Abstract
HIV-1 Nef plays an essential role in enhancing virion infectivity by antagonizing the host restriction molecule SERINC5. Because Nef is highly polymorphic due to the selective forces of host cellular immunity, we hypothesized that certain immune-escape polymorphisms may impair Nef’s ability to antagonize SERINC5 and thereby influence viral fitness in vivo. To test this hypothesis, we identified 58 Nef polymorphisms that were overrepresented in HIV-infected patients in Japan sharing the same HLA genotypes. The number of immune-associated Nef polymorphisms was inversely correlated with the plasma viral load. By breaking down the specific HLA allele-associated mutations, we found that a number of the HLA-B*51:01-associated Y120F and Q125H mutations were most significantly associated with a reduced plasma viral load. A series of biochemical experiments showed that the double mutations Y120F/Q125H, but not either single mutation, impaired Nef’s ability to antagonize SERINC5 and was associated with decreasing virion infectivity and viral replication in primary lymphocytes. In contrast, other Nef functions such as CD4, CCR5, CXCR4 and HLA class I downregulation and CD74 upregulation remained unchanged. Taken together, our results suggest that the differential ability of Nef to counteract SERINC5 by naturally occurring immune-associated mutations was associated with the plasma viral load in vivo.
Subject terms: Retrovirus, Viral immune evasion, Viral pathogenesis, Virus-host interactions, HIV infections
Introduction
Nef is an accessory protein of HIV-1 and other primate immunodeficiency viruses that is crucial for efficient virus replication in infected individuals and for virus pathogenicity1,2. Despite its small size of about 27–35 KDa, Nef performs a striking number of functions, including downregulation of the viral entry receptors (CD4, CCR5 and CXCR4) and HLA class I molecules and upregulation of HLA class II invariant chain (CD74) from the cell surface as well as stimulation of viral replication in CD4+ T cells3–8. Nef also enhances the infectivity of progeny virions9–11 mediated in part by counteracting host serine incorporator (SERINC) 3 and 5, which molecules restrict HIV-1 infectivity, of which SERINC5 is the most potent12,13. SERINC5 is incorporated into the membranes of progeny virions in virus-producing cells and antagonizes fusion with target cells14. Nef inhibits this process by internalizing SERINC5 from the surface of the virus-producing cells12.
Despite being one of HIV-1’s most variable proteins, Nef nevertheless possesses several functionally-important, highly conserved motifs. Motifs responsible for each of Nef’s functions have been identified in mutagenesis studies on laboratory-adapted HIV-1 strains3–5,15–19. For instance, the introduction of mutations to the highly conserved FPD motif (Phe121-Pro122-Asp123) in Nef was shown to result in disruption of Nef’s ability to counteract SERINC3/5, thus decreasing the infectivity of progeny virions12,20. However, it is unclear whether highly diverse naturally-occurring (patient-derived) Nef sequences also display differential abilities to counteract SERINC3/5; and if so, it remains elusive whether this ability of patient-derived Nef influences viral fitness in vivo. Because Nef is a dominant target of host cellular immunity21,22, certain immune-escape polymorphisms might affect the Nef function and plasma viremia. Indeed, recent literature demonstrates that two CTL escape mutations, K94E and H116N, observed in elite controllers impair to some extent Nef’s ability to internalize SERINC523. In the present study, we analyzed HLA-associated Nef polymorphisms inversely associated with the plasma viral load in 375 HIV-infected individuals in Japan and further investigated how these polymorphisms affected Nef’s functions.
Results
Nef HLA-associated polymorphisms in chronically HIV-1 subtype B infected subjects
We first sought to identify and characterize HLA-associated Nef polymorphisms in 375 treatment-naïve, chronically HIV-1 subtype B-infected subjects in Japan, a unique population of HLA class I alleles and predominantly HIV-1 subtype B epidemic. A total of 108 HLA class I alleles, defined by four-digit resolution, were observed at frequencies consistent with previous literature24–26. Among these, 49 alleles (including 12 HLA-A, 23 HLA-B, and 14 HLA-C) were observed in at least 10 individuals and thus included in the statistical analyses of HLA-associated polymorphisms. HLA-associated polymorphisms were identified by using a phylogenetically corrected logistic-regression model that corrects the potential confounders, which include HLA linkage disequilibrium between host HLA class I alleles, evolutional relationship between the viral sequences, and viral codon covariation21,27. The identified HLA-associated amino acid residues in Nef were classified as adapted and non-adapted associations when amino acid enriched or depleted in the presence of a particular HLA28. At a threshold of a false-discovery rate (q value) of < 0.2, we identified a total of 112 HLA-associated Nef polymorphisms comprising 58 adapted and 54 non-adapted associations occurring at 55 out of 206 codons (Table S1). These numbers were largely consistent with a previously published study showing that a total of 104 HLA-associated Nef polymorphisms occurred at 45 codons in a cohort of an HIV-infected Japanese population (N = 306)26. For example, both studies consistently demonstrated that Nef codon 81 was associated with HLA-B*35:01 and HLA-B*39:01, and that Nef codon 135 was associated with HLA-A*24:02, the most prevalent HLA class I allele in this population. However, there were some differences in HLA-Nef codon associations; e.g., 2 Nef polymorphisms at codons 120 and 125 were associated with HLA-B*51:01 in this study, whereas no HLA-B*51:01-associated Nef polymorphism was identified by Chikata et al.26.
