Abstract
Molecular pathology and personalized medicine are still being evolved in Saudi Arabia, and genetic testing for the detection of mutations as cancer markers have not been established in the diagnostics laboratories in Saudi Arabia. The aim of the present study was to determine the prevalence of isocitrate dehydrogenase (IDH1 and IDH2) mutations and epidermal growth factor receptor variant (EGFRv)III transcript expression in Saudi Arabian patients with glioma. Out of 117 brain tumors tested by reverse transcription-quantitative PCR for EGFRvIII, 41 cases tested positive. In the glioblastoma (GBM) category, 28/55 tumors were positive, in astrocytoma tumors 5/22, and in oligodendrogliomas 4/13 cases were positive respectively. EGFRvIII transcript was sequenced by capillary electrophoresis to demonstrate the presence of EGFRvIII-specific junction where exons 2–7 were deleted. In the present study 106 tumors were sequenced for IDH1 exon-4 mutations using the capillary sequencing method. The most common substitution missense mutation c.395G>A was found in 16 tumors. In the case of adamantinomatous craniopharyngioma, a novel missense mutation in c.472C>T was detected in IDH2 gene. Using next-generation sequencing (NGS), 74 tumors were sequenced for the IDH1 gene, and a total of 8 missense variants were identified in 36 tumors in a population of Saudi Arabia. The missense mutation (c.395G>A) was detected in 29/36 of tumors. A novel intronic mutation in c.414+9T>A was found in 13 cases in the IDH1 gene. In addition, one case exhibited a novel synonymous mutation in c.369A>G. Eleven tumors were found to have compound mutations in the IDH1 gene. In IDH2 gene, out of a total of 16 variants found in 6 out of 45 tumors, nine were missense, five were synonymous and one was intronic. This is the first report from Saudi Arabian laboratories analyzing glioma tumors for EGFRvIII expression, and the first study from Saudi Arabia to analyze IDH mutations in gliomas using the capillary and NGS methods.
Keywords: RT-qPCR, personalized medicine, isocitrate dehydrogenase, molecular diagnostics, capillary sequencing, next-generation DNA sequencing, glioma, molecular pathology, brain cancer, Saudi Arabia
Introduction
Recent advances in the areas of medical genetics, molecular diagnostics and molecular pathology have significantly helped provide a mechanistic understanding of various diseases (1). Personalized medicine is a new and innovative field of healthcare, especially in cancer (2). This field of medicine focuses on the detection of genomic signatures in an individual, and helps accurately forecast a person's susceptibility to a disease and their prognosis, as well as develop a treatment strategy, that became known as targeted therapy (3).
Gliomas are very aggressive forms of brain cancer and the most common form of primary brain tumors; despite many advances in the molecular biology and genetics of gliomas, these tumors remain incurable (4,5). World Health Organization (WHO) grade-IV gliomas, also known as glioblastomas multiforme (GBMs), account for >50% of gliomas (6). Among all central nervous system (CNS) malignancies, GBM has a very poor prognosis (7). According to a published report by the Central Brain Tumor Registry of the United States in a cohort analysis of glioblastoma tumors from the National Program of Cancer Registries between 2001 and 2015, the 1-year survival rates reported were 62.5, 71.8, 58.6, 47.4 and 31.2%, and the 5-year survival rates were 20.8, 21.9, 9.3, 5.9 and 3.9% in age groups ranging 0–19, 20–44, 45–54, 55–64 and 65–74 years, respectively (8).
The isocitrate dehydrogenase (IDH) mutation is an early event in brain tumorigenesis; >70% of grade-II and -III gliomas have an IDH gene mutation (9,10). IDH mutations have been found to be strongly associated with 1p/19q codeletion and O(6)-methyl guanine methyl transferase (MGMT) promoter methylation, but are mutually exclusive with epidermal growth factor receptor (EGFR) amplification and loss of chromosome 10 (11,12). Other common mutations present in GBM include methylation of promoters in the p14ARF, CDKN2A and RB1 genes, and overexpression and mutations in the platelet-derived growth factor receptor, amplification of MDM2, CDKN2A gene mutation, overexpression of or mutations in the oncogene EGFR, as well as mutations and deletions in tumor suppressor genes phosphatase and tensin homologue (PTEN) and tumor protein p53 (13–15). Among all these genes altered in GBM, the EGFR gene was recognized as a convincing target for treatment development, since it is overexpressed, amplified and/or mutated in ≥40-50% of GBMs (16,17). In addition, out of the EGFR rearrangements that result in truncated isoforms, EGFR transcriptional variant III (EGFRvIII) is the most common mutational variant, expressed in 25–64% of GBMs (18). This mutant product contains an in-frame deletion of 801 base pairs of the coding sequence due to the deletion of exons 2–7 (19). This rearrangement gives rise to a ligand-independent kinase activation that persists downstream of the Ras-MAP kinase and PI3K pathways, promoting cell proliferation (20).
IDH1 and IDH2 are homologous metabolic isozymes, and the mutation of these genes generates a neomorphic enzyme, which can lead to the abnormal accumulation of 2-hydroxyglutarate (2-HG) and promote tumorigenesis. The IDH1 protein is localized in the cytoplasm and the peroxisomes (21). In the cytoplasm, the role of the IDH1 protein is to provide NADPH, when the pentose phosphate pathway is impaired, whereas IDH2 is localized in the mitochondria and catalyzes the same reaction as IDH1. The mutation of the IDH1 and IDH2 genes play an important role in gliomas by converting α-ketoglutarate (α-KG), which is produced in tricarboxylic acid (TCA) cycle, to an oncometabolite, 2-HG (22). Glioma tissue with an IDH1 mutation produces less α-KG and is known to contain high levels of 2-HG (23). This metabolite stimulates cellular proliferation through the degradation of hypoxia-inducible factor (HIF)-α, and inhibits α-KG-dependent dioxygenases (24). The conversion of α-KG to 2-HG decreases intracellular NADPH levels, which contributes to oncogenesis by creating a prooxidant state that benefits the development of glioma.
There is an abundance of clinical data associated with mutation detection, genetic profiling and its use in targeted therapy, prognosis prediction and survival of various types of cancer, including gliomas in the literature; however, such data from the Kingdom of Saudi Arabia is sporadic. Commercial molecular diagnostics laboratories are not ready to provide several cancer genetic tests in Saudi Arabia. Several drugs aimed at inhibiting EGFR signaling have already yielded good results in lung and colorectal cancer; however, for gliomas, clinical trials are still on-going, and considerable efforts are currently being made to develop immunotherapy targeting EGFRvIII for glioma (25,26). Reverse transcription-quantitative (RT-q) PCR for EGFRvIII became a basic requirement for monitoring the efficacy of this treatment, and for selecting the patients for the clinical trials. The molecular detection of IDH1 and IDH2 mutations has become standard practice in several institutions, and has been proven useful for the clinical management of gliomas. The aim of the present study was to determine the prevalence of IDH1 and IDH2 mutations and EGFRvIII transcript expression status in Saudi Arabian patients with glioma. Sequencing for IDH1 and IDH2 mutations in brain tumor samples was performed using the next-generation DNA sequencing (NGS) and capillary methods. In addition, the EGFRvIII expression was analyzed in brain tumor samples, by an in-house RT-qPCR assay developed for the first time in the Kingdom of Saudi Arabia. This type of investigations will help transfer the clinical assays to the diagnostics laboratories in the Kingdom, so that this approach can be utilized to become independent in molecular diagnostics and established oncology testing for clinical use.
Materials and methods
Chemicals, reagents and equipment
All NGS reagents for Ion Proton sequencing were purchased from Thermo Fisher Scientific, Inc. Sequencing reagents POP-7 (cat. no. 4393708) and BigDye v3.1 (cat. no. 4336923), TaqMan Fast universal PCR master mix (cat. no. 4352042), EGFRvIII-specific TaqMan MGB probe, GAPDH-specific TaqMan MGB probe and formamide were obtained from Applied Biosystems (Thermo Fisher Scientific, Inc.). A QuantiTect reverse transcription kit was purchased from Qiagen AB (cat. no. 204443). HPLC-purified EGFRvIII-specific primers, HPLC-purified GAPDH-specific primers, and IDH1 and IDH2 primers were from Integrated DNA Technologies, Inc. Platinum Taq DNA polymerase (cat. no. 10966-018). PCRx enhancer system (cat. no. 11495-017) was from Invitrogen (Thermo Fisher Scientific, Inc.). Real-time PCR instrument (ABI 7500 Fast; Thermo Fisher Scientific, Inc.), capillary sequencing instrument (3500 Genetic analyzer), speed vac (Eppendorf), NanoDrop 2000C, microAmp optic adhesive film for PCR plate (7500 Fast) and fast optical 96 well reaction plate (7500 Fast; cat. no. 4346907) were also purchased from Thermo Fisher Scientific, Inc.
Clinical specimens and ethics statement
This study was performed in accordance with the principles of the Declaration of Helsinki, and was approved by the Institutional review board bioethics committee of King Abdullah Medical City (KAMC), Makkah, Kingdom of Saudi Arabia (approval no. 14-140). Before starting the study, a written informed consent was obtained from all patients or the parent or guardian, if the patient was a minor. Formalin-fixed, paraffin-embedded (FFPE) tumor samples were collected for analysis from the Al-Noor Specialty Hospital Makkah, KAMC Makkah, and King Khalid University Hospital, King Saud University, Riyadh. In the present study, 165 CNS tumors were included; the majority of these (146/165) were gliomas and some were non-gliomas. Non-glioma tumors included one atypical choroid plexus papilloma, one primitive neuroectodermal tumor (PNET), one synovial sarcoma, two atypical teratoid rhabdoid tumors (ATRTs; 10 h and 2 years of age at the time of diagnosis), two adamantinomatous craniopharyngiomas, three medulloblastomas, four meningiomas (including one spinal meningioma Psammomatous type) and five hemangioblastomas. The mean age of the patients was 39 years (age range, 10 h to 83 years). The patients included 92 males and 73 females; the mean age was 42 and 36 years for males and females, respectively. The histological classification of tumors was based on the criteria set by the WHO (27). The computed tomography or magnetic resonance imaging data of all patients were reviewed by the consultant radiologist to confirm the diagnosis. One limitation that needs to be acknowledged in the present analysis of tumor tissue is obtaining a brain biopsy or FFPE tissue from the healthy individuals to use as a control; this was difficult, unless there is a pathological condition the patients cannot be operated for the brain tissue. For this reason, RNA and DNA isolated from cell lines were used as the controls.
