Abstract
Osteosarcoma is a common primary bone cancer that there are currently no effective treatment strategies for. Forkhead box M1 (FoxM1) is key in the development of osteosarcoma, and microRNA (miR)-216b serves an antitumor role by targeting FoxM1. Moreover, thiostrepton (TST), a natural thiazole antibiotic, induces antitumor effects and specifically targets FoxM1. Therefore, the present study investigated whether thiostrepton and miR-216b synergistically inhibited osteosarcoma cells by targeting FoxM1. The MTT assay, reverse transcription-quantitative PCR, a dual-luciferase reporter assay and flow cytometry were performed. Compared with the human osteoblast cell line hFOB1.19, miR-216b expression was significantly downregulated in the osteosarcoma cell lines U2OS, MG63 and Saos-2. By contrast, FoxM1 expression was significantly upregulated in osteosarcoma cell lines compared with the hFOB1.19 cell line. The results indicated that miR-216b targeted the 3′-untranslated region of FoxM1. Moreover, the results suggested that miR-216b cooperated with TST to decrease cell cytotoxicity and increase cell apoptosis. In addition, miR-216b cooperated with TST to increase Bax expression and decrease Bcl-2 expression. In conclusion, the combination of TST and miR-216b synergistically promoted osteosarcoma cell cytotoxicity and apoptosis by targeting FoxM1. Therefore, the present study suggested that the combination of TST and miR-216b may serve as a promising therapeutic strategy for osteosarcoma.
Keywords: thiostrepton, microRNA-216b, osteosarcoma, forkhead box M1
Introduction
Osteosarcoma is a common type of primary bone cancer, with an incidence of 4–5 per million individuals and a 60–70% 5-year survival rate (1). In recent years, increasing research has been conducted to develop clinical therapeutic strategies for osteosarcoma, including chemotherapy, radiotherapy and surgery; however, the survival rate of patients with osteosarcoma remains low (2). Therefore, identifying the molecular mechanisms underlying the occurrence and progression of osteosarcoma is important for the development of effective therapeutic agents.
MicroRNAs (miRNAs/miRs) are small, endogenous non-coding RNAs that contain 20–25 nucleotides. By post-transcriptionally regulating the silencing of genes in the 3′-untranslated region (3′-UTR), miRNAs are involved in biological processes, including tumor occurrence, growth and progression (3). miR-216 is downregulated in numerous types of tumors, which suggests that miR-216 may serve as a promising prognostic indicator (4,5). By knocking down syndecan binding protein in breast cancer or Cyclin T2 in gastric cancer, miR-216 effectively inhibits cancer cell proliferation and migration (6,7). By targeting FoxM1, miR-216 suppresses proliferation, migration and invasion in osteosarcoma cells, as well as in liver and cervical cancer cells (8–10). In addition, miR-216 is closely linked to improved outcomes in patients with cervical cancer (10).
Thiostrepton (TST) is a natural cyclic oligopeptide antibiotic of the thiopeptide class, which is derived from streptomyces (11). TST was first studied in breast cancer as a FoxM1 and proteasome inhibitor in 2008 (12). In vivo and in vitro evidence has demonstrated the significant anticancer effects of TST (11). By targeting FoxM1 protein and the proteasome, TST induces cell apoptosis in ovarian and lung cancer, as well as in other types of tumor cells (13–15). As a novel anticancer target, TST has been extensively studied in tumor research (11). Additionally, by targeting FoxM1, TST is involved in the progression of Ewing's sarcoma and synoviosarcoma (16–18). The present study investigated FoxM1 as a common target of TST and miR-216b, and the co-influences of TST and miR-216b on osteosarcoma cell behaviors.
Materials and methods
Cell culture
Human osteosarcoma cell lines (U2OS, MG63 and Saos-2) and the human osteoblast cell line (hFOB1.19) were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were cultured in Dulbecco's Modified Eagle's medium (Gibco, Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin with 5% CO2 at 37°C. Cells in the logarithmic growth phase were used for subsequent experiments.
Cell transfection
Cells were seeded (5×105 cells/well) into 6-well plates. Subsequently, cells were transfected with miR-216b mimic (Shanghai GenePharma Co., Ltd.) or mimic-negative control (NC) (Shanghai GenePharma Co., Ltd.). The miR-216b mimic sequence was 5′-AAAUCUCUGCAGGCAAAUGUGA-3′, and the mimic-NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. miR-216b mimic or mimic-NC were transfected at a final concentration of 50 nM using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following culture for 6 h at 37°C, the medium was replaced. At 48 h post-transfection, cells were used for subsequent experiments.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® (Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the M-MLV reverse transcription kit (cat. no. RR047A; Takara Bio, Inc.). Relative miR-216b expression was determined using the miRNA Real-Time PCR Assay kit (Aidlab Biotechnologies, Ltd.). Relative FoxM1 expression was determined using SYBR Premix Ex Taq™ II with Tli RNaseH (cat. no. RR820A, Takara Bio, Inc.) and an ABI PRISM 7500 system (Thermo Fisher Scientific, Inc.). The following primers were used for qPCR: miR-216b forward, 5′-AAAUCUCUGCAGGCAAAUGUGA-3′ and reverse, 5′-ACAUUUGCCUCCAGAGAUUUUU-3′; β-actin forward, 5′-AAACTGGAACGGTGAAGGTG-3′ and reverse, 5′-AGTGGGGTGGCTTTTAGGAT-3′; FoxM1 forward, 5′-TGCCCAGCAGTCTCTTACCT-3′ and reverse, 5′-CTACCCACCTTCTGGCAGTC-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. miRNA and mRNA expression levels were quantified using the 2−ΔΔCq method (19) and normalized to the internal reference genes U6 and β-actin, respectively.
