC, Breast cancer; BCa, Bladder Cancer; BCLC, Barcelona clinic liver cancer; CA-724, Carbohydrate antigens-724; CC, Cervical cancer; CCND1, Cyclin D1; CCNE1, Cyclin E1; CDK4, Cyclin dependent kinase 4; CEA, Carcinoembryonic antigen; circRNAs, Circular RNAs; CLL, Chronic lymphocytic leukemia; CML, Chronic lymphocytic leukemia; CNNM3, CBS domain divalent metal cation transport mediator 3; CRC, Colorectal cancer; CSE1L, Chromosome segregation 1 like; CXCL6, Chemokine (C-X-C Motif) ligand 6; DFS, Disease free survival; DHX9, DExH-Box Helicase 9; DLBCL, Diffuse large B-cell lymphoma; dsRBDs, Double stranded RNA-binding domains; E2F5, E2F transcription factor 5; EGFR, Epidermal growth factor receptor; EMT, Epithelial-mesenchymal transition; ER, Estrogen receptor; ESCC, Esophageal squamous cell cancer; ESRP1, Epithelial Splicing Regulatory Protein 1; EYA4, EYA transcriptional coactivator and phosphatase 4; EZH2, Enhancer of zeste homolog 2; FASN, Fatty acid synthase; FBXW7, F-Box and WD repeat domain containing 7; FIGO, Federation of gynecology and obstetrics; FLT3, Fms-like tyrosine kinase 3; FMNL2, Formin like protein 2; FOXO3, Forkhead Box O3; FZD3, Frizzled class receptor 3; GAGE1, G-antigen 1; GBM, Glioblastoma; GC, Gastric cancer; GFRA1, GDNF family receptor alpha 1; HBV, Hepatitis B virus; HCC, Hepatocellular carcinoma; HNRNPL, Heterogeneous nuclear ribonucleoprotein; HNSCC, Head and Neck Squamous cell cancer; HPGD, Hydroxyprostaglandin dehydrogenase; HPSE, Heparanase; HSCC, Hypopharyngeal squamous cell carcinoma; HuR, Hu-antigen R; HUVEC, Human umbilical vein epithelial cells; ICS, Intronic complementary sequences; IGHV, Immunoglobulin heavy-chain variable region; IL6R, interleukin 6 receptor; IRESs, Internal Ribososmal Entry Sites; KIF4A, Kinesin family member 4A; LASP1, LIM and SH3 protein 1,LC, Lung cancer; LFS, Leukemia-free survival; LIF, Leukemia inhibitory factor; LSCC, Laryngeal squamous cell carcinoma; LUAD, Lung adenocarcinoma; m6A, N6-methyladenosine; MACC1, Metastasis associated in colon cancer 1; MG, β -microglobulin; MM, Multiple myeloma; MMP13, Matrix metalloproteins 13; NF1, Neurofibromin 1; NF90, Nuclear factor 90; NF110, Nuclear factor 110; NFKBIA, NFKB Inhibitor Alpha; NPM1, Nucleophosmin; NSCLC, Non small cell lung carcinoma; ORFs, Open reading Frames; OS, Overall survival; OSC, Osteosarcoma; OSCC, Oral squamous cell carcinoma; PCK1, Phosphoenolpyruvate carboxykinase 1; PDK1, 3-phosphoinositide-dependent protein kinase; PFS, Progression-free survival; PIK3CD, Phosphotidylinositol-4,5-Bisphosphate 3-kinase catalytic subunit delta; PLK1, Polo like kinase 1; PRKACB, Protein kinase CAMP-activated catalytic subunit Beta; PTBP1, polyprimidine tract binding protein 1; QKI, Quaking; RBM20, RNA-binding motif protein 20; RBPs, RNA-binding proteins; RFC3, Replication factor C subunit 3; RFS, Recurrance free survival; RT-qPCR, Real-time quantitative polymerase chain reaction; shRNAs, short hairpin RNAs; siRNAs, Small interfering RNAs; SOCS3, Suppressor of cytokine signaling 3; T1F1γ, Transcription intermediatory factor 1-gamma; TET1, Ten-eleven translocation methyclcytosine dioxygenase 1; TERT, Telomerase reverse transcriptase; TGFBR1, Transforming growth factor-β receptor type 1; TIMP3, TIMP Metallopeptidase inhibitor 3; TMZ, temozolomide; TNBC, Triple negative breast cancer; TNKS, Tankyrase; TNM, Tumor; Node and Metastasis; U1snRNP, U1 small nuclear ribonucleoprotein particle; ULK1, Uridine kinase-like protein 1; USP28, Ubiquitin specific peptidase 28; VEGFA, Vascular endothelial growth factor A; YAP, Yes-associated protein 1; ZEB1, Zinc finger E-box binding homeobox 1.
References
- Abdelmohsen K., Panda A. C., Munk R., Grammatikakis I., Dudekula D. B., De S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369. 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., et al. (2015). Rolling circle translation of circular RNA in living human cells. Sci. Rep. 5:16435. 10.1038/srep16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abulizi R., Li B., Zhang C. G. (2019). Circ_0071662, a novel tumor biomarker, suppresses bladder cancer cell proliferation and invasion by sponging miR-146b-3p. Oncol. Res. 41, 1056–1064. 10.3727/096504019X15740729375088 [DOI] [PubMed] [Google Scholar]
- Ahmed I., Karedath T., Andrews S. S., Al I. K., Mohamoud Y. A., Querleu D., et al. (2016). Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 7, 36366–36381. 10.18632/oncotarget.8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhoondi S., Sun D., Von Der Lehr N., Apostolidou S., Klotz K., Maljukova A., et al. (2007). FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67, 9006–9012. 10.1158/0008-5472.CAN-07-1320 [DOI] [PubMed] [Google Scholar]
- Aktaş T., Ilik I. A., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., et al. (2017). DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119. 10.1038/nature21715 [DOI] [PubMed] [Google Scholar]
- Andrews S. J., Rothnagel J. A. (2014). Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204. 10.1038/nrg3520 [DOI] [PubMed] [Google Scholar]
- Ashwal-fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). Article circRNA biogenesis competes with Pre-mRNA splicing. Mol. Cell. 56, 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Bach D. H., Lee S. K., Sood A. K. (2019). Circular RNAs in cancer. Mol. Ther. Nucleic Acids 16, 118–129. 10.1016/j.omtn.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J. H., Zhang Q., Li F., Chan T. M., Lin X., Kim Y., et al. The landscape of microRNA, Piwi-interacting RNA, circular RNA in human saliva. Clin Chem. (2015) 61:221–230. 10.1373/clinchem.2014.230433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H., Lei K., Huang F., Jiang Z., Zhou X. (2019). Exo-circRNAs: a new paradigm for anticancer therapy. Mol. Cancer 18:56. 10.1186/s12943-019-0986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Zhang Y., Han B., Yang L., Chen X., Huang R., et al. (2018). Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood–brain barrier integrity. J. Neurosci. 38, 32–50. 10.1523/JNEUROSCI.1348-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L., Zhi X., Ma L., Zhang J., Chen P. (2018). Biochemical and biophysical research communications Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532 e 3p/FOXO4 axis. Biochem. Biophys. Res. Commun. 505, 346–352. 10.1016/j.bbrc.2018.09.073 [DOI] [PubMed] [Google Scholar]
- Black D. L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336. 10.1146/annurev.biochem.72.121801.161720 [DOI] [PubMed] [Google Scholar]
- Cai J., Chen Z., Zuo X. (2019). circSMARCA5 functions as a diagnostic and prognostic biomarker for gastric cancer. Dis. Markers. 2019:2471652 10.1155/2019/2473652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Miao C., Sang L., Huang Y., Zhang R., Sun L. (2020). Circ _ 0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 243:117255. 10.1016/j.lfs.2020.117255 [DOI] [PubMed] [Google Scholar]
- Cao Q., Shi Y., Wang X., Yang J., Mi Y., Zhai G. (2019). Circular METRN RNA hsa _ circ _ 0037251 promotes glioma progression by sponging miR- 1229-3p and regulating mTOR expression. Sci. Rep. 9:19791. 10.1038/s41598-019-56417-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Ma W., Ke Z., Xie F. (2018). CircRNA hsa _ circ _ 100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17, 2080–2090. 10.1080/15384101.2018.1515553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wang X., Fu S., Wang S., Fu Y., Zhang J. (2020). Circular RNA circ-CDYL sponges miR-1180 to elevate yes- associated protein in multiple myeloma. Exp. Biol. Med. 245, 925–932. 10.1177/1535370220918191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Li X. N., Ye C. X., Chen Z. L., Wang Z. J. (2020). Circular RNA circHUWE1 is upregulated and promotes cell proliferation, migration and invasion in colorectal cancer by sponging mir-486. Oncotargets Ther. 13, 423–434. 10.2147/OTT.S233338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490. 10.1038/s41580-020-0243-y [DOI] [PubMed] [Google Scholar]