RNA isolation
The FFPE samples (5–10 sections of 5-µm thickness) were collected in Eppendorf tubes, and the paraffin was dissolved using 1.0 ml xylene (2X) at 55°C for 5 min. The supernatant was discarded by pipetting, and 1.0 ml ethanol (2X) at room temperature was added to neutralize the residual xylene. The supernatant was then removed by pipetting, and the pellets were dried at 65°C for 3 min; if any remaining ethanol was present, the heating continued until all residual ethanol had evaporated. The pellet was resuspended in 240 µl proteinase K digestion buffer and treated with 10–20 µl proteinase K. This incubation was carried out at 55°C for 2–3 h or overnight, and then the incubations were carried out at 80°C for 15 min, to reverse the RNA crosslinking. Further steps were carried out as described in the QIAamp FFPE RNA kit (cat. no. 73504). Finally, RNA was eluted in 50–100 µl of RNAase-free water and stored at −80°C in aliquots for future use.
Primers and probe design for EGFRvIII and GAPDH
Junction-specific (exon 1 and 8 junctions) RT-qPCR primers for EGFRvIII were previously published (Table SI) (28). As this sequence is not present in genomic DNA, only the cDNA target was amplified. Sequencing PCR primers for EGFRvIII were designed using NCBI-Primer Blast. These primers were used to validate the positive cases found by RT-qPCR for EGFRvIII. To monitor the genomic DNA contamination, for GAPDH mRNA, intron spanning primers were used (Table SI). The forward primer was in exon 2, and reverse primer in exon 4, and the probe is in exon 3. The probe was a sense probe, which binds to the reverse strand.
RT-qPCR
The final concentration of the reverse and forward primers was 25 µM. The primers used were as follows: EGFRvIII forward, 5′-GGCTCTGGAGGAAAAGAAAGGTAATT-3′ and reverse, 5′-CCGTCTTCCTCCATCTCATAGC-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GATGGGATTTCCATTGATGAC-3′. MGB probes were prepared at a final concentration of 10 µM, and stored in aliquots at −20°C or lower, protected from light. The sequences of TaqMan-MGB probes used were: 6-FAM-5′-TGACAGATCACGGCTC-NFQ-3′ for EGFRvIII; and VIC-5′-TCACCAGGGCTGCTT-NFQ-3′ for GAPDH. Total RNA was used in a two-step RT-qPCR system. In this system, each sample was evaluated for the EGFRvIII and GAPDH genes using MGB probes. First the reverse transcription reaction was performed in 20 µl with 1 µg of RNA at 42°C for 30 mins, as described in the QuantiTect reverse transcription kit manual. The reverse transcription reaction was stopped by incubation for 3 mins at 95°C. This c-DNA was stored at −20°C for future use. qPCR was then performed using MGB probes and TaqMan Fast universal PCR master mix on a 7500 Fast RT-qPCR machine (ABI 7500; Applied Biosystems; Thermo Fisher Scientific, Inc.). A total of 2 µl cDNA was used for qPCR in 20 µl of TaqMan Fast universal PCR master mix containing 1 µl of 25 µM forward and reverse primers and 1 µl of 10 µM specific probe. The amplification protocol was as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. Mean Ct values of duplicate wells were used for data analysis. The relative expression of EGFRvIII compared with the reference gene (GAPDH) was calculated as ∆Ct (∆-threshold cycle), by subtracting the Ct value of GAPDH from that of EGFRvIII as previously described (29).
Cell lines used
The human U-87 MG ATCC® HTB-14 glioblastoma (origin unknown) cell line was originally obtained from the American Type Culture Collection (ATCC). U-87 MG cells harbouring the DR-GFP DNA repair reporter (U87 MG/DR-GFP) served as EGFRvIII-negative control cell line (30). As EGFRvIII-positive cells, U87 MG/DR-GFP were infected with EGFRvIII retrovirus WZL-hygro-EGFRvIII construct and selected for hygromycin resistance. The generation of these clones has been previously described (30–32). The DR-GFP plasmid was a gift from Dr Maria Jasin (Memorial Sloan Kettering Cancer Center, New York, NY, USA; cat. no. 26475; Addgene, Inc.) and WZL-hygro plasmid was a gift from Dr Scott Lowe (Memorial Sloan-Kettering Cancer Center, New York, NY, USA; cat. no. 18750; Addgene, Inc.). RNA and DNA isolated from these cell lines were used as the positive and negative control in RT-qPCR. Additionally, RNA isolated from immortalized human myelogenous leukaemia cell line K562 (cat. no. 04379012001) part of c-DNA synthesis kit from Roche was used in RT-qPCR as a negative control.
Validation of RT-qPCR results by sequencing the PCR product
The RT-qPCR test was validated with Sanger sequencing for EGFRvIII-positive samples using the Platinum Taq DNA polymerase (Invitrogen; Thermo Fisher Scientific, Inc.) and GC enhancer system (Promega Corporation). The following thermocycling conditions were used: 95°C for 3 min, followed by 40 cycles at 95°C for 30 sec, 54°C for 30 sec and 68°C for 30 sec, and a final extension step at 68°C for 5 min. The products were purified and sequenced in both directions for confirmation. A separate set of primers flanking the deletion site resulting in a 238-bp PCR product (113 bp upstream and 29 bp in downstream of 96 bp RT-qPCR product) was used for this sequencing. The sequencing primers are shown in Table SI.
Sanger sequencing for IDH1 and IDH2
The FFPE samples (5–10 sections of 5-µm thickness), were collected in a 1.5- or 1.7-ml Eppendorf tube. DNA extraction was performed using QIAamp DNA FFPE tissue kit (Qiagen) following the protocol described by the manufacturer (cat. no. 56404). Investigation of mutations in exon 4 of IDH1 and IDH2 were performed as previously reported, using primers to flank variants in the IDH1 (GenBank sequence NM_005896) and IDH2 (GenBank sequence NM_002168) genes (Table SI) (33). For IDH1, a ~196-bp partial exon 4 sequence containing hot spots was amplified using a forward primer within exon 4 and a reverse primer within intron 4, this product size was 213 bp. For IDH2, a 288-bp sequence with the entire exon 4 containing R140 and R172 hot spots was amplified using forward primer within intron 3 and a reverse primer within intron 4. For PCR 30–50 ng genomic DNA was amplified. The quality of the PCR product was checked by electrophoresis on agarose (1.2%) gel, and images were captured on the Bio-Rad's gel documentation system. PCR products were purified using Agencourt AM-Pure XP kit (Beckman Coulter, Inc.). All primers were tagged with M13 sequences, and M13 primers were used in sequencing PCR (BigDye reaction). BigDye reaction products were purified using the BigDye xTerminator kit (cat. no. 4376484) containing BigDye xTerminator and SAM solutions (Life Technologies; Thermo Fisher Scientific, Inc.). The sequencing was then performed on an ABI 3500 Genetic analyzer (8-capillary sequencer).
NGS analysis on Ion Proton
DNA (10 ng) was used for NGS analysis. DNA was sequenced using Ion PI v3 chip on Ion Proton instrument. Libraries were prepared using Ion AmpliSeq primer pools by Ion AmpliSeq™ cancer panel v.1 (cat. no. 4471262), Ion AmpliSeq™ Cancer Hotspot Panel v2 (cat. no. 4475346), or Comprehensive cancer panel genes (cat. no. 4477685) (34–37). Ion AmpliSeq 2.0 library and Ion PI Hi-Q OT2 200 kits were used for library and template preparation, respectively. Sequencing was performed using an Ion PI Hi-Q sequencing 200 kit, and the library was tagged with Ion Xpress™ Barcode Adapters. Sixteen tumour samples were pooled per chip run when libraries were prepared using Ion AmpliSeq™ cancer panel v.1 and Ion AmpliSeq™ Cancer Hotspot Panel v2, and 8 samples were pooled when comprehensive cancer panel primers were used. Following sequencing, amplicon sequences were aligned to the human reference genome GRCh37 (hg19) in the target regions of the cancer panel using the Torrent Suite v.5.0.2 software (Life Technologies; Thermo Fisher Scientific, Inc.). Variants call format (vcf) file was generated by running the Ampliseq™ Variant Caller plug-in v5.2 within the Ion Torrent Suite software. The vcf file data were analysed using Ion Reporter v5.6 (Thermo Fisher Scientific, Inc.). True mutations were considered based on a Phred score of >100, and mutation calling was considered significant by Ion Reporter software at P<0.05 (36,37).
Results
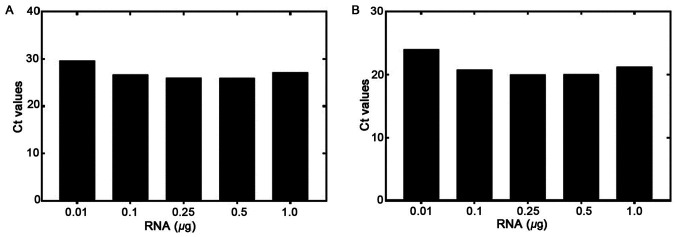
The RT-qPCR sensitivity was tested using various concentrations of RNA isolated from the EGFRvIII-positive U87 MG/DR-GFP cells. RNA was diluted from 1.0 to 0.01 µg and used for the RT reaction. RT-qPCR was then performed to detect the levels of EGFRvIII and GAPDH (Fig. 1A and B, respectively). The GAPDH Ct at a starting amount of 0.01 µg RNA was <30 cycles, demonstrating that RNA quality and not quantity was a more important determinant of assay success. The amplification plots for both genes are shown in Figs. S1–S3. The Ct values ranges from 29.69 to 25.97 for EGFRvIII, and 24.5 to 20.0 for GAPDH, from 0.01 to 1.0 µg RNA, respectively. The difference between the Cts of the two genes (EGFRvIII and GAPDH) was <7. This showed that the amount of starting RNA, over a 100-fold range, was not a significant factor determining assay results.
Figure 1.
Sensitivity of the RT-qPCR assay with the dilution of EGFRvIII-positive RNA. Amplification plots for EGFRvIII panel (A) and for GAPDH panel (B) RNA was diluted from 1.0–0.01 µg and used in RT reaction, then the qPCR was performed for EGFRvIII and GAPDH respectively. The Ct values are plotted against the RNA concentration. RT-qPCR, reverse transcription-quantitative PCR; EGFRv, epidermal growth factor receptor variant.