MTT assay
The MTT assay was performed to assess cell cytotoxicity. Transfected cells were seeded (3×103 cells/well) into 96-well plates and cultured at 37°C overnight. Subsequently, cells were treated with different concentrations (1, 2, 3 and 5 µM) of TST (cat. no. MB13332; Meilunbio) at 37°C for 48 h. Cells were incubated with 5 mg/ml MTT solution at 37°C for 4 h. Subsequently, DMSO was added to dissolve the formazan crystals. The optical density (OD) of each well was measured at a wavelength of 490 nm. Cell cytotoxicity (%) = (average OD490 of the control group-average OD490 of the experimental group)/(average OD490 of the control group-average OD490 of the blank group) ×100.
Western blotting
Cells were washed twice in cold PBS and total protein was extracted using RIPA lysate (cat. no. P0013C; Beyotime Institute of Biotechnology) containing protease inhibitor (Merck KGaA). Total protein was quantified using the bicinchoninic acid protein assay kit (cat. no. P0012S; Beyotime Institute of Biotechnology). Proteins (40 µg) were separated via 10% SDS-PAGE and transferred onto PVDF membranes, which were blocked in 5% skim milk at room temperature for 1 h. The membranes were incubated the following primary antibodies at 4°C: Anti-FoxM1 (cat. no. A2493; ABclonal Biotech Co., Ltd.), anti-Bax (cat. no. A0207; ABclonal Biotech Co., Ltd.), anti-Bcl-2 (cat. no. 2870; Cell Signaling Technology, Inc.) and anti-tubulin (cat. no. 2144; Cell Signaling Technology). Subsequently, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (cat. no. A0208; Beyotime Institute of Biotechnology) at room temperature for 1 h. Protein bands were visualized using ECL plus reagent (Thermo Fisher Scientific, Inc.) and a Chemo XRS+ luminometer (Bio-Rad Laboratories). Protein expression levels were quantified using Quantity One software (Bio-Rad Laboratories) with tubulin as the loading control.
Renilla luciferase activity
The binding sites between FoxM1 and miR-216b were predicted using TargetScan software (version 7.2.0; www.TargetScan.org/vert_72). The 1.8 kb binding sequences in the 3′UTR of FoxM1 were cloned into the pmiR-GLO vector (Promega Corporation) to construct FoxM1 3′-UTR wild-type (wt). Using the GeneTailer site-directed mutagenesis kit (Invitrogen; Thermo Fisher Scientific, Inc.), binding sequences were mutated to generate FoxM1 3′-UTR mutant (mut). U2OS and MG63 cells were seeded into 96-well plates until the confluence reached 70%, and then the cells were co-transfected by Lipofectamine® 2000 with FoxM1 3′-UTR wt or FoxM1 3′-UTR mut and miR-216b mimic or mimic-NC according to the protocol. Following transfection for 48 h, relative luciferase activity was measured using the Dual-Luciferase Reporter assay kit (Promega Corporation).
Apoptosis analysis (early and late apoptosis)
At 48 h post-transfection/TST treatment, cells were washed twice with cold PBS. Cell apoptosis was detected using the Annexin APC/7AAD kit (Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's protocol. Annexin V-positive cells were defined as apoptotic cells. Apoptotic cells were analyzing using a FACSCalibur flow cytometer (BD Biosciences) and CellQuest Pro software (version 5.1; BD Biosciences).
Statistical analysis
Statistical analyses were performed using SPSS software (version 17.0; IBM Corp.). Data are expressed as the mean ± standard deviation. All data conformed to normal distribution. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's or Bonferroni's post hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate and repeated three times.
Results
Opposite expression patterns of FoxM1 and miR-216b in osteosarcoma cells
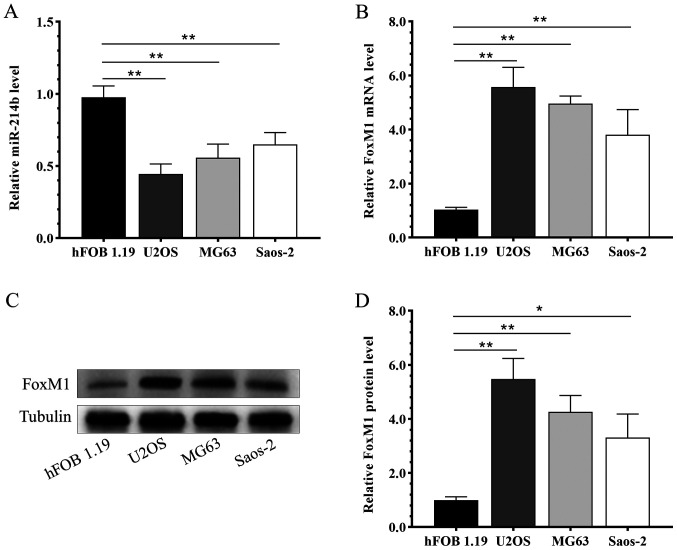
Compared with the human osteoblast cell line hFOB1.19, miR-216b was significantly downregulated in osteosarcoma cell lines U2OS, MG63 and Saos-2 (Fig. 1A). By contrast, the mRNA (Fig. 1B) and protein (Fig. 1C and D) expression levels of FoxM1 were significantly upregulated in the osteosarcoma cell lines U2OS, MG63 and Saos-2 compared with the hFOB1.19 cell line.
