Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the deadliest cancer types with a poor prognosis due to the lack of symptoms in the early stages and a delayed diagnosis. The present study aimed to identify the risk factors significantly associated with prognosis and to search for novel effective diagnostic modalities for patients with early-stage ESCC. mRNA and methylation data of patients with ESCC and the corresponding clinical information were downloaded from The Cancer Genome Atlas (TCGA) database, and the representation features were screened using deep learning autoencoder. The univariate Cox regression model was used to select the prognosis-related features from the representation features. K-means clustering was used to cluster the TCGA samples. Support vector machine classifier was constructed based on the top 75 features mostly associated with the risk subgroups obtained from K-means clustering. Two ArrayExpress datasets were used to verify the reliability of the obtained risk subgroups. The differentially expressed genes and methylation genes (DEGs and DMGs) between the risk subgroups were analyzed, and pathway enrichment analysis was performed. A total of 500 representation features were produced. Using K-means clustering, the TCGA samples were clustered into two risk subgroups with significantly different overall survival rates. Joint multimodal representation strategy, which showed a good model fitness (C-index=0.760), outperformed early-fusion autoencoder strategy. The joint representation learning-based classification model had good robustness. A total of 1,107 DEGs and 199 DMGs were screened out between the two risk subgroups. The DEGs were involved in 70 pathways, the majority of which were correlated with metastasis and proliferation of various cancer types, including cytokine-cytokine receptor interaction, cell adhesion molecules PPAR signaling pathway, pathways in cancer, transcriptional misregulation in cancer and ECM-receptor interaction pathways. The two survival subgroups obtained via the joint representation learning-based model had good robustness, and had prognostic significance for patients with ESCC.
Keywords: esophageal squamous cell carcinoma, differentially expressed genes, autoencoder strategy, joint representation learning, multi-omics classification prediction
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the deadliest cancer types and accounts for ~90% of all incidental esophageal cancers in China (1,2). Although the causes of ESCC have been increasingly clarified and various treatment strategies have been applied in recent decades, there remains a lack of effective ESCC therapies (1). Usually, ESCC has a poor prognosis as most patients lack symptoms at an early stage and are diagnosed too late to achieve curative treatment (3). In China, the survival rate of patients with ESCC with a late diagnosis is <10%, but if it is diagnosed at an early stage, the survival rate can be as high as 85% (4). Therefore, the understanding of the pathogenesis of ESCC, the identification of the risk factors that are significantly associated with prognosis, and the search for novel effective diagnostic modalities for early-stage ESCC are urgently required (5).
It is widely accepted that abnormal molecular alterations, including aberrant gene expression and promoter methylation, are usually associated with the pathogenesis of ESCC, a multifactorial disease. It has been reported that genes involved in cell cycle and apoptosis regulation are mutated in 99% of ESCC cases, and mutations in the epigenetic modulators are associated with prognosis, with potential therapeutic implications (6). Furthermore, epigenetic alterations, particularly DNA methylation, serve a crucial role in cancer, and aberrant promoter island methylation of tumor suppressor genes has been established as a common epigenetic mechanism underlying tumorigenesis (7,8). Considering that the overall survival (OS) time of cancer patients is usually associated with a variety of risk factors, it is increasingly recognized that incorporating multiple variables into cancer prediction models would be more accurate than an estimation based on a single predictor (9). Recently, several risk assessment models have been developed to identify the high-risk patients and have proved to be useful in estimating the likelihood of patients with a specific set of risk factors suffering from diseases of interest (10,11). However, the majority of the existing analytical tools only analyze individual groups independently. It is a difficult and urgent issue to integrate data from multiple groups with different technical protocols to acquire more useful prognosis-related information (12).
Deep learning (DL), a new category of machine learning methods, has been successfully applied in solving numerous structural analysis problems (13,14). A DL computation framework has been used to successfully predict survival in several cancer types, based on large-scale omics data, including liver cancer, pan-cancer, and kidney renal clear cell carcinoma (15–17). However, no autoencoder-based DL models have been applied to multi-omics data of various cancer types. Autoencoder is an unsupervised feed-forward neural network which may be built using different strategies, including early-fusion autoencoder and joint multimodal representation (15).
The Cancer Genome Atlas (TCGA) database is an application platform for large-sample genome sequencing analysis of 33 types of cancer. In the present study, large quantities of mRNA and methylation data of patients with ESCC were downloaded from TCGA, along with their clinical information. The representation features were screened using an autoencoder framework. Univariate Cox proportional hazards (PH) analysis was used to screen out the features that were significantly associated with prognosis. According to these features, K-means grouping was used to cluster the prognosis-related features, and its robustness was evaluated. Furthermore, two ArrayExpress datasets was were used to verify the reliability of the clinical prognosis-related features. The present study aimed to develop a reliable stratification method for identifying the ESCC patients with high risk of mortality and to provide potential clinical diagnostic biomarkers for patients with ESCC.
Materials and methods
Data collection and preprocessing
Methylation beta-value of 202 esophageal cancer (ESCA) cases generated on Illumina Human Methylation 450 platform and FPKM values of 196 cases generated from RNA-Seq Illumina HiSeq 2000 platform were downloaded from TCGA database (18) (https://gdc-portal.nci.nih.gov/), and their clinical data were also downloaded. A total of 194 samples possessing RNA-seq and methylation array data were obtained, including 99 ESCC samples and 95 esophageal adenocarcinoma samples. Ninety-six ESCC samples had overall survival (OS) information and were used as the training set (TCGA set).
Raw data were preprocessed using the following steps: i) the probes that were missing in >50% of samples were filtered out; ii) the methylation data were initially annotated using Illumina Human Methylation 450 kanno.ilmn12. hg19 package (19) of R software, and the mean beta values of multiple methylation sites in the promoter region were subsequently calculated and used as promoter methylation values. The promoter region referred to all CpG islands within 1,500 bp ahead of the transcription start site (TSS); iii) the samples that were missing across >20% of the promoter features were filtered out; iv) the missing values of the two-omics data were filled with impute package of R software (version 3.5.2) (20); and v) the gene features with zero values across all samples were filtered out.