Number of Nef HLA-adapted polymorphisms inversely associated with plasma viral load
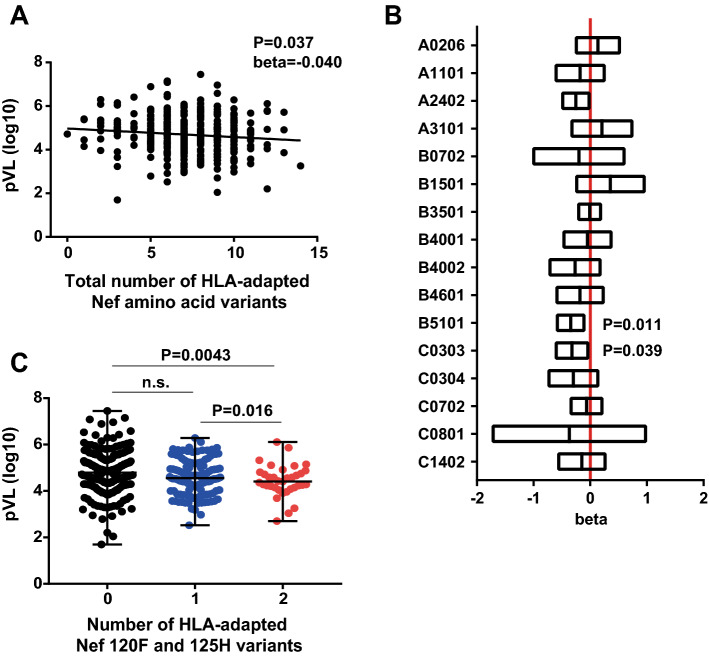
We then sought to investigate the relationship between the presence of HLA-associated amino acid variants in Nef and the plasma viral RNA load (pVL) and CD4 count of the patients. Amino acid variants within a given Nef sequence were counted as HLA-adapted amino acid variants if they had been identified as being HLA-adapted associations in this study, regardless of the HLA class I alleles expressed by the patient. For example, Nef-8C is an HLA-C*15:02-associated adapted polymorphism (Table S1); and, as such, any Nef sequence harboring Cys at codon 8 was counted as having an HLA-adapted amino acid variant at this site. A weak but statistically significant inverse association was observed between pVL and the total number of HLA-adapted Nef amino acid variants (beta = − 0.040, p = 0.037; Fig. 1A). In contrast, we observed no significant association between the CD4 count and the total number of HLA-adapted Nef amino acid variants (p = 0.10). Although the overall pVL association was weak, these results may raise the interesting hypothesis that selection of certain HLA-driven variants in Nef could modulate viremia in this population.
Figure 1.
Association between the number of Nef HLA-associated adapted polymorphisms and plasma viral load. (A) Results of regression analysis between the total number of HLA-adapted Nef amino acid variants and log10-transformed plasma viral load (pVL) in chronically HIV-1 subtype B infected subjects in Japan (n = 375). (B) Regression coefficient and 95% CI obtained from the regression analysis in which pVL and the number of HLA-adapted Nef variants were used as dependent and independent variables for HLA-matched subjects. (C) Difference in plasma viral load between the subjects harbouring HLA-adapted Nef 120F and 125H mutations regardless of the HLA class I alleles expressed by the subject. Statistical analysis was performed by using the Mann–Whitney U-test. n.s. not significant.
We postulated that the observed weakness of association between HLA-adapted Nef variants and a lower pVL could be dominantly attributed to only certain HLA-adapted variants. To address this issue, we stratified the HLA-adapted Nef variants associated with 22 prevalent HLA class I alleles (> 10% frequency) in this cohort and tested for association with the pVL values by performing regression analysis; however, only 16 HLA class I alleles were associated with HLA-adapted Nef variants (Fig. 1B). Statistically significant inverse associations were observed between pVL versus the number of HLA-B*51:01-adapted variants (beta = − 0.35, p = 0.011) and the number of HLA-C*03:03-adapted variant (beta = − 0.32, p = 0.039; Fig. 1B). Because there were 2 HLA-B*51:01-adapted variants (120F and 125H) and an HLA-C*03:03-adapted variant (85F) (Table S1), we further examined the association of pVL with the number of the 120F and 125H variants or the number of the 85F variant, regardless of the HLA class I alleles expressed by the subject. Subjects infected with plasma viruses encoding both Nef 120F and 125H mutations exhibited statistically significantly lower pVL compared to those encoding the consensus amino acid residues (Y120 and Q125) or a subset of the mutations (p < 0.02, Mann–Whitney test; Fig. 1C). In contrast, subjects infected with plasma viruses encoding the 85F mutation did not show any significant difference in pVL compared to those encoding the consensus residue (p > 0.05, Mann–Whitney test). One might be concerned that HLA-B*51:01 is a protective allele in this population and that patients expressing either of them had superior immune-mediated viral control; and indeed 120YFPDWQNY125 has been shown to be a CTL epitope presented by HLA-B*51:0129,30. However, no significant association was observed between pVL and this allele (p > 0.05, Mann–Whitney test). Rather, because F121 and D123 are highly conserved residues important for certain Nef functions31–34, these results suggest that modulation of pVL may be attributable to altered Nef functions mediated by the HLA-adapted Nef Y120F and Q125H variants.