Figure 1.
Opposite expression patterns of FoxM1 and miR-216b in osteosarcoma cells. The expression level of (A) miR-216b and (B) FoxM1 mRNA in osteoblast and osteosarcoma cells. FoxM1 protein expression levels were (C) determined by western blotting and (D) semi-quantified. *P<0.05, **0.05≤P<0.01. FoxM1, forkhead box M1; miR, microRNA.
miR-216b targets the 3′-UTR of FoxM1
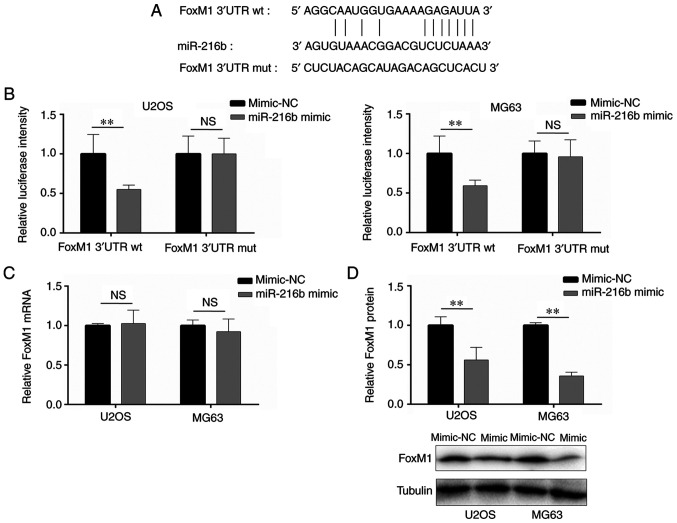
Based on a previous report (6) and online prediction, the results indicated that miR-216b bound to the 3′-UTR of FoxM1 (Fig. 2A). Compared with the mimic-NC group, miR-216b overexpression significantly decreased the luciferase activity of FoxM1 3′-UTR wt, whereas miR-216b overexpression did not significantly alter the luciferase activity of FoxM1 3′-UTR mut (Fig. 2B), indicating that miR-216b bound to the 3′-UTR of FoxM1. Additionally, the regulatory effects of miR-216b on FoxM1 expression were examined via RT-qPCR and western blotting. FoxM1 mRNA expression levels were not significantly altered by miR-216b overexpression compared with the mimic-NC group (Fig. 2C). However, miR-216b overexpression significantly decreased FoxM1 protein expression levels in U2OS and MG63 cells compared with the mimic-NC group (Fig. 2D).
Figure 2.
FoxM1 is a target of miR-216b. (A) The binding sites between miR-216b and FoxM1. (B) The interaction between miR-216b and the 3′-UTR of FoxM1 was detected by performing a dual-luciferase reporter assay. The (C) mRNA and (D) protein expression levels of FoxM1 following miR-216b overexpression. **0.05≤P<0.01. FoxM1, forkhead box M1; miR, microRNA; 3′-UTR, 3′-untranslated region; wt, wild-type; mut, mutant; NS, not significant.
Effects of TST and miR-216b on osteosarcoma cell cytotoxicity and apoptosis
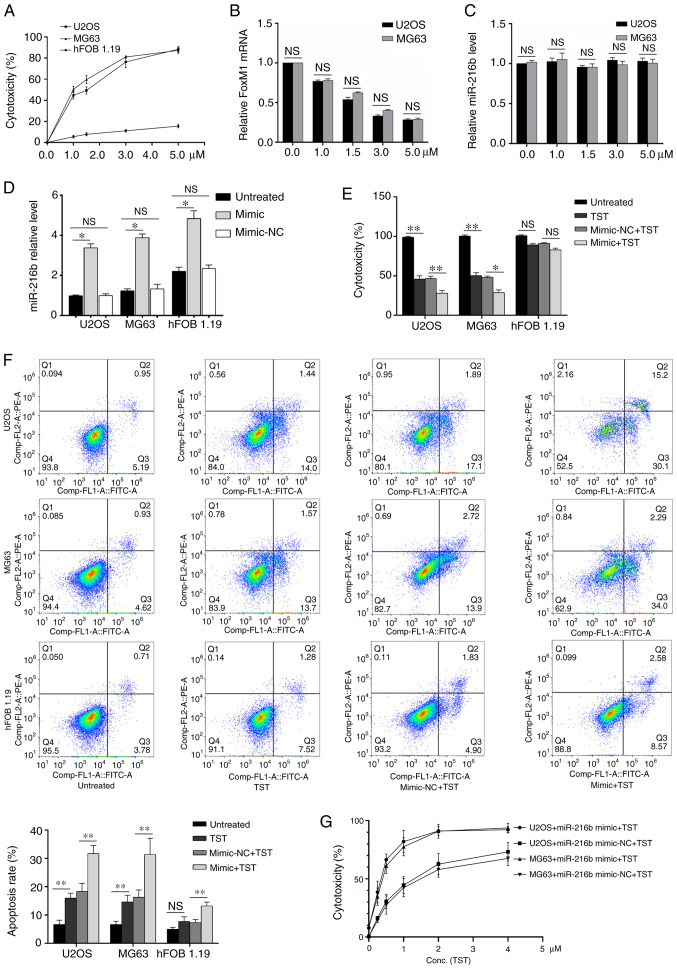
TST increased U2OS and MG63 cell cytotoxicity in a dose-dependent manner (Fig. 3A). FoxM1 mRNA expression levels were decreased by TST treatment in U2OS and MG63 cells in a dose-dependent manner compared with the control group (0 µM TST; Fig. 3B). By contrast, compared with the control group (0 µM TST), miR-216b expression levels were not notably altered by TST treatment (Fig. 3C). miR-216b overexpression significantly