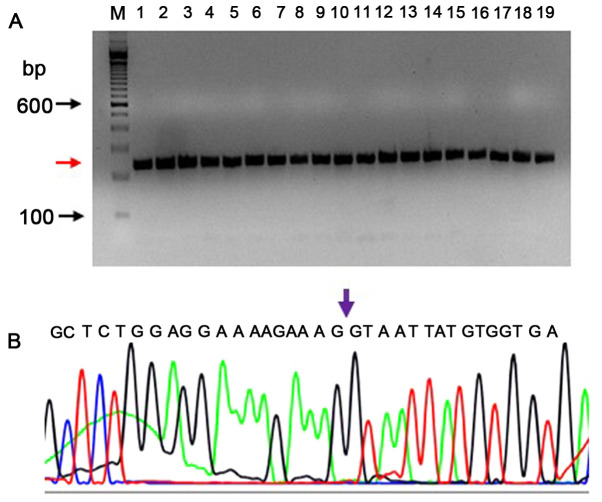
Amplification plots for RNA from K-562 cells, used as a negative control, are shown in Fig. S3B. The RT reaction was performed with 1.0 µg RNA, which was followed by the qPCR reaction for EGFRvIII and GAPDH respectively. There was no amplification of the EGFRvIII transcript, whereas GAPDH was amplified (Ct value 22.78). Similarly, in Fig. S4 demonstrates the EGFRvIII and GAPDH amplification plots of DNA isolated from positive control cells (panel A) and negative control cells (panel B). qPCR results confirmed that only DNA from the positive control cells amplified both the EGFRvIII and GAPDH genes (Ct values 21.23 and 16.85, respectively), whereas in negative control cells EGFRvIII was not amplified, but only GAPDH amplification was detected (Ct value 19.43). In Fig. S5 the verification of the EGFRvIII-specific product by PCR is shown using known positive and negative controls on agarose gel. PCR products of 238 bp (red arrow) are formed by DNA from positive cells (lanes 2–10) and no EGFRvIII-specific PCR products (lanes 11–19) were formed in the negative controls. The forward primer is a junctional primer spanning exons 1–8 (5′-CAGTATTGATCGGGAGAGCC-3′) and amplifies only the exon 2–7 deletion mutant of EGFR (EGFRvIII); lane denoted as ‘M’ is a 100 bp DNA ladder.
As shown in Table I, out of a total of 117 brain tumors tested for EGFRvIII by RT-qPCR, 41 (35%) tumors were positive (Table I). Out of 55 glioblastoma tumors, which included low-grade gliomas and GBMs, 28 tumors (50.9%) were positive for EGFRvIII expression (Table SII). In addition, 22.7% (5/22) of astrocytoma tumors, which included pilocytic, anaplastic and diffuse types, 30.8% (4/13) of oligodendrogliomas and anaplastic oligodendrogliomas, and 14.8% (4/27) of other brain tumors, including meningiomas, craniopharyngiomas, medulloblastomas, hemangioblastomas, gliosarcomas, ependymomas and gangliogliomas were also positive (Table SIII–SV).
Table I.
Epidermal growth factor receptor variant III expression in different brain tumors in Saudi Arabian population.
| Tumor type | Total tested | Positive (n) | Percentage |
|---|---|---|---|
| Glioblastoma | 55 | 28 | 50.9 |
| Oligodendroglioma | 13 | 4 | 30.8 |
| Astrocytoma | 22 | 5 | 22.7 |
| Others | 27 | 4 | 14.8 |
| Total | 117 | 41 | 35.0 |
The EGFRvIII transcript was sequenced by capillary electrophoresis to demonstrate the presence of an EGFRvIII-specific junction where exons 2–7 were deleted. The EGFRvIII expression was confirmed by sequencing the PCR product on an Applied Biosystem's sequencing instrument (3500 Genetic analyzer). The sequencing PCR primers were different from those used in RT-qPCR, in order to confirm a larger product which extends from downstream to upstream of the deletion. The RT-qPCR product spanned 20 bp upstream and 76 bp downstream of the deletion. The sequencing PCR product was 238-bp long (113 bp upstream and 29 bp downstream of the 96 bp RT-qPCR product). A total of 26 DNA samples that were found to be positive by RT-qPCR were tested to confirm the deletion sequence by Sanger sequencing. The RNA samples were reverse transcribed, and as this PCR product had 64.28% of GC content, a PCRx enhancer system was used with Platinum Taq DNA polymerase. All (26/26) samples exhibited the correctly-sized product (238 bp; a representative PCR gel picture is shown in Fig. 2A). Using the BigDye v3.1 method, the sequences were verified for the deletion of exons 2–7. A representative electropherogram confirming the presence of an exon 1 and exon 8 at the junction is shown in Fig. 2B.
Figure 2.
Validation of EGFRvIII transcript by sequencing the PCR product. (A) PCR products of 238 bp (red arrow) are shown for positive samples. The forward primer is a junctional primer spanning exons 1–8 (5′-CAGTATTGATCGGGAGAGCC-3′) and amplifies only the exon 2–7 deletion mutant of EGFR (EGFRvIII). This 238 bp product was sequenced by Sanger sequencing; a representative electropherogram is shown (B) The reverse primer is 5′-GCCCTTCGCACTTCTTACAC-3′. The arrow mark shown in the electropherogram confirms the presence of the junction at exon 1 and exon 8 in the sequencing PCR product. Lane denoted as (M) is a DNA ladder. EGFRv, epidermal growth factor receptor variant.
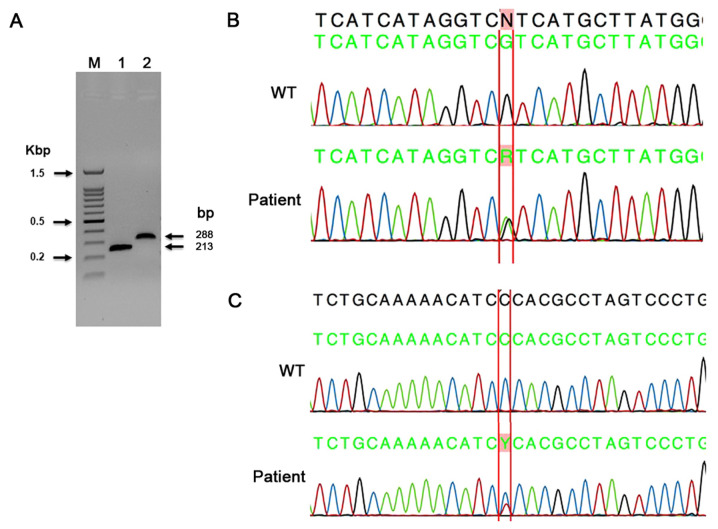
In the present study, 106 tumors were sequenced for IDH1 exon-4 mutations using the capillary sequencing method. A summary of all mutations found by capillary sequencing is shown in Table II. The primers used in the present study allowed for an interrogation of all three active-site arginine residues for both IDH1 (R100, R109 and R132) and IDH2 (R140, R149 and R172), as previously reported (38). In the present study, the substitution missense mutation in c.395G>A; p. (Arg132His) was found in the majority of the tumors. This mutation was identified in 16 tumor samples, including 4 oligodendrogliomas, 3 anaplastic oligodendrogliomas and 1 pilocytic astrocytoma. In addition, 2 astrocytomas, 2 GBMs, 1 diffuse astrocytoma and 1 grade-II ependymoma were also positive for this mutation (Table II). A representative image of the PCR gel of IDH1 and IDH2 PCR product sizes (Fig. 3A), and a representative electropherogram of a known missense mutation in IDH1 c.395G>A; p. (Arg132His) are shown in Fig. 3B. In the present study, 45 tumors were sequenced for IDH2 exon-4 mutations using the capillary sequencing method. The most common IDH2 mutation (Arg172Lys) was not found in any of the present cases. In a case of adamantinomatous craniopharyngioma, a novel missense mutation in IDH2 c.472C>T; p. (Pro158Ser) was detected (Fig. 3C). This novel mutation had not been reported in CNS tumors so far.
Table II.
Summary of IDH1 and IDH2 mutations detected by capillary sequencing.
| Serial number | Lab code | Sex | Age, years | Diagnosis | IDH1 | IDH2 |
|---|---|---|---|---|---|---|
| 1 | 15-N | F | 47 | Glioblastoma multiforme | c.395G>A; p. (Arg132His) | ND |
| 2 | HALK-27 | M | 59 | Glioblastoma | c.395G>A; p. (Arg132His) | ND |
| 3 | 50-N | F | 6 | Oligodendroglioma (WHO grade-II) | c.395G>A; p. (Arg132His) | NEG |
| 4 | HALK-10 | F | 17 | Anaplastic oligodendroglioma | c.395G>A; p. (Arg132His) | NEG |
| 5 | HALK-25 | F | 25 | Oligodendroglioma | c.395G>A; p. (Arg132His) | NEG |
| 6 | HALK-30 | F | 51 | Oligodendroglioma | c.395G>A; p. (Arg132His) | NEG |
| 7 | HALK-35 | M | 33 | Anaplastic oligodendroglioma | c.395G>A; p. (Arg132His) | ND |
| 8 | HALK-44 | F | 27 | Anaplastic oligodendroglioma | c.395G>A; p. (Arg132His) | NEG |
| 9 | KAMC-41 | M | 42 | Oligodendroglioma | c.395G>A; p. (Arg132His) | ND |
| 10 | HALK-11 | M | 1 | Diffuse astrocytoma | c.395G>A; p. (Arg132His) | NEG |
| 11 | KAMC-33 | M | 36 | Pilocytic astrocytoma | c.395G>A; p. (Arg132His) | ND |
| 12 | KAMC-37 | M | 58 | Pilocytic astrocytoma | c.395G>A; p. (Arg132His) | ND |
| 13 | KAMC-50 | F | 9 | Astrocytoma | c.395G>A; p. (Arg132His) | ND |
| 14 | KAMC-51 | M | 6 | Astrocytoma | c.395G>A; p. (Arg132His) | ND |
| 15 | 13-N | M | 19 | Pilocytic astrocytoma (WHO grade-I) | c.395G>A; p. (Arg132His) | ND |
| 16 | 49-N | M | 35 | Craniopharyngioma | NEG | c.472C>T; p. (Pro158Ser) |
| 17 | KAMC-28 | F | 39 | Ependymoma (WHO grade-II) | c.395G>A; p. (Arg132His) | ND |
IDH, isocitrate dehydrogenase; WHO, World Health Organization; ND, not done; NEG, negative.