Viral replication capacity of HIV-1 Nef variants in primary CD4+ cells
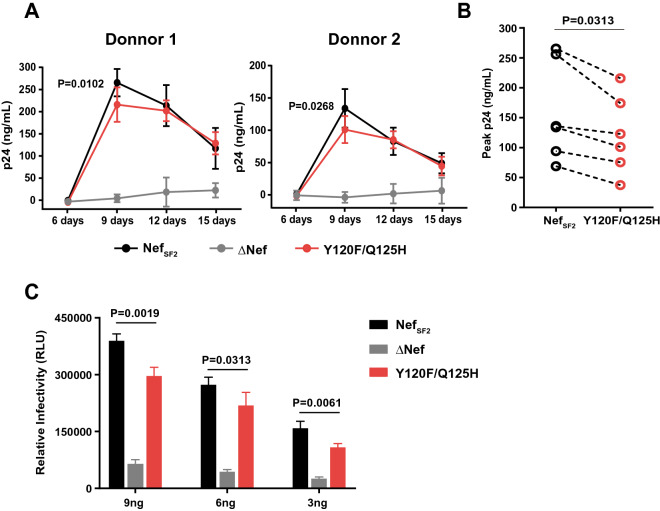
We first tested whether the Y120F and Q125H mutations in Nef impaired viral replication capacity in primary CD4+ cells. We prepared PBMC from 2 HIV-negative donors, exposed them to HIV-NefSF2, HIV∆Nef or HIV-NefY120F/Q125H at Day 0, and measured time-course changes in p24 Gag antigen secreted into the culture supernatant (as a measure of viral replication) until Day15. As expected, HIV-NefSF2, as compared to HIV∆Nef, exhibited a substantial increase in p24 Gag antigen at Day 9 in PBMC from both donors. HIV-NefY120F/Q125H exhibited a substantial increase in p24 Gag antigen, but the peak level reached at Day 9 was significantly reduced compared to that of HIV-NefSF2 in PBMC from both donors (p < 0.03; Fig. 2A). We conducted this assay with 4 additional HIV-negative donors. The data demonstrated that the peak level of p24 Gag antigen of HIV-NefY120F/Q125H was significantly lower than that of NefSF2 (Wilcoxon matched-pairs test, p = 0.0313; Fig. 2B), indicating that the Nef Y120F and Q125H mutations in combination impaired viral replication.
Figure 2.
Effects of Nef Y120F/Q125H mutations on viral replication in PBMC. (A) PBMC (106 cells) prepared from 2 HIV-negative donors were infected for 6 h with HIV-NefSF2, HIVΔNef or HIV-Nef120F/125H that had been produced in HEK293T cells at 10 ng of p24 Ag, and then continuously cultured at 37 °C in fresh culture medium for an additional 15 days. Culture supernatants were collected and replaced with fresh medium every 3 days. To monitor viral replication, we quantified the concentration of p24 Ag in the culture supernatant by use of ELISA. HIV-NefSF2, HIVΔNef, and HIV-Nef120F/125H replication kinetics are shown in each panel. P-values were calculated by using the unpaired t-test at Day 9 (HIV-NefSF2 vs. HIV-Nef120F/125H). (B) A series of the same experiments was done by using PBMC prepared from 4 additional HIV-negative donors. The peak p24 Ag values were plotted and statistically analyzed by using the Wilcoxon matched-pairs signed rank test. (C) TZM-bl cells (1 × 104 cells) were exposed to HIV-NefSF2, HIVΔNef or HIV-Nef120F/125H prepared as above at 9, 6, and 3 ng of p24 Ag. Twenty-four hr later, the reporter cells were lysed and β-galactosidase activity generated as a consequence of infection was measured using a chemiluminescence substrate. Data shown are mean ± SD from 3 or 4 independent experiments. P-values were determined by ANOVA with multiple comparisons.
The observed difference in Nef’s ability to stimulate viral replication could be attributed by viral infectivity of the viral inocula used, because Nef is known to enhance virion infectivity35 and the Y120F and Q125H mutations may affect this Nef’s function. Virion infectivity was tested using TZM-bl reporter cells as target cells and the same preparations of viral inocula used in the replication assays. As expected, HIV-NefSF2, as compared to HIV∆Nef, exhibited a substantial increase in infectivity, and the level of infectivity was increased proportionally to the amount of the viral inocula used (Fig. 2C). NefY120F/Q125H exhibited a substantial increase in infectivity, but ~ 20% reduced level compared to HIV-NefSF2 regardless of the amount of inocula used (p < 0.04; Fig. 2C). These results suggested that the mutations impair Nef’s ability to enhance viral infectivity and stimulate viral replication in primary PBMC.