- Chen L. L., Yang L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang J., He Y., Wang Y. (2018). hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol. Ther. Nucleic Acids 13, 55–63. 10.1016/j.omtn.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen S., Li T., Zhao Q., Xiao B., Guo J. (2017). Clinica chimica acta using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 466, 167–171. 10.1016/j.cca.2017.01.025 [DOI] [PubMed] [Google Scholar]
- Chen S.-P., Liu B.-X., Xu J., Pei X.-F., Liao Y.-J., Yuan F., et al. (2015). MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer 15:706. 10.1186/s12885-015-1738-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Wang L., Zhang J. (2020). Circular RNA 0016788 displays as a biomarker for tumor progression and poor prognosis in surgical hepatocellular carcinoma patients. J. Clin. Lab. Anal. 34:e23300. 10.1002/jcla.23300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. (2020). Circ_0067934 correlates with poor prognosis and promotes laryngeal squamous cell cancer progression by sponging miR-1324. Eur. Rev. Med. Pharmacol. Sci. 24, 4320–4327. 10.26355/eurrev_202004_21013 [DOI] [PubMed] [Google Scholar]
- Conn S. J., Pillman K. A., Toubia J., Conn V. M., Salmanidis M., Phillips C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Cortés-López M., Miura P. (2016). Focus: epigenetics: emerging functions of circular RNAs. Yale J. Biol. Med. 89, 527–537. [PMC free article] [PubMed] [Google Scholar]
- Deng G., Mou T., He J., Chen D., Lv D., Liu H., et al. (2020). Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J. Exp. Clin. Cancer Res. 39, 1–16. 10.1186/s13046-019-1487-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N., Li L., Gao J., Zhou J., Wang Y., Wang C. (2018). Biochemical and biophysical research communications Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem. Biophys. Res. Commun. 495, 189–196. 10.1016/j.bbrc.2017.11.028 [DOI] [PubMed] [Google Scholar]
- Ding C., Yi X., Wu X., Bu X., Wang D., Wu Z., et al. (2020). Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 479, 1–12. 10.1016/j.canlet.2020.03.002 [DOI] [PubMed] [Google Scholar]
- Dong W., Bi J., Liu H., Yan D., He Q., Zhou Q., et al. (2019). Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol. Cancer 18, 1–16. 10.1186/s12943-019-1025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Li D., Guo X., Li P., Li X., Tong S., et al. (2019). Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif. Cells Nanomed. Biotechnol. 47, 4120–4130. 10.1080/21691401.2019.1671857 [DOI] [PubMed] [Google Scholar]
- Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., et al. (2017). FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 8:14741. 10.1038/ncomms14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zhang L., Wu J., Khadka B., Fang Z., Gu J., et al. (2019). CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J. Exp. Clin. Cancer Res. 38:54. 10.1186/s13046-019-1071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Wang Q. B., Zhi Y., Ren W. H., Li S. M., Zhao C. Y., et al. (2020). Down-regulation of hsa_circ_0092125 is related to the occurrence and development of oral squamous cell carcinoma. Int. J. Oral Maxillof. Surg. 49, 292–297. 10.1016/j.ijom.2019.07.014 [DOI] [PubMed] [Google Scholar]
- Gao M., Xiao H., Hang D., Jiang S., Fu Y., Gong L. (2019). hsa_circ_0007841: a novel potential biomarker and drug resistance for multiple myeloma. Front. Oncol. 9:1261. 10.3389/fonc.2019.01261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang J., Zheng Y., Zhang J., Chen S., Zhao F. (2016). Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 7:12060. 10.1038/ncomms12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Lin X., Zhang Y., Li Q., Guo Y., Fang C., et al. (2020). Exosomal circular RNA sorting mechanisms and their function in promoting or inhibiting cancer. Oncol. Lett. 19, 3369–3380. 10.3892/ol.2020.11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Bao Y., Zhang W., Deng L., Su D., Zheng H. (2020). Circular RNA hsa_circ_0014130 inhibits apoptosis in non–small cell lung cancer by sponging miR-136-5p and upregulating BCL2. Mol. Cancer Res. 18, 748–756. 10.1158/1541-7786.MCR-19-0998 [DOI] [PubMed] [Google Scholar]
- Gu X., Wang G., Shen H. U. I., Fei X. (2018). Hsa_circ_0033155 : a potential novel biomarker for non - small cell lung cancer. Exp. Ther. Med. 10, 3220–3226. 10.3892/etm.2018.6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J., Bezzi M., Jeong J. C., Paffenholz S. V., Berry K., Naldini M. M., et al. (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165, 289–302. 10.1016/j.cell.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Guo J. N., Li J., Zhu C. L., Feng W. T., Shao J. X., Wan L., et al. (2016). Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Oncotargets Ther. 9, 7451–7458. 10.2147/OTT.S123220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Zhang J., Zhang D., Cao S., Li G. (2017). Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget 8, 48169–48177. 10.18632/oncotarget.18327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Li J., Wang H., Su X., Hou J., Gu Y., et al. (2017). Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66, 1151–1164. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- Han W., Wang L., Zhang L., Wang Y., Li Y. (2019). Biochemical and biophysical research communications circular RNA circ-RAD23B promotes cell growth and invasion by miR-593 e 3p/CCND2 and miR-653 e 5p/TIAM1 pathways in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 510, 462–466. 10.1016/j.bbrc.2019.01.131 [DOI] [PubMed] [Google Scholar]
- Hanada K., Akiyama K., Sakurai T., Toyoda T., Shinozaki K., Shiu S. H. (2009). sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics 26, 399–400. 10.1093/bioinformatics/btp688 [DOI] [PubMed] [Google Scholar]
- Hang D., Zhou J., Qin N., Zhou W., Ma H., Jin G., et al. (2018). A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 7, 2783–2791. 10.1002/cam4.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- He F., Zhong X., Lin Z., Lin J., Qiu M., Li X., et al. (2020). Plasma exo-hsa_circRNA_0056616 : a potential biomarker for lymph node metastasis in lung adenocarcinoma. J. Cancer 11, 4037–4046. 10.7150/jca.30360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Huang Z., He M., Liao J., Zhang Q., Wang S., et al. (2020a). Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer 19:17. 10.1186/s12943-019-1120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Lv X., Zeng Z. (2020b). A potential disease monitoring and prognostic biomarker in cervical cancer patients: the clinical application of circular RNA_0018289. J. Clin. Lab. Anal. 34:e23340. 10.1002/jcla.23340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Yan D., Dong W., Bi J., Huang L., Yang M., et al. (2020). circRNA circFUT8 upregulates kru factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol. Ther. Oncolytics 16, 172–187. 10.1016/j.omto.2019.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Liu P., Xie X., Zhou Y., Liao Q., Xiong W., et al. (2017). CircGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 36, 1–12. 10.1186/s13046-017-0614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Ju H., Li N., Jiang Y., Zhao W., Song T., et al. (2020). Association between hsa_circ_0006156 expression and incidence of gastric cancer. Eur. Rav. Med. Pharmacol. Sci. 24, 3030–3036. 10.26355/eurrev_202003_20667 [DOI] [PubMed] [Google Scholar]
