Abstract
As the number of infections and deaths caused by the recent COVID-19 pandemic is increasing dramatically day-by-day, scientists are rushing towards developing possible countermeasures to fight the deadly virus, SARS-CoV-2. Although many efforts have already been put forward for developing potential vaccines; however, most of them are proved to possess negative consequences. Therefore, in this study, immunoinformatics methods were exploited to design a novel epitope-based subunit vaccine against the SARS-CoV-2, targeting four essential proteins of the virus i.e., spike glycoprotein, nucleocapsid phosphoprotein, membrane glycoprotein, and envelope protein. The highly antigenic, non-allergenic, non-toxic, non-human homolog, and 100% conserved (across other isolates from different regions of the world) epitopes were used for constructing the vaccine. In total, fourteen CTL epitopes and eighteen HTL epitopes were used to construct the vaccine. Thereafter, several in silico validations i.e., the molecular docking, molecular dynamics simulation (including the RMSF and RMSD studies), and immune simulation studies were also performed which predicted that the designed vaccine should be quite safe, effective, and stable within the biological environment. Finally, in silico cloning and codon adaptation studies were also conducted to design an effective mass production strategy of the vaccine. However, more in vitro and in vivo studies are required on the predicted vaccine to finally validate its safety and efficacy.
Keywords: COVID-19, SARS-CoV-2, In silico, Immunoinformatics, Vaccine designing
1. Introduction
The Coronavirus Disease-2019 or COVID-19 is caused by a virus known as the Severe Acute Respiratory Syndrome Coronavirus- 2 (SARS-CoV-2). This disease was originated in Wuhan City of Hubei Province of China in December 2019. But now its spread is abruptly accelerated on a worldwide scale from its origin [1,2]. Coronaviruses (CoVs) are a group of immensely diversified RNA viruses with some general features i.e. enveloped and positive-sense viruses, containing ssRNA (single-stranded RNA). They are responsible for several systemic manifestations of the respiratory, hepatic, neurological as well as enteric systems. Actually, the severity of their infection varies across species [3,4]. To date, seven strains of Human Coronaviruses (HCoVs) i.e., HCoV-OC43, HCoV-HKU1, HCoV-229E, HCoV-NL63, SARS-CoV, MERS-CoV, and SARS-CoV-2 are discovered that can cause mild to severe respiratory diseases in humans [5]. But most importantly, the SARS-CoV, MERS-CoV, and SARS-CoV-2, these three HCoVs have appeared during the last two decades [6,7]. Although the SARS-CoV and MERS-CoV epidemics didn't have much impact worldwide; however, the recent COVID-19 pandemic causing SARS-CoV-2 has already caused much sufferings and claimed the lives of millions of people, let alone infecting millions of people around the world. As of October 22, 2020, the COVID-19 has disseminated in around 217 countries and territories all over the world. Not only that, it has put an end to 1,137,804 lives with almost 41 million infected cases [8].
Perhaps, the development and evaluation of specified drugs for the treatment of COVID-19 disease are going to take a couple of years. Regardless of this, repurposing a broad range of existing host-directed therapies are in course of investigation at present [9]. The available therapeutics are mainly used for the treatment of the symptomatic conditions caused by SARS-CoV-2 infection. Currently, different types of anti-viral agents i.e., remdesivir, lopinavir, ritonavir etc., antibiotics (such as azithromycin), neuraminidase inhibitors, RNA synthesis inhibitors, and plasma therapy are being used for treatment [10], [11]. However, studies have reported that most of these therapeutics showed many adverse effects on the health of the individuals on whom they were tested [11,12]. As the current therapies can only alleviate the symptoms associated with the infection, to reduce the transmission or even to eradicate this infectious virus, the development of an effective vaccination strategy has become a must-do work [13]. Currently, at least 40 amongst the prestigious pharmaceutical and research institutions from numerous countries have demonstrated their engagement in developing COVID-19 vaccines passionately after the sequenced genome of the virus has been unraveled and released officially on January 11, 2020 [14]. And some institutions have already started efficacy evaluation of their potential candidates in animals as well as clinical trials [15], [16]. Again, the Phase I clinical trial of the most current vector-based vaccine, LV-SMENP DC engaging 100 patients was performed on March 24, 2020. But the completion date of the study is estimated to be December 31, 2024 (NCT04276896) [17]. Furthermore, a conventional vaccination strategy has also been displayed using whole inactivated or live-attenuated virus vaccines. Researchers from the University of Hong Kong have come up with a live influenza vaccine that expresses proteins of SARS-CoV-2 [[18], [19]]. Besides these, “Codon de-optimization” technology has been developed by Codagenix. Basically, this technology attenuates viruses and the company is now probing for COVID-19 vaccination strategies [20]. Nonetheless, the main drawback of attenuated vaccines is that secondary mutations can promote virulence during reversion and can give rise to more serious conditions. In contrast with such vaccines, the prime advantage of subunit vaccines is that they are safe to use because the constituents of such vaccines are only the recombinant proteins or synthetic peptides from any target infectious pathogen. These vaccines do not include the whole infectious agent. Therefore, when it comes to subunit vaccines, the probability of inducing any adverse effect after administration is lesser [21]. Such type of vaccines are more important to combat the highly infectious and pathogenic viruses like the SARS-CoV-2. Currently, research is going on to develop potential subunit vaccines against some other CoVs such as SARS-CoV, MERS-CoV etc. If an attenuated or whole-inactivated type vaccine has been developed against the SARS-CoV-2, then with its high mutation rate and infectivity, there could a chance that the attenuated virus might revert to its virulence form [22,23]. Therefore, with relatively much less adverse effects on human health, scientists should give more priority to develop subunit vaccines against the SARS-CoV-2.
In this study, an effective subunit vaccine was designed against various isolates of SARS-CoV-2 from different countries around the world, utilizing the immunoinformatics approach. In immunoinformatics, the novel antigens of a pathogen or virus are identified by dissecting its genomic data and then different tools of in silico biology are used for vaccine development by analyzing its genome [[24], [25], [26]]. In our study, a blueprint of epitope-based vaccine was designed which might produce substantial immune response towards SARS-CoV-2, isolated from different countries around the world, targeting the spike glycoprotein, nucleocapsid phosphoprotein, membrane glycoprotein, and envelope protein of the virus. The protein sequences of SARS-CoV-2, isolated from Bangladesh, was used as the model to construct the vaccine. Only those epitopes which were found to be 100% conserved in some other selected countries were used for vaccine construction. As a result, the designed vaccine is also expected to be effective in different regions around the world.
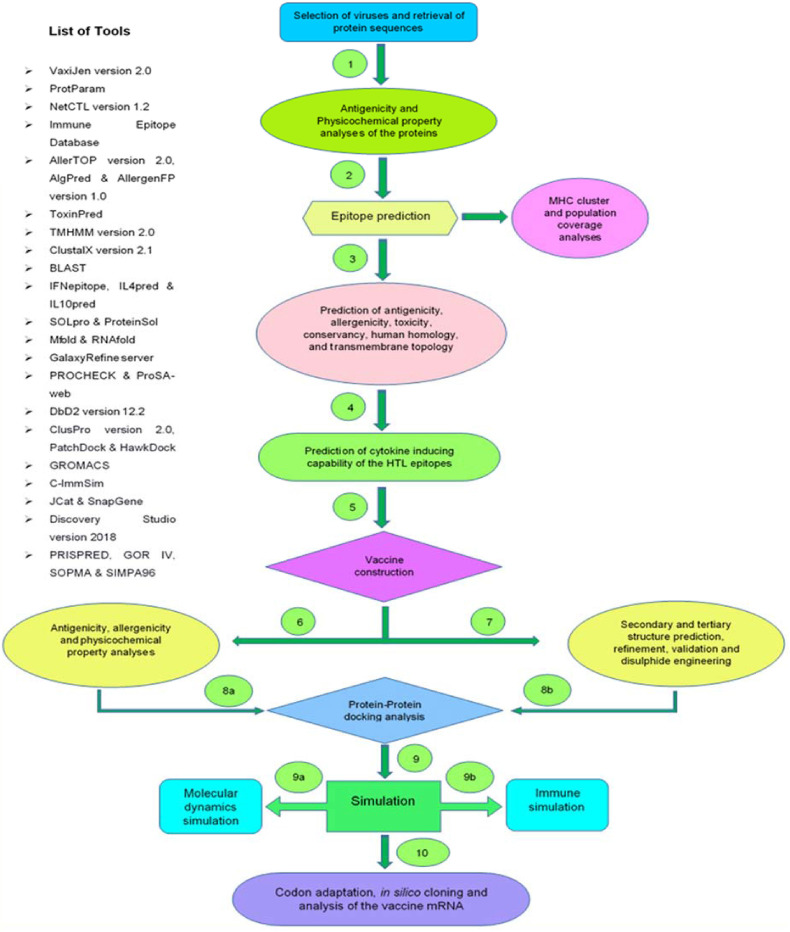
The spike glycoproteins of SARS-CoV-2 promote and facilitate the entry of the virus into the host cells and these proteins are the prime target of antibodies. The nucleocapsid phosphoprotein is vital for packaging the viral genome into a helical ribonucleocapsid (RNP) and it plays an elementary role during viral self-assembly. Also, the membrane and envelope proteins are important for viral entry, replication, budding, and particle assembly within the host cells [12], [27]. Therefore, these four proteins were used as potential targets in this study to design the vaccine with the purpose of interfering the viral life cycle. Fig. 1 represents the step-by-step procedure used in this study to design the vaccine.
Fig. 1.
The step-by-step procedure adapted in the vaccine designing study.
2. Materials and methods
2.1. Strain identification and retrieval of the protein sequences
The SARS-CoV-2 virus isolated from Bangladesh was identified and four target proteins of the virus i.e. spike glycoprotein, nucleocapsid phosphoprotein, membrane glycoprotein, and envelope protein were retrieved from the National Center for Biotechnology Information or NCBI (https://www.ncbi.nlm.nih.gov/) database.
2.2. Antigenicity prediction and physicochemical property analysis of the proteins
The antigenicity of the retrieved protein sequences of SARS-CoV-2 was predicted by the online tool, VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), using the prediction accuracy threshold of 0.4 because the 0.4 threshold has been proved to increase the prediction accuracy [28]. The algorithm of this server uses an alignment-free approach for determining the antigenicity of query peptides or proteins, which is solely based on auto cross covariance (ACC) transformation method. In this method, the general ACC calculations of the query peptides or proteins are made solely based on their physicochemical properties [[28], [29], [30]]. Thereafter, various physicochemical properties of the selected antigenic protein sequences were predicted by the ExPASy's online tool ProtParam (https://web.expasy.org/protparam/), where all the parameters were also kept at their default values [31].
2.3. Prediction of T-cell epitopes
An effective multi-epitope subunit vaccine must comprise of cytotoxic T-lymphocytic (CTL) and helper T-lymphocytic (HTL) epitopes so that after the administration, the vaccine would be able to stimulate the immune cells and generate substantial immune responses [32,33]. The MHC class-I or CTL epitopes of the selected protein sequences were predicted using the online epitope prediction server, NetCTL 1.2 (http://www.cbs.dtu.dk/services/NetCTL/) [34]. This server uses the NetCTL 1.2 statistical method for predicting the possible epitopes from a given query protein sequence. This statistical method takes into account the probability of proteasomal cleavage, TAP (transporter associated with antigen processing) transport efficiency, and MHC class-I binding, while providing quite specific and sensitive predictions. When analyzed by the Receiver Operating Characteristic (ROC) method, this method generated better results than EpiJen, WAPP, and many other epitope prediction methods [34]. The MHC class-II epitopes were predicted using another online epitope prediction server, Immune Epitope Database or IEDB (https://www.iedb.org/). The IEDB server houses a huge amount of data on antibody and T cell epitopes, experimented in humans, non-human primates, and other animal species in terms of infectious disease, allergy, auto-immunity and transplantation [35,36]. The MHC class-II restricted CD4+ HTL epitopes were obtained for the full HLA set, using the IEDB recommended 2.22 prediction method [36,37]. The top MHC class-I and MHC class-II epitopes which were found to be common for all the selected reference HLA alleles were taken for further analyses.