Effects of Nef variants on counteraction of SERINC3/5-mediated inhibition of HIV-1 infectivity
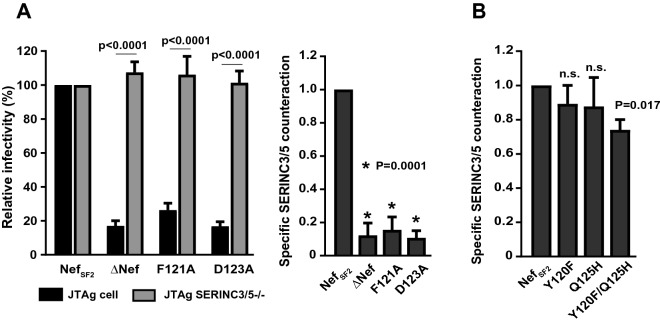
SERINC3 and 5 (SERINC3/5) molecules were recently revealed as being inhibitors of HIV-1 virion infectivity and counteracted by HIV-1 Nef12,13. So we wanted to examine whether the Y120F and Q125H mutations would impair the ability of Nef to counteract SERINC3/5. Before testing this directly, we first undertook the following control experiments. We transfected JTAg cells with pNL43-NefSF2 and pNL43-∆Nef plasmids, harvested the virus-containing supernatant, and then exposed TZM-bl cells to it. By measuring the luminescence intensity generated from HIV-infected TZM-bl cells, we assessed the virion infectivity. Relative infectivity was calculated as luminescence intensity obtained by HIV-NefSF2 normalized to 100%. As expected, infectivity of HIV-NefSF2 was much enhanced as compared to that of HIV∆Nef (Fig. 3A). Moreover, when we transfected JTAg cells that had been engineered to knock out both SERINC3/5 (JTAg-SERINC3/5−/−) with the same pNL43-NefSF2 and pNL43-∆Nef plasmids, both virus preparations showed comparable infectivity (Fig. 3A), clearly confirming that SERINC3/5 were inhibitors of virion infectivity counteracted by Nef. We defined the specific ability of Nef to counteract SERINC3/5 by calculating infectivity of viral particles secreted from parental JTAg cells divided by that of JTAg-SERINC3/5−/− cells, such that values > 1.0 and < 1.0 indicated increased or decreased ability of Nef to counteract SERINC3/5 compared to NefSF2, respectively (Fig. 3A). Testing of the F121A and D123A mutations in NefSF2 showed nearly complete disruption of Nef’s ability to counteract SERINC3/5 (Fig. 3A), confirming previously reported findings12. We then tested the NefSF2 harboring the Y120F, Q125H, and Y120F/Q125H mutations for SERINC3/5 counteraction. No substantial effects were observed with the single mutations; whereas the double Y120F/Q125H mutation showed ~ 20% reduced ability to counteract SERINC3/5 (Fig. 3B).
Figure 3.
Effects of Nef Y120F and Q125H mutations on counteraction of SERINC3/5-mediated infectivity inhibition. (A) Infectivity of viruses that were produced from JTAg cells and JTAg-SERINC3/5−/− cells (left panel). Those cells were transduced with HIV-1 NL43 proviral constructs lacking Nef (ΔNef) or carrying NefSF2 and the mutants (F121A and D123A), and then TZM-bl cells were exposed to the resultant viruses to determine viral infectivity. Results are expressed as the mean of triplicate assessments, normalized to the control strain, NL4.3-NefSF2. Statistical analysis was performed by using the paired t test. Ability of NefSF2 and the indicated mutants (F121A and D123A) to counteract SERINC3/5 was determined by dividing the relative infectivity of viral particles secreted from JTAg cells by that from JTAg-SERINC3/5−/− cells (right panel). (B) Specific SERINC3/5 counteraction function of NefSF2 and the indicated mutants (Y120F, Q125H, Y120F/Q125H). Data shown are the mean results ± SD from 3 independent experiments. Statistical analysis was performed by ANOVA with multiple comparisons vs NefSF2. n.s. not significant.