- Hirsch S., Blätte T. J., Grasedieck S., Cocciardi S., Rouhi A., Jongen-lavrencic M., et al. (2017). Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologia 102, 2039–2047. 10.3324/haematol.2017.172866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Zhu H., Zhao S., Wang K., Zhang N., Tian Y., et al. (2019). The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 10:950. 10.1038/s41419-019-2183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K., Lin Y., Gupta S. K., Chang N., Yen L., Sun H. S., et al. (2017). Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 77, 2339–2351. 10.1158/0008-5472.CAN-16-1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Han Q., Gu Y., Ma J., Mcgrath M., Qiao F. (2018). Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics 10, 723–732. 10.2217/epi-2017-0142 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhao Y., Shi C., Ren P., Wei B., Guo Y., et al. (2019). A circular RNA from APC inhibits the proliferation of diffuse large B-cell lymphoma by inactivating Wnt/β -catenin signaling via interacting with TET1 and miR-888. Aging 11, 8068–8084. 10.18632/aging.102122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E., Fu J., Yu Q., Xie P., Yang Z., Ji H. (2020). CircRNA hsa circ 0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics 12, 587–603. 10.2217/epi-2019-0404 [DOI] [PubMed] [Google Scholar]
- Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. (2015). cir-ITCH Plays an inhibitory role in colorectal cancer by regulating the Wnt/β -catenin pathway. PLoS ONE 10:e0131225. 10.1371/journal.pone.0131225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Chen M., Pan J., Yu W. (2018). Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem. Biophys. Res. Commun. 500, 511–517. 10.1016/j.bbrc.2018.04.131 [DOI] [PubMed] [Google Scholar]
- Huang M., He Y., Liang L., Huang Q., Zhu Z. (2017). Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J. Gastroenterol. 23, 6330–6338. 10.3748/wjg.v23.i34.6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. X., Zhang Q., Hu H., Jin Y., Zeng A. L., Xia Y. B., et al. (2020). A novel circular RNA circFN1 enhances cisplatin resistance in gastric cancer via sponging miR-182-5p. J. Cell. Biochem. 10.1002/jcb.29641 [DOI] [PubMed] [Google Scholar]
- Huang X. Y., Huang Z. L., Huang J., Xu B., Huang X. Y., Xu Y. H., et al. (2020). Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 39, 1–16. 10.1186/s13046-020-1529-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Huang Z. L., Xu Y. H., Zheng Q., Chen Z., Song W., et al. (2017). Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/MIR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 7:5428. 10.1038/s41598-017-05432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H. T., Orejuela M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K. A. I., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Qiu C., Wang M., Mao N., Wu S., Dai Y. (2018). Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem. Biophys. Res. Commun. 497, 122–126. 10.1016/j.bbrc.2018.02.036 [DOI] [PubMed] [Google Scholar]
- Jin Y. D., Ren Y. R., Gao Y. X., Zhang L., Ding Z. (2019). Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 3311–3319. 10.26355/eurrev_201904_17693 [DOI] [PubMed] [Google Scholar]
- Jing Z., Jian R., Sun R., Li Y. (2020). Circular RNA hsa_circ_0002052 promotes osteosarcoma via modulating miR - 382/STX6 axis. Hum. Cell. 33, 810–818. 10.1007/s13577-020-00335-9 [DOI] [PubMed] [Google Scholar]
- Kelly S., Greenman C., Cook P. R., Papantonis A. (2015). Exon skipping is correlated with exon circularization. J. Mol. Biol. 427, 2414–2417. 10.1016/j.jmb.2015.02.018 [DOI] [PubMed] [Google Scholar]
- Kong S., Yang Q., Tang C., Wang T., Shen X., Ju S. (2019). Identification of hsa_circ_0001821 as a novel diagnostic biomarker in gastric cancer via comprehensive circular RNA profiling. Front. Genet. 10:878. 10.3389/fgene.2019.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Yang L., Wei W., Lyu N., Zou Y., Gao G. (2019). CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics 11, 1163–1176. 10.2217/epi-2019-0093 [DOI] [PubMed] [Google Scholar]
- Kou P., Zhang C., Lin J., Wang H. (2019). Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol. Lett. 17, 2091–2098. 10.3892/ol.2018.9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen L. S., Ebbesen K. K., Hansen T. B. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- Kun-peng Z., Chun-lin Z., Jian-ping H., Lei Z. (2018a). A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int. J. Biol. Sci. 14, 1513–1520. 10.7150/ijbs.27523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kun-peng Z., Xiao-long M., Chun-lin Z. (2018b). Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 14, 321–330. 10.7150/ijbs.24360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E., Parker R., Lasda E., Parker R. O. Y. (2014). Circular RNAs : diversity of form and function. RNA 20, 1829–1842. 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B., Tian Z., Fan W., Ni B. (2019). Circular RNA: a novel biomarker and therapeutic target for human cancers. Int. J. Med. Sci. 16, 292–301. 10.7150/ijms.28047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. (2019a). Circ - U2AF1 promotes human glioma via derepressing neuro - oncological ventral antigen 2 by sponging hsa - miR - 7 - 5p. J. Cell Physiol. 234, 9144–9155. 10.1002/jcp.27591 [DOI] [PubMed] [Google Scholar]
- Li J., Ni S., Zhou C., Ye M. (2018b). The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manage. Res. 10, 2777–2784. 10.2147/CMAR.S172388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J., Chen Z., Chen Y., Jin M. (2018a). Hsa_circ_0079530 promotes cell proliferation and invasion in non-small cell lung cancer. Gene 665, 1–5. 10.1016/j.gene.2018.04.059 [DOI] [PubMed] [Google Scholar]
- Li J., Zhen L., Zhang Y., Zhao L., Liu H., Cai D., et al. (2017). Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco. Targets Ther. 10, 3521–3529. 10.2147/OTT.S136347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. (2018). Hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac. Cancer 9, 924–930. 10.1111/1759-7714.12761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Pei Y., Wang W., Liu F., Zheng K., Zhang X. (2020). Extracellular nanovesicles-transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J. Cell. Mol. Med. 24, 2202–2214. 10.1111/jcmm.14877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhong C., Jiao J., Li P., Cui B., Ji C., et al. (2017). Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int. J. Mol. Sci. 18:597. 10.3390/ijms18030597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. (2019b). Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR - 671 - 5p and CDR1. J. Cell. Physiol. 234, 13807–13819. 10.1002/jcp.28061 [DOI] [PubMed] [Google Scholar]
- Li X., Liu C. X., Xue W., Zhang Y., Jiang S., Yin Q. F., et al. (2017). Coordinated circRNA biogenesis and function with article coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67, 214–227.e7. 10.1016/j.molcel.2017.05.023 [DOI] [PubMed] [Google Scholar]