Two validation datasets
E-GEOD-53624 (https://www.ebi.ac.uk/arrayexpress/experiments/E-GEOD-53624/;A- GEOD- 18109 platform) and E-GEOD-53625 (https://www.ebi.ac.uk/arrayexpress/experiments/E-GEOD-53625/;A-GEOD- 18109 platform) datasets consisting of RNA expression profile data of ESCC were downloaded from ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/), which included 119 and 179 cases, respectively. The probes and samples were filtered and preprocessed according to the steps stated above steps. The average eigenvalue of all probes corresponding to a single RNA was selected as the eigenvalue of the RNA. The two datasets were used as validation datasets in the present analysis. As no methylation dataset with clinical prognostic information could be found, only the two RNA datasets were used for verification.
Clinical characteristics of the TCGA set and of the two validation datasets are presented in Table I.
Table I.
Clinical characteristics of three datasets.
| Clinical index | TCGA cohort (n=96) | E-GEOD-53624 (n=119) | E-GEOD-53624 (n=179) |
|---|---|---|---|
| Age, mean ± SD | 58.29±10.24 | 59.03±8.93 | 59.35±9.03 |
| Sex, female/male | 14/82 | 21/98 | 33/146 |
| OS, years, mean ± SD | 1.25±0.96 | 3.09±2.02 | 3.02±1.91 |
| OS status, alive/dead | 63/33 | 46/73 | 73/106 |
| DFS, mean years ± SD) | 1.11±1.02 | – | – |
| DFS status, 0/1/- | 59/32/5 | – | – |
| Stage, I/II/III/IV/- | 7/55/27/4/3 | 8/44/67/0/0 | 10/77/92/0/0 |
| Pathological T, T1/T2/T3/T4/- | 8/31/50/4/3 | 8/20/62/29 | 12/27/110/30 |
| Pathologic N, N0/N1/N2/N3- | 54/29/6/3/4 | 54/42/13/10/0 | 83/62/22/12/0 |
| Pathological M, M0/M1/- | 83/4/9 | – | – |
| Eastern cancer oncology group, 0/1/2/3/- | 3/28/5/3/57 | – | – |
TCGA, The Cancer Genome Atlas; OS, overall survival; SD, standard deviation; DFS, disease-free survival; T, tumor; N; node; M, metastasis.
Autoencoder construction using two strategies
Autoencoder is an unsupervised feedforward neural network (15). In this technique, according to the reconstruction error between input layer X (input layers) and output layer X ‘(output layers), the depth neural network is trained to reproduce input layer X. For example, if you use the reconstructed square difference, the train model could be used to optimize the following loss function: L(X, X′=||X–X′||2. A key feature of Autoencoder is to learn a useful feature representation of data, usually compressed data. The input layer X is firstly transformed to the middle layer Z, and the mapping from layer X to Z is completed by the encoder part of the network. For example, if the encoder contains only one neural network layer, Z=α(WeX + be), where, α is a nonlinear activation function, and We and be are linear weights and deviations, the second part of the Autoencoder network is the decoder, from layer Z to reconstructed output layer X′: X′=α(WdZ + bd).
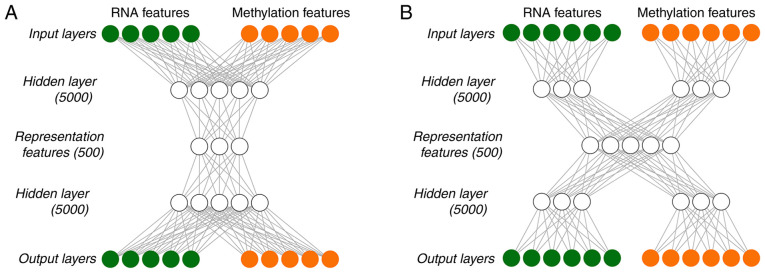
In the present study, autoencoder was built using two different strategies. One was early-fusion autoencoder (Fig. 1A), which was the traditional autoencoder used as the benchmark model, and the other one was a joint multimodal representation strategy (Fig. 1B). Early-fusion involves fusing data from multiple sources into a single feature vector, which is then used as an input for the DL algorithm (Fig. 1A). The fused data was original or preprocessed data from sensors. If data fusion is performed without feature extraction, it will be very challenging. For example, the sampling rates of different sensors may vary. If a data source generates discrete data and another data source provides a continuous data stream, simultaneous interpretation of data from multiple sources may be unavailable. In order to alleviate certain problems associated with fusion of original data, higher-level characterization features were extracted from each mode, which may be common learning representations in DL, and were then fused at a specified level.
Figure 1.
Analysis flow chart. (A) Early-fusion antoencoder strategy, (B) joint multimodal representation strategy.
As DL essentially involves learning the hierarchical representation from the original data, it produces an intermediate-level fusion. A common autoencoder of intermediate-level fusion is joint multimodal representation strategy (Fig. 1B). A neural network transforms the original input into a higher-level representation. Each layer usually alternates linear and non-linear operations, scales, shifts and tilts the input, resulting in a new representation of the original data. In a multimodal environment, when all modes are transformed into representations, different representations may be fused into a separate hidden layer, and consequently a joint multimodal representation strategy may be learned. The majority of the work of deep multimodal fusion adopts the intermediate fusion method.
Tanh was used as the activation function of each layer in the in-DL process. To train Autoencoder, a gradient descent algorithm with 128 epochs and a 20% dropout was used. The Hidden layernodes and Hidden layer presentation nodes were set to be 5,000 and 500, respectively. The TCGA data were integrated and recoded by implementing Autoencoder with Python lasagne library, and 500 representation features were eventually obtained.