2.4. Antigenicity, allergenicity, toxicity, and transmembrane topology determination
In the study, a few criteria were set to select the most promising epitopes from all the epitopes predicted by the NetCTL 1.2 and IEDB servers. Only those epitopes that were found to be highly antigenic, non-allergenic, non-toxic, 100% conserved (among the selected sequences from different countries around the world), and non-homolog to the human proteome, were considered as the most promising or best-selected epitopes and only these epitopes were used in the vaccine construction process. The antigenicity of the selected epitopes was determined using the VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) server again, keeping the prediction accuracy parameter threshold 0.4 to get better predictions. Thereafter, the allergenicity of the selected epitopes was predicted using two different online tools, AllerTOP v2.0 (https://www.ddg-pharmfac.net/AllerTOP/) and AllergenFP v1.0 (http://ddg-pharmfac.net/AllergenFP/) to get better prediction accuracy. However, the results generated by AllerTOP v2.0 were given priority since the server has better prediction accuracy of 88.7% than AllergenFP server (87.9%) [38,39]. Both these severs use ACC transformation method to generate their predictions, however, the AllerTop 2.0 server classifies the query proteins by k-nearest neighbor algorithm (kNN, k = 1), on the other hand, the AllergenFP server uses the Tanimoto coefficient to determine the allergenicity or non-allergenicity of the query proteins. Both servers are developed by machine learning method, based on a training set containing 2427 known allergens and 2427 known non-allergens from different species [38,39].
After that, the toxicity prediction of the epitopes was conducted using the ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/) [40]. Like many online tools used in this study, this server gives predictions based on machine learning technique, developed by using a positive training data set containing 1805 sequences and two negative data sets: one set contains 3593 negative sequences from Swissprot and the other one has 12,541 negative sequences from TrEMBLE. Some independent data sets are also frequently used by the server. During toxicity prediction, the Support Vector Machine (SVM) method was used and all the parameters were kept default. The SVM method showed excellent performance in terms of accuracy when evaluated by the Matthew's correlation coefficient (MCC) [40]. And finally, the transmembrane topology of the selected epitopes was determined using the transmembrane topology of protein helices determinant, TMHMM v2.0 server (http://www.cbs.dtu.dk/services/TMHMM/) [41].
2.5. Conservancy and human homology prediction of the epitopes
The conservancy analysis of the selected epitopes was performed using the ‘epitope conservancy analysis’ module of the IEDB server (https://www.iedb.org/conservancy/) [42]. To further confirm the results generated by the conservancy analysis of IEDB server, the multiple sequence alignment (MSA) of the epitopes with the target proteins from different countries, was carried out by the ClustalX 2.1 tool [43]. The tool compiles different statistical methods within a single module which can be used to generate the MSA profile of target peptides or proteins. The epitopes, that were found to be 100% conserved among the selected isolates of SARS-CoV-2 from different countries across the world, were considered for vaccine construction (along with some mentioned criteria) because this will ensure the efficacy of the designed vaccine over the selected isolates. Thereafter, the homology of the epitopes to the human proteome was determined to find out any epitope that had homology with the human proteome. Only the non-homolog epitopes were taken into consideration to prevent any type of autoimmune response [44]; Adianingsih and Kharisma, 2019). The protein Basic Local Alignment Search Tool (BLAST) module (blastP) of the BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) tool was used in the human homology determination, where Homo sapiens (taxid: 9606) was used as the comparing organism, keeping all other parameter default. An e-value cut-off of 0.05 was set in the experiment and the epitopes that had no hits below the e-value inclusion threshold were selected as non-homologous peptides [45].
2.6. Cytokine inducing ability prediction of the epitopes
The helper T-cells generate several types of cytokines including IFN-gamma (interferon-gamma), IL-4 (interleukin-4), and IL-10 (interleukin-10) to activate and stimulate different types of immune cells i.e. cytotoxic T-cells, macrophages, B-cells etc. [46]. So, prediction of the cytokine inducing capability of the HTL or MHC class-II epitopes is an important criterion for vaccine construction. The IFN-gamma inducing capability of the predicted HTL epitopes was determined using IFNepitope (http://crdd.osdd.net/raghava/ifnepitope/) server. The Hybrid (Motif + SVM) prediction approach was used for determining the IFN-gamma inducing ability which is one of the highly accurate approaches for the prediction of IFN-gamma inducing capability of the epitopes. Furthermore, IL-4 and IL-10 inducing properties of the HTL epitopes were determined using IL4pred (https://webs.iiitd.edu.in/raghava/il4pred/index.php) and IL10pred (http://crdd.osdd.net/raghava/IL-10pred/) servers, respectively [47], [48], [49]. SVM method was used in both IL4pred and IL10pred predictions, keeping the threshold at default values.
2.7. Population coverage analysis
To design a multi-epitope vaccine, it is a crucial prerequisite to consider the distribution of specific HLA alleles among different populations and ethnicities around the world because the expression of different HLA alleles may vary from one population to another. The IEDB population coverage tool (http://tools.iedb.org/population/) was used to determine the population coverage of the most promising epitopes across different HLA alleles in different regions around the world [42].
2.8. Cluster analysis of the MHC alleles
Cluster analysis of the MHC alleles was carried out in order to identify of the relationship of the MHC class-I and class-II alleles used in the study. The cluster analysis of the MHC alleles was conducted using the online tool MHCcluster 2.0 (http://www.cbs.dtu.dk/services/MHCcluster/) [50]. During the analysis, all the parameters of the server were kept default and all the used HLA super-type representatives (MHC class-I), and HLA-DR representatives (MHC class-II) were selected.
2.9. Vaccine construction
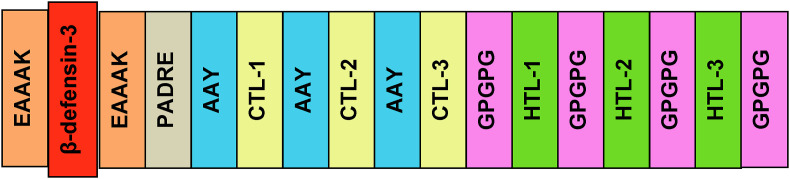
In this step, the most promising epitopes from 2.4 subsection were conjugated with one another for constructing a possible vaccine. Human beta-defensin-3, as an adjuvant sequence, was connected to the epitopes by EAAAK linkers. Adjuvants are important to enhance the antigenicity and immunogenicity of the constructed vaccines [51,52]. The pan HLA-DR epitope (PADRE) sequence was also associated with the adjuvant and epitopes. The PADRE sequence provokes the immune response by improving the capacity of the CTL epitopes of the vaccines [53,54]. Thereafter, the AAY and GPGPG linkers were used to conjugate the CTL and HTL epitopes. The EAAAK linkers are proved to provide a partition of the domains of a bifunctional fusion protein [55], whereas the GPGPG linkers are used to generate the junctional epitopes and also to enhance the immune processing and presentation [56]. And the AAY linker is also used widely in the in silico vaccine designing experiments because this linker conjugates the epitopes effectively and efficiently . Fig. 2 represents a graphical illustration of how the vaccine was designed in this study.
Fig. 2.
Schematic representation of the possible vaccine construct with linkers (EAAAK, AAY, and GPGPG), PADRE sequence, adjuvant (human beta-defensin-3), and epitopes (CTL-1, CTL-2, CTL-3, as well as HTL-1, HTL-2, HTL-3, and so on) in sequential and appropriate manner.
2.10. Antigenicity, allergenicity, and physicochemical property analyses
The antigenicity of the constructed vaccine was predicted by the online server VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), keeping the prediction threshold set at 0.4 [28]. Thereafter, both AlgPred (http://crdd.osdd.net/raghava/algpred/) and AllerTop v2.0 (https://www.ddg-pharmfac.net/AllerTOP/) servers were used to determine the allergenicity of the vaccine construct 57. Multiple Expression motifs for Motif Elicitation (MEME)/Motif Alignment & Search Tool (MAST) prediction approach was used in the allergenicity prediction of the vaccine by the AlgPred server. This server has also been developed based on machine learning technique using protein data sets for training. The algorithm of the server has been trained by clustering proteins into five different training sets and this clustering is done on the basis of similarity matrix using BLAST [58]. After that, various physicochemical properties of the vaccine were analyzed by the online server, ProtParam (https://web.expasy.org/protparam/) [31]. Finally, the solubility of the vaccine construct was determined by the SOLpro module of the SCRATCH protein predictor (http://scratch.proteomics.ics.uci.edu/) and further clarified by the Protein-sol server (https://protein-sol.manchester.ac.uk/). Both of these servers use machine learning techniques to provide much reliable predictions. During prediction, all the parameters were kept at their default values [59,60].
2.11. Secondary and tertiary structure prediction of the vaccine construct
After the physiochemical property analysis of the vaccine, it was subjected to the secondary structure prediction. Several online tools i.e., PRISPRED (http://bioinf.cs.ucl.ac.uk/psipred/) (using PRISPRED 4.0 prediction method), GOR IV (https://npsa-prabi.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_gor4.html), SOPMA (https://npsa-prabi.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_sopma.html), and SIMPA96 (https://npsaprabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_simpa96.html) were used to run the prediction keeping all the parameters default [61,62]. After that, the tertiary or 3D structure of the vaccine was predicted using RaptorX (http://raptorx.uchicago.edu/) online server. The server predicts the tertiary or 3D structure of a protein using an efficient template-based method [63].
2.12. 3D structure refinement and validation
The 3D structures generated by computational methods may not provide the true or native structures of the proteins. Therefore, the 3D structure refinement is performed to enhance the resolution of the computationally predicted models so that they can closely resemble the native protein structures. The generated 3D structure of the designed vaccine was refined by the GalaxyRefine module of the GalaxyWEB server (http://galaxy.seoklab.org/), which uses CASP10 tested refinement method for protein structure refinement [64,65]. After refining the structure, validation process of the vaccine construct was carried out by analyzing the Ramachandran plot, generated by PROCHECK (https://servicesn.mbi.ucla.edu/PROCHECK/) server [66,67] and z-score provided by another online tool, ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php). A z-score within the range of the z-scores of all the experimentally determined protein chains in the PDB database corresponds to a higher quality of a query protein [68].
2.13. Vaccine protein disulfide engineering
The vaccine protein disulfide engineering was conducted by the online tool, Disulfide by Design −2 v12.2 (http://cptweb.cpt.wayne.edu/DbD2/) [69]. The server determines the possible sites within a protein structure which have the greater possibility to form the disulfide bonds. During the prediction, the intra-chain, inter-chain, and Cβ for glycine residue were selected. The χ3 and Cα−Cβ-Sγ angles were kept at −87° or +97°± 5 and 114.6°±10, respectively and the amino acid pairs with less than 2.2 kcal/mol bond energy were selected for mutation into cysteine residues to form the disulfide bonds among themselves. The 2.2 kcal/mol was used as a threshold in this study because almost 90% of the naturally formed disulfide bonds have energy value of less than 2.2 kcal/mol [69,70].