- Li X., Wang J., Zhang C., Lin C., Zhang J., Zhang W., et al. (2018a). Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 246, 166–179. 10.1002/path.5125 [DOI] [PubMed] [Google Scholar]
- Li X., Yang L., Chen L. (2018b). Review the biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 71, 428–442. 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Z., Jiang H., Li Q., Wang R., Pan H., et al. (2018c). Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell. Physiol. Biochem. 51, 2324–2340. 10.1159/000495876 [DOI] [PubMed] [Google Scholar]
- Li X-N., Wang Z., Ye C., Zhao B., Li Z., Yang Y. (2018). RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J. Exp. Clin. Cancer Res. 37:325. 10.1186/s13046-018-1006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Liu Y. R., Zhou J. H., Li W., Guo H. H., Ma H. P. (2019). Enhanced expression of circular RNA hsa_circ_000984 promotes cells proliferation and metastasis in non-small cell lung cancer by modulating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci., 23, 3366–3374. 10.26355/eurrev_201904_17700 [DOI] [PubMed] [Google Scholar]
- Li Y, Hu J., Li L., Cai S., Zhang H., Zhu X., et al. (2018). Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem. Biophys. Res. Commun. 503, 2089–2094. 10.1016/j.bbrc.2018.07.164 [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., et al. (2017). CircHIPK 3 sponges miR- 558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 18, 1646–1659. 10.15252/embr.201643581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhou Y. E., Yang G., He S., Qiu X., Zhang L., et al. (2019). Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin. Chim. Acta 492, 37–44. 10.1016/j.cca.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Liang D., Wilusz J. E. (2014). Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28, 2233–2247. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Ling Y., Mehrpour M., Saw P. E., Liu Z., Tan W., et al. (2020). Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 19:65. 10.1186/s12943-020-01152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H. F., Zhang X. Z., Liu B. G., Jia G. T., Li W. L. (2017). Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 7, 1566–1576. [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu Y., Bian Z., Zhang J., Zhang R., Chen X., et al. (2018). Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 Kip1 axis. Mol. Cancer 17:151. 10.1186/s12943-018-0902-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wu Y., Wang S., Jiang J., Zhang C., Jiang Y., et al. (2019). Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer 19:937. 10.1186/s12885-019-6088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Hou K., Ji H., Mi S., Yu G., Hu S., et al. (2019). Overexpression of circular RNA circ-CDC45 facilitates glioma cell progression by sponging miR-516b and miR-527 and predicts an adverse prognosis. J. Cell. Biochem. 121, 690–697. 10.1002/jcb.29315 [DOI] [PubMed] [Google Scholar]
- Liu J., Liu T., Wang X., He A. (2017). Circles reshaping the RNA world: from waste to treasure. Mol. Cancer. 16, 1–12. 10.1186/s12943-017-0630-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang X. (2020). Hsa_circRNA_101237 : a novel diagnostic and prognostic biomarker and potential therapeutic target for multiple myeloma. Cancer Manage. Res. 12, 2109–2118. 10.2147/CMAR.S241089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Liu K., Zhang L., Cai J., Yao H., Bai Y., Zhang Z. (2018). Circ_0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur. Rev. Med. Pharmocol. Sci. 22, 8248–8256. 10.26355/eurrev_201812_16519 [DOI] [PubMed] [Google Scholar]
- Liu T., Song Z., Gai Y. (2018). Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem. Biophys. Res. Commun. 503, 1503–1509. 10.1016/j.bbrc.2018.07.070 [DOI] [PubMed] [Google Scholar]
- Liu W., Ma W., Yuan Y., Zhang Y., Sun S. (2018). Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering. Biochem. Biophys. Res. Commun. 500, 864–851. 10.1016/j.bbrc.2018.04.172 [DOI] [PubMed] [Google Scholar]
- Liu X. X., Yang Y. E., Liu X., Zhang M. Y., Li R., Yin Y. H., et al. (2019). A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 17:50. 10.1186/s12967-019-1800-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Fu L., Qian X., Dou L., Cang S. (2020). Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330-5p/LASP1 axis. Arch. Biochem. Biophys. 689:108434. 10.1016/j.abb.2020.108434 [DOI] [PubMed] [Google Scholar]
- Lu J., Wang Y-H., Huang X., Xie J.-W., Wang J.-B., Lin J., et al. (2020). circ-CEP85L suppresses the proliferation and invasion of gastric cancer by regulating NFKBIA expression via miR-942-5p. J. Cell. Physiol. 235, 6287–6299. 10.1002/jcp.29556 [DOI] [PubMed] [Google Scholar]
- Lu J., Zhang P., Xie J., Wang J., Lin J., Chen Q., et al. (2019a). Circular RNA hsa_circ_0006848 related to ribosomal protein L6 acts as a novel biomarker for early gastric cancer. Dis. Markers 2019:3863458. 10.1155/2019/3863458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Zhang P., Xie J., Wang J., Lin J., Chen Q., et al. (2019b). Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 33:e22726. 10.1002/jcla.22726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Wu Y., Zeng B., Sun J., Li Y., Luo J., et al. (2020). CircEHMT1 inhibits metastatic potential of breast cancer cells by modulating miR-1233-3p/KLF4/MMP2 axis. Biochem. Biophys. Res. Commun. 526, 306–313. 10.1016/j.bbrc.2020.03.084 [DOI] [PubMed] [Google Scholar]
- Luo Y. H., Yang Y. P., Chien C. S., Yarmishyn A. A., Ishola A. A., Chien Y., et al. (2020). Plasma level of circular RNA hsa_circ_0000190 correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers 12:1740. 10.3390/cancers12071740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. H., Zhu X. Z., Huang K. W., Zhang Q., Fan Y. X., Yan P. W., et al. (2017). Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed. Pharmacother. 96, 892–898. 10.1016/j.biopha.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Luo Z., Rong Z., Zhang J., Zhu Z., Yu Z., Li T., et al. (2020). Circular RNA circCCDC9 acts as a miR-6792- 3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol. Cancer 19:86. 10.1186/s12943-020-01203-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Han C., Xia W., Li X., Fang P., Yin R., et al. (2020). circ5615 functions as a ceRNA to promote colorectal cancer progression by upregulating TNKS. Cell Death Dis. 11:356. 10.1038/s41419-020-2514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang L., Li Y. (2019). circEIF4G2 modulates the malignant features of cervical cancer via the miR - 218/HOXA1 pathway. Mol. Med. Rep. 19, 3714–3722. 10.3892/mmr.2019.10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Memczak S., Papavasileiou P., Peters O., Rajewsky N. (2015). Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 10:e0141214. 10.1371/journal.pone.0141214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Chen S., Han J. X., Qian B., Wang X. R., Zhong W. L., et al. (2018). Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 78, 4150–4162. 10.1158/0008-5472.CAN-17-3009 [DOI] [PubMed] [Google Scholar]
- Meng Q., Li Y., Kong C., Gao X., Jiang X. (2020). Circ_0000388 exerts oncogenic function in cervical cancer cells by regulating miR-337-3p/ TCF12 axis. Cancer Biother. Radiopharm. 10.1089/cbr.2019.3159 [DOI] [PubMed] [Google Scholar]