Selection of transformed features and K-means clustering
For the representation features obtained using joint multimodal representation or early-fusion autoencoder, the coefficient of variation of each feature was analyzed using survival package of R language (https://cran.r-project.org/web/packages/survival/index.html), from which the significant transformed features with a coefficient of variation of <0.1 were screened out. Next, the remaining features underwent a univariate Cox-PH analysis, using survival package of R language. Subsequently, according to the representation features with P<0.05, K-means clustering was applied to the 96 samples of the TCGA set using R nbclust package (https://cran.r-project.org/web/packages/NbClust/index.html) (21), which can set several evaluation parameters and automatically generate the optimal clustering number. In the present study, Silhouette index and Calski-Harabasz criterion were calculated to generate the optimal clustering number, and the classification label of each sample was consequently obtained. Kaplan-Meier survival curves of different survival groups were drawn, and concordance index (C-index) and log-rank P-values were calculated. When the optimal clustering number was generated, the survival risk subgroups of samples were also obtained. In the subsequent parts, the ‘risk subgroups’ were used instead of the ‘optimal clustering number’.
Data grouping and robustness evaluation
In order to evaluate the robustness of the obtained risk subgroups, a cross-validation (CV)-like procedure was used to partition the TCGA set as follows: TCGA data was split into training set/test set (60/40%) to have enough test samples to generate the evaluation indicators. Specifically, the 96 ESCC samples were randomly divided into 5-folds, and then 2-folds were used as the test set and the remaining 3-folds were used as the training set. This way, a total of 10 new combinations were obtained. For each of the 10 new combinations, the 60% samples (training set) were used to construct a model and predict the risk subgroups in the test set. Finally, C-index and Brier scores were used to evaluate the robustness of the grouping model. Data grouping was implemented using caret package of R language.
C-index refers to the proportion of the samples whose predicted survival times are ordered correctly. A C-index score of ~0.70 suggests that the model performs well, while a score of ~0.50 suggests random background. In the present study, C-index was calculated using survcomp package of R language (https://www.bioconductor.org/packages/release/bioc/html/survcomp.html) (22).
Brier score is another scoring function to measure the accuracy of probability prediction. It ranges between 0 and 1, with a greater score indicating greater inaccuracy. In survival analysis, Brier score measures the average difference between the observed survival and the estimated survival over a given period of time. Brier score was calculated using sbrier.score2proba function of survcomp package.
Supervised classification
After determining the risk subgroups using K-means clustering, RNAs and methylation features of the TCGA set were analyzed using analysis of variance (ANOVA). Based on the ANOVA F value, the top 75 RNAs and 75 methylation features that were most relevant to the risk subgroups were selected, respectively, to construct support vector machine (SVM) classifiers for predicting the validation set. Additionally, C-index and Brier score values were calculated.
The SVM classifiers were constructed using penalize SVM package of R (https://cran.r-project.org/web/packages/penalizedSVM/index.html) (23). The package used 5-fold CV to perform a grid search for the optimal hyperparameters for the SVM model and constructed SVM models.
Verification of the risk subgroups in two independent datasets
In order to verify the robustness of the two risk subgroups obtained from K-means clustering in predicting the survival risk, two independent RNA-seq validation sets (E-GEOD-53624 and E-GEOD-53625) were used. Firstly, the probes were converted into the corresponding gene names according to the platform probe annotation. When there were several probes for the same RNA, the average eigenvalues of these probes was selected as the eigenvalue of the RNA. Secondly, the common genes between the TCGA set and the validation sets were selected, respectively. Thirdly, the common genes were scaled twice via Median scale normalization and Robust scale normalization, as follows:
Median absolute deviation (mad) is a robust evaluation method for the variability of a univariate sample. For a set of eigenvectors,
Regarding robust scale normalization, the values between 1/4 and 3/4 quantiles in the training set were used to calculate the mean and the standard deviation of each gene feature. This normalization method eliminated the effect of the outliers and was helpful to calculate robust means and standard deviations.
The two normalizations were completed using R software. Following the normalizations, the top 75 RNA features in the training set were selected based on ANOVA F values and were used to construct the SVM models. The risk subgroups in the validation sets were predicted using the SVM model. Kaplan-Meier survival curves of different risk subgroups were drawn, and C-index and log-rank P-values were calculated.
Bioinformatics analysis
Differential expression analysis was performed between the two risk subgroups of the TCGA set. For RNA-seq data of the TCGA set, differentially expressed genes (DEGs) between the two risk subgroups were analyzed using DESeq2 package of R (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) (24), and |logFC|>0.585 and an FDR <0.05 were selected as the threshold values for identifying significant DEGs. For methylation data of the TCGA set, differential methylation genes (DMGs) between the two risk subgroups were identified using moderate t-test test and limma package of R (https://bioconductor.org/packages/release/bioc/html/limma.html) (25), with |beta difference|>0.1 and an FDR<0.05 as the threshold for identifying significant DMGs.
In order to obtain the functional pathways involving the significant DEGs, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using KEGG orthology-based annotation system (KOBAS) (26). An FDR <0.05 was selected as the threshold for the significantly enriched pathways.
Statistical analysis
Firstly, the risk subgroups and clinical characteristics of patients from the TCGA set were subjected to univariate Cox regression to evaluate their associations with survival. The significant variables with a log-rank P<0.05 were then included in multivariate Cox regression analysis to identify independent prognostic factors.