2.14. Protein-protein docking
In protein-protein docking, the vaccine construct was docked against multiple TLRs. It's highly important for a vaccine to have a good binding affinity with the TLRs because TLR proteins generate potential immune responses after recognizing the vaccines which mimic the original viral infections. Thus, they facilitate to produce a strong immunity towards a particular virus or pathogen [71]. In this study, the vaccine construct was docked with TLRs i.e. TLR-1 (PDB ID: 6NIH), TLR-2 (PDB ID: 3A7C), TLR-3 (PDB ID: 2A0Z), TLR-4(PDB ID: 4G8A), TLR-8 (PDB ID: 3W3M), and TLR9 (PDB ID: 4QDH). The protein-protein docking was conducted using three different online tools to improve the prediction accuracy. Initially, the ClusPro v2.0 (https://cluspro.bu.edu/login.php) was used for docking where the lower energy score corresponds to the better binding affinity [[72], [73], [74], [75]]. The algorithm of the ClusPro v2.0 server calculates the energy score based on the following equation:
| E = 0.40Erep+(−0.40Eatt) + 600Eelec +1.00EDARS [72,73] |
Then the docking was again performed using PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) server and the results were refined by FireDock server (http://bioinfo3d.cs.tau.ac.il/FireDock/php.php). The algorithm of the PatchDock server is inspired by object recognition and image segmentation techniques used in artificial intelligence. When performing the docking analysis, the algorithm follows three major stages: molecular shape representation, surface patch matching, and filtering and scoring [[76], [77], [78]]. The FireDock server is mainly used in eliminating the problems associated with protein-protein docking solutions by flexible refinement. The server is based on fast rigid-body docking algorithms that refine and score the docked solutions according to an energy function in a very short amount of time (about 3.5 s per candidate solution). Thereafter, the final round of docking was performed by the HawkDock server (http://cadd.zju.edu.cn/hawkdock/) along with the Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) study [79]. The HawkDock server works on the rigid-body docking protocol of the ATTRACT algorithm, which predicts different binding poses of proteins residing in protein-protein interaction (PPI) networks. Then the scoring function of the server recognizes the near-native binding poses of the proteins with the help of the ATTRACT score. Again, the MM-GBSA analysis of the server also allows the users to analyze the key residues in the PPIs and re-rank the top docked models for reliable prediction [79,80]. Then the vaccine docked with the TLR (with the highest binding affinity) was visualized using Discovery Studio Visualizer [81].
2.15. Conformational B-cell epitope prediction
The humoral immunity, along with the cell mediated immunity, is very important to fight the pathogens inside the body. The humoral immunity of the body depends on the B-cells that produce antibodies when they encounter an antigen. Therefore, the constructed vaccine should have effective conformational B-cell epitopes so that it can provide more potent immunity. The conformational B-cell epitopes of the constructed vaccine protein were predicted by IEDB ElliPro tool (http://tools.iedb.org/ellipro/), keeping all the parameters default [82].
2.16. Molecular dynamics (MD) simulation
For observing the state changes and effects of the environment on the crystalized structure of the vaccine protein as well as protein-TLR complex structure, MD simulation was carried out. The GROMACS (GROningen MAchine for Chemical Simulations) a Linux based command-line program had been used for this purpose. Two separate simulations were run for crystalized and complex protein structures. For conducting the two simulations, first the crystalized and complex protein ‘pdb’ files were cleaned to remove any environmental substrates generated by the server. After that, the pdb2gmx was run using OPLS-AA (Optimized Potential for Liquid Simulation-All Atom) force field to generate the topology for both the structures. The structures were positioned in the center of a 2 nm sized cube, being 1 nm from each edge during their simulations. Solvation was performed by filling the box with water molecules spatially placed with a force constant of 1000 kJ mol−1 nm−2. Then the energy minimization had been conducted to stabilize the structures. From Supplementary Fig. S1, we can see that the potential energy of the structure had quickly reduced below the order of 106 for the protein complex. From this, we inferred that the complex system was stable enough to conduct further simulations. NVT (Number Volume Temperature) equilibration had been performed for 100 ps to stabilize the temperature. Afterwards, the NPT (Number Pressure Temperature) equilibration was done for 100 ps and pressure as well as density were calculated. The crystallized protein structure was also prepared in the same way. Then the resulting stabilized structures had been subjected to MD simulation of 20 ns The RMSD, RMSF of backbone of the energy minimized structures was predicted and the radius of gyration was also calculated for the two structures. All plots and simulation graphs had been analyzed using the Xmgrace and QtGrace tool. Finally, the two simulations were compared with each other.
2.17. Immune simulation
The immune simulation study of the constructed vaccine was carried out to predict its immunogenicity and immune response profile using the C-ImmSim server (http://150.146.2.1/CIMMSIM/index.php). The server predicts the real-life-like immune interactions by means of machine learning techniques and position-specific scoring matrix (PSSM) [83]. All the parameters except the time steps were kept at their default values during the experiment. However, the time steps were kept at 1, 84, and 170, and the number of simulation steps was set at 1050. So, three injections would require four weeks apart because the recommended interval between two doses of most of the commercial vaccines is proved to be four weeks [84]. The Simpson's Diversity index, D was calculated from the figures.
2.18. Codon adaptation and in silico cloning
Codon adaptation and in silico cloning are two essential steps in vaccine designing by immunoinformatics. An amino acid can be encoded by more than one codon in different organisms because the cellular machinery of an organism can be completely different from another organism, this phenomenon is known as codon bias. Therefore, codon adaptation is performed to predict the suitable codon that efficiently encodes a specific amino acid in a particular organism. In the codon adaptation study, the vaccine protein was reverse translated to the possible DNA sequence which was expected to encode the amino acids of the designed vaccine protein [84], [85]. The Java Codon Adaptation Tool or JCat server (http://www.jcat.de/) was used for the codon adaptation study of the constructed vaccine [86]. In the server, the prokaryotic E. coli strain K12 was selected as the target organism and rho-independent transcription terminators, prokaryotic ribosome binding sites, and EaeI and StyI cleavage sites of restriction enzymes were avoided at the server. Finally, the SnapGene restriction cloning software was used for inserting the newly adapted DNA sequence between the EaeI (position: from 172nd to 177th base pair) and StyI (position: from 206th to 211th base pair) restriction sites of the pETite vector [87]. The pETite vector plasmid contains ubiquitin-like modifier or SUMOtag and 6X-His tag which facilitates the solubilization and viable affinity purification of the recombinant protein [88].
2.19. Analysis of the vaccine mRNA
The mRNA secondary structure prediction was carried out using two different online tools i.e., Mfold (http://unafold.rna.albany.edu/?q=mfold) and RNAfold (http://rna.tbi.univie.ac.at/cgibin/RNAWebSuite/RNAfold.cgi). Both servers predict the mRNA secondary structures thermodynamically and generate minimal free energies for the structures [[89], [90], [91], [92]]. To analyze the mRNA folding and secondary structure of the vaccine, the optimized DNA sequence from the JCat server was converted to the possible RNA sequence by the DNA<−>RNA- > Protein tool (http://biomodel.uah.es/en/lab/cybertory/analysis/trans.htm). Then the RNA sequence was pasted in the Mfold and RNAfold servers for lower minimum free energy prediction using the default parameters.
3. Results
3.1. Identification, selection, and retrieval of viral protein sequences
The four proteins i.e. spike glycoprotein (GenBank ID: QLF97699.1), nucleocapsid phosphoprotein (GenBank ID: QLF97707.1), membrane glycoprotein (GenBank ID: QLF97702.1), and envelope protein (GenBank ID: QLF97701.1) of the SARS-CoV-2 of Bangladeshi isolate were selected and then the protein sequences were retrieved from the NCBI database.
3.2. Antigenicity prediction and physicochemical property analysis
All the query proteins were found to be antigenic in the physicochemical property analysis. Again, all of them had a similar half-life of 30 h in the mammalian cell culture system but only the nucleocapsid phosphoprotein was predicted to be unstable, having instability index over 40. All the proteins were also found to have quite high aliphatic indexes. Furthermore, the spike glycoprotein had the lowest GRAVY value of −0.077. The results of the physicochemical property analysis are listed in Table 1 .
Table 1.
The antigenicity and physicochemical property analyses of the selected viral proteins. AN; antigenicity, pI; isoelectric point, II; instability index, AI; aliphatic index, GRAVY; grand average of hydropathicity.
| Name of the protein | AN | pI | Total number of negatively charged residues | Total number of positively charged residues | Ext. coefficient | Half-life | II | AI | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| Spike glycoprotein | Antigenic | 6.32 | 109 | 103 | 148,960 | 30 h (mammalian reticulocytes, >20 h (yeast, >10 h (Escherichia coli | Stable (32.86) | 84.67 | −0.077 |
| Nucleocapsid phosphoprotein | Antigenic | 10.09 | 36 | 61 | 43,890 | 30 h (mammalian reticulocytes, >20 h (yeast, >10 h (Escherichia coli | Unstable (55.81) | 52.53 | −0.980 |
| Membrane Glycoprotein | Antigenic | 9.51 | 13 | 21 | 52,160 | 30 h (mammalian reticulocytes, >20 h (yeast, >10 h (Escherichia coli | Stable (39.14) | 120.86 | −0.446 |
| Envelope Protein | Antigenic | 8.57 | 3 | 5 | 6085 | 30 h (mammalian reticulocytes, >20 h (yeast, >10 h (Escherichia coli | Stable (38.68) | 144.00 | −0.827 |
3.3. Epitope prediction and filtering the most promising epitopes
The MHC class-I and class-II epitopes were predicted from the target protein sequences for constructing the vaccine. The epitopes that were found to follow all the previously mentioned criteria, were finally selected for vaccine construction considering as the most promising epitopes (Table 2 ). The 100% conservancy of the epitopes among the selected isolates from different regions of the world ensured that this possible vaccine should be effective in controlling the SARS-CoV-2 infections in various regions around the world along with Bangladesh. Supplementary Table S1 lists the potential epitopes of spike glycoprotein, Supplementary Table S2 lists the potential epitopes of nucleocapsid phosphoprotein, and Supplementary Table S3 and Supplementary Table S4 list the potential T-cell epitopes of membrane glycoprotein and envelope protein, respectively. Furthermore, Supplementary Table S5 lists the proteins from different countries (with their GenBank IDs) which were used in the conservancy analysis and Supplementary Fig. S2 represents the result of the MSA analysis of the most promising epitopes with the selected protein sequences from different regions of the world.
Table 2.
List of the epitopes finally selected for vaccine construction (selection criteria: antigenicity, non-allergenicity, non-toxicity, 100% conservancy and non-homolog to the human proteome).
| Protein name | MHC class-I epitopes | MHC class-II epitopes |
|---|---|---|
| Spike Glycoprotein | VLPFNDGVY | QSLLIVNNATNVVIK |
| WTAGAAAYY | VLSFELLHAPATVCG | |
| GAAAYYVGY | VVLSFELLHAPATVC | |
| QLTPTWRVY | – | |
| STECSNLLL | – | |
| VLKGVKLHY | – | |
| Nucleocapsid phosphoprotein | SSPDDQIGY | IAQFAPSASAFFGMS |
| SPDDQIGYY | GTRNPANNAAIVLQL | |
| DLSPRWYFY | – | |
| LSPRWYFYY | – | |
| Membrane Glycoprotein | LVGLMWLSY | IKLIFLWLLWPVTLA |
| ATSRTLSYY | VGLMWLSYFIASFRL | |
| AGDSGFAAY | KLIFLWLLWPVTLAC | |
| Envelope Protein | VSLVKPSFY | LLFLAFVVFLLVTLA |
| – | VLLFLAFVVFLLVTL | |
| – | LFLAFVVFLLVTLAI | |
| – | AFVVFLLVTLAILTA | |
| – | VNSVLLFLAFVVFLL | |
| – | NSVLLFLAFVVFLLV | |
| – | SVLLFLAFVVFLLVT | |
| – | FLLVTLAILTALRLC | |
| – | FLAFVVFLLVTLAIL | |
| – | LAFVVFLLVTLAILT |
3.4. Cytokine inducing ability prediction of the epitopes
IFN-gamma, IL-4, and IL-10 inducing capacity prediction of the HTL epitopes showed that many of the selected HTL epitopes had the capability of inducing at least one of these cytokine. Moreover, all the most promising epitopes were also found to have at least one cytokine production capability (Supplementary Table S1, S2, S3, and S4).