Functional effects of Nef variants in the context of patient-derived Nef sequences
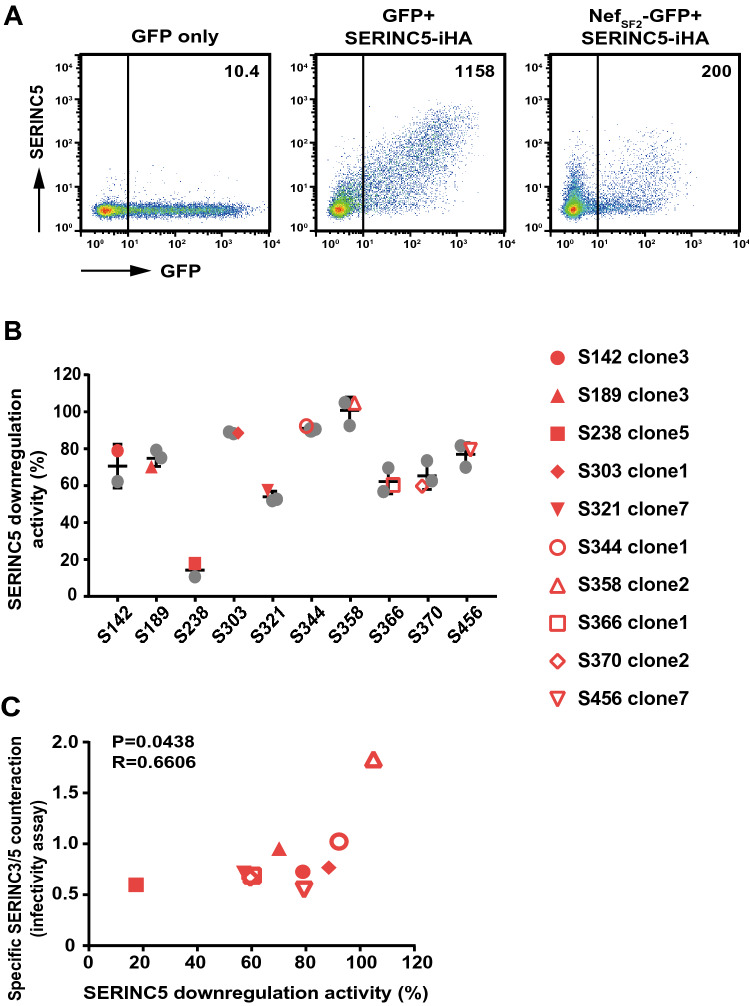
Mutational effects on Nef functionality are often dependent on backbone sequences or genetic lineages. We first analyzed the sequence of amplified nef gene fragments after they had been cloned into a plasmid (average of 8 clones per subject) in 10 out of 12 HLA-B*51:01+ patients whose autologous viruses had the Y120F/Q125H mutations. Nef clones clustered closely with their respective bulk plasma HIV RNA sequences in the phylogenetic tree (Fig. S1A). In 6 patients, all sequenced clones had both Y120F/Q125H mutations; whereas in the other patients, a minority of the Nef clones had the single Y120F mutation (Fig. S1B). Next, by the use of a transfection-based assay, 2 or 3 Nef sequences harboring the Y120F/Q125H mutations were tested for Nef’s ability to downregulate SERINC5 from the cell surface. SERINC5-iHA expression was increased when JTAg-SERINC3/5−/− cells were transfected with DNA encoding SERINC5-iHA, whereas co-transfection with DNA encoding SERINC5-iHA and NefSF2-GFP resulted in substantial reduction in cell surface expression of SERINC5-iHA (Fig. 4A), confirming Nef’s ability to downregulate SERINC523,36,37. All patient-derived Nef clones tested were functional with respect to the SERINC5-iHA downregulation function, but the activity level was different to a relatively small extent within a host, but to a large extent across hosts (Fig. 4B). A nef clone from each patient (shown by the red plots in Fig. 4B and their amino acid sequences are given in Fig. S2) was subcloned into pNL43, and infectivity potential of the recombinant viruses harboring patient-derived Nef clones was determined. We observed a weak but statistically significant correlation in Nef functions between SERINC5-iHA downregulation and the specific counteraction of SERINC3/5 in the infectivity assay (Spearman R = 0.6606, p = 0.0438; Fig. 4C).
Figure 4.
Effects of patient-derived Nef clones on SERINC5 downregulation and counteraction of SERINC3/5-mediated infectivity inhibition. (A) Representative flow cytometry plots display the expression of surface SERINC5 in JTAg-SERINC3/5−/− cells that had been co-transfected with genes encoding GFP and Nef SF2-GFP together with SERINC5-iHA or an empty vector. The resultant cells were analyzed for cell-surface SERINC5 expression (as HA) and GFP. MFI values for SERINC5-iHA in the GFP+ subset are shown. (B) Nef clones isolated from the indicated HIV-infected subjects (2 or 3 clones per patient) were analyzed for their ability to downregulate SERINC5-iHA. Results are expressed as relative downregulation activity, normalized to the control strain, NefSF2. Plots indicated in red were used for functional analyses in panel C and Fig. 5. (C) Correlation analysis between Nef’s ability to counteract SERINC3/5 (for viral infectivity) and to downregulate cell surface expression of SERINC5. Nef clones tested are indicated in (B). Statistical analysis was done with the Spearman test.