increased miR-126b expression levels in U2OS, MG63 and hFOB1.19 cells compared with the untreated cells (Fig. 3D). Treatment with 1 µM TST significantly decreased osteosarcoma cell cytotoxicity (Fig. 3E) and significantly increased osteosarcoma cell apoptosis (Fig. 3F) compared with untreated osteosarcoma cells. miR-216b overexpression enhanced the regulatory effects of TST on osteosarcoma cell cytotoxicity and apoptosis (Fig. 3E and F). Subsequently, osteosarcoma cells were treated with miR-126 mimic and different concentrations of TST. The results indicated that there was a notable synergistic effect between miR-216b and TST on osteosarcoma cell cytotoxicity (Fig. 3G).
Figure 3.
Effects of TST and miR-216b on cell cytotoxicity and apoptosis. (A) Effect of TST on U2OS and MG63 cell (A) cytotoxicity, (B) FoxM1 mRNA expression and (C) miR-216b expression. (D) miR-216b expression levels following transfection with miR-216b mimic or mimic-NC. The effects of TST (1 µM) and miR-216b mimic on U2OS and MG63 cell (E) cytotoxicity of miR-216b and TST treatment in normal and cancer cells; and (F) apoptosis. (G) Relationship between TST concentration and its potency. The combined effect of TST (1 µM) and miR-216b mimic on U2OS and MG63 cell cytotoxicity. *P<0.05, **0.05≤P<0.01. TST, thiostrepton; miR, microRNA; FoxM1, forkhead box M1; NC, negative control; NS, not significant.
miR-216b and TST co-regulate apoptosis-associated genes in osteosarcoma cells
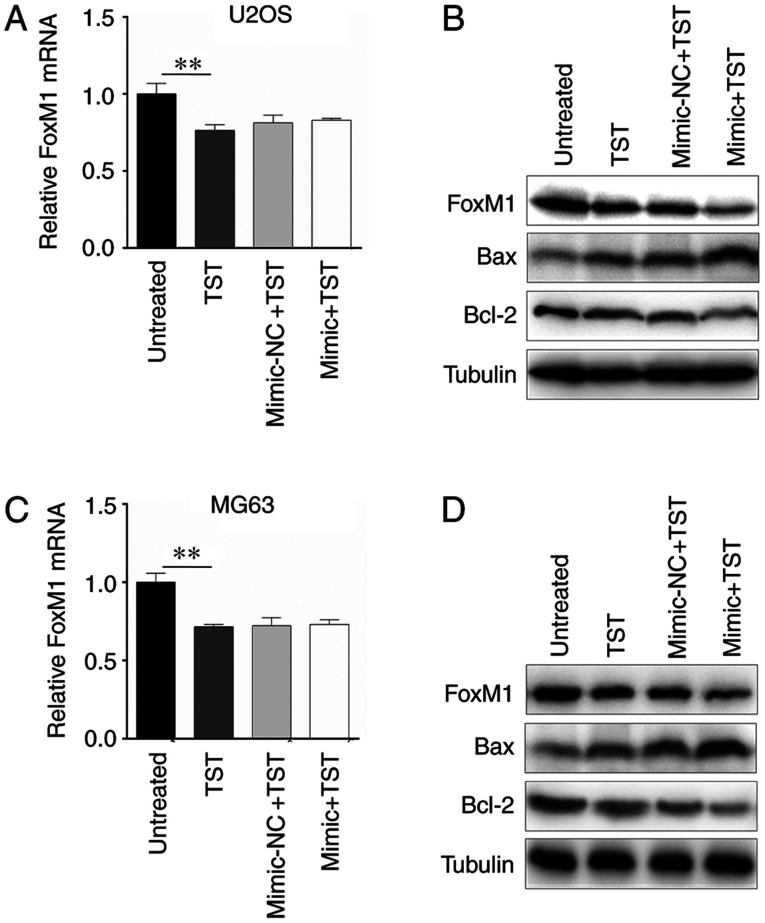
The results indicated that 1 µM TST notably downregulated the mRNA and protein expression levels of FoxM1 in U2OS cells compared with untreated cells (Fig. 4A and B). However, miR-216b overexpression only amplified the inhibitory effect of TST on FoxM1 protein expression (Fig. 4B). In addition, protein expression levels of apoptosis-associated genes Bax and Bcl-2 were detected. The western blotting results indicated that TST treatment markedly increased Bax expression and decreased Bcl-2 expression in U2OS cells compared with untreated cells (Fig. 4B); however, TST-induced alterations to Bax and Bcl-2 expression were enhanced by miR-216b overexpression. Similar results were obtained in MG63 cells (Fig. 4C and D).