Figure 3.
A representative PCR gel and electropherograms for IDH1 and IDH2 sequencing. (A) Representative image of gel IDH1 and IDH2 exon 4 PCR products Lane denoted as (M) is a 100 bp ladder. Lanes 1 and 2 are IDH1 and IDH2 PCR products showing 213 and 288 bp product size. (B) Representative electropherogram of a known missense mutation in IDH1 c.395G>A; p. (Arg132His). Upper sequence is WT, lower sequence is tumor sample. Letter ‘R’ represent nucleotide G changed to G/A heterozygous. (C) Representative electropherogram showing a novel missense mutation in IDH2 c.472C>T; p. (Pro158Ser). Upper sequence is WT, lower sequence is tumor sample. Letter ‘Y’ represents nucleotide C changed to T/C heterozygous. IDH, isocitrate dehydrogenase; WT, wild type.
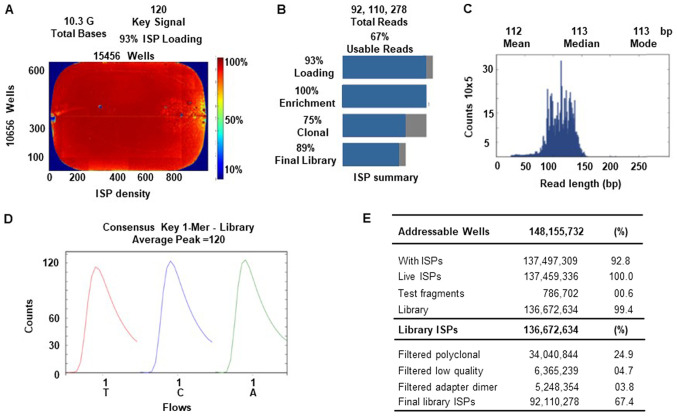
An Ion Torrent Suite v.5.0.2-generated Ion PI chip run report metrics of NGS sequencing is shown in Fig. 4. The Ion sphere particle (ISP) density image shows the semi-conductor chip loading across the wells that contain live ISPs; the total number of bases reported in the output file were 10.3 G (Fig. 4A). An ISP summary showing loading, enrichment, clonality and final library quality is shown in Fig. 4B. In the histogram, the first row shows 93% of loaded wells, of which 7% were empty, 100% were wells with a predicted number of live library ISPs (enrichment), 75% were clonal ISPs and 25% were polyclonal ISPs (ISPs carrying clones from >2 templates). The mean sequencing read length (trimmed lengths) of all reads was 112 bp (Fig. 4C). In Fig. 4D and E, the key signal (i.e. percentage of live ISPs readings for flows of the bases T, C and A in the library key), and the numerical values of addressable wells and library details are shown respectively.
Figure 4.
Torrent Suite software generated Ion PI chip run report metrics. (A) The ISP density image is showing percent loading across the semi-conductor chip, key signal and total bases read. (B) ISP summary showing loading, enrichment, clonality and final library quality. (C) Read length histogram showing trimmed lengths of all reads in base pairs. (D) Key signal details. (E) Addressable wells, and library ISP details. ISP, ion sphere particle.
A summary of IDH1 and IDH2 mutations identified by the NGS method is shown in Tables II–V. A total of 74 tumors were sequenced for the IDH1 gene by NGS, and 8 missense variants were found in 36 tumors in a Saudi Arabian population (Tables III–V). A codon 132 mutation (c.395G>A) was detected in 29/36 (80.5%) tumor samples by the NGS method. Other missense mutations in codons c.352C>T, c.368G>A c.369A>G, c.380C>T, c.394C>A and c.709T>C, each were found in 1 tumor, and 2 tumors were found to contain another missense mutation in c.356G>A. A novel intronic mutation (c.414+9T>A) was found in 13 tumors in the IDH1 gene by the NGS method (Tables III and IV). As shown in Tables III and IV, 8 tumors were found to have synonymous mutations (rs11554137) in c.315C>T; p. (Gly105Gly), and 1 to have a novel synonymous mutation in c.369A>G; p. (Gly123Gly). A total of 11 tumors were found to have compound mutations in the IDH1 gene (Table IV). One case of astrocytoma and a case of GBM tumor showed a compound, an intronic and a synonymous mutation (c.414+9T>A; and c.395G>A). Also, a PNET case contains a compound mutation in exons 2 and 5 respectively (Table IV). Compound variants were also found in each case of pilocytic astrocytoma, gliosarcoma and grade-II ependymoma tumors. The allele frequency, Phred score and P-values for all variants are shown in Tables III–V. One case of ATRT contained a synonymous mutation p. (Ser332Ser), and an intronic variant in IDH2 gene (Table V). A total of 6/45 of the screened cases had 16 mutations in the IDH2 gene (Table V). Nine of these variants were missense mutations, 5 were synonymous and 1 was intronic. However, all these IDH2 mutations were present as compound mutations with IDH1 gene mutations. One case of glioma had a mutation in exon 4 of the IDH1 gene and 6 compound mutations in exon 4 of the IDH2 gene also (Table V). However, the allele frequencies were low for these mutations.
Table V.
Summary of the IDH1 and IDH2 compound mutations detected by next-generation sequencing.
| Serial number | Lab code | Sex | Age, years | Diagnosis | Variant type | Gene name | Frequency,% | Phred score | Coverage | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | KAMC-15 | F | 62 | GBM | c.395G>A; p. (Arg132His) | IDH1 | 19.41 | 2940.93 | 1999 | 0.00001 |
| 2 | KAMC-15 | F | 62 | GBM | c.368G>A, p. (Gly123Glu) | IDH1 | 3.3 | 85.9895 | 2000 | 0.00001 |
| 3 | KAMC-15 | F | 62 | GBM | c.453C>T; p. (Pro151Pro)a | IDH2 | 7.96 | 359.68 | 1986 | 0.00001 |
| 4 | KAMC-15 | F | 62 | GBM | c.420G>A; p. (Arg140Arg)a | IDH2 | 6.01 | 176.401 | 1997 | 0.00001 |
| 5 | KAMC-15 | F | 62 | GBM | c.410G>A; p. (Gly137Glu) | IDH2 | 9.59 | 548.867 | 1991 | 0.00001 |
| 6 | KAMC-20 | M | 73 | Glioma | c.395G>A; p. (Arg132His) | IDH1 | 12.81 | 1488.5 | 1999 | 0.00001 |
| 7 | KAMC-20 | M | 73 | Glioma | c.500C>T; p. (Pro167Leu)a | IDH2 | 3.45 | 24.9168 | 1999 | 0.00322 |
| 8 | KAMC-20 | M | 73 | Glioma | c.487G>A; p. (Gly163Ser)a | IDH2 | 5.65 | 148.411 | 2000 | 0.00001 |
| 9 | KAMC-20 | M | 73 | Glioma | c.483C>T; p. (Gly163Gly)a | IDH2 | 4.05 | 49.1264 | 2000 | 0.00001 |
| 10 | KAMC-20 | M | 73 | Glioma | c.476G>A; p. (Arg159His) | IDH2 | 5.81 | 160.295 | 1998 | 0.00001 |
| 11 | KAMC-20 | M | 73 | Glioma | c.475C>T; p. (Arg159Cys) | IDH2 | 4.4 | 67.3793 | 1999 | 0.00001 |
| 12 | KAMC-20 | M | 73 | Glioma | c.404C>T; p. (Pro135Leu)a | IDH2 | 5.25 | 119.363 | 2000 | 0.00001 |
| 13 | KAMC-44 | M | 53 | GBM | c.395G>A; p. (Arg132His) | IDH1 | 26.03 | 4639.97 | 1998 | 0.00001 |
| 14 | KAMC-44 | M | 53 | GBM | c.368G>A; p. (Gly123Glu) | IDH1 | 5.05 | 246.285 | 2000 | 0.00001 |
| 15 | KAMC-44 | M | 53 | GBM | c.519C>T; p. (His173His)a | IDH2 | 13.44 | 1063.23 | 1942 | 0.00001 |
| 16 | KAMC-44 | M | 53 | GBM | c.459C>G; p. (Ile153Met)a | IDH2 | 12.71 | 954.66 | 1944 | 0.00001 |
| 17 | 71-N | M | 35 | Astrocytoma-(grade-II) | c.395G>A; p. (Arg132His) | IDH1 | 37.84 | 8160.23 | 1998 | 0.00001 |
| 18 | 71-N | M | 35 | Astrocytoma-(grade-II) | c.448G>A; p. (Glu150Lys)a | IDH2 | 5.15 | 112.741 | 2000 | 0.00001 |
| 19 | 71-N | M | 35 | Astrocytoma (grade-II) | c.409G>A; p. (Gly137Arg) | IDH2 | 3.75 | 36.0244 | 2000 | 0.00025 |
| 20 | 62-N | M | 63 | GBM | c.512G>A; p. (Gly171Asp) | IDH2 | 2.2 | 22.7072 | 2000 | 0.00536 |
| 21 | 62-N | M | 63 | GBM | c.395G>A; p. (Arg132His) | IDH1 | 6.46 | 416.312 | 1997 | 0.00001 |
| 23 | 91-N | M | 10b | ATRT grade-IV | c.996C>T; p. (Ser332Ser)a | IDH2 | 51.34 | 2138.71 | 411 | 0.00001 |
| 24 | 91-N | M | 10b | ATRT grade-IV | c.116-65T>Ca | IDH2 | 44.07 | 2328.36 | 565 | 0.00001 |
Novel mutation.
Hours. IDH, isocitrate dehydrogenase; ATRT, atypical teratoid rhabdoid tumor; GBM, glioblastoma; WHO, World Health Organization; M, male; F, female.
Table III.