3.5. Population coverage analysis
The population coverage analysis showed that the MHC class-I and class-II epitopes covered 90.42% and 93.23%, respectively and the combined MHC class-I and class-II covered 84.56% of the world population. India was found to possess the highest percentage of population coverage of both the MHC class-I epitopes (93.49%) and MHC class-II epitopes (91.20%). And the highest percentage of population coverage of the MHC class-I and class-II epitopes in combination was obtained by China (85.67%) (Supplementary Fig. S3).
3.6. Cluster analysis of the MHC alleles
The cluster analysis of the possible MHC class-I and MHC class-II alleles, that may interact with the predicted epitopes of the selected proteins, was carried out using the online tool MHCcluster 2.0. The tool demonstrates the relationship of the clusters of the alleles in a phylogenetic manner. Supplementary Figure S4 illustrates the result of the experiment where the red zone indicates a strong interaction and the yellow zone represents a weaker interaction.
3.7. Vaccine construction
The vaccine has been constructed using the most promising epitopes which could be used to fight against the selected viral isolates effectively. EAAAK, AAY, and GPGPG linkers were used at their appropriate positions during conjugating the epitopes. The newly constructed vaccine candidate were designated as: CV (Table 3 ).
Table 3.
Vaccine constructed against the SARS-CoV-2. The bolded letters represent the linker sequences.
| Name of the vaccine | Vaccine Construct |
|---|---|
| “CV” vaccine | EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKAKFVAAWTLKAAAAAYVLPFNDGVYAAYWTAGAAAYYAAYGAAAYYVGYAAYQLTPTWRVYAAYSTECSNLLLAAYVLKGVKLHYAAYSSPDDQIGYAAYSPDDQIGYYAAYDLSPRWYFYAAYLSPRWYFYYAAYLVGLMWLSYAAYATSRTLSYYAAYAGDSGFAAYAAYVSLVKPSFYGPGPGQSLLIVNNATNVVIKGPGPGVLSFELLHAPATVCGGPGPGVVLSFELLHAPATVCGPGPGIAQFAPSASAFFGMSGPGPGGTRNPANNAAIVLQLGPGPGIKLIFLWLLWPVTLAGPGPGVGLMWLSYFIASFRLGPGPGKLIFLWLLWPVTLACGPGPGLLFLAFVVFLLVTLAGPGPGVLLFLAFVVFLLVTLGPGPGLFLAFVVFLLVTLAIGPGPGAFVVFLLVTLAILTAGPGPGVNSVLLFLAFVVFLLGPGPGNSVLLFLAFVVFLLVGPGPGSVLLFLAFVVFLLVTGPGPGFLLVTLAILTALRLCGPGPGFLAFVVFLLVTLAILGPGPGLAFVVFLLVTLAILTGPGPG |
3.8. Antigenicity, allergenicity, and physicochemical property analyses of the vaccine Candidate
The CV candidate vaccine was predicted to be a potent antigen as well as a non-allergen. In the physicochemical property analysis of the vaccine, a high (basic) theoretical pI, half-life of 1 h in the mammalian cells, and more than 10 h in the E. coli cell culture system were predicted. Moreover, the vaccine construct was also found to be quite stable as well as soluble upon overexpression in E. coli cell culture system. Furthermore, it had an aliphatic index of 113.03 and a quite low GRAVY value of −0.843 (Table 4 ).
Table 4.
Antigenicity, allergenicity and physicochemical property analysis of the vaccine construct. AN; antigenicity, AG; allergenicity, pI; isoelectric point, II; instability index, AI; aliphatic index, GRAVY; grand average of hydropathicity.
| Name of the protein | AN | AG | pI | Total number of negatively charged residues | Total number of positively charged residues | Ext. coefficient | Half-life | II | AI | GRAVY | Solubility |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CV | Antigenic | Non-allergenic | 9.24 | 14 | 30 | 117,745 | 1 h (mammalian reticulocytes, 20 min (yeast, >10 h (Escherichia coli | Stable (32.45) | 113.03 | - 0.843 | Soluble (SolPro: 0.760, Protein-Sol: 0.668) |
3.9. Secondary and tertiary structure prediction of the vaccine candidate
In the secondary structure analysis of the vaccine candidate, the amino acid percentage of α-helix, β-strand, and coil structure of the vaccine protein was predicted using four different online servers. The vaccine had the highest percentage of the amino acids in the coil structure, considering the results from all the servers. The amino acid percentages of the vaccine are given with a tabular representation (Supplementary Table S6 and Supplementary Fig. S5).
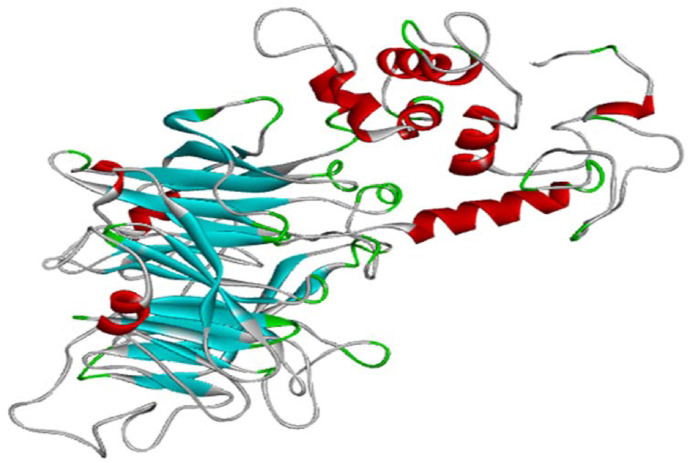
The 3D structure of the CV vaccine construct was predicted by the online server RaptorX. The vaccine had 5 domains with quite low p-value of 3.56e-08, which declared that the quality of the prediction 3D structure was quite good because according to the documentation of the RaptorX server, the smaller p-value represents the higher quality the model [26], [93]. The homology modeling of the vaccine construct was performed using 1KJ6A as template from the Protein Data Bank. The 3D structure of CV is illustrated in Fig. 3 .
Fig. 3.
The tertiary structure of the CV vaccine.
3.10. Protein 3D structure refinement and validation of the vaccine candidate
The protein structure generated by the RaptorX server was subjected to the refinement process using the Galaxy-web server, which was then analyzed by Ramachandran plot generated by the PROCHECK server and the z-score predicted by the ProSA-web server. The Ramachandran plot analysis depicted that CV vaccine had 86.2% of the amino acids in the most favored region, 11.7% of the amino acids in the additional allowed regions, 0.8% of the amino acids in the generously allowed regions, and 1.3% of the amino acids in the disallowed regions. Moreover, CV had the z-score of −5.75 which lies within the range of experimentally proven X-ray crystal structures of proteins from the Protein Data Bank (PDB) (Supplementary Fig. S6).
3.11. Vaccine protein disulfide engineering
In protein disulfide engineering, only those amino acid pairs were selected which had a bond energy value of less than 2.2 kcal/mol. The CV generated four pairs of amino acids with bond energy less than 2.2 kcal/mol i.e. 93 Ala and 431 Phe, 369 Tyr and 396 Cys, 435 Thr and 528 Ala, 459 Gly and 468 Phe. The selected amino acid pairs formed the mutant version (with disulfide bonds) of the original vaccine in the DbD2 server (Supplementary Figure S7). Since CV was predicted to have four possible pairs of amino acid residues with the capability to form potential disulfide bonds, therefore, it can be considered as a quite stable vaccine construct.
3.12. Protein-protein docking study
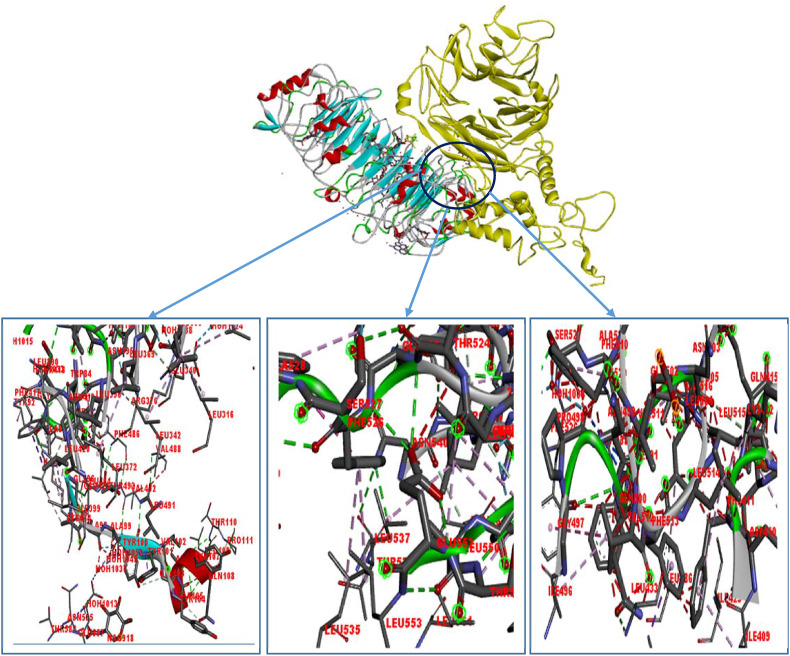
The protein-protein docking study was performed to analyze the ability of the designed vaccine to interact with different TLRs which would occur during an actual immune response. CV vaccine showed a very high binding affinity when docked by ClusPro 2.0. For better prediction, it was further docked using two other servers i.e. PatchDock and HawkDock servers, respectively. ClusPro 2.0 server showed the highest binding affinity with TLR-8 (−1261.2 kcal/mol). Again, when analyzed by PatchDock and FireDock server, CV vaccine again showed the best global energy score with TLR-8 (−32.29 kcal/mol). Furthermore, the docking and MM-GBSA analyses by HawkDock server also predicted the highest binding affinity of the vaccine protein with TLR-8 (Table 5 ). Since all of the servers predicted very good binding affinities of the CV vaccine with the TLR-8, therefore, the visualization of the interaction (Fig. 4 ) and molecular dynamics (MD) simulation was performed only for the CV-TLR-8 docked complex.
Table 5.
Results of molecular docking study of the CV vaccine interacted with the TLR-8. MM-GBSA; Molecular Mechanics-Generalized Born Surface Area.
| Target TLRs (with PDB IDs) | ClusPro energy score (the lowest energy in kcal mol−1) | Global energy (PatchDock server) | HawkDock score (the lowest score) | MM-GBSA (binding free energy, in kcal mol−1) |
|---|---|---|---|---|
| TLR-1 (6NIH) | −1131.7 | −8.30 | −5701.54 | −43.28 |
| TLR-2 (3A7C) | −1021.3 | −24.33 | −4102.30 | −66.69 |
| TLR-3 (2A0Z) | −923.5 | −11.73 | −3708.21 | −49.22 |
| TLR-4 (4G8A) | −955.1 | −2.92 | −5568.56 | −65.08 |
| TLR-8 (3W3M) | −1261.2 | −32.29 | −5819.85 | −112.41 |
| TLR-9 (4QDH) | −1139.0 | −6.02 | −3988.37 | −109.86 |
Fig. 4.
The interaction between TLR-8 (receptor protein on left in variable color) and the CV vaccine (ligand protein on right in yellow color). Here the interacting amino acid pairs are: Leu 342 (receptor)-Glu 340 (ligand), Ala 99 (receptor)– Glu 555 (ligand), Ala 97 (receptor)- Ile 496 (ligand), Phe 486 (receptor)- Leu 372 (ligand), Leu 342 (receptor)- Val 488 (ligand), Leu 535 (receptor)-Phe 526 (ligand), Tyr 563 (receptor)-Asp 543 (ligand), Lys 333 (receptor)-Asp 543 (ligand), Ile 409 (receptor)- Leu 514 (ligand), Lys 412 (receptor)- Leu 515 (ligand), Leu 433 (receptor)- Ile 496 (ligand). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.13. Conformational B-cell epitope prediction
The conformational B-cell epitope prediction of the CV vaccine candidate determined four potential regions of the vaccine with scores ranging from 0.537 to 0.774 and covering total 329 amino acids (Supplementary Table S7).