Results
Risk subgroups of ESCC
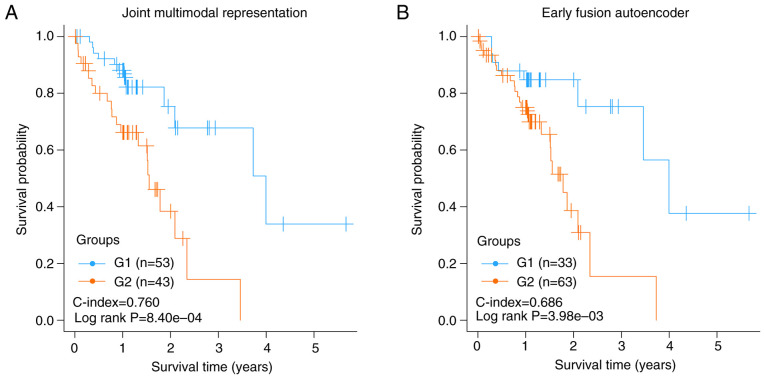
A total of 96 ESCC cases, including coupled RNA-seq and DNA methylation data, were obtained from the TCGA database. Following data preprocessing, 16,772 genes from RNA-seq and 20,112 genes from DNA methylation data were acquired. Early-fusion autoencoder and joint multimodal representation framework were constructed using sklearn library for deep learning of the two-omics data, and 500 representation features were generated using each of the two strategies. After filtering out the features with a CV <0.1, the remaining features underwent univariate Cox-PH analysis using OS data. The resulting representation features significantly associated with OS (P<0.005) were used for K-means clustering of all ESSC samples of the TCGA set. Two risk subgroups (G1 and G2) were obtained using K-means clustering, and the detailed grouping information of the TCGA set is shown in Table SI. There were significant differences in the OS time between G1 and G2 subgroups. The C-index and log-rank P-value using joint multimodal representation strategy were 0.760 and 8.40×10−4, respectively (Fig. 2A), while the two metrics using early-fusion autoencoder strategy were 0.686 and 3.98×10−3, respectively (Fig. 2B). The results showed that joint multimodal representation strategy was superior to early-fusion autoencoder strategy.
Figure 2.
Kapan-Meier diagrams of the risk subgroups obtained by using different strategies. (A) Kaplan-Meier graphs of the risk subgroups obtained using joint multimodal representation strategy, (B) Kaplan-Meier graphs of the risk subgroups obtained using early fusion autoencoder strategy.
The two risk subgroups were independent prognostic factors
In order to evaluate whether the two risk subgroups obtained using the joint multimodal representation-based classification model may be used as independent prognostic factors, a univariate Cox regression model was used to analyze these risk subgroups, as well as the clinical factors of patients. The risk subgroup G2 was significantly associated with prognosis with a hazard ratio (HR) of 3.465, a 95% CI of 1.618–7.421, and a significant P-value of 1.38×10−3 (Table II). Pathologic_N, stage, sex, additional pharmaceutical therapy and additional radiation therapy were significantly associated with prognosis, with P-values of 6.03×10−3, 1.07×10−3, 4.53×10−3, 4.25×10−2 and 3.16×10−2, respectively (Table II). Furthermore, in multivariate Cox regression analysis, only the risk subgroup G2 was found to be an independent predictor of prognosis (HR=2.469, 95% CI=1.061–5.747, P-value=3.60×10−2, Table II), indicating that the risk subgroup G2 was a significant prognostic factor independent of pathologic_N, stage, sex, additional pharmaceutical and radiation therapy.
Table II.
Univariate and multivariate cox regression analysis of clinical factors in two risk subgroups.
| Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinical features | HR | 95% CI | Z | P-value | HR | 95% CI | Z | P-value |
| Group | 8.40×10−4 | |||||||
| G1 | 1.000 | – | – | – | 1.000 | – | – | – |
| G2 | 3.465 | 1.618–7.421 | 3.198 | 1.38×10−3 | 2.469 | 1.061–5.747 | 2.097 | 0.036 |
| Pathological N | 6.03×10−3 | |||||||
| N0 | 0.217 | 0.091–0.519 | −3.434 | 5.95×10−4 | 0.682 | 0.200–2.326 | −0.611 | 0.541 |
| N1 | 0.339 | 0.135–0.851 | −2.304 | 2.12×10−2 | 0.549 | 0.183–1.652 | −1.066 | 0.286 |
| Stage | 1.07×10−3 | |||||||
| I+II | 0.067 | 0.018–0.248 | −4.052 | 5.08×10−5 | 0.171 | 0.028–1.034 | −1.923 | 0.055 |
| III++IV | 0.160 | 0.043–0.593 | −2.745 | 6.00×10−3 | 0.221 | 0.042–1.163 | −1.782 | 0.075 |
| Sex | 4.53×10−3 | |||||||
| Female | 1.000 | – | – | – | 1.000 | – | – | – |
| Male | 5.365 | 1.246–23.094 | 2.256 | 2.41×10−2 | 4.704 | 0.991–22.322 | 1.949 | 0.051 |
| Additional pharmaceutical therapy | 4.25×10−2 | |||||||
| No | 1.017 | 0.348–2.977 | 0.031 | 0.975 | 0.471 | 0-Inf | 0.000 | 1.000 |
| Yes | 0.000 | 0.000-Inf | −0.004 | 0.997 | 0.000 | 0-Inf | −0.001 | 0.999 |
| Additional radiation therapy | 3.16×10−2 | |||||||
| No | 0.979 | 0.34–2.822 | −0.038 | 0.969 | 2.805 | 0-Inf | 0.000 | 1.000 |
| Yes | 0.000 | 0.000-Inf | −0.004 | 0.997 | 0.000 | 0-Inf | −0.001 | 1.000 |
HR, hazards ratio; CI, confidence interval; N, node.
Robustness evaluation of the two risk subgroups
The robustness of the two risk subgroups obtained using the joint multimodal representation-based model was evaluated using an SVM classification model, which was built with the two risk subgroups as labels using a CV procedure. The 96 TCGA samples were split into 10 combinations of training and test sets (60/40%). The geometric means of the C-indexes and Brier scores were 0.77±0.04 and 0.13±0.03 for the training set, and 0.75±0.06 and 0.14±0.04 for the test set, respectively (Table III).