Figure 4.
miR-216b and TST co-regulate Bax/Bcl-2 expression via FoxM1. FoxM1 (A) mRNA and (B) protein expression levels in U2OS cells. FoxM1 (C) mRNA and (D) protein expression levels in MG63 cells. **0.05≤P<0.01. miR, microRNA; TST, thiostrepton; FoxM1, forkhead box M1.
Discussion
As a cancerous bone tumor, osteosarcoma primarily affects the metaphysis of long bones, especially the regions around the knee, and it is highly prevalent in children and teenagers (1). Currently, the therapeutic efficacy of osteosarcoma treatment strategies is not ideal. Limb-preserving surgery is widely used for the treatment of osteosarcoma; however, even with radical amputation, 10% of patients experience local relapse and 40–50% suffer from lung metastases (20–22). Moreover, large bone defects caused by surgery do not heal easily, which seriously affects the limb function of affected patients (1). Therefore, understanding the pathogenesis and etiology of osteosarcoma is important for the development of highly specific and less toxic targeted drugs. Osteosarcoma progression is complicated, involving multiple signaling pathways, vital factors, and regulatory miRNAs and their target genes (23,24).
FoxM1 is part of the forkhead box family of transcription factors, containing a winged helix in the DNA-binding region that consists of 100 amino acids (25). FoxM1 expression is upregulated in numerous different human tumor types, which indicates its vital involvement in tumor progression (25,26). In the prognostic landscape of >18,000 genes across 39 human cancer types, the FoxM1 regulatory network is the main prognostic indicator for poor prognosis, in cancers such as lung cancer, colon cancer, liver cancer, head and neck cancer, brain cancer and others (27). Nevertheless, the exact mechanisms underlying the anticancer functions of FoxM1 are not completely understood. One potential theory suggested that FoxM1 transcription activation enhances tumorigenicity, including promoting tumor cell proliferation (28). Another theory proposed that FoxM1 supports different carcinogenetic signaling pathways, including the Wnt, SMAD3 and NF-κB signaling pathways, by interacting with other proteins (29). A relevant study indicated that FoxM1 is upregulated in osteosarcoma cells and triggers cell proliferation, migration and invasion (8). FoxM1 knockdown via RNA interference induces tumor cell proliferation and anchors independent growth (30). Meanwhile, FoxM1 silencing can inhibit tumor cell migration, invasion and tumorigenesis (28). Based on the aforementioned studies, it has been suggested that FoxM1 may serve as a novel osteosarcoma drug target. The present study indicated that FoxM1 expression was upregulated in osteosarcoma cell lines compared with the human osteoblast cell line hFOB1.19.
The diagnostic and prognostic potential of miRNAs for tumors has recently been reported, with a number of miRNAs being used a tumor biomarkers and targeted drugs (31). Moreover, miRNAs are involved in osteosarcoma-associated signaling pathway regulation, which provides a novel strategy for identifying the molecular mechanisms underlying osteosarcoma (32). The miRNAs and drug resistance relationships in osteosarcoma have been identified, which further broadens the knowledge of the structure and functions of miRNAs in the disease (33). It has been predicted that miRNAs will reverse chemotherapy resistance in future analyses (34). miR-216b is a member of the miR-216 cluster and is located on human chromosome 2p16.1 (35). A previous study demonstrated the close relationship between miR-216b and different types of human cancer such as breast cancer, cervical cancer and colorectal cancer (35). miR-216b inhibits proliferation and promotes apoptosis in pancreatic cancer and clear cell renal cell carcinoma by downregulating KRAS (36,37). In colorectal cancer, miR-216b suppresses tumors by activating the JAK2/STAT3 signaling pathway (38). miR-216b is downregulated in non-small cell lung cancer (NSCLC) tissues and cell lines. Multivariate Cox regression analysis indicated that miR-216b is an independent prognostic factor for patients with NSCLC (39). By targeting FoxM1 and SOX9, miR-216b regulates NSCLC proliferation and invasion (39,40). miR-216b is also downregulated in breast cancer samples, which suppresses breast cancer cell proliferation by targeting the purinergic receptor P2X7 (41). A recent study reported that the miR-216b/FoxM1 axis inhibits osteosarcoma proliferation, migration and metastasis (8). In the present study, miR-216b was downregulated in osteosarcoma cell lines compared with the hFOB1.19 cell line, and the results indicated that miR-216b regulated FoxM1 protein expression by binding to the 3′UTR.
TST has been widely applied in the clinic as a natural cyclic oligopeptide antibiotic of the thiopeptide class, which is derived from streptomyces (11). TST exerts a specific targeted inhibitory effect on FoxM1, but whether or not FoxM1 can be downregulated by TST poses little impact on the activity of other transcription factors (42–44). Previous studies have demonstrated that TST can inhibit proliferation and induce apoptosis in different types of malignant tumors by downregulating FoxM1, thereby indicating that TST displays strong anticancer activity (45,46) TST has a limited toxic effect on normal cells, and in vivo experiments also verified that TST treatment at an effective anticancer concentration presents few toxic effects (46–48). The combined treatment of TST, cisplatin and Bortezomib significantly improves the therapeutic sensitivity of breast cancer (49,50). Therefore, the aforementioned studies indicated that TST may serve as a potential anticancer treatment by targeting FoxM1. Previous studies have proposed that TST, as a FoxM1 inhibitor, may serve as a novel therapeutic agent for Ewing's sarcoma (17,18). The results of the present study indicated that TST reduced osteosarcoma cell cytotoxicity and negatively regulated FoxM1 mRNA expression levels.