3.14. MD simulation of CV-TLR-8 docked complex
The CV-TLR-8 protein complex was selected for the MD simulation. One chain had a mass of 85724.286 amu, charge −10.00 e, and 751 residues. The other chain had a mass of 62965.010 amu, charge 15.00 e, and 601 residues. A total of 92,459 water molecules was added to the system after solvation from which 5 were replaced by CL ions during ionization to neutralize the system's charge. Energy minimization had been completed in 2383 steps when the steepest descent had converged and the force had reached <1000 kJ/mol. The average potential energy was calculated to be −5.2177795e+06 kJ/mol with a maximum force of 9.6646100e+02 on atom 8176. From the temperature equilibration plot of Supplementary Fig. S8 (a), it is clear that the target minimization value of 300 K remained stable over the remainder of the equilibration and fluctuated by only ±1.37 K. The pressure value was also predicted which showed fluctuations around 0 bar with a range of ±100 bar and the average pressure had been found to be - 1.64581 bar. Similarly, density had also been calculated over 100 ps and the average density had been found as 1067.77 kgm3 (Supplementary Fig. S8). The density values are mostly stable over time, indicating that the system had been well equilibrated.
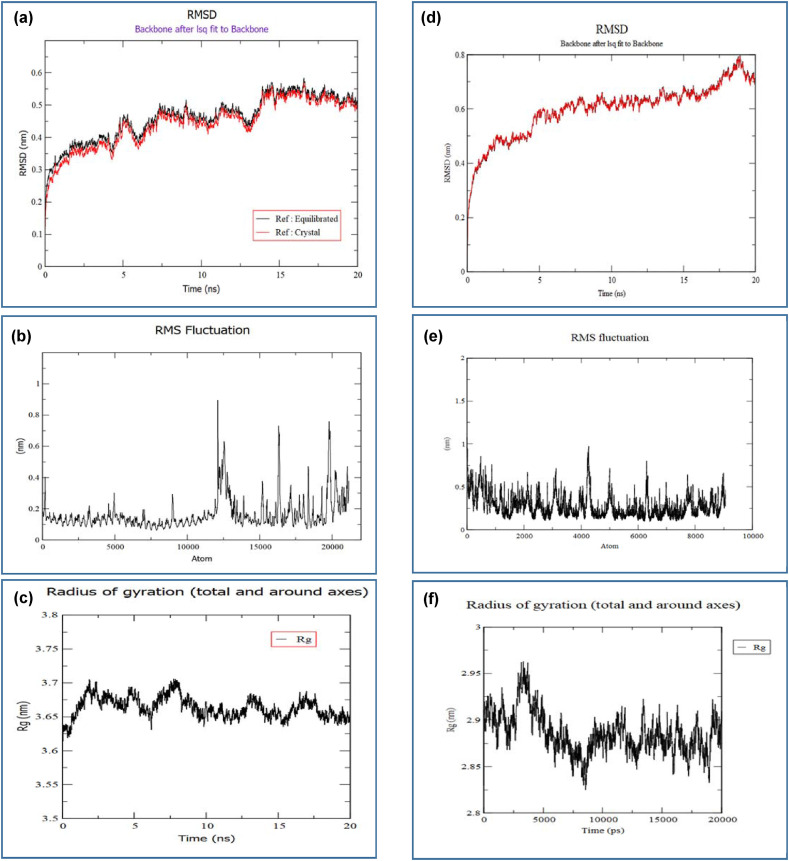
Trajectory analysis was conducted and thereafter the RMSD, RMSF, and the radius of gyration were also calculated after completion of the 20 ns simulation (Fig. 5 ). A plot of the RMSD backbone has been depicted in Fig. 5a which revealed that RMSD levels had gone up to ~0.6 nm. The black line of the graph refers to the RMSD relative to the structure present in the minimized, equilibrated system and the red line is the RMSD relative to the crystal structure. Since both these plots are almost highly correlated, so it can be declared that the structure remained quite stable during the experiment. The RMSF and radius of gyration graphs also point to the fact that the CV-TLR-8 complex was quite stable during the experiment (Fig. 5b & c). Like the complex structure, the single vaccine crystal structure also generated very good results in the MD simulation. In the RMSD graph of Fig. 5d, it can be noticed that the RMSD value of the crystal structure had gone up to ~0.8 nm, which is quite normal for such single crystallized structure. Again, from the RMSF and radius of gyration graphs of the structure also pointed towards the fact that the crystallized structure didn't have any significant region with the possibility to undergo deformation, so it can be deduced that the crystalized vaccine CV might be quite stable in the biological environment (Fig. 5e & f). So, overall, both structures (complex and crystallized vaccine structure) showed quite acceptable and sound performance in the MD simulation study.
Fig. 5.
The results of the MD simulation of the CV-TLR-8 complex and also the only crystal structure of the vaccine. Here, (a) RMSD plot of backbone of the complex showing the complex structure had maintained a stable structure with minimum fluctuations. (b) RMS Fluctuations of all the atoms of the complex at their average positions. The peaks and dips in the graph denote the flexibility of the corresponding region in the molecular structure. (c) Radius of gyration of the protein complex. (d) RMSD plot of backbone of the crystal structure showing the structure had maintained a stable structure with minimum fluctuations. (e) RMS Fluctuations of all the atoms of the crystal structure at their average positions. (f) Radius of gyration of the crystalized vaccine structure.
3.15. Immune simulation
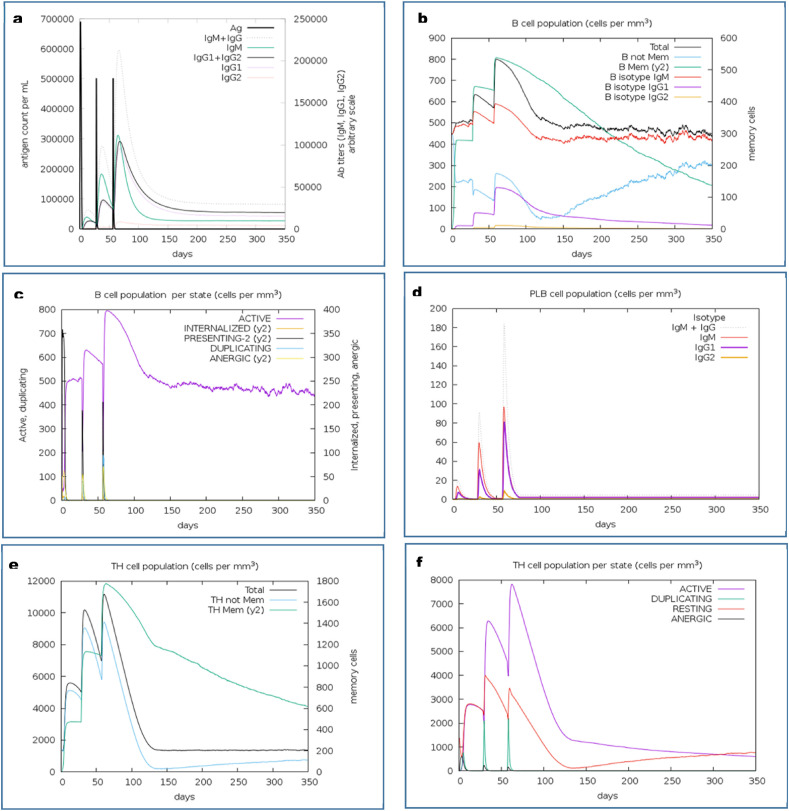
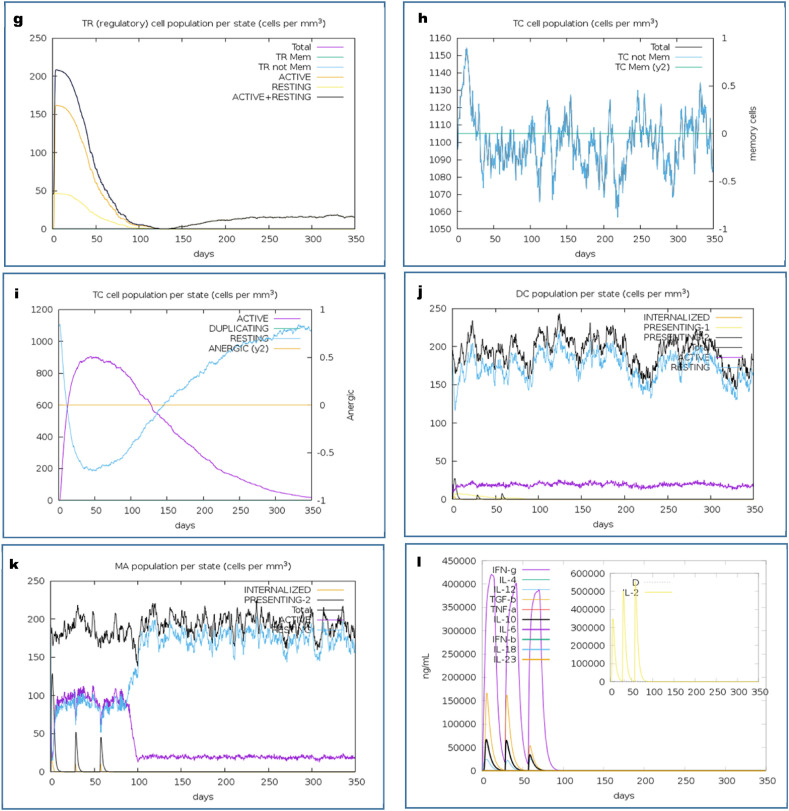
The immune simulation of the CV vaccine candidate was analyzed by the C-ImmSimm server which determines the stimulation of adaptive immunity as well as the immune interactions of the epitopes with their specific targets [46]. The immune simulation study expressed that, after administrating the three doses of the vaccine, the primary immune response against the vaccine was found to be stimulated significantly as indicated by the gradual increase in the levels of different immunoglobulins (Fig. 6 a). Again, the successive augmentation in the concentrations of active B-cell (Fig. 6b and c), plasma B-cell (Fig. 6d), helper T-cell (Fig. 6e and f), regulatory T-cell, and cytotoxic T-cell (Fig. 6g, h and i) were also predicted, which represents the capability of the vaccine to create a very potent secondary immune response. Furthermore, the increase in the concentration of macrophages and dendritic cells indicated a very potent presentation of antigen by these antigen-presenting cells (APCs) (Fig. 6j and k). The CV vaccine was also predicted to produce different types of cytokines (Fig. 6l). Henceforth, the overall immune simulation study revealed that, the CV vaccine might be able to stimulate strong immunogenic response after its administration.
Fig. 6.
C-IMMSIMM representation of the immune simulation of the best predicted vaccine, CV. (a) The immunoglobulin and immunocomplex response to the CV vaccine inoculations (lines colored in black) and specific subclasses are indicted by colored lines, (b) Rise in the B-cell population over the course of the three injections, (c) Increment in the B-cell population per state over the course of vaccination, (d) Increase in the plasma B-cell population over the course of the injections, (e) Enhancement of the helper T-cell population over the course of the three injections, (f) Increment in the helper T-cell population per state over the course of the vaccination, (g) Elevation in the regulatory T lymphocyte over the course of the three injections, (h) Increment in the cytotoxic T lymphocyte population over the course of the injections, (i) Increase in the active cytotoxic T lymphocyte population per state over the course of the three injections, (j) Rise in the active dendritic cell population per state over the course of the three injections, (k) Increment in the macrophage population per state over the course of the injections, (l) Augmentation in the concentrations of different types of cytokines over the course of the three injections.