Table III.
C-index and Brier score of the SVM classifier for robustness evaluation of the risk subgroups using CV procedure.
| Dataset | 10-fold cv | C-index | Brier score |
|---|---|---|---|
| Training | JMR (60%) | 0.77±0.04 | 0.13±0.03 |
| Methylation only | 0.72±0.10 | 0.14±0.03 | |
| RNA only | 0.74±0.05 | 0.13±0.03 | |
| Test | JMR (40%) | 0.75±0.06 | 0.14±0.04 |
| Methylation only | 0.65±0.17 | 0.16±0.05 | |
| RNA only | 0.73±0.11 | 0.14±0.04 |
JMR, joint multimodal representation.
Regarding single-omics data, the performance of the two-omics model was also good. According to the ANOVA F values, the top 75 RNAs (Table SII) and the top 75 methylation genes (Table SIII) were selected to construct the SVM classification model, respectively. Using methylation data only, the training set generated a C-index of 0.72±0.10 and a Brier score of 0.14±0.03, while the test set achieved a C-index of 0.65±0.17 and a Brier score of 0.16±0.05 (Table III). Using RNA-seq data only, the C-index and Brier score of the training set were 0.74±0.05 and 0.13±0.03, while the two metrics of the test set were 0.73±0.11 and 0.14±0.04, respectively (Table III). These results showed that our classification model had good robustness to predict the survival subgroups.
Validation of the two risk subgroups in two independent RNA-seq datasets
The joint multimodal representation-based classification model on two independent datasets of RNA-seq data (E-GEOD-53624 and E-GEOD-53625), which had 119 and 179 samples, respectively, was validated. There were 5,776 and 5,776 common RNAs between the two independent validation datasets with the TCGA set, respectively. According to the results of risk subgroup classification and ANOVA F values, the top 75 genes from the common RNAs were screened to construct an SVM model and to predict the two independent datasets.
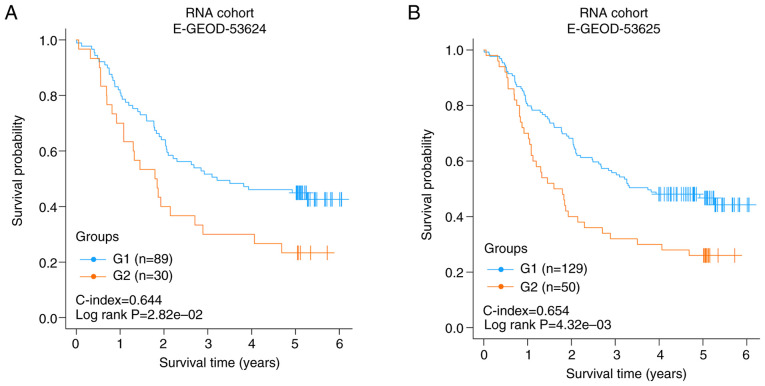
Each validation set was divided into two risk subgroups using the SVM model. In E-GEOD-53624 or E-GEOD-53625, the two risk groups had a significantly different OS time (C-index=0.644, log rank P-value=2.82×10−2, Fig. 3A; C-index=0.654, log rank P-value=4.32×10−3, Fig. 3B). The results of the two independent validation sets suggested that the two risk subgroups obtained using the joint multimodal representation-based model had good stability.
Figure 3.
Verification of the classification model in the two independent validation sets. (A) Kaplan-Meier graphs of the risk subgroups obtained in E-GEOD-53624, (B) Kaplan-Meier graphs of the risk subgroups obtained in E-GEOD-53625.
Failed validation of the two risk subgroups of esophageal cancer samples
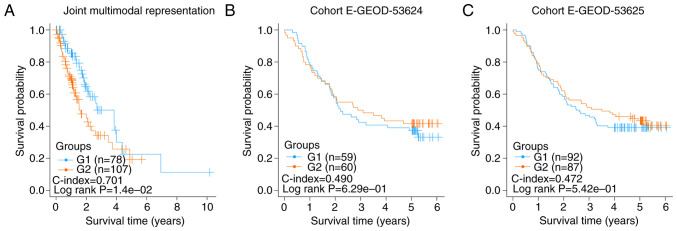
A total of 185 esophageal cancer samples with OS information were downloaded from TCGA database. From the RNA-seq and DNA methylation data of these samples, 500 representation features were produced using the autoencoder based on joint multimodal representation strategy. Consequently, two risk subgroups (G1a and G2) were obtained using the same procedure as described earlier. The C-index and log-rank P-value were 0.701 and 1.40×10−2, respectively (Fig. 4A).
Figure 4.
Risk subgroups of all esophageal cancer samples using the model based on joint multimodal representation strategy. (A) Kaplan-Meier graphs of the risk subgroups of all esophageal cancer samples obtained using joint multimodal representation strategy, (B) Kaplan-Meier graphs of the risk subgroups obtained in E-GEOD-53624, (C) Kaplan-Meier graphs of the risk subgroups obtained in E-GEOD-53625.
Similarly, according to the results of the risk subgroups classification and ANOVA F values, the top 75 genes were screened to construct SVM models and predict in E-GEOD-53624 and E-GEOD-53625 datasets using the aforementioned procedure. The C-index and log-rank P-value of E-GEOD-53624 were 0.490 and 6.29×10−1, respectively (Fig. 4B), and those of E-GEOD-53625 were 0.472 and 5.42×10−1, respectively (Fig. 4C). These insignificant results showed that the two risk subgroups of the esophageal cancer samples could not be successfully verified in E-GEOD-53624 and E-GEOD-53625, indicating that the risk factors of esophageal adenocarcinoma and ESCC may be different.