- Moore M. J., Proudfoot N. J. (2009). Pre-mRNA processing reaches back totranscription and ahead to translation. Cell 136, 688–700. 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Motegi A., Sood R., Moinova H., Markowitz S. D., Liu P. P., Myung K. (2006). Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175, 703–708. 10.1083/jcb.200606145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeli P., Pourhanifeh M. H., Karimzadeh M. R., Shabaninejad Z., Movahedpour A., Tarrahimofrad H., et al. (2020). Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit. Rev. Oncol. Hematol. 145:102854. 10.1016/j.critrevonc.2019.102854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A. (2019). Twist1 activated circRNA-10720 is a new player in hepatocellular carcinoma metastasis. Transl. Cancer Res. 8, S135–S140. 10.21037/tcr.2018.12.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W., Graveley B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Li Y., Huang Y., Li X., Zhu Y., Long Y., et al. (2019). CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J. Cell. Physiol. 234, 10458–10469. 10.1002/jcp.27714 [DOI] [PubMed] [Google Scholar]
- Pan B., Qin J., Liu X., He B., Wang X., Pan Y., et al. (2019). Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front. Genet. 10:1096. 10.3389/fgene.2019.01096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Li T., Jiang Y., Pan C., Ding Y., Huang Z., et al. (2018). Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell. Biochem. 119, 440–446. 10.1002/jcb.26201 [DOI] [PubMed] [Google Scholar]
- Panda A., Gorospe M. (2018). Detection and analysis of circular RNAs by RT-PCR. Bio. Protoc. 8:e2775. 10.21769/BioProtoc.2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño C., Haenni A. L., Urcuqui-Inchima S. (2015). NF90 isoforms, a new family of cellular proteins involved in viral replication? Biochimie 108, 20–24. 10.1016/j.biochi.2014.10.022 [DOI] [PubMed] [Google Scholar]
- Peng Y., Pu K., Su H., Zhang J., Zheng Y., Ji R. (2020). Circular RNA hsa_circ_0010882 promotes the progression of gastric cancer via regulation of the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 1142–1151. 10.26355/eurrev_202002_20165 [DOI] [PubMed] [Google Scholar]
- Ping L., Jian-jun C., Chu-shu L., Guang-hua L., Ming Z. (2019a). Blood Cells, Molecules and Diseases Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol. Dis. 75, 41–47. 10.1016/j.bcmd.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Ping L., Jian-Jun C., Chu-Shu L., Guang-Hua L., Ming Z. (2019b). High circ_100053 predicts a poor outcome for chronic myeloid leukemia and is involved in imatinib resistance. Oncol. Res. 10.3727/096504018X15412701483326. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Piwecka M., GlaŽar P., Hernandez-Miranda L. R., Memczak S., Wolf S. A., Rybak-Wolf A., et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- Qin M., Liu G., Huo X., Tao X., Sun X., Ge Z., et al. (2016). Hsa-circ-0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers 16, 161–169. 10.3233/CBM-150552 [DOI] [PubMed] [Google Scholar]
- Qin M., Wei G., Sun X. (2018). Circ-UBR5: an exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem. Biophys. Res. Commun. 503, 1027–1034. 10.1016/j.bbrc.2018.06.112 [DOI] [PubMed] [Google Scholar]
- Qiu B. Q., Wu Y. B., Xiong D., Zhang X. M. (2019). CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non - small cell lung cancer by regulating galectin - 1 - AKT/ERK1/2 signaling. J. Cell. Physiol. 234, 11256–11264. 10.1002/jcp.27783 [DOI] [PubMed] [Google Scholar]
- Qiu L., Huang Y., Li Z., Dong X., Chen G., Xu H., et al. (2019). Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol. Oncol. 13, 441–455. 10.1002/1878-0261.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., et al. (2018). The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 78, 2839–2852. 10.1158/0008-5472.CAN-17-2808 [DOI] [PubMed] [Google Scholar]
- Qu D., Yan B., Xin R., Ma T. (2018). A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am. J. Cancer Res. 8, 1387–1402. [PMC free article] [PubMed] [Google Scholar]
- Ren C., Zhang Z., Wang S., Zhu W., Zheng P., Wang W. (2020). Circular RNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer through upregulating ZEB1 via sponging multiple miRNAs. Biol. Chem. 401, 487–496. 10.1515/hsz-2019-0350 [DOI] [PubMed] [Google Scholar]
- Rong D., Lu C., Zhang B., Fu K., Zhao S., Tang W., et al. (2019). CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer 18:25. 10.1186/s12943-019-0958-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7:e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer L., Bär C., Thum T. (2019). Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 27, 1350–1363. 10.1016/j.ymthe.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabaninejad Z., Vafadar A., Movahedpour A., Ghasemi Y., Namdar A., Fathizadeh H., et al. (2019). Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J. Ovarian Res. 12:84. 10.1186/s13048-019-0558-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X., Li G., Liu H., Li T., Liu J., Zhao Q., et al. (2016). Comprehensive circular RNA profiling reveals that hsa-circ-0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine 95, 1–7. 10.1097/MD.0000000000003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Chen L., Lu R., Zhang X., Xiao B., Ye G., et al. (2017). Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 39:1010428317699125. 10.1177/1010428317699125 [DOI] [PubMed] [Google Scholar]
- Shi J. (2017). Novel insights into circular RNAs in clinical application of carcinomas. Onco Targets Ther. 10, 2183–2188. 10.2147/OTT.S134403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Xu D., Shi P., He B., Li Z., Ji Y., et al. (2019). Upregulated circ RNA HSA_CIRC_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma. Cancer Manag. Res. 11, 1997–2006. 10.2147/CMAR.S195546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Liang Y., Sang Y., Li Y., Zhang H., Chen B., et al. (2020). circHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Mol. Ther. Nucleic Acid 20, 518–533. 10.1016/j.omtn.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. Z., Li J. F. (2018). Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem. Biophys. Res. Commun. 495, 2369–2375. 10.1016/j.bbrc.2017.12.050 [DOI] [PubMed] [Google Scholar]
- Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.-H., et al. (2015). Exon circularization requires canonical splice signals. Cell Rep. 10, 103–111. 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Sun H., Tang W., Rong D., Jin H., Fu K., Zhang W., et al. (2017). Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 21, 299–306. 10.3233/CBM-170379 [DOI] [PubMed] [Google Scholar]
- Sun Y. M., Wang W. T., Zeng Z. C., Chen T. Q., Han C., Pan Q., et al. (2019). circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 134, 1533–1546. 10.1182/blood.2019000802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Sun D., Pu W., Gou Q., Guo C., Gong Y., et al. (2018). Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol. Cancer 17:138. 10.1186/s12943-018-0887-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Huang X., Wang J., Yang L., Kong Y., Gao G., et al. (2019). circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer 18:23. 10.1186/s12943-019-0946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Fu K., Sun H., Rong D., Wang H., Cao H. (2018). CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer 17:137. 10.1186/s12943-018-0888-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Xue R., Weng S., Wu J., Fang Y., Wang Y., et al. (2015). BIRC6 promotes hepatocellular carcinogenesis: interaction of BIRC6 with p53 facilitating p53 degradation. Int. J. Cancer 136, E475–E487. 10.1002/ijc.29194 [DOI] [PubMed] [Google Scholar]