IDH1 missense and intronic mutation status in brain tumors of Saudi Arabian population analyzed by next-generation sequencing.
| Serial number | Lab code | Sex | Age, years | Diagnosis | IDH1 | Exon | Allele frequency, % | Phred score | Coverage | P-value | IDH2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | KAMC-12 | F | 51 | GBM | c.395G>A; p. (Arg132His) | 4 | 9.66 | 901.25 | 1988 | 0.00001 | NEG |
| 2 | 83-N | M | 25 | Ependymoma (WHO grade-II) | c.395G>A; p. (Arg132His) | 4 | 17.72 | 2519.02 | 1981 | 0.00001 | NEG |
| 3 | HALK-36 | M | 13 | Pilocytic astrocytoma | c.394C>A; p. (Arg132Ser) | 4 | 35.72 | 4976.78 | 1996 | 0.00001 | ND |
| 4 | HALK-19 | F | 40 | Gliosarcoma | c.395G>A; p. (Arg132His) | 4 | 10.16 | 986.822 | 1998 | 0.00001 | NEG |
| 5 | 61-N | M | 7 | Medulloblastoma | c.315C>T; p. (Gly105Gly) | 4 | 9.6 | 350.583 | 2000 | 0.00001 | NEG |
| 6 | 63-N | M | 63 | GBM | c.352C>T; p. (Pro118Ser) | 4 | 6.26 | 388.295 | 1934 | 0.00001 | NEG |
| 7 | 65-N | M | 13 | GBM-grade-IV | c.315C>T; p. (Gly105Gly) | 4 | 59.53 | 11659.2 | 1999 | 0.00001 | NEG |
| 8 | 46-N | F | 38 | GBM-grade-IV | c.356G>A, p. (Arg119Gln) | 4 | CC=0.82 | 49.4243 | 1941 | 0.00001 | NEG |
| 9 | 47-N | M | 62 | GBM-grade-IV | c.356G>A, p. (Arg119Gln) | 4 | CC=0.51 | 69.8561 | 1967 | 0.00001 | NEG |
| 10 | 51-N | M | 73 | GBM-grade-IV | c.315C>T; p. (Gly105Gly) | 4 | 90.91 | 328.334 | 44 | 0.00001 | ND |
| 11 | HALK-13 | M | 7 | Pilocytic astrocytoma | c.395G>A; p. (Arg132His) | 4 | 5.50 | 298.817 | 2000 | 0.00001 | NEG |
| 12 | HALK-37 | M | 5 | Pilocytic astrocytoma | c.395G>A; p. (Arg132His) | 4 | 3.71 | 116.617 | 1997 | 0.00001 | NEG |
| 13 | HALK-42 | F | 4 | Pilocytic astrocytoma | c.395G>A; p. (Arg132His) | 4 | 3.05 | 67.7037 | 1999 | 0.00001 | NEG |
| 14 | HALK-46 | M | 32 | Ganglioglioma | c.395G>A; p. (Arg132His) | 4 | 3.85 | 129.056 | 1999 | 0.00001 | NEG |
| 15 | KAMC-2 | M | 67 | Oligodendroglioma | c.395G>A; p. (Arg132His) | 4 | 19.35 | 2918.86 | 1995 | 0.00001 | NEG |
| 16 | KAMC-47 | F | 75 | GBM | c.395G>A; p. (Arg132His) | 4 | 9.00 | 792.927 | 1988 | 0.00001 | NEG |
| 17 | HALK-25 | F | 25 | Oligodendroglioma | c.395G>A; p. (Arg132His) | 4 | 43.06 | 9872.2 | 1995 | 0.00001 | NEG |
| 18 | HALK-30 | F | 51 | Oligodendroglioma | c.395G>A; p. (Arg132His) | 4 | 40.07 | 8890.1 | 1999 | 0.00001 | NEG |
| 19 | HALK-35 | M | 33 | Anaplastic oligodendroglioma | c.395G>A; p. (Arg132His) | 4 | 35.97 | 7562.31 | 1996 | 0.00001 | NEG |
| 20 | 40-N | F | 59 | Meningioma-(WHO grade-I) | c.414+9T>Aa | Intronic | 75 | 1001.64 | 200 | 0.00001 | ND |
| 21 | 73-N | F | 65 | Meningioma-(WHO grade-I) | c.414+9T>Aa | Intronic | 72.28 | 469.808 | 101 | 0.00001 | ND |
| 22 | 75-N | M | 65 | Meningioma-(WHO grade-I) | c.414+9T>Aa | Intronic | 80.9 | 526.674 | 89 | 0.00001 | ND |
| 23 | 72-N | M | 14 | GBM | c.414+9T>Aa | Intronic | 77.07 | 829.327 | 157 | 0.00001 | ND |
| 24 | 78-N | M | 83 | GBM- (WHO grade-VI) | c.414+9T>Aa | Intronic | 84.92 | 816.555 | 126 | 0.00001 | ND |
| 25 | 79-N | M | 3 | Meningioma-(WHO grade-I) | c.414+9T>Aa | Intronic | 84.62 | 333.176 | 52 | 0.00001 | ND |
| 26 | 80-N | F | 31 | Hemangioblastoma | c.414+9T>Aa | Intronic | 79.1 | 376.734 | 67 | 0.00001 | ND |
| 27 | KAMC-52 | M | 55 | GBM-(WHO grade-VI) | c.414+9T>Aa | Intronic | 68.18 | 273.568 | 66 | 0.00001 | ND |
| 28 | 76-N | F | 72 | GBM- (WHO grade-VI) | c.414+9T>Aa | Intronic | 66.67 | 160.187 | 39 | 0.00001 | ND |
| 29 | HALK-2 | M | 45 | GBM | c.414+9T>Aa | Intronic | 78.85 | 576.863 | 104 | 0.00001 | ND |
| 30 | HALK-3 | M | 56 | GBM | c.414+9T>Aa | Intronic | 79.05 | 583.85 | 105 | 0.00001 | ND |
| 31 | KAMC-10 | F | 60 | Astrocytoma | c.395G>A; p. (Arg132His) | 4 | 8.68 | 742.359 | 1994 | 0.00001 | NEG |
| 32 | KAMC-31 | M | 28 | Astrocytoma | c.395G>A; p. (Arg132His) | 4 | 7.67 | 588.959 | 1995 | 0.00001 | NEG |
| 33 | KAMC-50 | F | 9 | Astrocytoma | c.395G>A; p. (Arg132His) | 4 | 43.54 | 6950 | 1998 | 0.00001 | NEG |
| 34 | HALK-41 | M | 51 | GBM | c.395G>A; p. (Arg132His) | 4 | 10.19 | 995.207 | 1993 | 0.00001 | NEG |
Novel mutation. IDH, isocitrate dehydrogenase; GBM, glioblastoma; WHO, World Health Organization; M, male; F, female; ND, not done; NEG, negative.
Table IV.
IDH1 compound mutation status in brain tumors of Saudi Arabian population analyzed by next-generation sequencing.
| Serial number | Lab code | Sex | Age, years | Diagnosis | IDH1 | Exon | Allele frequency, % | Phred score | Coverage | P-value | IDH2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HALK-45 | F | 38 | Gliosarcoma | c.395G>A; p. (Arg132His) | 4 | 5.01 | 242.779 | 1996 | 0.00001 | NEG |
| 2 | HALK-45 | F | 38 | Gliosarcoma | c.315C>T; p. (Gly105Gly) | 4 | 30.18 | 5818.57 | 1998 | 0.00001 | NEG |
| 3 | HALK-48 | M | 68 | GBM | c.395G>A; p. (Arg132His) | 4 | 3.66 | 113.809 | 1995 | 0.00001 | NEG |
| 4 | HALK-48 | M | 68 | GBM | c.315C>T; p. (Gly105Gly) | 4 | 33.35 | 6742.06 | 1994 | 0.00001 | NEG |
| 5 | HALK-39 | M | 55 | Low grade glioma | c.369A>G; p. (Gly123Gly)a | 4 | 5.26 | 119.741 | 1998 | 0.00001 | NEG |
| 6 | HALK-39 | M | 55 | Low grade glioma | c.315C>T; p. (Gly105Gly) | 4 | 4 | 143.241 | 2000 | 0.00001 | NEG |
| 7 | 84-N | M | 9 | Pilocytic astrocytoma | c.395G>A; p. (Arg132His) | 4 | 18.82 | 2798.99 | 1998 | 0.00001 | NEG |
| 8 | 84-N | M | 9 | Pilocytic astrocytoma | c.368G>A; p. Gly123Glu) | 4 | 3 | 65.2141 | 2000 | 0.00001 | NEG |
| 9 | KAMC-27 | M | 42 | GBM | c.395G>A; p. (Arg132His) | 4 | 20.92 | 2191.56 | 1319 | 0.00001 | NEG |
| 10 | KAMC-27 | M | 42 | GBM | c.394G>A; p. (Arg132Ser) | 4 | 33.75 | 6868.22 | 1994 | 0.00001 | NEG |
| 11 | HALK-44 | F | 27 | Anaplastic oligodendroglioma | c.315C>T; p. (Gly105Gly) | 4 | 43.57 | 10052 | 1997 | 0.00001 | NEG |
| 12 | HALK-44 | F | 27 | Anaplastic Oligodendroglioma | c.395G>A; p. (Arg132His) | 4 | 36.95 | 7849.08 | 1992 | 0.00001 | NEG |
| 13 | 92-N | F | 8 | PNET | c.709T>C; p. (Ser237Pro)a | 5 | 47.22 | 472.971 | 108 | 0.00001 | NEG |
| 14 | 92-N | F | 8 | PNET | c.395G>A; p. (Arg132His) | 4 | 0.79 | 21.675 | 252 | 0.0068 | NEG |
| 15 | HALK-27 | M | 59 | GBM | c.414+9T>Aa | Intronic | 70.93 | 384.357 | 86 | 0.00001 | ND |
| 16 | HALK-27 | M | 59 | GBM | c.395G>A; p. (Arg132His) | 4 | 49 | 809.429 | 400 | 0.00001 | ND |
| 17 | 77-N | M | 14 | Astrocytoma | c.414+9T>Aa | Intronic | 85.29 | 221.041 | 34 | 0.00001 | ND |
| 18 | 77-N | M | 14 | Astrocytoma | c.395G>A; p. (Arg132His) | 4 | 42.5 | 69.0889 | 40 | 0.00001 | ND |
| 19 | KAMC-39 | F | 39 | Oligoastrocytoma | c.395G>A; p. (Arg132His) | 4 | 6.41 | 411.865 | 1996 | 0.00001 | NEG |
| 20 | KAMC-39 | F | 39 | Oligoastrocytoma | c.315C>T p. (Gly105Gly) | 4 | 6.21 | 384.588 | 1998 | 0.00001 | NEG |
| 21 | 54-N | F | 14 | Medulloblastoma | c.380C>T; p. (Pro127Leu) | 4 | 3.97 | 45.0045 | 1992 | 0.00003 | NEG |
| 22 | 54-N | F | 14 | Medulloblastoma | c.315C>T p. (Gly105Gly) | 4 | 34.1 | 4605.54 | 2000 | 0.00001 | NEG |
Novel mutation. IDH, isocitrate dehydrogenase; GBM, glioblastoma; WHO, World Health Organization; M, male; F, female; ND, not done; NEG, negative.