3.16. Codon adaptation, in silico cloning, and analysis of the vaccine mRNA structure
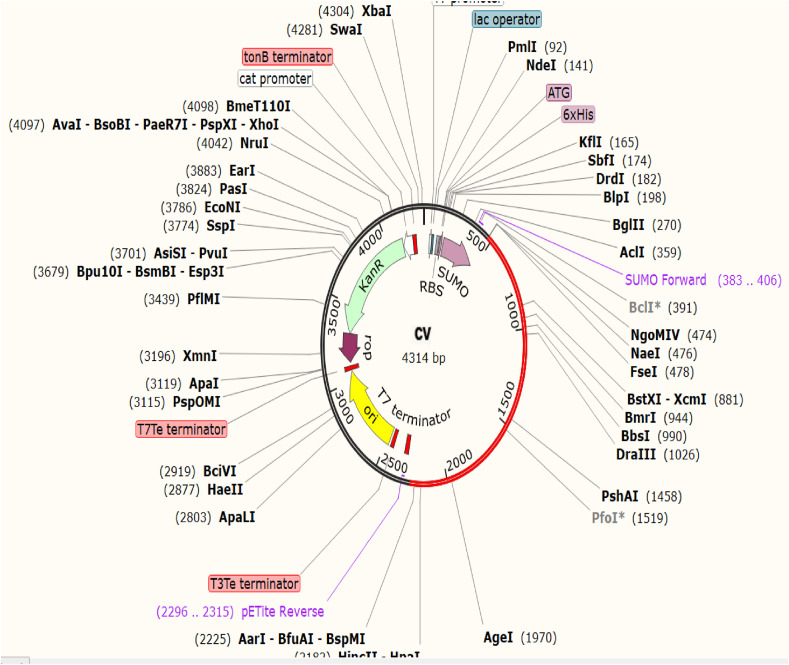
For in silico cloning and plasmid construction, at first the protein sequence of the CV vaccine was adapted by the JCat server. The codon adaptation index (CAI) value of CV was found to be 0.903 which indicated that the DNA sequence contained a higher proportion of the codons that are most likely to be used in the target organism E. coli strain K12 (codon bias) for efficient production the CV vaccine (Supplementary Fig. S9) [90,92]. A good GC content of 56.79% was also recorded for the adapted sequence. After codon adaptation, the predicted DNA sequence of CV vaccine was inserted into the pETite vector plasmid. The plasmid contains sequences of the SUMO tag and 6X His tag, for this reason, these sequences are expected to facilitate the purification of the CV vaccine during the downstream processing [94] (Fig. 7 ). Thereafter, the secondary structure of the CV mRNA was predicted by Mfold and RNAfold servers. The Mfold server generated a minimum free energy score of −517.50 kcal/mol, which was in agreement with the prediction of the RNAfold server (−531.10 kcal/mol). Supplementary Fig. S10 depicts the vaccine mRNA structure predicted by the RNAfold server.
Fig. 7.
Constructed pETite vector plasmid with the CV vaccine insert (marked in red color). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Vaccines are widely produced pharmaceutical products that are currently being administered worldwide to combat various pathogenic infections. Developing vaccines by conventional means is a time consuming process which sometimes takes several years to achieve desired results [95]. However, the finesse of modern technology and the accessibility of the genomic data of about all the pathogens have saved the time of vaccine development and made it possible to develop the novel peptide-based “subunit vaccines”, which are comprised of only the antigenic protein segments of the target pathogen and thus the toxic or allergenic parts of an antigen can be eliminated during vaccine designing and development [96]. Again, vaccine designing through these computation-based methods also saves time and cost of designing and developing processes to a greater extent [97,98]. In this study, the methods of immunoinformatics were exploited to design an epitope-based polyvalent vaccine against the SARS-CoV-2 virus, isolated from Bangladesh, targeting the spike glycoprotein, nucleocapsid phosphoprotein, membrane glycoprotein, and envelope protein of the virus.
After identifying the target proteins, their antigenicity and physicochemical properties were analyzed. In the antigenicity test, all the selected proteins were found to be antigenic which should aid in the antigenic response of the vaccine. The theoretical pI refers to the pH at which a protein contains no integrated charge. All the proteins except the spike glycoprotein were found to possess basic theoretical pI (pH more than 7.0), which should be quite achievable. The aliphatic index of a protein determines the relative volume of the amino acids in its side chains occupied by the aliphatic amino acids i.e. valine, alanine, etc. and higher aliphatic index of a protein represents its more thermostable state [57], [99], [100]. All the proteins had quite high aliphatic indexes, so they were considered to be quite thermostable. The GRAVY value determines the hydrophilic and hydrophobic nature of a compound. The negative GRAVY value represents the hydrophilic characteristics, whereas the positive value indicates the hydrophobic characteristics of a compound [101]. With the negative GRAVY value, all the proteins were found to be hydrophilic in nature so they should be easily soluble in water. All the proteins generated sound and satisfactory results in the physicochemical property analysis.
After analyzing the physicochemical properties, the possible T-cell epitopes were determined to construct the vaccine. The cytotoxic T-cells recognize the antigens, whereas the helper T-cells are involved in activating the B-cell, macrophages, and even the cytotoxic T-cells [32,33,102]. Furthermore, the cell mediated immune response also provides a life-long immunity by secreting antiviral cytokines and by recognizing and destroying the infected cells [100,103]. A set of filters were set to select the most promising T-cell epitopes from all the epitopes predicted by the servers. Only the highly antigenic (so that the epitopes would generate sufficient immunogenic response), non-allergenic (so that the epitopes won't be able to cause any unwanted allergenic reaction), non-toxic, 100% conserved, and non-homolog (to the human proteome to prevent autoimmunity from setting in after the administration of the designed vaccine) epitopes were considered for constructing the vaccine. Again, cytokines i.e. the IFN-gamma, IL-10, and IL-4 are very important to establish a network among the cells of the immune system during immunogenic responses as they activate and mediate the proper functioning of the immune cells [46]. Therefore, the prediction of the cytokine producing ability of the HTL epitopes is important before the vaccine construction. Almost all the HTL epitopes as well as the all the most promising epitopes were predicted to be the inducer of at least one cytokine (among IFN-gamma, IL-10, and IL-4). This capability would largely contribute to the immunogenic activities of the constructed vaccine. Again, the population coverage analysis demonstrated that very good portions of the world population, about 90.42% and 93.23%; respectively, as well as the population from different countries (with India and China containing the highest percentage of population individually), have occupied the selected MHC-I and MHC-II epitopes and their alleles. With this excellent result, it can be deduced that our designed vaccine should be effective on more than 90% people of the world population in combating the SARS-CoV-2 infection. Again, in the cluster analysis of the MHC-I alleles, quite good correlation between HLA-A02:01, HLA-A02:06, HLA-A29:02, and HLA-A01:01 was observed. On the other hand, the HLA-A11:01 and HLA-A03:01 showed relatively less correlation with the other MHC-I alleles. However, a very good correlation among all the selected MHC-II alleles i.e., DRB5:0101, DRB1:1501, DRB1:0401, DRB3:0101, and DRB1:0301 was found. Therefore, it can be concluded that in the cluster analysis, all the MHC alleles showed sound and satisfactory performance.
The most promising epitopes obtained by filtering from the previous step, were conjugated together using different linkers i.e., EAAAK, AAY, and GPGPG at appropriate positions. During vaccine construction, the EAAAK linker was added to the start or N-terminal region of the vaccine to protect it from degradation [104]. The final vaccine candidate was designated as “CV”. In the antigenicity and allergenicity analyses, the CV vaccine was found to be antigenic as well as non-allergenic. Hence, the vaccine might generate robust immune response as well as cause no allergic reaction within the body. Moreover, in the physicochemical property analysis of the vaccine, it was found to be basic with a high theoretical pI, which should be achievable. The aliphatic index refers to the protein's thermal stability and thus, the higher the aliphatic index of a protein is, the more thermostable state it acquires [105]. Therefore, the CV vaccine could be considered as a thermostable vaccine protein due to having a high aliphatic index (114.48). Again, the vaccine candidate was found to have negative GRAVY value which revealed that it might be hydrophilic. And along with the hydrophilic nature of the vaccine, it was also found to have a half-life of more than 10 h in E. coli, for this reason, the mass production and purification of the vaccine in the E. coli cell culture system should be much easier. Furthermore, the vaccine protein was found to be soluble upon over-expression in E. coli by both of the servers (SolPro and Protein-Sol). While using E. coli as a host, solubility is an important factor to be considered during the production of recombinant proteins. This is because if a recombinant protein is not soluble, then that protein may become non-functional due to lack of proper folding or it may form insoluble inclusion bodies. And the E. coli culture system is one of the most widely used and recommended systems for mass production of recombinant proteins [106]. Therefore, in this study, we have also recommended E. coli as the host for mass production of our designed vaccine candidate. And the instability index (31.49) of the vaccine protein dictated that it might be quite stable in the biological environment because a compound with instability index less than 40 is considered to be stable [100,107]. Considering all these aspects of the physicochemical property analysis, it can be considered that the predicted vaccine might be quite effective and responsive as a potential vaccine candidate.
After the physicochemical property analysis, the secondary and tertiary structures of the CV vaccine were determined and thereafter, the protein refinement and validation steps were also performed. The refined vaccine protein was predicted to have very good amount of the amino acids in the favored regions of the Ramachandran plot analysis and the protein also had a remarkable z-score which pointed towards a good quality structure of the protein. Thereafter, the disulfide engineering of the vaccine was performed where four amino acid pairs with less than 2.2 kcal/mol bond energy were selected for mutation into cysteine residues to form the disulfide bonds among themselves. Therefore, with these four pairs of amino acids capable of forming disulfide bonds, CV can be considered as a stable vaccine candidate.
The protein-protein docking of the CV vaccine candidate with different TLRs was performed using several online tools. The docking step is one of the necessary steps in the vaccine designing experiment because it determines the possible interaction of the constructed CV vaccine with different TLRs, which might occur during immune response. The docking experiment, performed by all the servers showed that the CV vaccine had good capability to interact with all the selected TLRs, with the highest score generated by TLR-8. Therefore, after the docking study, the MD simulation study of the CV-TLR-8 was carried out because CV vaccine generated the best docking results with the TLR-8. MD simulation is performed to simulate a biological environment for the vaccine and analyze the physical movements and interactions of the atoms of a protein complex with environment molecules for a fixed length of time, which provides a view of the dynamic evolution of the system. The results of an MD simulation show how stable the vaccine complex (in this case, CV-TLR-8) is in terms of changing pressure, temperature, and motion. In our experiment, a low average potential energy of −5.2177795e+06 kJ/mol, an average temperature fluctuation of only ±1.3 K, and also quite desired RMSD, RMSF as well as the radius of gyration were obtained which pointed towards the fact that the energy minimized CV-TLR-8 structure might be quite stable in the biological environment. The MD simulation also predicted that the crystalized vaccine CV structure should also be stable within the biological environment with quite acceptable values in the experiment.
Thereafter, the immune simulation study of the candidate vaccine was performed which predicted that the vaccine might generate an immune response which is consistent with the typical and natural immune system. After the vaccine was administrated, the primary immune response was found to be stimulated, which might stimulate the secondary immune response in later stages. The CV vaccine was found to stimulate both humoral and cell-mediated mediated immune responses as indicated by the increase in the levels of the cytotoxic T-cells, helper T-cells, memory B-cells, plasma B-cells, and different antibodies [108,17]. Again, the rise in the concentration of APCs like the macrophages and dendritic cells also indicated a very good antigen presentation. Moreover, the augmentation in the cytokine profile was also reported which represents the possibility of the vaccine to produce effective immune response against the viral invasion [[109], [110], [111]]. The negligible Simpson index (D) predicted a diverse immune response of the vaccine CV [83]. Overall, the CV vaccine was found to be effective and satisfactory in the results of the immune simulation study.
Finally, the codon adaptation and in silico cloning were performed to design a recombinant plasmid which might be used for the mass production of the CV vaccine in the E. coli strain K12. In the codon adaptation study of the vaccine, the obtained results were found to be satisfactory with the CAI value of 0.903 and the GC content of 56.79%, because any CAI value over 0.80 and the GC content within 30%–70% are considered to be good scores [85], [111], [112], [113]. Following that, the optimized CV DNA sequence was inserted into the pETite plasmid. During stability prediction of the vaccine mRNA secondary structure, both Mfold and RNAfold servers generated negative and much lower minimal free energies of −517.50 kcal/mol and −531.10 kcal/mol, respectively. Therefore, with these lower minimal free energies, it can be declared that the predicted CV vaccine might be quite stable upon transcription in vivo [114].