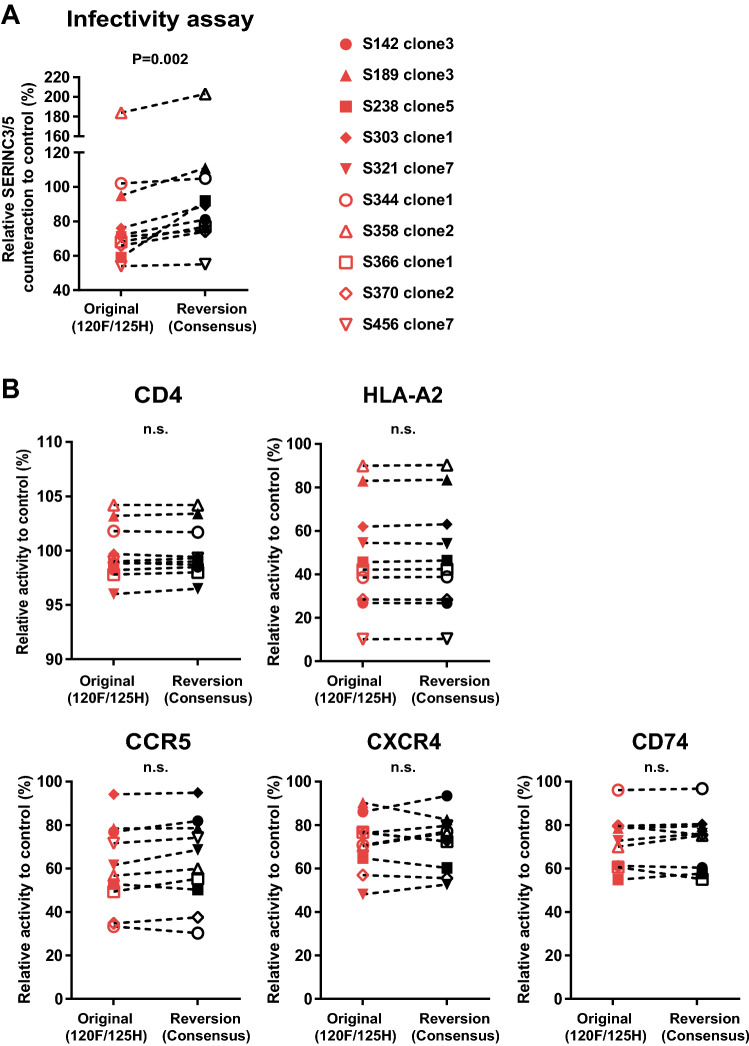
Finally, to validate the effects of the Y120F/Q125H mutations on Nef function in the context of patient-derived Nef sequences, we reverted each Nef clone to the consensus Y120 and Q125 to generate paired mutant/revertant constructs. The pair-wise comparison showed that Nef’s ability to counteract SERINC3/5 was restored when the reversions, from 120F/125H to Y120/Q125, were introduced (Wilcoxon matched-paired test, P = 0.002; Fig. 5A). In contrast, Nef’s ability to downregulate CD4, CCR5, CXCR4 and HLA-A*02 and upregulate CD74 was not influenced by the reversions (Fig. 5B). These results indicate that the HLA-B*51:01-adapted Y120F/Q125H mutations selectively impaired Nef’s ability to counteract SERINC3/5 but that CD4, CCR5, CXCR4 and HLA downregulation functions and CD74 upregulation function remained unaffected.
Figure 5.
Effects of Y120F and Q125H mutations in patient-derived Nef clones on counteraction of SERINC3/5-mediated infectivity inhibition and modulation of cell surface receptor expression. Patient-derived Nef clones (n = 10) harboring HLA-B*51:01-adapted 120F and 125H mutations (see legend to Fig. 4B) were subcloned into HIV-1 pNL43 or Nef-GFP fusion plasmids, and their corresponding Y120 and Q125 reversion mutants were constructed. Pair-wise analyses of Nef’s ability to counteract SERINC3/5 (A) as well as downregulate CD4, CCR5, CXCR4 and HLA class I, and upregulate CD74 (B) were conducted. Data represent indicated Nef functions by parental and revertant clones. Data shown are the mean results ± SD from 3 or 4 independent experiments. Statistical analysis was performed by the Wilcoxon matched-pairs signed rank test. n.s. not significant.
Discussion
We have demonstrated here that a number of naturally-occurring Nef variants (Y120F and Q125H) associated with a certain HLA class I allele (HLA-B*51:01) inversely correlated with the plasma viral load in treatment-naïve HIV-infected patients harboring this HLA allele. In addition, a set of in vitro functional analyses of the Nef variants demonstrated that these 2 mutations in combination selectively impaired Nef’s ability to antagonize the restriction function of SERINC5, associating with decreased viral replication and virion infectivity while preserving other Nef functions including downregulation of HLA class I, CD4, CCR5, and CXCR4 as well as upregulation of CD74. Our findings indicate that certain HLA-associated Nef variants were associated with a decreased plasma viral load and impaired Nef’s ability to antagonize SERINC5 function.
It has been well documented that, by performing mutational analyses, highly conserved Nef residues, including PxxP72, D123, and DD174, 175 are responsible for Nef’s important functions such as HLA downregulation, dimerization, and CD4 downregulation, respectively19,33,38–41. In addition, analyses of patient-derived Nef sequences have revealed the contribution of naturally-occurring polymorphisms at Nef’s more variable sites on Nef functions of patient-derived sequences42–47. In our study, the immune-escape mutations, Y120F, Q125H, and the double mutations (Y120F/Q125H), exhibited the prevalence of 39.2, 13.1, and 9.3%, respectively; in this study cohort in Japan, the values are much higher than those of subtype B sequences in Los Alamos Database, which are 13.6, 5.2, and 1.6%, respectively. This suggests a unique population of HLA class I alleles of this cohort in Japan, compared to the regions with subtype B epidemic; and implicates that the double mutations could not be prevalent without HLA-driven selective pressure. However, the double mutations are relatively prevalent in subtype D (11.9%) in the data from Los Alamos database, compared to subtype A and C (3.9 and 1.3%, respectively). Further detailed studies are needed to clarify the effects of the Y120F and Q125H mutations on Nef functions of different subtype backbones.