The EGFRvIII expression status in the IDH1 and IDH2 mutated cases is shown in the Table SVI. Five of the tumors contain both IDH1 and IDH2 mutations along with EGFRvIII expression also. Out of 36 tumors with IDH1 mutations, 14 case showed EGFRvIII expression, and 22 cases were negative for EGFRvIII. In GBM tumor category 5/14 tumors were positive and 9 were negative. Furthermore, in oligodendrogliomas including anaplastic type 2/8, and in astrocytoma's including pilocytic and diffuse type, 5/6 were positive for EGFRvIII respectively. In medulloblastoma 1/2 tumor was positive; and each case of adamantinomatous craniopharyngioma, ganglioglioma, and two case of gliosarcoma tumors have IDH1 mutations but they were negative for EGFRvIII.
Discussion
Studies describing the prevalence of IDH mutations and its associations with overall survival and tumor progression in the Saudi Arabian population are limited. The fields of genomic medicine and targeted therapy are new in the Kingdom of Saudi Arabia. Mutations in the IDH1 and IDH2 genes have been shown to be predictive markers for favorable clinical outcomes in gliomas; it has also been shown that PARP inhibitors enhance the radio sensitization of glioma cells with an IDH mutation (38,39). The clinical trials on IDH1 inhibitors, such as Ivosidenib (TIBSOVO®), have recently yielded successful results for acute myeloid leukemia (AML) cases with an IDH1 gene mutation; however, this approach remains unclear for gliomas. As IDH-mutant astrocytomas have a more favorable survival, they may require a less aggressive treatment approach. Tumors without IDH mutations may have other genetic abnormalities characteristic of GBM that predict an aggressive clinical course and require an intensified treatment protocol. Several studies have shown that the IDH1 exon 4 mutations in grade-II and -III gliomas and secondary glioblastomas are in fact common (70%), and are less frequent in primary glioblastomas (29.4%) (9,35,40,41). In the present study, 71% (5/7) of anaplastic oligodendrogliomas, 75% (6/8) of astrocytomas and 31% (5/16) of pilocytic astrocytomas contained this exon 4 missense mutation, which was in agreement with previous reports (35,41,42). However, in our glioblastoma tumors, which included grade-IV GBM, the mutation detection rate was 43% (14/32) which is higher than that found in previous studies (15,42,43). The most common Arg132His mutation accounted for 92.7% of all exon 4 mutations in the IDH1 gene, with other mutations in this codon such as Arg132Cys, Ser, Gly, Leu and Val being very rare (9). In the present study, medulloblastoma (1/2) and grade-II ependymoma (1/1) tumors were found to have the missense mutation, and 2 of these medulloblastoma tumors also contained the synonymous mutations. Previous studies have reported that in meningiomas, medulloblastomas and ependymal tumors, IDH mutations were absent (42,44). Previously, a case of anaplastic grade-III ependymoma with a c.395G>A mutation was reported (37). Few studies have reported IDH1 codon 132 (Arg132His) mutations in PNET cases (42). In the present study, a novel mutation in c.709T>C; p. (Ser237Pro) was detected in exon 5 of 1 PNET case, which had not been reported in the COSMIC or ExAC databases; a codon 132 mutation was also present in this tumor. In the majority of cases, the arginine in position 132 was replaced by histidine (Arg132His) in IDH1, and in IDH2 the arginine residue at amino acid codon 172 or codon 140 was mutated (45). Mutations affected in these codons, which belongs to an evolutionary conserved region of the isocitrate binding site. Using the capillary and NGS methods, it was also found in the present study that c.395G>A was the major mutation in this Saudi Arabian population. This mutation in IDH1 (COSM28746) had a FATHMM score of 0.94 and Polyphen score of 0.127, suggesting it is a deleterious mutation; this mutation is also listed in the ClinVar database as pathogenic. Other IDH1 missense mutations found in the present studied cases, such as c.394G>A, c.368G>A, and c.356G>A, were also described in the COSMIC database as pathogenic. The novel IDH1 synonymous mutation c.369 A>G, found in low-grade glioma, had not previously been reported in the SNP and COSMIC databases; however, another synonymous variant, c.315C>T, was reported in the ClinVar database as a benign one. Recently, a study describing the prevalence of IDH mutations reported from Saudi Arabian patients with glioma; Alassiri et al (46) have shown that the MGMT promoter methylation, and IDH1 mutations and their associations with survival. However, in their study, the IDH1 was analyzed by IHC/qPCR targeting limited codons, and MGMT methylation was performed at the Mayo clinic. Alissiri et al, have reported that out of 65 cases screened for the IDH mutation, 6 (9.2%) tested positive. In another study this mutation rate was 33.87% by droplet digital PCR, 27.42% RT-qPCR, and 30.65% by Sanger direct sequencing (47). By NGS method this mutation rate was found to be ~47%, and by Sanger sequencing this was 15%. Recent advances are transferring many of the molecular tests to the NGS platforms for its sensitivity and accuracy in mutation detection (48). The synonymous IDH1 SNP (rs11554137; c.315C>T) mutations found in the present study were also shown to have an adverse prognosis in Egyptian adult patients with AML (49). This synonymous mutation, p. (Gly105Gly), and the intronic c.414+9T>A variants were found mostly is meningioma and glioblastoma tumors, but few cases of medulloblastoma, gliosarcoma, hemangioblastoma and anaplastic oligodendroglioma also contained these variants.
It has been shown that mutations in the IDH1 and IDH2 genes are mutually exclusive in gliomas, and mutations in IDH2 are mutually exclusive with PTEN, P53 and ATRX mutations (10,41,50). As compared with the IDH1 gene, mutations in the IDH2 gene are less prevalent in gliomas (40,45). The IDH2 missense mutation in c.410G>A; and c.476G>A are pathogenic mutations; these two variants were reported only twice each in the COSMIC database, in cutaneous melanoma, and in bladder urothelial carcinoma, and endometrial carcinomas respectively. Other IDH2 missense mutations in c.409G>A and c.475C>T were also reported in 1 sebaceous neoplasm and 1 bladder carcinoma, respectively, and they are pathogenic. This is the first report describing these 4 variants in brain tissue. Four of the IDH2 missense mutations detected in the present study (in c.404C>T, c.448G>A, c.459C>G, c.487G>A and c.512C>T) and the 5 synonymous variants (in c.420G>A, c.453C>T, c.483C>T and c.519C>T), as well as a synonymous mutation in c.996C>T and an intronic variant in c.116-65T>C were not reported in the COSMIC database; these novel mutations in the IDH2 gene were identified in the present study. In 1 GBM, the IDH2 synonymous mutation detected in this study in c.519C>T; p. (His173His) had also been reported in an atypical choroid plexus papilloma tumor (36). In the present study, no missense mutations were detected in IDH2 amino acid codons 172 and 140; however, all IDH2 mutations found were in exon 4, with the exception of c.996C>T; p. (Ser332Ser), which was present in exon 8 of an ATRT case. The c.116-65T>C variant detected in this case was present in intron 1. Only 1 synonymous mutation was present in codon 140 [c.420G>A; p. (Arg140Arg)] in exon 4. The novel exon 4 mutation in c.472C>T; p. (Pro158Ser) detected in the IDH2 gene by capillary sequencing was not reported in CNS tumors, and has only been reported in a case of pancreatic ductal adenocarcinoma so far (51).
Previous investigation revealed that EGFRvIII expression predominated in primary glioblastomas and in high grade GBMs but was rare in secondary glioblastomas. Furthermore, IDH1 mutation are rare in primary glioblastomas and they are common in secondary glioblastomas (52,53). By NGS analysis it was observed that 32% (12/37) of GBMs that includes glioblastomas also, and in 69% (11/16) of astrocytoma's tumors including pilocytic, anaplastic, and diffuse type, and 63% (5/8) of oligodendrogliomas (all grades) showed IDH1 c.395G>A mutations. One patient had right frontal lobe tumor with a history of GBM grade-IV, who was operated for decompression by excision biopsy. This tumor was tested negative for EGFRvIII, nine months post surgically this case had a recurrence and the tumor became positive for EGFRvIII. Immunohistochemistry results also positive for EGFR, FISH results showed EGFR amplification, and Ki 67 proliferation index was high in this tumor, suggesting EGFRvIII expression may change in a subset of patients at recurrence (54). This case had IDH1 mutation in c.352C>T preoperatively, and post-operatively showed mutations in both IDH1 in c.395G>A and in c.512G>A of IDH2 gene. Patients with meningiomas generally have a good prognosis, the present study identified that one grade-I secretary meningioma case doesn't contain IDH1 mutation and it is EGFRvIII-negative. In a total of 22 astrocytomas, two anaplastic tumors (grade-III) including one recurrent, two pilocytic (grade-I), and one diffuse type (grade-II) were positive for EGFRvIII. All these tumors were also positive for IDH1 mutation. One case of astrocytoma grade-II was negative for EGFRvIII, but this tumor had IDH1 c.395G>A, and IDH2 mutations in c.448G>A, and in c.409G>A which are pathogenic. Of a total of 13 oligodendrogliomas, four tumors that includes two anaplastic (grade-III) types are positive for both EGFRvIII and IDH1. In an adamantinomatous craniopharyngioma grade-I case which is negative for IDH1 mutation and EGFRvIII and no evidence of tumor recurrence after craniotomy. A case of ependymoma (grade-II) had an IDH1 exon-4 mutation and positive for EGFRvIII, another medulloblastoma (grade-IV) case had an IDH1 different missense mutation in c.380C>T and positive for EGFRvIII.
It has been postulated that the EGFRvIII detection may help distinguish patients with glioma who will respond to TKIs therapy, which makes this mutant an interesting target for immunotherapy too (55). These clinical trials showed promise in early phase II clinical trials; however, at later stages, the results were not promising (56,57). Such molecular targeted approaches for clinical trials have made the detection of EGFRvIII a priority, at least for the time being (58). Thus, few molecular laboratories are providing RT-qPCR for EGFRvIII; however, the utility of this assay is limited to clinical trials, since the targeted therapy for this molecule is at an early stage. Immunochemical detection and southern blotting are not very sensitive and accurate compared to molecular techniques like PCR and RT-qPCR. Frozen tissue is not routinely available due to transportation delays, and the instability of RNA causes difficulties; therefore, the development of molecular tests for EGFRvIII detection in the FFPE tissue is a convenient approach (28,29).