- Tang X., Liu S., Ding Y., Guo C., Guo J., Hua K., et al. (2020). Serum circular FoxO3a serves as a novel prognostic biomarker in squamous cervical cancer. Cancer Manag. Res. 12, 2531–2540. 10.2147/CMAR.S243329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. Y., Zhao P., Zou T. N., Duan J. J., Zhi R., Yang S. Y., et al. (2017). Circular RNA hsa-circ-0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 36, 901–908. 10.1089/dna.2017.3862 [DOI] [PubMed] [Google Scholar]
- Tao X., Shao Y., Lu R., Ye Q., Xiao B., Ye G., et al. (2020). Pathology - research and practice clinical signi fi cance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol. Res. Pract. 216:152763. 10.1016/j.prp.2019.152763 [DOI] [PubMed] [Google Scholar]
- Tay F. C., Lim J. K., Zhu H., Hin L. C., Wang S. (2015). Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug Deliv. Rev. 81, 117–127. 10.1016/j.addr.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Tian J., Xi X., Wang J., Yu J., Huang Q., Ma R., et al. (2019). CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag. Res. 11, 5413–5423. 10.2147/CMAR.S199436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu F., Guo X., Wu H., He Z., Wang F., Sun A., Dai X. (2020). Circ-0001313/miRNA-510-5p/AKT2 axis promotes the development and progression of colon cancer. Am J. Transl. Res. 12, 281–291. [PMC free article] [PubMed] [Google Scholar]
- Vo J. N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176, 869–881.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang J., Cao X., Xu L., Chen L. (2018). Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed. Pharmacother. 98, 775–782. 10.1016/j.biopha.2018.01.015 [DOI] [PubMed] [Google Scholar]
- Wang G., Liu W., Zou Y., Wang G., Deng Y., Luo J., et al. (2019). Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a–MET pathway. EBioMedicine 40, 432–445. 10.1016/j.ebiom.2018.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li H. (2018). CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur. Rev. Med. Pharmacol. Sci. 22, 3053–3060. 10.26355/eurrev_201805_15063 [DOI] [PubMed] [Google Scholar]
- Wang J., Li X., Lu L., He L., Hu H., Xu Z. (2018). Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J. Clin. Lab. Anal. 32, 1–7. 10.1002/jcla.22379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Luo J., Liu G., Li X. (2020). Circular RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation and migration via the miR-382-5p/PTEN axis. Biochem. Biophys. Res. Commun. 527, 503–510. 10.1016/j.bbrc.2020.03.165 [DOI] [PubMed] [Google Scholar]
- Wang J. X., Liu Y., Jia X. J., Liu S. X., Dong J. H., Ren X. M., et al. (2019). Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA–miR-486-3p-FLNA axis. Cancer Cell Int. 19:196. 10.1186/s12935-019-0924-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Gan T. Y., Li N., Liu C. Y., Zhou L. Y., Gao J. N., et al. (2017). Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 24, 1111–1120. 10.1038/cdd.2017.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Long H., Zheng Q., Bo X., Xiao X., Li B. (2019). Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 18, 1–12. 10.1186/s12943-019-1046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tong X., Zhou Z., Wang S., Lei Z., Zhang T., et al. (2018). Circular RNA hsa-circ-0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer 17, 1–18. 10.1186/s12943-018-0889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu H., Liu Z., Yang L., Zhou J., Cao X., et al. (2020). Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet. 240, 33–39. 10.1016/j.cancergen.2019.11.001 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang S., Chen X., Li N., Li J., Jia R., et al. (2018a). CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 78, 4812–4825. 10.1158/0008-5472.CAN-18-0532 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang S., Chen X., Li N., Li J., Jia R., et al. (2018b). EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer 17, 1–12. 10.1186/s12943-018-0911-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang S. (2020). Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. Onco. Targets Ther. 13, 1941–1951. 10.2147/OTT.S240642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., He H., Wang F. (2019). Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene 720:144099. 10.1016/j.gene.2019.144099 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu J., Huang B. O., Xu Y. M., Li J., Huang L. F., et al. (2015). Mechanism of alternative splicing and its regulation. Biomed. Rep. 3, 152–158. 10.3892/br.2014.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang L., Wang W., Guo X. (2020). Overexpression of circular RNA hsa_circ_0001038 promotes cervical cancer cell progression by acting as a ceRNA for miR-337-3p to regulate cyclin-M3 and metastasis-associated in colon cancer 1 expression. Gene 733:144273. 10.1016/j.gene.2019.144273 [DOI] [PubMed] [Google Scholar]
- Wang Y.-M., Huang L., Li D., Shao J., Xiong S., Wang C., et al. (2017). Hsa_circ_0101996 combined with hsa_circ_0101119 in peripheral whole blood can serve as the potential biomarkers for human cervical squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 10, 11924–11931. [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wei P., Wei D., Cao S., Liu H., Chen L., et al. (2020). Effect of up-regulation of circMATR3 on the proliferation, metastasis, progression and survival of hypopharyngeal Carcinoma. J. Cell. Mol. Med. 24, 4687–4697. 10.1111/jcmm.15134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Wei W., Xu H., Wang Z., Gao W., Wang T., et al. (2019). Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 27, 139–145. 10.3233/CBM-182029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., et al. (2017). Circular RNA ciRS-7 — A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 23, 3918–3929. 10.1158/1078-0432.CCR-16-2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. M., Wen X., Han X. R., Wang S., Wang Y. J., Shen M., et al. (2018). Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol. Cell. Biol. 38, e00259–18. 10.1128/MCB.00259-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Liu D., Yang Y. (2019). Enhanced expression of circular RNA circ-DCAF6 predicts adverse prognosis and promotes cell progression via sponging miR-1231 and miR-1256 in gastric cancer. Exp. Mol. Pathol. 110:104273. 10.1016/j.yexmp.2019.104273 [DOI] [PubMed] [Google Scholar]
- Wu Z., Shi W., Jiang C. (2018). Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem. Biophys. Res. Commun. 502, 465–471. 10.1016/j.bbrc.2018.05.184 [DOI] [PubMed] [Google Scholar]
- Wu Z., Sun H., Liu W., Zhu H., Fu J., Yang C., et al. (2019). Circ-RPL15 : a plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia 34, 919–923. 10.1038/s41375-019-0594-6 [DOI] [PubMed] [Google Scholar]
- Xia L., Wu L., Bao J., Li Q., Chen X., Xia H., Xia R. (2018). Biochemical and biophysical research communications circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/b -catenin pathway. Biochem. Biophys. Res. Commun. 503, 385–390. 10.1016/j.bbrc.2018.06.045 [DOI] [PubMed] [Google Scholar]