The highly conserved D123 residue (> 99% prevalence in subtype A, B, C and D) is encompassed by the immune-escape mutation sites of Y120F and Q125H. Several studies demonstrate that introduction of mutations at this residue results in impairment of multiple Nef functions including downregulation of CD4 and HLA as well as enhancement of viral infectivity and replication31–33,48,49. In addition, D123A mutant impairs Nef’s ability to counteract SERINC3/5 as shown in this study. Crystal structural analysis of the Nef core domain demonstrates that D123 involves a dimerization interface toward R10548. Indeed, D123N mutation prevents Nef core dimer formation when bound to Hck SH3 of Src-family kinases50. It is thus interesting to see whether and to what extent the immune-escape mutations at the neighboring residues of Y120F and Q125H play a secondary role in Nef dimerization.
Some limitations of our study merit mention. Although we investigated Nef clones isolated from 375 treatment-naïve HIV-infected patients in Japan, this panel did not capture the entirety of subtype B Nef genetic diversity. Also, the cohort of 375 individuals did not capture the entirety of host genetic diversity at HLA class I loci in this population. Several key Nef functions, including enhancing viral replication and virion infectivity, as well as downregulation of SERINC5, CD4, CCR5, CXCR4, and HLA class I molecules and upregulation of CD74, were tested in in vitro assays using Nef variants harboring HLA-associated mutations; but we could not rule out a role for other known or unknown Nef functions. For instance, SERINC3 is also known to restrict HIV-1 infectivity12,13,51,52, albeit much less extent compared to SERINC5, and counteracted by HIV-1 Nef. The Nef mutations tested here may differently affect the counteraction functions against SERINC3 and 5. This issue was not specifically addressed here. Due to the limited availability of PBMC samples from the HIV-infected donors, HLA class I-restricted immune responses to Nef variants could not be experimentally addressed. Also, because of the cross-sectional sampling of patients’ specimens in this study, intra-host changes of nef sequences over time could not be tested. Despite these limitations, our study provides strong evidence that naturally-occurring variations in Nef-mediated SERINC5 counteraction function may contribute, at least to some extent, to clinical outcomes in HIV-1 infections. Our results highlight the conflicting fitness effects of Nef arising by the interplay between antiviral immunity and intrinsic restriction by the host.
Materials and methods
Study subjects
Plasma samples were collected from 446 HIV-1 subtype B chronically infected and treatment-naïve patients who were monitored at the National Center for Global Health and Medicine, Tokyo and Institute of Medical Science, University of Tokyo, Japan from 1996 to 2012. The HLA class I typing of these patients was done by using a high-resolution sequence-based typing protocol as previously described25. In a subset of the cohort (N = 375), the data for CD4 counts (median: 307 [IQR: 190.3 to 400.5]/mm3) and plasma viral load (median: 45,000 [IQR: 16,000 to 165,000] copies/ml) were available. This study was approved by the Human Research Ethics Committee of the National Center for Global Health and Medicine and the Institutional Review Board of the University of Tokyo, and was conducted at Joint Research Center for Human Retrovirus Infection, Kumamoto University, Kumamoto, Japan, according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all study participants.
Sequence analysis of autologous nef genes
After precipitation of HIV-1 particles by ultracentrifugation (50,000 rpm, 15 min) of patients’ plasma, the viral RNA was purified by use of a QIAamp Viral RNA Min kit (Qiagen) followed by the synthesis of cDNA carried out with a Cloned AMV First-strand cDNA Synthesis Kit (Invitrogen Corp, Carlsbad, CA), as previously describe24. Through nested PCR, DNA fragments spanning the nef gene were amplified with the set of primers previously described53. The resultant PCR product was purified and directly sequenced with an automated sequencer (ABI 3500/3500XL; Applied Biosystems, Carlsbad, CA). The sequence data were analyzed and aligned by using Seqscape software version 2.7 and Gene cutter tool in the Los Alamos sequence database (https://www.hiv.lanl.gov/) with respect to a reference HXB2 strain. To facilitate a consistent codon numbering scheme (based on the NefHXB2 reference strain), we pairwise-aligned all nef sequences to nefHXB2 by using an in-house algorithm based on the HyPhy platform54 and insertions stripped out.
Cloning and plasmid construction
Patient-derived nef genes amplified as above were cloned into a plasmid by using a Zero Blunt TOPO PCR Cloning kit (Invitrogen). A median of 8 nef clones was sequenced per patient. To examine functionality of nef gene products, control (strain SF2) and the patient-derived Nef sequences were subcloned in the pcDNA3.1-GFP plasmid55 and HIV-1NL43 proviral construct as previously described43,56,57. Defined mutations of interest were then introduced by using overlapping PCR22,58. All control, patient-derived and mutation-introduced plasmid constructs were re-confirmed by DNA sequencing of the entire nef region.