In The Cancer Genome Atlas (TCGA) samples, this mutated form of EGFRvIII was reported to be present in 24% of GBMs, and this was reported in 50–60% of EGFR-overexpressing GBMs (45,59). In the present study this deletion was found in 41 (35%; n=117) tumors that includes all brain tumors. Only 51% of GBMs, 23% of astrocytomas, including diffuse and pilocytic astrocytomas, and 31% of anaplastic oligodendrogliomas were positive for EGFRvIII. In addition, 1 case of each from medulloblastoma, atypical ganglioglioma, reactive gliosis and ependymoma were positive. Previous studies found that nearly 34.5% of tumors, all grades of glioma, including astrocytoma and pilocytic astrocytoma tumors, express EGFRvIII, a percentage high enough for therapeutic targeting (60). Previous studies have also reported the EGFRvIII expression in ependymomas, medulloblastomas, ATRT grade-IV, oligoastrocytomas grade-III and anaplastic astrocytoma's (61,62). The present data from a Saudi population is in concordance to these published reports.
NGS screening for the diagnosis of glioma is already provided in many genetics' laboratories. The NGS method detected more mutations than the capillary method in this study, suggesting the advantage of the NGS screening technique on all exons of IDH genes. That may have been the reason compound mutations were not detected in previous studies that used capillary sequencing. The IDH2 primers were not included in Ion AmpliSeq™ cancer panel v.1, but in Ion AmpliSeq™ Cancer Hotspot Panel v2, and Comprehensive cancer primer panels IDH2 gene was included (34–37). The samples screened with Ion AmpliSeq™ cancer panel v.1 primers are marked as ‘ND’ in the Tables for the IDH2 gene. The NGS method detected mutations with a high accuracy, as evidenced by the P-values (0.00001) and the high Phred quality score of all variants, indicating high confidence in the variants found in this tumor. Many of the IDH mutations found in this study had low frequencies, which indicates they are somatic variants; however, they all had a Phred score of ~100 and high suggest they are true variants. All mutations were verified in various databases (COSMIC, ExAc and dbSNP) to confirm whether the variants were novel in the IDH1 and IDH2 genes.
Molecular genetic testing for the detection of mutations as cancer markers have not been established in the diagnostics laboratories of Saudi Arabia. Considering the lack of molecular services to test cancer markers in the Kingdom, EGFRvIII RT-qPCR tests in brain tumors were developed. This is the first report from Saudi Arabian laboratories analyzing gliomas for EGFRvIII expression, and the second report in a Saudi Arabian population showing the prevalence of IDH mutations in gliomas. It is also the first study from Saudi Arabia to analyze IDH mutations in glioma cases using the capillary and NGS methods (46). In certain circumstances, the differential diagnosis of CNS lesions based on histology alone can be difficult. For example, distinguishing between reactive gliosis and diffuse glioma, or between oligodendroglioma and other similar entities, can be challenging. In some of these cases, the IDH status was proven to be an extremely useful diagnostic biomarker, and immunochemical detection methods are not very sensitive and accurate compared with molecular techniques, such as Sanger sequencing, a gold standard in molecular pathology for mutation detection (63,64).
The aim of the present study was to determine the prevalence of IDH1 and IDH2 mutations and EGFRvIII transcript expression in Saudi Arabian patients with glioma. As these methods are not available in this region, the aim of the present study was to transfer these tests to specialty hospitals for patient's use. This type of research will help transfer the clinical assays to the diagnostics laboratories in the Kingdom, so that this approach can be utilized to become independent in molecular diagnostics and established oncology testing for clinical use. Finally, one limitation that needs to be acknowledged in the present study is the lack of functional studies. Retrospective, comparative survival analysis correlating with glioma characteristics and types, surgery type, the specifics of radiotherapy and the adjuvant therapies were not performed in the present study. In the International Cancer Genome Consortium (https://dcc.icgc.org) data portal IDH1 and EGFR genes are in top 20 mutated cancer genes with high functional impact somatic mutations. IDH1 and EGFR are most recurrently mutated cancer driver genes in GBM_TCGA dataset (https://www.intogen.org). Poor prognostic markers included genetic changes in the EGFR mutations in this group (65), and among most driver genes, IDH mutations are good prognostic factor in diffuse gliomas (66,67).
In conclusion, the present study analyzed the IDH mutations by NGS and capillary methods. The p. (Arg132His) mutation in IDH1 was the predominant mutation in the Saudi Arabian population. Several novel IDH1 and IDH2 mutations were reported in this study. The novel IDH1 missense mutation in c.709T>C, a novel synonymous mutation c.369 A>G, and a novel intronic mutation (c.414+9T>A) was not reported previously. Also, in IDH2 gene four missense mutations, six synonymous variants, and an intronic variant were novel variants found. In the present study, missense mutations in IDH2 amino acid codons 172 and 140 were not detected. The exon 4 mutation in IDH2 (c.472C>T) was not reported previously in CNS tumors. In this study, 35% brain tumors were positive for EGFRvIII expression. In the GBM category, 51% were positive, and in astrocytoma and in anaplastic oligodendroglioma 23 and 31% were positive, respectively. The RT-qPCR test was validated by Sanger sequencing in EGFRvIII-positive samples.
Development of molecular testing helps the clinician to prescribe the correct medication, as this approach is giving good results to treat different cancer types effectively, saves money, avoids purchasing the diagnostic kits, and this will prevent sending the samples to companies outside the Kingdom. The development and application of a genetic test for brain tumors, such as IDH1 and IDH2 mutations, is significant in this regard because these tests have prognostic and therapeutic values.
Supplementary Material
Acknowledgements
The authors would like to thank Mrs. Rowa Abbas Bakhsh and Mr. Mohammed Bader Al-Hamad (Histopathology Division, Al-Noor Specialty Hospital, Makkah, Saudi Arabia) for their technical help. The authors would also like to thank Dr Emad A. Felemban and Dr Ahmed Shawky (Science and Technology Unit, Umm Al-Qura University, Makkah, Saudi Arabia) for their continuous support.
Glossary
Abbreviations
- AML
acute myeloid leukemia
- ATRT
atypical teratoid rhabdoid tumor
- ClinVar
clinical variant
- CNS
central nervous system
- COSMIC
Catalogue Of Somatic Mutations In Cancer
- Ct
cycle threshold
- ddPCR
droplet digital PCR
- EGFRvIII
epidermal growth factor receptor variant III
- ExAc
The Exome Aggregation Consortium
- FDA
The Food and Drug Administration
- HG
hydroxyglutarate
- HIF
hypoxia-inducible factor
- IDH
isocitrate dehydrogenase
- ISP
Ion sphere particle
- KAMC
King Abdullah Medical City
- KG
ketoglutarate
- KSA
Kingdom of Saudi Arabia
- MGMT
O(6)-methyl guanine methyl transferase
- NGS
next-generation DNA sequencing
- PNET
primitive neuroectodermal tumor
- RT-qPCR
reverse transcription-quantitative PCR
- TCGA
The Cancer Genome Atlas
- TKI
tyrosine kinase inhibitor
Funding
The present study was supported by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdul Aziz City for Science and Technology (grant no. 12-MED 2961-10), to Dr MM. Taher.
Availability of data and materials
The IDH2 novel variant [(c.472C>T; p. (Pro158Ser)] sequence file was deposited in SRA (Sequence Read Archive) database with access numbers SRA: PRJNA644191; BioSample access number: SAMN15452876.