Bioinformatics analysis of the risk subgroups in TCGA ESCC samples
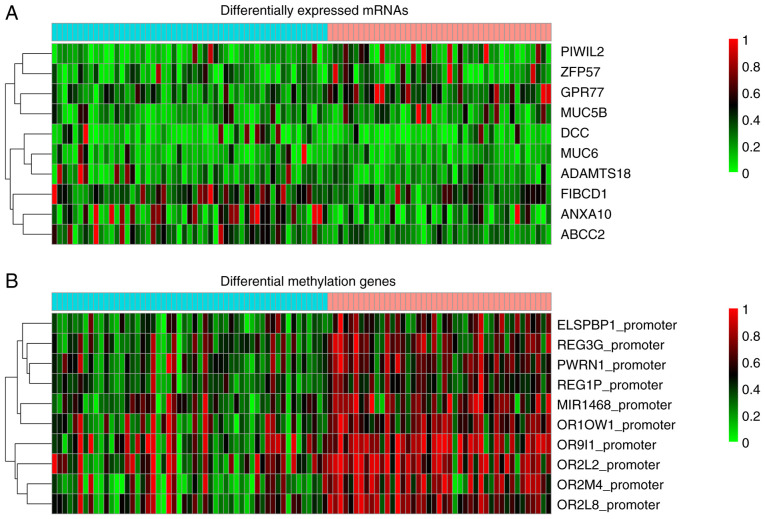
Differential gene expression between the risk subgroup G1 and G2 of the TCGA set was analyzed using DESeq2 package of R software, and the genes with |logFC|>0.585 and an FDR<0.05 were considered significant. A total of 1,107 DEGs were screened out, consisting of 773 upregulated and 334 downregulated genes in the risk subgroup G2. Their detailed information is presented in Table SIV. The top 10 genes, including PIWIL2, ZFP57, GPR77, MUC5B, DCC, MUC6, ADAMTS18, FIBCD1, ANXA10 and ABCC2, are presented in Fig. 5A. The significant DMGs between the two subgroups were analyzed using limma package and moderate t-test test. An FDR <0.05 and |delta methylation|>0.1 were selected as the threshold values of significance. A total of 199 DMGs, including 195 upregulated and 4 downregulated DMGs were screened out and their detailed information are presented in Table SV. The top 10 DMGs were comprised of ELSPBP1_promoter, REG3G_promoter, PWRN1_promoter, REG1P_promoter, MIR1468_promoter, OR10W1_promoter, OR9I1_promoter, OR2L2_promoter, OR2M4_promoter and OR2L8_promoter and are presented in Fig. 5B.
Figure 5.
Heatmap of (A) top 10 differentially expressed mRNAs and (B) top 10 differential methylation genes between risk subgroup G1 and G2.
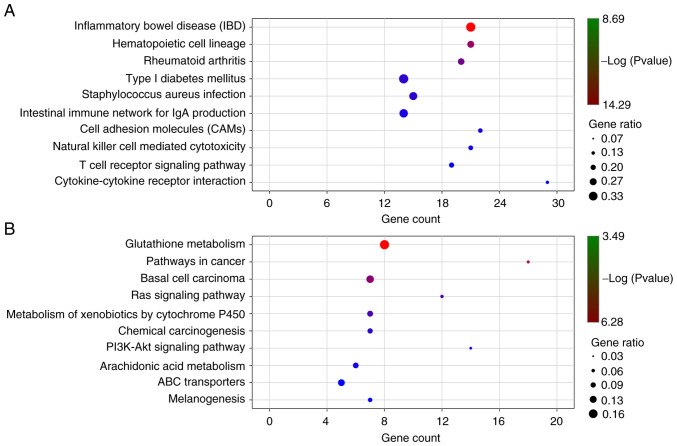
KEGG enrichment pathways of the significant DEGs were analyzed with a P-value <0.05 using KOBAS. There were 46 significant pathways for the upregulated DEGs (Fig. 6A and Table SVI) and 24 significant pathways for the downregulated DEGs (Fig. 6B and Table SVII), including cytokine-cytokine receptor interaction, cell adhesion molecules (CAMs) cAMP signaling pathway, PPAR signaling pathway, pathways in cancer, basal cell carcinoma, Ras signaling pathway, PI3K-Akt signaling pathway, transcriptional misregulation in cancer, mTOR signaling pathway and ECM-receptor interaction.
Figure 6.
KEGG pathway enrichment analysis of significant upregulated and downregulated genes. (A) top 10 pathways of significant upregulated genes, and (B) top 10 pathways of significant downregulated genes.
Discussion
It is important to identify the risk factors significantly associated with prognosis and to search for novel effective diagnostic modalities for early-stage ESCC. In recent years, cancer has been widely accepted as a genetic disease, and aberrant expression of various mRNAs and aberrant promoter island methylation of tumor suppressor genes have been considered as the common epigenetic mechanisms underlying the pathogenesis of ESCC (6,27,28). Due to the rapid development of bioinformatics, multi-omics data, including whole genome gene expression and methylation, are increasingly used in cancer research (17). In the present study, a large quantity of RNA-seq and methylation data of patients with ESCC and their clinical information was integrated to screen gene features for the following construction of an Autoencoder framework.