In the present study, since FoxM1 is the common target of TST and miR-216b, osteosarcoma cells were treated with different TST concentrations and subsequently, miR-216b expression levels were detected. miR-216b expression levels were not significantly altered by TST compared with the control group. The present study also indicated that 1 µM TST displayed an inhibitory effect on osteosarcoma cytotoxicity. Subsequently, miR-216b mimic- or mimic-NC-transfected osteosarcoma cells were treated with 1 µM TST. Cytotoxicity was decreased by TST treatment in miR-216b-overexpression osteosarcoma cells compared with the control group. The combination of miR-216b mimic and TST indicated that there was a notable synergistic effect between miR-216b and TST. Furthermore, TST significantly decreased FoxM1 mRNA expression levels, which were not altered by miR-216b overexpression, compared with the control group. However, miR-216b synergistically downregulated FoxM1 protein expression levels in TST-treated osteosarcoma cells. It has been suggested that TST and miR-216b independently target FoxM1, which combined with the results of the present study indicated that TST and miR-216b may display synergistic effects on osteosarcoma progression via FoxM1.
Jin et al (51) reported that TST treatment downregulated FoxM1, matrix metallopeptidase 9 and Bcl-2 expression in a dose-dependent manner in endometriosis rat samples. By downregulating cyclin D1 and cyclin E1, TST stops cell cycle progression at the early S phase; moreover, TST suppresses laryngeal squamous cell carcinoma (LSCC) DNA synthesis in a dose- and time-dependent manner (52). Similarly, TST induces LSCC apoptosis in a dose- and time-dependent manner via cytochrome C release, Bcl-2 downregulation, and upregulation of Bax, p53, cleaved Caspase-9, cleaved Caspase-3 and cleaved poly(ADP-ribose) polymerase 1 (52). The present study assessed cell apoptosis by detecting Bax and Bcl-2 expression levels. miR-216b overexpression combined with TST treatment increased Bax expression and decreased Bcl-2 expression compared with the control group, thereby promoting osteosarcoma cell apoptosis.
Collectively, to the best of our knowledge, the present study indicated for the first time that miR-216b was downregulated in osteosarcoma cells compared with hFOB1.19 cells, and its expression was not altered by TST treatment. Moreover, the results indicated that TST and miR-216b targeted FoxM1, synergistically inhibiting cytotoxicity and stimulating apoptosis in osteosarcoma cells. Therefore, the combined used of TST and miR-216b may serve as a promising therapeutic strategy for osteosarcoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by Shanghai Municipal Health Commission (grant no. 201740160) and the Chongming District of Shanghai Science and Technology Project (grant no. CKY2017-18).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XC and JG substantially contributed to the conception and designed the work. WX and JS mainly completed the acquisition, analysis and interpretation of data for the work. HL, YL and XL were also involved in the acquisition, analysis and interpretation of data for the work. HL and YL drafted the work and revised it critically for important intellectual content. XL agreed to be accountable for the work in ensuring that questions related to the integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wu J, Sun H, Li J, Guo Y, Zhang K, Lang C, Zou C, Ma H. Increased survival of patients aged 0–29 years with osteosarcoma: A period analysis, 1984–2013. Cancer Med. 2018;7:3652–3661. doi: 10.1002/cam4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hattinger CM, Patrizio MP, Magagnoli F, Luppi S, Serra M. An update on emerging drugs in osteosarcoma: Towards tailored therapies? Expert Opin Emerg Drug. 2019;24:153–171. doi: 10.1080/14728214.2019.1654455. [DOI] [PubMed] [Google Scholar]
- 3.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Wang M, Wang N, Wang J, Qi L, Mao P. Downregulation of microRNA-216b contributes to glioma cell growth and migration by promoting AEG-1-mediated signaling. Biomed Pharmacother. 2018;104:420–426. doi: 10.1016/j.biopha.2018.05.048. [DOI] [PubMed] [Google Scholar]
- 5.Azevedo-Pouly AC, Sutaria DS, Jiang J, Elgamal OA, Amari F, Allard D, Grippo PJ, Coppola V, Schmittgen TD. miR-216 and miR-217 expression is reduced in transgenic mouse models of pancreatic adenocarcinoma, knockout of miR-216/miR-217 host gene is embryonic lethal. Funct Integr Genomics. 2017;17:203–212. doi: 10.1007/s10142-016-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jana S, Sengupta S, Biswas S, Chatterjee A, Roy H, Bhattacharyya A. miR-216b suppresses breast cancer growth and metastasis by targeting SDCBP. Biochem Biophys Res Commun. 2017;482:126–133. doi: 10.1016/j.bbrc.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Zhang L, Song Q, Chen Z. MicroRNA-216b regulates cell proliferation, invasion and cycle progression via interaction with cyclin T2 in gastric cancer. Anticancer Drugs. 2020;31:623–631. doi: 10.1097/CAD.0000000000000915. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Guo Z, Yu H, Fan L. miR-216b inhibits osteosarcoma cell proliferation, migration, and invasion by targeting Forkhead Box M1. J Cell Biochem. 2019;120:5435–5443. doi: 10.1002/jcb.29213. [DOI] [PubMed] [Google Scholar]