The genome based technologies for developing subunit vaccines will continue to dominate the field of immunology and vaccinology since it allows the researchers to have an opportunity to develop vaccines by optimizing their target antigens [115], [116]. The field of immunoinformatics is still an emerging field and it has a long way to go. Research is going on to develop potential subunit vaccines against almost all the infectious diseases and in many cases positive results are observed with minimal harmful effects, which is why such subunit vaccines are gaining more attention and acceptance among the common population. The monovalent subunit vaccine, RTS,S/AS01E (Mosquirix™) and the Hepatitis B surface antigen (HBsAg) have gained market access and mass acceptance recently. Exploiting the immunoinformatics approach during development, these two vaccines are used as preventative measures against malaria and Hepatitis B virus, respectively. These two subunit vaccines have showed positive results in many studies and therefore, such vaccines will certainly pave the way of developing potential subunit vaccines to fight various infectious pathogens [117], [118], [119], [120].
Overall, this study suggests the designed CV vaccine as a safe and effective epitope based subunit vaccine to fight against the SARS-CoV-2 virus, based on the strategies employed in the study. However, results obtained from such computational studies are regarded only as “predictions” and therefore, we can't be 100% sure that these results will be similar in the biological environment. Although the techniques and tools of modern bioinformatics can provide predictions with very high accuracy, but still in vivo and in vitro studies are required to finally confirm the outcomes of such in silico studies. Therefore, we suggest wet-lab based studies to finally validate the safety and efficacy of our designed vaccine. If positive findings are achieved in the wet-lab based studies, then our experiment should definitely open new avenues to design a subunit vaccine against the SARS-CoV-2 virus.
5. Conclusion
The recent COVID-19 pandemic has caused the sufferings of millions of people worldwide, while claiming the lives of hundreds of thousands of people. Although researchers are rushing to develop potential counter-measure to fight the virus, however, till now, all the efforts have shown potential drawbacks. Different types of antiviral therapies are also being tried all over the world, but their outcomes are also raising serious questions about their usage. Moreover, the vaccines that are now under development or various stages of trials, are inactivated or live-attenuated vaccines, which can have serious consequences if any unwanted reversal back to the virulence form occurs. Therefore, in this study, a blue-print of a potential vaccine against the SARS-CoV-2 has been designed targeting four distinctive proteins i.e., spike glycoprotein, nucleocapsid phosphoprotein, membrane glycoprotein, and envelope protein. During the vaccine construction, only those epitopes which were found to be highly antigenic (so that the vaccine would be able to generate robust immune response), non-allergenic (so that the vaccine won't cause any harmful reaction within the body), non-toxic, non-human homolog, and 100% conserved epitopes (so that vaccine would be effective against different isolates around the world) were used. Numerous in silico validations conducted in the study also indicated that, the designed vaccine might be quite safe and effective as well as well responsive to use. If satisfying results are achieved in the wet-lab based studies, then this epitope-based vaccine candidate might provide a relatively cheap and effective option to reach the entire world to combat the COVID-19 pandemic.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to the COVID Research Cell (CRC), Wazed Miah Science Research Centre, Jahangirnagar University, for providing the support to conduct this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100478.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Islam H., Rahman A., Masud J., Shweta D.S., Araf Y., Ullah M.A., Sium S.M., Sarkar B. A generalized overview of SARS-CoV-2: where does the current knowledge stand? Electron J Gen Med. 2020;17(6) doi: 10.29333/ejgm/8258. [DOI] [Google Scholar]
- 2.Ullah M.A., Araf Y., Sarkar B., Moin A.T., Reshad R.A., Hasanur M.D. 2020. Pathogenesis, diagnosis and possible therapeutic options for COVID-19. [DOI] [Google Scholar]
- 3.Chan J.F., Lau S.K., Woo P.C. The emerging novel Middle East respiratory syndrome coronavirus: the "knowns" and "unknowns. J Formos Med Assoc. 2013;112(7):372–381. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses -drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y., Li C., Chen L., Xu B., Zhou Y., Cao L., Shang Y., Fu Z., Chen A., Deng L., Bao Y. A novel human coronavirus OC43 genotype detected in mainland China. Emerg Microb Infect. 2018;7(1):1–4. doi: 10.1038/s41426-018-0171-5. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1? Accessed on: 22 October, 2020.
- 9.Amanat F., Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amanat F., Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu R., Wang L., Kuo H.C., Shannar A., Peter R., Chou P.J., Li S., Hudlikar R., Liu X., Liu Z., Poiani G.J. An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Rep. 2020 doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos I.D., Grosche V.R., Bergamini F.R., Sabino-Silva R., Jardim A.C. Antivirals against coronaviruses: candidate drugs for SARS-CoV-2 treatment? Front Microbiol. 2020;11:1818. doi: 10.3389/fmicb.2020.01818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehman M., Tauseef I., Aalia B., Shah S.H., Junaid M., Haleem K.S. Therapeutic and vaccine strategies against SARS-CoV-2: past, present and future. Future Virol. 2020 Jul;15(7):471–482. doi: 10.2217/fvl-2020-0137. [DOI] [Google Scholar]
- 14.Zhang N., Li C., Hu Y., Li K., Liang J., Wang L., Du L., Jiang S. Current development of COVID-19 diagnostics, vaccines and therapeutics. Microb Infect. 2020 doi: 10.1016/j.micinf.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moin A.T., Sakib M.N., Araf Y., Sarkar B., Ullah M.A. 2020. Combating COVID-19 pandemic in Bangladesh: a memorandum from developing country. Preprints. [DOI] [Google Scholar]
- 16.Kaiser Permanente Washington Health Research Institute, Kaiser Permanente, Washington Health Research Institute . 2020. Kaiser Permanente launches first coronavirus vaccine trial. Seattle: 16 March. [Internet] Retrieved: 23 March 2020. [Google Scholar]
- 17.Carvalho L.H., Sano G.I., Hafalla J.C., Morrot A., De Lafaille M.A.C., Zavala F. IL-4-secreting CD4+ T cells are crucial to the development of CD8+ T-cell responses against malaria liver stages. Nat Med. 2002;8(2):166–170. doi: 10.1038/nm0202-166. [DOI] [PubMed] [Google Scholar]
- 18.Cheung E. South China Morning Post; 2020. China coronavirus: Hong Kong researchers have already developed vaccine but need time to test it, expert reveals.https://www.scmp.com/news/hong-kong/health-environment/article/3047956/china-coronavirus-hongkong-researchers-have Accessed 28 Feb 2020. [Google Scholar]
- 19.Inovio Pharmaceuticals Inovio collaborating with Beijing advaccine to advance INO-4800 vaccine against new coronavirus in China. 2020. http://ir.inovio.com/news-and-media/news/press-release-details/2020/IVI-INOVIO-and-KNIH-to-Partner-with-CEPI-in-Phase-12-Clinical-Trial-of-INOVIOs-COVID-19-DNA-Vaccine-in-South-Korea/default.aspx
- 20.Shieber J. Tech Crunch; 2020. Codagenix raises $20 million for a new flu vaccine and other therapies.https://techcrunch.com/2020/01/13/codagenix-raises-20-million-for-a-new-flu-vaccine-and-othertherapies/ 16 August. [Google Scholar]
- 21.Sarkar B., Islam S.S., Zohora U.S., Ullah M.A. Virus like particles-A recent advancement in vaccine development. J Microbiol Soc. 2019 Dec;55(4):327–343. doi: 10.7845/kjm.2019.9089. [DOI] [Google Scholar]
- 22.Khalaj-Hedayati A. Protective immunity against SARS subunit vaccine candidates based on spike protein: lessons for coronavirus vaccine development. J Immunol Res. 2020:2020. doi: 10.1155/2020/7201752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong L.C., Khan A.M. Vaccine target discovery. 2019. [DOI]
- 25.Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3(5):445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar B., Ullah M.A., Johora F.T., Taniya M.A., Araf Y. Immunoinformatics-guided designing of epitope-based subunit vaccine against the SARS Coronavirus-2 (SARS-CoV-2) Immunobiology. 2020 doi: 10.1016/j.imbio.2020.151955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Futur Med. 2019;14(4):275–286. doi: 10.2217/fvl-2018-0144. [Internet] Apr 1 [cited 2020 Jun 10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doytchinova I.A., Flower D.R. Bioinformatic approach for identifying parasite and fungal candidate subunit vaccines. Open Vaccine J. 2008 Sep;1(1):4. [Google Scholar]
- 30.Ullah A., Sarkar B., Islam S.S. Exploiting the reverse vaccinology approach to design novel subunit vaccine against ebola virus. Immunobiology. 2020:151949. doi: 10.1016/j.imbio.2020.151949. [DOI] [PubMed] [Google Scholar]
- 31.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. The proteomics protocols handbook. Humana press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [DOI] [Google Scholar]
- 32.Chaudhri G., Quah B.J., Wang Y., Tan A.H., Zhou J., Karupiah G., Parish C.R. T cell receptor sharing by cytotoxic T lymphocytes facilitates efficient virus control. Proc Natl Acad Sci Unit States Am. 2009;106(35):14984–14989. doi: 10.1073/pnas.0906554106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J., Paul W.E. CD4 T cells: fates, functions, and faults. Blood. J Am Soc Hematol. 2008;112(5):1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Large-scale validation of methods for cytotoxic, lymphocyte epitope prediction T.-, Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. BMC Bioinf. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkar B., Ullah M.A., Araf Y. A systematic and reverse vaccinology approach to design novel subunit vaccines against dengue virus type-1 and human Papillomavirus-16. Inf Med Unlocked. 2020:100343. doi: 10.1016/j.imu.2020.100343. [DOI] [Google Scholar]
- 36.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P., Sidney J., Kim Y., Sette A., Lund O., Nielsen M., Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinf. 2010;11(1):568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitrov I., Flower D.R., Doytchinova I. April. AllerTOP-a server for in silico prediction of allergens. BMC Bioinf. 2013;14(6) doi: 10.1186/1471-2105-14-s6-s4. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 40.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R. Consortium, OSDD; raghava, GPS in silico approach for predicting toxicity of peptides and proteins. PloS One. 2013;8 doi: 10.1371/journal.pone.0073957. e73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Bui H.H., Sidney J., Li W., Fusseder N., Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinf. 2007;8(1):361. doi: 10.1186/1471-2105-8-361nehka. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Kharisma V.D., Ansori A.N. Construction of epitope-based peptide vaccine against SARS-CoV-2: immunoinformatics study. J Pure Appl Microbiol. 2020;14(suppl 1):999–1005. doi: 10.22207/JPAM.14.SPL1.38. [DOI] [Google Scholar]
- 45.Mehla K., Ramana J. Identification of epitope-based peptide vaccine candidates against enterotoxigenic Escherichia coli: a comparative genomics and immunoinformatics approach. Mol Biosyst. 2016;12(3):890–901. doi: 10.1039/c5mb00745c. [DOI] [PubMed] [Google Scholar]