Authors' contributions
KV, FAA, and MMT conceived and designed the study. RAJ, HK and EMB performed the pathologic diagnoses. MMT, GD, NMB and MA performed the mutational analyses and acquired and interpreted the data. KQ, EMB and THN drafted the manuscript and analyzed the clinical data. KV, MMT, FAA and NMB revised it critically and gave final approval of the version to be published. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional review board bioethics committee of King Abdullah Medical City, Makkah, Kingdom of Saudi Arabia (approval no. 14-140). Written informed consent was obtained from all patients or the parent or guardian, if the patient was a minor.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Claussnitzer M, Cho JH, Collins R, Cox NJ, Dermitzakis ET, Hurles ME, Kathiresan S, Kenny EE, Lindgren CM, MacArthur DG, et al. A brief history of human disease genetics. Nature. 2020;577:179–189. doi: 10.1038/s41586-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobel ME, Bagg A, Caliendo AM, Ladanyi M, Zehnbauer B. The evolution of molecular genetic pathology: Advancing 20th-Century diagnostic methods into potent tools for the new millennium. J Mol Diagn. 2008;10:480–483. doi: 10.2353/jmoldx.2008.080113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shendure J, Findlay GM, Snyder MW. Genomic medicine-progress, pitfalls, and promise. Cell. 2019;177:45–57. doi: 10.1016/j.cell.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R, Reifenberger G. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 5.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schüler D, Probst-Hensch NM, Maiorka PC, et al. Genetic pathways to glioblastoma: A population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 6.Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. Glioblastoma multiforme: A review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 7.Delgado-Lopez PD, Corrales-Garcıa EM. Survival in glioblastoma: A review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 8.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 10.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, Wiemels JL, Nelson HH, Karagas MR, Wrensch MR, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, Frenay M, Tijssen CC, Lacombe D, Idbaih A, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: A report of the European organization for research and treatment of cancer brain tumor group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 13.Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee ShU. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev. 2017;18:3–9. doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. L Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 17.Herbst RS, Shin DM. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors. A new paradigm for cancer therapy. Cancer. 2002;94:1593–1611. doi: 10.1002/cncr.10372. [DOI] [PubMed] [Google Scholar]
- 18.Pelloski CE, Ballman KV, Furth AF, Zhang L, Lin E, Sulman EP, Bhat K, McDonald JM, Yung WK, Colman H, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 19.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP (+)-dependent isocitrate dehydrogenase. J Biol Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 22.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: Mechanisms models, and clinical opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 23.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi BD, Maus MV, June CH, Sampson JH. Immunotherapy for glioblastoma: Adoptive T-cell strategies. Clin Cancer Res. 2019;25:2042–2048. doi: 10.1158/1078-0432.CCR-18-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An Z, Aksoy O, Zheng T, Fan QW, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene. 2018;37:1561–1575. doi: 10.1038/s41388-017-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 28.Steffensen KD, Waldstrøm M, Olsen DA, Corydon T, Lorentzen KA, Knudsen HJ, Jeppesen U, Brandslund I, Jakobsen A. Mutant epidermal growth factor receptor in benign, borderline, and malignant ovarian tumors. Clin Cancer Res. 2008;14:3278–3282. doi: 10.1158/1078-0432.CCR-07-4171. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimoto K, Dang J, Zhu S, Nathanson D, Huang T, Dumont R, Seligson DB, Yong WH, Xiong Z, Rao N, et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 30.Golding SE, Rosenberg E, Khalil A, McEwen A, Holmes M, Neill S, Povirk LF, Valerie K. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J Biol Chem. 2004;279:15402–15410. doi: 10.1074/jbc.M314191200. [DOI] [PubMed] [Google Scholar]
- 31.Golding SE, Morgan RN, Adams BR, Hawkins AJ, Povirk LF, Valerie K. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol Ther. 2009;8:730–738. doi: 10.4161/cbt.8.8.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lammering G, Hewit TH, Holmes M, Valerie K, Hawkins W, Lin PS, Mikkelsen RB, Schmidt-Ullrich RK. Inhibition of the type III epidermal growth factor receptor variant mutant receptor by dominant-negative EGFR-CD533 enhances malignant glioma cell radiosensitivity. Clin Cancer Res. 2004;10:6732–6743. doi: 10.1158/1078-0432.CCR-04-0393. [DOI] [PubMed] [Google Scholar]
- 33.Patel KP, Barkoh BA, Chen Z, Ma D, Reddy N, Medeiros LJ, Luthra R. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J Mol Diagn. 2011;13:678–686. doi: 10.1016/j.jmoldx.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehrotra M, Duose DY, Singh RR, Barkoh BA, Manekia J, Harmon MA, Patel KP, Routbort MJ, Medeiros LJ, Wistuba II, Luthra R. Versatile ion S5XL sequencer for targeted next generation sequencing of solid tumors in a clinical laboratory. PLoS One. 2017;12:e0181968. doi: 10.1371/journal.pone.0181968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballester LY, Fuller GN, Powell SZ, Sulman EP, Patel KP, Luthra R, Routbort MJ. Retrospective analysis of molecular and immunohistochemical characterization of 381 primary brain tumors. J Neuropathol Exp Neurol. 2017;76:179–188. doi: 10.1093/jnen/nlw119. [DOI] [PubMed] [Google Scholar]
- 36.Taher MM, Hassan AA, Saeed M, Jastania RA, Nageeti TH, Alkhalidi H, Dairi G, Abduljaleel Z, Athar M, Bouazzaoui A, et al. Next generation DNA sequencing of atypical choroid plexus papilloma of brain: Identification of novel mutations in a female patient by Ion Proton. Oncol Lett. 2019;18:5063–5076. doi: 10.3892/ol.2019.10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butt M, Alyami S, Nageeti T, Saeed M, AlQuthami K, Bouazzaoui A, Athar M, Abduljaleel Z, Al-Allaf F, Taher M. Mutation profiling of anaplastic ependymoma grade III by Ion Proton by next generation DNA sequencing. F1000Res. 2019;8:613. doi: 10.12688/f1000research.18721.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Kwintkiewicz J, Liu Y, Tech K, Frady LN, Su YT, Bautista W, Moon SI, MacDonald J, Ewend MG, et al. Chemosensitivity of IDH1-mutated gliomas due to an impairment in PARP1-mediated DNA repair. Cancer Res. 2017;77:1709–1718. doi: 10.1158/0008-5472.CAN-16-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 42.Hayden JT, Frühwald MC, Hasselblatt M, Ellison DW, Bailey S, Clifford SC. Frequent IDH1 mutations in supratentorial primitive neuroectodermal tumors (sPNET) of adults but not children. Cell Cycle. 2009;8:1806–1807. doi: 10.4161/cc.8.11.8594. [DOI] [PubMed] [Google Scholar]
- 43.Mukasa A, Takayanagi S, Saito K, Shibahara J, Tabei Y, Furuya K, Ide T, Narita Y, Nishikawa R, Ueki K, Saito N. Significance of IDH mutations varies with tumor histology, grade, and genetics in Japanese glioma patients. Cancer Sci. 2012;103:587–592. doi: 10.1111/j.1349-7006.2011.02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellai M, Piazzi A, Caldera V, Monzeglio O, Cassoni P, Valente G, Schiffer D. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J Neurooncol. 2011;105:345–357. doi: 10.1007/s11060-011-0596-3. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–5571. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alassiri AH, Alkhaibary A, Al-Sarheed S, Alsufani F, Alharbi M, Alkhani A, Aloraidi A. O6-methylguanine-DNA methyltransferase promoter methylation and isocitrate dehydrogenase mutation as prognostic factors in a cohort of Saudi patients with glioblastoma. Ann Saudi Med. 2019;39:410–416. doi: 10.5144/0256-4947.2019.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Zhao Y, Li JF, Guo CC, Chen FR, Su HK, Zhao HF, Long YK, Shao JY, To Ss, Chen ZP. IDH1 mutation detection by droplet digital PCR in glioma. Oncotarget. 2015;6:39651–39660. doi: 10.18632/oncotarget.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin H, Sa JK, Bae JS, Koo H, Jin S, Cho HJ, Choi SW, Kyoung JM, Kim JY, Seo YJ, et al. Clinical targeted next-generation sequencing panels for detection of somatic variants in gliomas. Cancer Res Treat. 2020;52:41–50. doi: 10.4143/crt.2019.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ali MAM, Ahmed EK, Assem MMA, Helwa R. The synonymous isocitrate dehydrogenase 1 315C>T SNP confers an adverse prognosis in egyptian adult patients with NPM1-/CEBPA-negative acute myeloid leukemia. Indian J Hematol Blood Transfus. 2018;34:240–252. doi: 10.1007/s12288-017-0852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang HY, Tang K, Liang TY, Zhang WZ, Li JY, Wang W, Hu HM, Li MY, Wang HQ, He XZ, et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J Exp Clin Cancer Res. 2016;35:86. doi: 10.1186/s13046-016-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wartenberg M, Cibin S, Zlobec I, Vassella E, Eppenberger-Castori S, Terracciano L, Eichmann MD, Worni M, Gloor B, Perren A, Karamitopoulou E. Integrated Genomic and Immunophenotypic classification of pancreatic cancer reveals three distinct subtypes with prognostic/predictive Significance. Clin Cancer Res. 2018;24:4444–4454. doi: 10.1158/1078-0432.CCR-17-3401. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 54.Wikstrand CJ, McLendon RE, Friedman AH, Bigner DD. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res. 1997;57:4130–4140. [PubMed] [Google Scholar]
- 55.Johnson LA, Scholler J, Ohkuri T, Kosaka A, Patel PR, McGettigan SE, Nace AK, Dentchev T, Thekkat P, Loew A, et al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med. 2015;7:275ra22. doi: 10.1126/scitranslmed.aaa4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heimberger AB, Sun W, Hussain SF, Dey M, Crutcher L, Aldape K, Gilbert M, Hassenbusch SJ, Sawaya R, Schmittling B, et al. Immunological responses in a patient with glioblastoma multiforme treated with sequential courses of temozolomide and immunotherapy: Case study. Neuro Oncol. 2008;10:98–103. doi: 10.1215/15228517-2007-046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Binder DC, Ladomersky E, Lenzen A, Zhai L, Lauing KL, Otto-Meyer SD, Lukas RV, Wainwright DA. Lessons learned from rindopepimut treatment in patients with EGFRvIII-expressing glioblastoma. Transl Cancer Res. 2018;7(Suppl 4):S510–S513. doi: 10.21037/tcr.2018.03.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwatra MM. A rational approach to target the epidermal growth factor receptor in glioblastoma. Curr Cancer Drug Targets. 2017;17:290–296. doi: 10.2174/1568009616666161227091522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kastenhuber ER, Huse JT, Berman SH, Pedraza A, Zhang J, Suehara Y, Viale A, Cavatore M, Heguy A, Szerlip N, et al. Quantitative assessment of intragenic receptor tyrosine kinase deletions in primary glioblastomas: Their prevalence and molecular correlates. Acta Neuropathol. 2014;127:747–759. doi: 10.1007/s00401-013-1217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Felsberg J, Hentschel B, Kaulich K, Gramatzki D, Zacher A, Malzkorn B, Kamp M, Sabel M, Simon M, Westphal M, et al. Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: Prognostic role and comparison between primary and recurrent tumors. Clin Cancer Res. 2017;23:6846–6855. doi: 10.1158/1078-0432.CCR-17-0890. [DOI] [PubMed] [Google Scholar]
- 61.Carvalho PO, Uno M, Oba-Shinjo SM, Rosemberg S, Wakamatsu A, da Silva CC, Teixeira MJ, Marie SK. Activation of EGFR signaling from pilocytic astrocytomas to glioblastomas. Int J Biol Markers. 2014;29:e120–e128. doi: 10.5301/JBM.5000045. [DOI] [PubMed] [Google Scholar]
- 62.Wong A, Mitra S, Del Vecchio CA, Skirboll S. Expression of EGFRvIII in brain tumor stem cells. J Clin Oncol. 2008;26(Suppl 15):S2002–S2002. doi: 10.1200/jco.2008.26.15_suppl.2002. [DOI] [Google Scholar]
- 63.Katsanis SH, Katsanis N. Molecular genetic testing and the future of clinical genomics. Nat Rev Genet. 2013;14:415–426. doi: 10.1038/nrg3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Di Resta C, Galbiati S, Carrera P, Ferrari M. Next-generation sequencing approach for the diagnosis of human diseases: Open challenges and new opportunities. EJIFCC. 2018;29:4–14. [PMC free article] [PubMed] [Google Scholar]
- 65.Liang A, Zhou B, Sun W. Integrated genomic characterization of cancer genes in glioma. Cancer Cell Int. 2017;17:90. doi: 10.1186/s12935-017-0458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jonsson P, Lin AL, Young RJ, DiStefano NM, Hyman DM, Li BT, Berger MF, Zehir A, Ladanyi M, Solit DB, et al. Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin Cancer Res. 2019;25:5537–5547. doi: 10.1158/1078-0432.CCR-19-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The IDH2 novel variant [(c.472C>T; p. (Pro158Ser)] sequence file was deposited in SRA (Sequence Read Archive) database with access numbers SRA: PRJNA644191; BioSample access number: SAMN15452876.