- 9.Zheng WW, Zhou J, Zhang CH, Liu XS. MicroRNA-216b is downregulated in hepatocellular carcinoma and inhibits HepG2 cell growth by targeting Forkhead box protein M1. Eur Rev Med Pharmacol Sci. 2016;20:2541–2550. [PubMed] [Google Scholar]
- 10.He S, Liao B, Deng Y, Su C, Tuo J, Liu J, Yao S, Xu L. miR-216b inhibits cell proliferation by targeting FOXM1 in cervical cancer cells and is associated with better prognosis. BMC Cancer. 2017;17:673. doi: 10.1186/s12885-017-3650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandu C, Ngounou Wetie AG, Darie CC, Steller H. Thiostrepton, a natural compound that triggers heat shock response and apoptosis in human cancer cells: A proteomics investigation. Adv Exp Med Biol. 2014;806:443–451. doi: 10.1007/978-3-319-06068-2_21. [DOI] [PubMed] [Google Scholar]
- 12.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7:2022–2032. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 13.Kongsema M, Wongkhieo S, Khongkow M, Lam EW, Boonnoy P, Vongsangnak W, Wong-Ekkabut J. Molecular mechanism of Forkhead box M1 inhibition by thiostrepton in breast cancer cells. Oncol Rep. 2019;42:953–962. doi: 10.3892/or.2019.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang TH, Wu ATH, Cheng TS, Lin KT, Lai CJ, Hsieh HW, Chang PM, Wu CW, Huang CF, Chen KY. In silico identification of thiostrepton as an inhibitor of cancer stem cell growth and an enhancer for chemotherapy in non-small-cell lung cancer. J Cell Mol Med. 2019;23:8184–8195. doi: 10.1111/jcmm.14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalathil D, Prasad M, Chelladurai M, John S, Nair AS. Thiostrepton degrades mutant p53 by eliciting an autophagic response in SW480 cells. J Cell Physiol. 2018;233:6938–6950. doi: 10.1002/jcp.26601. [DOI] [PubMed] [Google Scholar]
- 16.Maekawa A, Kohashi K, Kuda M, Iura K, Ishii T, Endo M, Nakatsura T, Iwamoto Y, Oda Y. Prognostic significance of FOXM1 expression and antitumor effect of FOXM1 inhibition in synovial sarcomas. BMC Cancer. 2016;16:511. doi: 10.1186/s12885-016-2542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen L, Joo J, Lee S, Wai D, Triche TJ, May WA. FOXM1 is an oncogenic mediator in Ewing Sarcoma. PLoS One. 2013;8:e54556. doi: 10.1371/journal.pone.0054556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengupta A, Rahman M, Mateo-Lozano S, Tirado OM, Notario V. The dual inhibitory effect of thiostrepton on FoxM1 and EWS/FLI1 provides a novel therapeutic option for Ewing's sarcoma. Int J Oncol. 2013;43:803–812. doi: 10.3892/ijo.2013.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Palmerini E, Torricelli E, Cascinu S, Pierini M, De Paolis M, Donati D, Cesari M, Longhi A, Abate M, Paioli A, et al. Is there a role for chemotherapy after local relapse in high-grade osteosarcoma? Pediatr Blood Cancer. 2019;66:e27792. doi: 10.1002/pbc.27792. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Zhang S. Survival of patients with primary osteosarcoma and lung metastases. J BUON. 2018;23:1500–1504. [PubMed] [Google Scholar]
- 22.Grimer RJ, Taminiau AM, Cannon SR, Surgical Subcommitte of the European Osteosarcoma Intergroup Surgical outcomes in osteosarcoma. J Bone Joint Surg Br. 2002;84:395–400. doi: 10.1302/0301-620X.84B3.0840395. [DOI] [PubMed] [Google Scholar]
- 23.Wang JY, Yang Y, Ma Y, Wang F, Xue A, Zhu J, Yang H, Chen Q, Chen M, Ye L, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. doi: 10.1016/j.biopha.2019.109627. [DOI] [PubMed] [Google Scholar]
- 24.Rickel K, Fang F, Tao J. Molecular genetics of osteosarcoma. Bone. 2017;102:69–79. doi: 10.1016/j.bone.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laoukili J, Stahl M, Medema RH. FoxM1: At the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halasi M, Gartel AL. FOX(M1) news-it is cancer. Mol Cancer Ther. 2013;12:245–254. doi: 10.1158/1535-7163.MCT-12-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gartel AL. FOXM1 in cancer: Interactions and vulnerabilities. Cancer Res. 2017;77:3135–3139. doi: 10.1158/0008-5472.CAN-16-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhat UG, Jagadeeswaran R, Halasi M, Gartel AL. Nucleophosmin interacts with FOXM1 and modulates the level and localization of FOXM1 in human cancer cells. J Biol Chem. 2011;286:41425–41433. doi: 10.1074/jbc.M111.270843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umeh-Garcia M, Simion C, Ho PY, Batra N, Berg AL, Carraway KL, Yu A, Sweeney C. A Novel bioengineered miR-127 prodrug suppresses the growth and metastatic potential of triple-negative breast cancer cells. Cancer Res. 2020;80:418–429. doi: 10.1158/0008-5472.CAN-19-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong HL, Tao Y, Mao XZ, Song DY, You D, Ni JD. MicroRNA-29a suppresses the invasion and migration of osteosarcoma cells by regulating the SOCS1/NF-κB signalling pathway through negatively targeting DNMT3B. Int J Mol Med. 2019;44:1219–1232. doi: 10.3892/ijmm.2019.