- 46.Luckheeram R.V., Zhou R., Verma A.D., Xia B. 2012. CD4+ T cells: differentiation and functions. Clinical and developmental immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhanda S.K., Vir P., Raghava G.P. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013 Dec;8(1):30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagpal G., Usmani S.S., Dhanda S.K., Kaur H., Singh S., Sharma M., Raghava G.P. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep. 2017;7 doi: 10.1038/srep42851. 42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhanda S.K., Gupta S., Vir P., Raghava G.P. Prediction of IL4 inducing peptides. Clin Dev Immunol. 2013 doi: 10.1155/2013/263952. 2013 Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomsen M., Lundegaard C., Buus S., Lund O., Nielsen M. MHCcluster, a method for functional clustering of MHC molecules. Immunogenetics. 2013;65(9):655–665. doi: 10.1007/s00251-013-0714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S., Nguyen M.T. Recent advances of vaccine adjuvants for infectious diseases. Immune Netw. 2015;15(2):51–57. doi: 10.4110/in.2015.15.2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meza B., Ascencio F., Sierra-Beltrán A.P., Torres J., Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: an in silico approach. Infect Genet Evol. 2017;49:309–317. doi: 10.1016/j.meegid.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Pandey R.K., Sundar S., Prajapati V.K. Differential expression of miRNA regulates T cell differentiation and plasticity during visceral leishmaniasis infection. Front Microbiol. 2016;7:206. doi: 10.3389/fmicb.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C.Y., Monie A., Pang X., Hung C.F., Wu T.C. Improving therapeutic HPV peptide based vaccine potency by enhancing CD4+ T help and dendritic cell activation. J Biomed Sci. 2010;17:88. doi: 10.1186/1423-0127-17-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14(8):529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 56.Saadi M., Karkhah A., Nouri H.R. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect Genet Evol. 2017;51:227–234. doi: 10.1016/j.meegid.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980;88(6):1895–1898. doi: 10.1093/oxfordjournals.jbchem.a133168. [DOI] [PubMed] [Google Scholar]
- 58.Saha S., Raghava G.P. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34(suppl_2):W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hebditch M., Carballo-Amador M.A., Charonis S., Curtis R., Warwicker J. Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magnan C.N., Randall A., Baldi P. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics. 2009;25(17):2200–2207. doi: 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- 61.Buchan D.W., Jones D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47(W1):W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 63.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat protoc. 2012;7:1511. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ko J., Park H., Heo L., Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40(W1):W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nugent T., Cozzetto D., Jones D.T. Evaluation of predictions in the CASP10 model refinement category. Proteins: Struct Funct Bioinf. 2014;82:98–111. doi: 10.1002/prot.24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laskowski R.A., MacArthur M.W., Thornton J.M. PROCHECK: validation of protein-structure coordinates. 2006. [DOI]
- 67.Morris A.L., MacArthur M.W., Hutchinson E.G., Thornton J.M. Stereochemical quality of protein structure coordinates. Proteins: Struct Funct Bioinf. 1992;12(4):345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 68.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(suppl_2):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craig D.B., Dombkowski A.A. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinf. 2013;14(1):346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petersen M.T.N., Jonson P.H., Petersen S.B. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- 71.Stern L.J., Calvo-Calle J.M. HLA-DR: molecular insights and vaccine design. Curr Pharmaceut Des. 2009;15:3249–3261. doi: 10.2174/138161209789105171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozakov D., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Vajda S. How good is automated protein docking? Proteins: Struct Funct Bioinf. 2013;81:2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohebbi A., Askari F.S., Ebrahimi M., Zakeri M., Yasaghi M., Bagheri H., Javid N. Susceptibility of the Iranian population to severe acute respiratory syndrome coronavirus 2 infection based on variants of angiotensin I converting enzyme 2. Future Virol. 2020 Aug;15(8):507–514. doi: 10.2217/fvl-2020-0160. [DOI] [Google Scholar]
- 75.Vajda S., Yueh C., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Kozakov D. New additions to the C lus P ro server motivated by CAPRI. Proteins: Struct Funct Bioinf. 2017;85:435–444. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Atapour A., Mokarram P., MostafaviPour Z., Hosseini S.Y., Ghasemi Y., Mohammadi S., Nezafat N. Designing a fusion protein vaccine against HCV: an in silico approach. Int J Pept Res Therapeut. 2019;25:861–872. doi: 10.1007/s10989-018-9735-4. [DOI] [Google Scholar]
- 77.Duhovny D., Nussinov R., Wolfson H.J. International workshop on algorithms in bioinformatics. Springer; Berlin, Heidelberg: 2002. Efficient unbound docking of rigid molecules; pp. 185–200. [DOI] [Google Scholar]
- 78.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(suppl_2):W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Biovia D.S., Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Richmond T.J. Dassault systèmes BIOVIA, discovery Studio visualizer, v. 17.2, san Diego: Dassault systèmes, 2016. J Chem Phys. 2000;10 0021-9991. [Google Scholar]
- 82.Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9(1):514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rapin N., Lund O., Bernaschi M., Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PloS One. 2010;5(4) doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castiglione F., Mantile F., De Berardinis P., Prisco A. How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Comput Math Methods Med. 2012 doi: 10.1155/2012/842329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khatoon N., Pandey R.K., Prajapati V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. DOI: doi.org/10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep. 2018;8(1):1–19. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biswal J.K., Bisht P., Mohapatra J.K., Ranjan R., Sanyal A., Pattnaik B. Application of a recombinant capsid polyprotein (P1) expressed in a prokaryotic system to detect antibodies against foot-and-mouth disease virus serotype O. J Virol Methods. 2015;215:45–51. doi: 10.1016/j.jviromet.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 89.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The vienna RNA websuite. Nucleic Acids Res. 2008;36(suppl_2):W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288(5):911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 91.Mathews D.H., Turner D.H., Zuker M. RNA secondary structure prediction. Curr Protoc Nucl Acid Chem. 2007;28(1):11–12. doi: 10.1002/0471142700.nc1102s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.2020. http://raptorx.uchicago.edu/documentation/ 22 October.
- 94.Carbone A., Zinovyev A., Képes F. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. doi: 10.1093/bioinformatics/btg272. doi.org/10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- 95.María R.R., Arturo C.J., Alicia J.A., Paulina M.G., Gerardo A.O. InTech; Rijeka, Croatia: 2017. The impact of bioinformatics on vaccine design and development. [Google Scholar]
- 96.Sarkar B., Ullah M.A., Araf Y., Das S., Hosen M.J. Blueprint of epitope-based multivalent and multipathogenic vaccines: targeted against the dengue and zika viruses. J Biomol Struct Dyn. 2020:1–21. doi: 10.1080/07391102.2020.1804456. [DOI] [PubMed] [Google Scholar]
- 97.Oli A.N., Obialor W.O., Ifeanyichukwu M.O., Odimegwu D.C., Okoyeh J.N., Emechebe G.O., Adejumo S.A., Ibeanu G.C. Immunoinformatics and vaccine development: an overview. ImmunoTargets Ther. 2020;9:13. doi: 10.2147/ITT.S241064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rappuoli R., Bottomley M.J., D'Oro U., Finco O., De Gregorio E. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J Exp Med. 2016;213:469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Enany S. Structural and functional analysis of hypothetical and conserved proteins of Clostridium tetani. J Inf Publ Health. 2014;7(4):296–307. doi: 10.1016/j.jiph.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 100.Sarkar B., Ullah M.A., Araf Y., Das S., Rahman M.H., Moin A.T. Designing novel epitope-based polyvalent vaccines against herpes simplex virus-1 and 2 exploiting the immunoinformatics approach. J Biomol Struct Dyn. 2020:1–21. doi: 10.1080/07391102.2020.1803969. [DOI] [PubMed] [Google Scholar]
- 101.Chang K.Y., Yang J.R. Analysis and prediction of highly effective antiviral peptides based on random forests. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saha C.K., Hasan M.M., Hossain M.S., Jahan M.A., Azad A.K. In silico identification and characterization of common epitope-based peptide vaccine for Nipah and Hendra viruses. Asian Pac J Trop Med. 2017;10(6):529–538. doi: 10.1016/j.apjtm.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 103.Cano R.L.E., Lopera H.D.E. El Rosario University Press; 2013. Introduction to T and B lymphocytes. In autoimmunity: from bench to bedside [internet] [PubMed] [Google Scholar]
- 104.Khan M., Khan S., Ali A., Akbar H., Sayaf A.M., Khan A., Wei D.Q. Immunoinformatics approaches to explore Helicobacter Pylori proteome (Virulence Factors) to design B and T cell multi-epitope subunit vaccine. Sci Rep. 2019;9(1):1–3. doi: 10.1038/s41598-019-49354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Panda S., Chandra G. Physicochemical characterization and functional analysis of some snake venom toxin proteins and related non-toxin proteins of other chordates. Bioinformation. 2012;8(18):891. doi: 10.6026/97320630008891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sørensen H.P., Mortensen K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Factories. 2005;4(1):1. doi: 10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Farhadi T. Effectiveness assessment of protein drugs and vaccines through in Silico analysis. Biomed Biotechnol Res J (BBRJ) 2018;2(2):106. doi: 10.4103/bbrj.bbrj_18_18. [DOI] [Google Scholar]
- 108.Almofti Y.A., Abd-elrahman K.A., Gassmallah S.A.E., Salih M.A. Multi epitopes vaccine prediction against severe acute respiratory syndrome (SARS) coronavirus using immunoinformatics approaches. Am J Microbiol Res. 2018;6(3):94–114. doi: 10.12691/ajmr-6-3-5. [DOI] [Google Scholar]
- 109.Hoque M.N., Istiaq A., Clement R.A., Sultana M., Crandall K.A., Siddiki A.Z., Hossain M.A. Metagenomic deep sequencing reveals association of microbiome signature with functional biases in bovine mastitis. Sci Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-49468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kambayashi T., Laufer T.M. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat Rev Immunol. 2014;14(11):719–730. doi: 10.1038/nri3754. [DOI] [PubMed] [Google Scholar]
- 111.Shey R.A., Ghogomu S.M., Esoh K.K., Nebangwa N.D., Shintouo C.M., Nongley N.F., Asa B.F., Ngale F.N., Vanhamme L., Souopgui J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019;9(1):1–18. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morla S., Makhija A., Kumar S. Synonymous codon usage pattern in glycoprotein gene of rabies virus. Gene. 2016;584(1):1–6. doi: 10.1016/j.gene.2016.02.047. [DOI] [PubMed] [Google Scholar]
- 113.Murina V., Kasari M., Takada H., Hinnu M., Saha C.K., Grimshaw J.W., Seki T., Reith M., Putrinš M., Tenson T., Strahl H. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J Mol Biol. 2019;431(18):3568–3590. doi: 10.1016/j.jmb.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hamasaki‐Katagiri N., Lin B.C., Simon J., Hunt R.C., Schiller T., Russek‐Cohen E., Komar A.A., Bar H., Kimchi‐Sarfaty C. The importance of mRNA structure in determining the pathogenicity of synonymous and non‐synonymous mutations in haemophilia. Haemophilia. 2017;23(1):e8–e17. doi: 10.1111/hae.13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Merten O.W. Virus contaminations of cell cultures–a biotechnological view. Cytotechnology. 2002;39(2):91–116. doi: 10.1023/A:1022969101804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tameris M.D., Hatherill M., Landry B.S., Scriba T.J., Snowden M.A., Lockhart S., Shea J.E., McClain J.B., Hussey G.D., Hanekom W.A., Mahomed H. MVA85A 020 Trial Study Team: safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013;381(9871):1021–1028. doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Adhikari U.K., Rahman M.M. Overlapping CD8+ and CD4+ T-cell epitopes identification for the progression of epitope-based peptide vaccine from nucleocapsid and glycoprotein of emerging Rift Valley fever virus using immunoinformatics approach. Infect Genet Evol. 2017;56:75–91. doi: 10.1016/j.meegid.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adianingsih O.R., Kharisma V.D. Study of B cell epitope conserved region of the Zika virus envelope glycoprotein to develop multi-strain vaccine. J Appl Pharmaceut Sci. 2019 Jan;9 doi: 10.7324/JAPS.2019.90114. 098-103. [DOI] [Google Scholar]
- 119.https://www.cdc.gov/hepatitis/index.htm. Accessed on: 22 October, 2020.
- 120.Oyarzún P., Kobe B. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum Vaccines Immunother. 2016;12(3):763–767. doi: 10.1080/21645515.2015.1094595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.