Autoencoder is a complex three-layer neural network, which may reconstruct multi-omics data to generate new features, and has achieved good performance in various fields (29). So far, an autoencoder-based model has been reported to be efficient and accurate in predicting the prognosis of multiple cancer types, using RNA-seq data (30). In the present study, two different strategies were used to construct an autoencoder, and the joint multimodal representation strategy was better than the early fusion autoencoder strategy. This was consistent with a previous study, in which it was believed that the joint multimodal representation strategy may alleviate the problems associated with the fusion of original data (31). The present study identified two risk subgroups of ESCC with significantly different survival by using the joint multimodal representation-based classification model. Wang et al determined and validated two molecular subtypes of ESCC by using consensus clustering, which have different functional implications, yet are not significantly different regarding OS time (32). Through comprehensive but analysis based on iCluster, three molecular subtypes of ESCC displaying a geographical trend were obtained, but the survival difference was not studied (33). Liu et al reported the identification of 3 subtypes possessing different clinical features, genomic complexity, p53 mutational status, and RNA expression; however they did not focus on survival comparison of the different subtypes (34). In contrast to these studies on the molecular subtypes of ESCC, the results of the present study may aid in improving the prognosis of patients with ESCC. Furthermore, risk subgroup G2 may be an independent prognostic factor for patients with ESCC. However, the obtained risk subgroups based on data of all esophageal cancer samples could not be verified in two independent validation sets, indicating that the risk factors of ESCC may be different from those of esophageal adenocarcinoma. As the accuracy of the autoencoder-based DL model in risk stratification of patients is significantly superior to that of similarity network (16), in the present study, an SVM model was constructed and it was proved that the present risk subgroup classification had good robustness and stability. The present autoencoder-based DL model may be helpful for cancer detection using gene expression data, and these highly interactive genes may be useful cancer biomarkers for the detection of ESCC, which requires further study.
In the present study, the differential expression between risk subgroups G1 and G2 of the TCGA set was analyzed, and 1,107 DEGs and 199 DMGs were screened out in the risk subgroup G2. Furthermore, KEGG enrichment pathways of the significant DEGs were analyzed. A total of 46 enrichment pathways, including cytokine-cytokine receptor interaction, cell adhesion molecules (CAMs), cAMP signaling pathway, and PPAR signaling pathway were identified for the upregulated DEGs, and 24 enrichment pathways, including pathways in cancer, basal cell carcinoma, Ras signaling pathway, PI3K-Akt signaling pathway, transcriptional misregulation in cancer, mTOR signaling pathway and ECM-receptor interaction were identified for the downregulated DEGs. The majority of these KEGG pathways are correlated with metastasis and proliferation of various cancer types, and may be important predictors (35–38). The results of the present study provided candidate genes for further functional research.
It should be noted that the present study is an extensive bioinformatics study based on published data. These results require further validation using in vitro or in vivo models. A TCGA study suggests that race is an important clinical factor for subtyping in esophageal cancer (33). Therefore, it is necessary to verify the obtained two risk subgroups of ESCC in different races in future studies.
In the present study, a joint multimodal representation strategy-based classification model that is able to robustly discriminate two subgroups of patients with ESCC with significantly different OS time was developed. Several cancer metastasis- and proliferation-related pathways were identified. This study provides more insights into the underlying molecular mechanisms of ESCC progression. Further studies are demanded to validate the feasibility of this prognostic model.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the Project of Jiang-su Province Health and Family (grant no. H2017035) and the Social Development Project of Jiangsu Provincial Science and Technology Department (grant no. BE2017759).
Availability of data and materials
All data used and/or analyzed in this study are available from the TCGA database (https://gdc-portal.nci.nih.gov/) or the EBI Array database (https://www.ebi.ac.uk/arrayexpress/).
Authors' contributions
JH and QZ designed the study. JY, XW, ML, YZ, XZ, JL and MZ performed all the statistical analyses. JY and XW finished the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lin DC, Wang MR, Koeffler HP. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology. 2018;154:374–389. doi: 10.1053/j.gastro.2017.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J, He YT, Zheng RS, Zhang SW, Chen WQ. Analysis of esophageal cancer time trends in China, 1989–2008. Asian Pac J Cancer Prev. 2012;13:4613–4617. doi: 10.7314/APJCP.2012.13.9.4613. [DOI] [PubMed] [Google Scholar]
- 3.Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14:33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GQ, Jiao GG, Chang FB, Fang WH, Song JX, Lu N, Lin DM, Xie YQ, Yang L. Long-term results of operation for 420 patients with early squamous cell esophageal carcinoma discovered by screening. Ann Thorac Surg. 2004;77:1740–1744. doi: 10.1016/j.athoracsur.2003.10.098. [DOI] [PubMed] [Google Scholar]
- 5.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154:360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 7.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 8.Yamada N, Yasui K, Dohi O, Gen Y, Tomie A, Kitaichi T, Iwai N, Mitsuyoshi H, Sumida Y, Moriguchi M, et al. Genome-wide DNA methylation analysis in hepatocellular carcinoma. Oncol Rep. 2016;35:2228–2236. doi: 10.3892/or.2016.4619. [DOI] [PubMed] [Google Scholar]