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patil SL, Palat A, Pan Y, Rajapakshe K, Mirchandani R, Bondesson M, Yustein JT, Coarfa C, Gunaratne PH. MicroRNA-509-3p inhibits cellular migration, invasion, and proliferation, and sensitizes osteosarcoma to cisplatin. Sci Rep. 2019;9:19089. doi: 10.1038/s41598-019-55170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Liu D, Chen Z. Potential therapeutic implications of miRNAs in osteosarcoma chemotherapy. Tumour Biol. 2017;39:1010428317705762. doi: 10.1177/1010428317705762. [DOI] [PubMed] [Google Scholar]
- 35.Jana S, Krishna M, Singhal J, Horne D, Awasthi S, Salgia R, Singhal SS. Therapeutic targeting of miRNA-216b in cancer. Cancer Lett. 2020;484:16–28. doi: 10.1016/j.canlet.2020.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Chen W, Cai H, Hu J, Wu B, Jiang Y, Chen X, Sun D, An Y. miR-216b inhibits pancreatic cancer cell progression and promotes apoptosis by down-regulating KRAS. Arch Med Sci. 2018;14:1321–1332. doi: 10.5114/aoms.2018.72564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Dong D, Jiang S, Zhang E, Zheng W, Mao L, Li W, Zhou J, Fan L, Cheng R, et al. miR-216b post-transcriptionally downregulates oncogene KRAS and inhibits cell proliferation and invasion in clear cell renal cell carcinoma. Cell Physiol Biochem. 2018;49:1755–1765. doi: 10.1159/000493621. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Liu X, He B, Pan Y, Sun H, Xu T, Hu X, Wang S. miR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. Am J Cancer Res. 2017;7:2051–2069. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Wang Y, Du X, Yao Y, Wang L, Jia Y. miR-216b suppresses cell proliferation, migration, invasion, and epithelial-mesenchymal transition by regulating FOXM1 expression in human non-small cell lung cancer. Onco Targets Ther. 2019;12:2999–3009. doi: 10.2147/OTT.S202523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Dong H, Dai H, Liu D, Wang Z. MicroRNA-216b regulated proliferation and invasion of non-small cell lung cancer by targeting SOX9. Oncol Lett. 2018;15:10077–10083. doi: 10.3892/ol.2018.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng L, Zhang X, Yang F, Zhu J, Zhou P, Yu F, Hou L, Xiao L, He Q, Wang B. Regulation of the P2X7R by microRNA-216b in human breast cancer. Biochem Biophys Res Commun. 2014;452:197–204. doi: 10.1016/j.bbrc.2014.07.101. [DOI] [PubMed] [Google Scholar]
- 42.Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PLoS One. 2009;4:e6593. doi: 10.1371/journal.pone.0006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011;3:725–731. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 44.Gartel AL. Thiostrepton, proteasome inhibitors and FOXM1. Cell Cycle. 2011;10:4341–4342. doi: 10.4161/cc.10.24.18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 46.Ahmed M, Uddin S, Hussain AR, Alyan A, Jehan Z, Al-Dayel F, Al-Nuaim A, Al-Sobhi S, Amin T, Bavi P, Al-Kuraya KS. FoxM1 and its association with matrix metalloproteinases (MMP) signaling pathway in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012;97:E1–E13. doi: 10.1210/jc.2011-1506. [DOI] [PubMed] [Google Scholar]
- 47.Halasi M, Schraufnagel DP, Gartel AL. Wild-type p53 protects normal cells against apoptosis induced by thiostrepton. Cell Cycle. 2009;8:2850–2851. doi: 10.4161/cc.8.17.9414. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Gartel AL. Micelle-encapsulated thiostrepton as an effective nanomedicine for inhibiting tumor growth and for suppressing FOXM1 in human xenografts. Mol Cancer Res. 2011;10:2287–2297. doi: 10.1158/1535-7163.MCT-11-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, Myatt SS, Lam EW. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M, Gartel AL. Combination with bortezomib enhances the antitumor effects of nanoparticle-encapsulated thiostrepton. Cancer Biol Ther. 2012;13:184–189. doi: 10.4161/cbt.13.3.18875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin P, Chen X, Yu G, Li Z, Zhang Q, Zhang JV. The clinical and experimental research on the treatment of endometriosis with thiostrepton. Anticancer Agents Med Chem. 2019;19:323–329. doi: 10.2174/1871520618666180108100211. [DOI] [PubMed] [Google Scholar]
- 52.Jiang L, Wu X, Wang P, Wen T, Yu C, Wei L, Chen H. Targeting FoxM1 by thiostrepton inhibits growth and induces apoptosis of laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2015;141:971–981. doi: 10.1007/s00432-014-1872-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.