- Xia P., Wang S., Ye B., Du Y., Li C., Xiong Z., et al. (2018). A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701.e7. 10.1016/j.immuni.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Xia W., Qiu M., Chen R., Wang S., Leng X., Wang J., et al. (2016). Circular RNA has-circ-0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci. Rep. 6:35576. 10.1038/srep35576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Tong H., Yuan X., Xiong C., Xu X., Zeng Y. (2020). CircFADS2 : a potential prognostic biomarker of colorectal cancer. Exp. Biol. Med. 245, 1233–1241. 10.1177/1535370220929965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao-Long M., Kun-Peng Z., Chun-Lin Z. (2018). Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J. Cancer 9:1856. 10.7150/jca.24619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaotong S., Jutong S., Hua H., Zhan Y., Haichao L. (2019). Hsa_circ_0070269 inhibits hepatocellular carcinoma progression through modulating miR-182/NPTX1 axis. Biomed. Pharmacother. 120:109497 10.1016/j.biopha.2019.109497 [DOI] [PubMed] [Google Scholar]
- Xie H., Ren X., Xin S., Lan X., Lu G., Lin Y., et al. (2016). Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 7, 26680–26691. 10.18632/oncotarget.8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Yu T., Jing X., Ma L., Fan Y., Yang F., et al. (2020). Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 19, 1–22. 10.1186/s12943-020-01208-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Li J., Li P., Li N., Zhang Y., Binang H., et al. (2020). RNA-Seq profiling of serum exosomal circular RNAs reveals circ-PNN as a potential biomarker for human colorectal cancer. Front. Oncol. 10:982. 10.3389/fonc.2020.00982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhang Y., Qi L., Ding L., Jiang H., Yu H. (2018). NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front. Mol. Neurosci. 11:225. 10.3389/fnmol.2018.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Z., Shao C. C., Wang X. J., Zhao X., Chen J. Q., Ouyang Y. X., et al. (2019). circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 10:175. 10.1038/s41419-019-1382-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhang M., Zheng X., Yi P., Lan C., Xu M. (2017). The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 143, 17–27. 10.1007/s00432-016-2256-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Yuan W., Yang X., Li P., Wang J., Han J., et al. (2018). Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21 PTEN expression. Mol. Cancer 17:19. 10.1186/s12943-018-0771-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Li X., Meng Q., Sun H., Wu S., Hu W., et al. (2020). CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol. Cancer 19, 1–15. 10.1186/s12943-020-1139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Yu Y., Yu X., Zhou J., Zhang Z., Ying S., et al. (2019). Downregulated expression of hsa_circ_0005556 in gastric cancer and its clinical significance. Dis. Markers 2019:2624586. 10.1155/2019/2624586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Qiu Z., Jiang Y., Dong L., Yang W., Gu C. (2019). Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget 7, 63449–63455. 10.18632/oncotarget.11523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110, 304–315. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Xie L., Han L., Qu X., Yang Y., Zhang Y., et al. (2017). Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics 7:3106. 10.7150/thno.19016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J. T., Zhao S. H., Liu Q. P., Lv M. Q., Zhou D. X., Liao Z.-J., et al. (2017). Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol. Res. Pract. 213, 453–456. 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Yao Y., Hua Q., Zhou Y., Shen H. (2019). CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β -catenin signaling pathway. Biomed. Pharmacother. 111, 1367–1375. 10.1016/j.biopha,.2018.12.120 [DOI] [PubMed] [Google Scholar]
- Ye D., Wang S., Huang Y., Chi P. (2019). PRIMARY RESEARCH A 3 - circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int. 19, 1–11. 10.1186/s12935-019-0995-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Gao G., Zou Y., Zheng S., Zhang L. (2019). circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol. Ther. Nucleic Acids 18, 88–98. 10.1016/j.omtn.2019.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y. Y., Lin J. (2018). Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J. Cell Physiol. 234, 3711–3719. 10.1002/jcp.27145 [DOI] [PubMed] [Google Scholar]
- Yin W. B., Yan M. G., Fang X., Guo J. J., Xiong W., Zhang R. P. (2018). Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin. Chim. Acta 487, 363–368. 10.1016/j.cca.2017.10.011 [DOI] [PubMed] [Google Scholar]
- You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. (2016). The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS ONE 11:e015847. 10.1371/journal.pone.0158347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yu X., Ding H., Yang L., Yu Y., Zhou J., Yan Z., et al. (2020). Reduced expression of circRNA hsa_circ_0067582 in human gastric cancer and its potential diagnostic values. J. Clin. Lab. Anal. 34:e23080 10.1002/jcla.23080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Lei L., Dong S., Liu D. (2020). Circular RNA hsa_circ_0068033 Acts as a diagnostic biomarker and suppresses the progression of breast cancer through sponging. Onco Targets Ther. 13, 1921–1929. 10.2147/OTT.S223542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Liu W., Zhang Y., Zhang Y., Sun S. (2018). CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem. Biophys. Res. Commun. 503, 870–875. 10.1016/j.bbrc.2018.06.089 [DOI] [PubMed] [Google Scholar]
- Zang Y., Li J., Wan B. (2020). circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646. J. Cell Mol. Med. 24, 2423–2433. 10.1111/jcmm.14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos P. G. (1996). Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. U.S.A. 93, 6536–6541. 10.1073/pnas.93.13.6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., et al. (2018). CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 9:417. 10.1038/s41419-018-0454-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zeng Y., Du W. W., Wu Y., Yang Z., Awan F. M., Li X., et al. (2017). A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7, 3842–3855. 10.7150/thno.19764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang C., Lin J., Wang H. (2018). Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cell. Physiol. Biochem. 51, 290–300. 10.1159/000495230 [DOI] [PubMed] [Google Scholar]
- Zhang G., Sun W., Zhu L., Feng Y., Wu L., Li T. (2019). Overexpressed circ_0029426 in glioblastoma forecasts unfavorable prognosis and promotes cell progression by sponging miR-197. J. Cell. Biochem. 120, 10295–10302. 10.1002/jcb.28313 [DOI] [PubMed] [Google Scholar]
- Zhang H., Deng T., Ge S., Liu Y., Bai M., Zhu K., et al. (2019). Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 38, 2844–2859. 10.1038/s41388-018-0619-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang H., Wang G., Ding C., Liu P., Wang R., Ding W., et al. (2017). Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 8, 61687–61697. 10.18632/oncotarget.18671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chang X., Zhai T., Yu J., Wang W., Du A., et al. (2020). A novel circular RNA, circ-ATAD1, contributes to gastric cancer cell progression by targeting miR-140-3p/YY1/PCIF1 signaling axis. Biochem. Biophys. Res. Commun. 525, 841–849. 10.1016/j.bbrc.2020.02.100 [DOI] [PubMed] [Google Scholar]