- 9.Riley RD, Hayden JA, Steyerberg EW, Moons KG, Abrams K, Kyzas PA, Malats N, Briggs A, Schroter S, Altman DG, et al. Prognosis research strategy (PROGRESS) 2: Prognostic factor research. PLoS Med. 2013;10:e1001380. doi: 10.1371/journal.pmed.1001380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iida M, Ikeda F, Hata J, Hirakawa Y, Ohara T, Mukai N, Yoshida D, Yonemoto K, Esaki M, Kitazono T, et al. Development and validation of a risk assessment tool for gastric cancer in a general Japanese population. Gastric Cancer. 2018;21:383–390. doi: 10.1007/s10120-017-0768-8. [DOI] [PubMed] [Google Scholar]
- 11.Pavlou M, Ambler G, Seaman SR, Guttmann O, Elliott P, King M, Omar RZ. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015;351:h3868. doi: 10.1136/bmj.h3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bengio Y. Learning Deep Architectures for AI. Foundations and Trends® in Machine Learning, 2009. https://doi.org/10.1561/2200000006
- 14.Papazafeiropoulos G, Vu QV, Truong VH, Luong MC, Pham VT. Prediction of buckling coefficient of stiffened plate girders using deep learning algorithm. In: Ha-Minh C, Dao D, Benboudjema F, Derrible S, Huynh D, Tang A, editors. CIGOS 2019, Innovation for Sustainable Infrastructure Springer. Springer; Singapore: Lecture Notes in Civil Engineering. Vol 54. [Google Scholar]
- 15.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24:1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirion OB, Chaudhary K, Huang S, Garmire LX. Multi-omics-based pan-cancer prognosis prediction using an ensemble of deep-learning and machine-learning models. medRxiv. doi: 10.1186/s13073-021-00930-x. doi: https://doi.org/10.1101/19010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie G, Dong C, Kong Y, Zhong JF, Li M, Wang K. Group lasso regularized deep learning for cancer prognosis from multi-omics and clinical features. Genes (Basel) 2019;10:240. doi: 10.3390/genes10030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu T, Chen D, Wang Y, Sun X, Li S, Miao S, Wo Y, Dong Y, Leng X, Du W, Jiao W. Identification of DNA methylation-driven genes in esophageal squamous cell carcinoma: A study based on the cancer genome atlas. Cancer Cell Int. 2019;19:52. doi: 10.1186/s12935-019-0770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen KD. IlluminaHumanMethylation450kanno. ilmn12. hg19: Annotation for illumina's 450k methylation arrays. R package version 0.2 12016. doi: 10.18129/B9.bioc.IlluminaHumanMethylation450kanno.ilmn12.hg19. [Google Scholar]
- 20.Hastie T, Tibshirani R, Narasimhan B, Chu G. Impute: Impute: Imputation for microarray data. R package version. 2019 [Google Scholar]
- 21.Malika C, Ghazzali N, Boiteau V, Niknafs A. NbClust: An R package for determining the relevant number of clusters in a data set. J Stat Software. 2014;61:1–36. [Google Scholar]
- 22.Schröder MS, Culhane AC, Quackenbush J, Haibe-Kains B. Survcomp: An R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker N, Werft W, Toedt G, Lichter P, Benner A. PenalizedSVM: A R-package for feature selection SVM classification. Bioinformatics. 2009;25:1711–1712. doi: 10.1093/bioinformatics/btp286. [DOI] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brembilla-Perrot B, de la Chaise Terrier A, Skeik L, Cherrier F, Pernot C. Factors predicting the response to an antiarrhythmic during an electrophysiologic study for ventricular tachycardia. Arch Mal Coeur Vaiss. 1987;80:1497–1503. (In French) [PubMed] [Google Scholar]
- 27.Chen J, Kwong DL, Cao T, Hu Q, Zhang L, Ming X, Chen J, Fu L, Guan X. Esophageal squamous cell carcinoma (ESCC): Advance in genomics and molecular genetics. Dis Esophagus. 2015;28:84–89. doi: 10.1111/dote.12088. [DOI] [PubMed] [Google Scholar]
- 28.Song G, Xu J, He L, Sun X, Xiong R, Luo Y, Hu X, Zhang R, Yue Q, Liu K, Feng G. Systematic profiling identifies PDLIM2 as a novel prognostic predictor for oesophageal squamous cell carcinoma (ESCC) J Cell Mol Med. 2019;23:5751–5761. doi: 10.1111/jcmm.14491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Yao H, Zhao S. Auto-encoder based dimensionality reduction. Neurocomputing. 2016;184:232–242. doi: 10.1016/j.neucom.2015.08.104. [DOI] [Google Scholar]
- 30.Xiao Y, Wu J, Lin Z, Zhao X. A semi-supervised deep learning method based on stacked sparse auto-encoder for cancer prediction using RNA-seq data. Comput Methods Programs Biomed. 2018;166:99–105. doi: 10.1016/j.cmpb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Qiao Y, Luo X, Li C, Tian H, Ma J. Heterogeneous graph-based joint representation learning for users and POIs in location-based social network. Information Processing Management. 2020;57:102151. doi: 10.1016/j.ipm.2019.102151. [DOI] [Google Scholar]
- 32.Wang F, Yan Z, Lv J, Xin J, Dang Y, Sun X, An Y, Qi Y, Jiang Q, Zhu W, et al. Gene expression profiling reveals distinct molecular subtypes of esophageal squamous cell carcinoma in Asian populations. Neoplasia. 2019;21:571–581. doi: 10.1016/j.neo.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women's Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University, et al, corp-author. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–174. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Snell JM, Jeck WR, Hoadley KA, Wilkerson MD, Parker JS, Patel N, Mlombe YB, Mulima G, Liomba NG, et al. Subtyping sub-saharan esophageal squamous cell carcinoma by comprehensive molecular analysis. JCI Insight. 2016;1:e88755. doi: 10.1172/jci.insight.88755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albelda S. Role of integrins and other cell adhesion molecules in tumor progression and metastasis. Lab Invest. 1993;68:4–17. [PubMed] [Google Scholar]
- 36.Chen YZ, Xue JY, Chen CM, Yang BL, Xu QH, Wu F, Liu F, Ye X, Meng X, Liu GY, et al. PPAR signaling pathway may be an important predictor of breast cancer response to neoadjuvant chemotherapy. Cancer Chemother Pharmacol. 2012;70:637–644. doi: 10.1007/s00280-012-1949-0. [DOI] [PubMed] [Google Scholar]
- 37.Dong C, Wang X, Xu H, Zhan X, Ren H, Liu Z, Liu G, Liu L. Identification of a cytokine-cytokine receptor interaction gene signature for predicting clinical outcomes in patients with colorectal cancer. Int J Clin Exp Med. 2017;10:9009–9018. [Google Scholar]
- 38.Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol Cancer. 2006;5:55. doi: 10.1186/1476-4598-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used and/or analyzed in this study are available from the TCGA database (https://gdc-portal.nci.nih.gov/) or the EBI Array database (https://www.ebi.ac.uk/arrayexpress/).