- Zhang L., Song X., Chen X., Wang Q., Zheng X., Wu C., Jiang J. (2019). Circular RNA CircCACTIN promotes gastric cancer progression by sponging MiR-331-3p and regulating TGFBR1 expression. Int. J. Biol. Sci. 15, 1091–1103. 10.7150/ijbs.31533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., et al. (2019). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814. 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., et al. (2018). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9:4475. 10.1038/s41467-018-06862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. F., Wei C. Y., Huang X. Y., Peng R., Yang X., Lu J. C., et al. (2019). Circular RNA circTRIM33–12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer 18, 1–15. 10.1186/s12943-019-1031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Xu J., Zhao J., Wang X. (2018). Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur. Rev. Med. Pharmacol. Sci. 22, 118–126. 10.26355/eurrev_201801_14108 [DOI] [PubMed] [Google Scholar]
- Zhang W., Yang S., Liu Y., Wang Y., Lin T., Li Y., et al. (2018). Hsa_circ_0007534 as a blood-based marker for the diagnosis of colorectal cancer and its prognostic value. Int. J. Clin. Exp. Pathol. 11, 1399–1406. [PMC free article] [PubMed] [Google Scholar]
- Zhang X. L., Xu L. L., Wang F. (2017). Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 41, 1056–1064. 10.1002/cbin.10826 [DOI] [PubMed] [Google Scholar]
- Zhang X. O., Dong R., Zhang Y., Zhang J. L., Luo Z., Zhang J., et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26, 1277–1287. 10.1101/gr.202895.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. O., Wang H. B., Zhang Y., Lu X., Chen L. L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu H., Li W., Yu J., Li J., Shen Z., et al. (2017). CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR - 630. Aging 9, 1585–1593. 10.18632/aging.101254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xue W., Li X., Zhang Y., Xue W., Li X., et al. (2016). The biogenesis of nascent circular RNAs article the biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624. 10.1016/j.celrep.2016.03.058 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao H. U. I., Zhang L. (2018). Identification of the tumor - suppressive function of circular RNA FOXO3 in non - small cell lung cancer through sponging miR - 155. Mol. Med. Rep. 3, 7692–7700. 10.3892/mmr.2018.8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yang T., Xiao J. (2018). EBioMedicine circular RNAs: promising biomarkers for human diseases. EBioMedicine 34, 267–274. 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Liu H., Liu H., Zhang X., Zhang M., Kolluri V. K. (2020). Downregulated expression of hsa_circ_0037515 and hsa_circ_0037516 as novel biomarkers for non-small cell lung cancer. Am. J. Transl. Res. 12, 162–170. [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Han Y., Liu Z., Zhao Z., Li Z., Jia K. (2018). circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci. Rep. 38:BSR20180570. 10.1042/BSR20180570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Dong G., Meng Q., Lin S., Li X. (2020). Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J. Cell. Biochem. 121, 4440–4449. 10.1002/jcb.29672 [DOI] [PubMed] [Google Scholar]
- Zhao S. Y., Wang J., Ouyang S. B., Huang Z. K., Liao L. (2018). Salivary circular RNAs Hsa-Circ-0001874 and Hsa-Circ-0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell. Physiol. Biochem. 47, 2511–2521. 10.1159/000491624 [DOI] [PubMed] [Google Scholar]
- Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., et al. (2020). Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 27, 919–933. 10.1038/s41418-019-0423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Chen H. (2020). Diagnostic signi fi cance of downregulated circMORC3 as a molecular biomarker of hypopharyngeal squamous cell carcinoma : a pilot study. Cancer Manag. Res. 12, 43–49. 10.2147/CMAR.S235888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Liu X., Xue Y., Gong W., Ma J., Xi Z., et al. (2017). TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J. Hematol. Oncol. 10, 1–19. 10.1186/s13045-017-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Huang M., Xing L., Yang R., Wang X., Jiang R., et al. (2020). The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol. Cancer 19:73. 10.1186/s12943-020-01183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Wang Y., Cheng Y., Wang W., Lu B., Zhu L., et al. (2018). Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem. Biophys. Res. Commun. 499, 1044–1049. 10.1016/j.bbrc.2018.03.221 [DOI] [PubMed] [Google Scholar]
- Zhong Z., Huang M., Lv M., He Y., Duan C. (2017). Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 403, 305–317. 10.1016/j.canlet.2017.06.027 [DOI] [PubMed] [Google Scholar]
- Zhou C., Molinie B., Daneshvar K., Pondick J. V., Wang J., Van Wittenberghe N., et al. (2017). Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20, 2262–2276. 10.1016/j.celrep.2017.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhang W. W., Peng F., Sun J. Y., He Z. Y., Wu S. G. (2018). Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag. Res. 10, 535–544. 10.2147/CMAR.S155923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Fan F. (2020). Genome-wide microarray analysis of circrnas revealed novel biomarkers for glioma treatment and their promoting effect on glioma progression. Onco Targets Ther. 13, 2739–2745. 10.2147/OTT.S241347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. H., Yang Y. C., Zhang R. Y., Wang P., Pang M. H., Liang L. Q. (2018). CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur. Rev. Med. Pharmacol. Sci. 22, 2297–2303. 10.26355/eurrev_201804_14818 [DOI] [PubMed] [Google Scholar]
- Zhou S., Wei J., Wang Y., Liu X. (2020). Cisplatin resistance-associated circRNA_101237 serves as a prognostic biomarker in hepatocellular carcinoma. Exp. Therapeut. Med. 19, 2733–2740. 10.3892/etm.2020.8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liu H., Wang W., Zhao H., Wang T. (2018). Hsa_circ_0102533 serves as a blood-based biomarker for non-small-cell lung cancer diagnosis and regulates apoptosis in vitro. Int. J. Clin. Exp. Pathol. 11, 4395–4404. [PMC free article] [PubMed] [Google Scholar]
- Zhu C. L., Sha X., Wang Y., Li J., Zhang M. Y., Guo Z. Y., et al. (2019). Circular RNA hsa-circ-0007142 is upregulated and targets miR-103a-2-5p in colorectal cancer. J. Oncol. 2019:9836819. 10.1155/2019/9836819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Xu Y., Chen Y., Yan F. (2017). Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed. Pharmacother. 88, 138–144. 10.1016/j.biopha.2016.12.097 [DOI] [PubMed] [Google Scholar]
- Zhu Q., Lu G., Luo Z., Gui F., Wu J., Zhang D., et al. (2018). CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem. Biophys. Res. Commun. 497, 626–632. 10.1016/j.bbrc.2018.02.119 [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Wei S., Chen Y., Chen Y., Fan X. (2017). Hsa_circ_0013958 : a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 284, 2170–2182. 10.1111/febs.14132 [DOI] [PubMed] [Google Scholar]
- Zhuo F., Lin H., Chen Z., Huang Z., Hu J. (2017). The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Oncotargets Ther. 10, 5187–5193. 10.2147/OTT.S147378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong L., Sun Q., Zhang H., Chen Z., Deng Y., Li D., et al. (2018b). Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed. Pharmacother. 102, 639–644. [DOI] [PubMed] [Google Scholar]
- Zong L., Sun Q., Zhang H., Chen Z., Deng Y., Li D., Zhang L. (2018a). Increased expression of circRNA_102231 in lung cancer and its clinical signifi cance. Biomed. Pharmacother. 102, 639–644. 10.1016/j.biopha.2018.03.084 [DOI] [PubMed] [Google Scholar]