Analysis of Nef-mediated modulation of cell surface molecules
Nef-mediated downregulation of cell surface CD4 and HLA-I molecules was assessed in CEM, a human CD4+ T cell line stably transfected with the gene encoding HLA-A*02:01 (CEM-A*02 cells). CEM-A*02 cells (2 × 106) were electroporated with 8 μg plasmid DNAs encoding GFP alone or Nef-GFP fusion proteins by electroporation in 0.4-mm cuvettes under the following Gene Pulser Xcell square-wave conditions: 250 V, 25 ms, and 1 pulse (Bio-Rad Laboratories, Inc.). Twenty-four hour later, the resultant cells were stained with brilliant violet-conjugated anti-human CD4 mAb and HLA-A2 serotype-specific mAb and 7-amino-actinomycin D (7-AAD) (all from BioLegend), as previously described46,47,57,59. Nef-mediated downregulation of cell surface CCR5 and CXCR4 molecules was assessed in TZM-bl cells. TZM-bl cells (1 × 105) were transfected with plasmid DNAs encoding GFP alone or Nef-GFP fusion proteins by Lipofectamine 2000 (Themo Fisher). Forty-eight hour later, the resultant cells were stained with phycoerythrin-conjugated anti-human CCR5 mAb, allophycocyanin-conjugated anti-human CXCR4 mAb (BioLegend), and 7-AAD, as previously described46. Nef-mediated upregulation of cell surface CD74 molecule was assessed in 721.221 cells. 721.221 cells (1 × 105) were exposed with the recombinant viruses harboring the patient-derived Nef sequences of interest. Forty-eight hour later, the resultant cells were stained with phycoerythrin-conjugated anti-CD74 (BioLegend), 7-AAD, and anti-p24 Gag-FITC (Beckman Coulter), as described57. Nef-mediated internalization of SERINC5 from the cell surface was assessed in JTAg cells with SERINC3/5 knocked out (denoted as JTAg-SERINC3/5−/−)12. JTAg-SERINC3/5−/− cells (3 × 106) were electroporated with 5 μg of pBJ5-SERINC5-internal HA tag (S5-iHA; kindly provided by H. Gottlinger)13 together with 5 μg of DNAs encoding GFP alone or Nef-GFP fusion proteins by using the Gene Pulser Xcell square-wave conditions described above. Twenty-four hr later, the resultant cells were stained with Alexa Fluor 647 anti-HA.11 and Zombie Aqua to remove dead cell fractions (all from BioLegend)36.
Note that in all systems described above, live cells were gated, and the mean fluorescence intensity (MFI) of CD4, CCR5, CXCR4, HLA-I, or CD74 in Nef-expressing cells (defined as the GFP+ or p24 Gag+ subset in the transduced cells, denoted MFI Nef+ in the below calculation) and non Nef-expressing cells (defined as the GFP− or p24 Gag− subsets in the transduced cells, denoted MFI Nef−) was analyzed by flow cytometry (FACS Verse: BD Biosciences). The following formula was used to calculate the CD4, CCR5, CXCR4 and HLA-I downregulation activity and CD74 upregulation activity of each Nef clone: (MFI Nef− − MFI Nef+)/MFI Nef− × 100. The MFI value of SERINC5-iHA downregulation activity for each Nef clone was normalized to the negative (GFP+/S5-iHA+) and positive (NefSF2-GFP+/S5-iHA+) controls by using the following formula: (MFInegative − MFIclone)/(MFInegative − MFIpositive) × 100. All Nef functional values were reported as the mean of a minimum of triplicate experiments.
Infectivity assay for SERINC3/5 activity
The recombinant viruses were produced by electroporation of JTAg or JTAg-SERINC3/5−/− cells with NL43-based proviral clones lacking or harboring various nef sequences. Twenty-four hour later, the virus-containing supernatant was harvested and quantified by assessing reverse transcriptase activity by using a one-step SYBR green I-based product-enhanced reverse transcriptase assay as described earlier60,61. TZM-bl reporter cells (NIH AIDS Research and Reference Reagent Program) were seeded into 96-well plates, exposed to the viruses for 24 h, and then lysed for measurement of β-galactosidase activity by using a Galacto-Star Reporter Assay System (Applied Biosystems) as described previously57. To obtain relative infectivity of the viruses, we divided the number of infected cells (as measured by luminescence value) by the amount of the input virus (as measured by reverse transcriptase activity), and then normalized it to the control strain, NL4.3-NefSF2. Furthermore, for quantification of Nef’s ability to counteract SERINC3/5, the relative infectivity value of the JTAg cell-derived virus was divided by that of the JTAg-SERINC3/5−/− cell-derived virus. Thereby, values > 1.0 and < 1.0 respectively indicated increased and decreased ability of Nef to counteract SERINC3/5. All Nef functional values were reported as the mean of a minimum of triplicate experiments.
Viral replication assay
HEK293T cells seeded on 6-well plates were transfected with NL43-based proviral clones harboring various nef sequences. Forty-eight hour later, virus-containing supernatants were harvested, quantified for the amount of p24Gag Ag by use of ELISA (ZeptoMetrix Corp.) and stored at − 80 °C until use. Freshly isolated PBMC from HIV-negative donors (106 cells) were exposed to the virus preparations (10 ng of p24Gag Ag) for 6 h, washed twice, and resuspended in culture medium (RPMI 1640, 10% fetal calf serum) as described previously22,57. Three days later, the PBMCs were stimulated with PHA. Culture supernatants were collected and replaced with fresh medium supplemented with human rIL-2 every 3 days. Viral replication was monitored by measuring p24Gag Ag in the culture supernatant by using ELISA over a 15-day period. ELISA values during the initial burst of viral replication (on day 9) were used as our measure of replication capacity. Results were expressed as the mean of quadruplicate assessments of each donor, normalized to control strain NL4.3-NefSF2.
Statistical analysis
Association between viral polymorphisms and host HLA class I alleles
The published phylogenetic dependency network model (PDN) was used to determine viral polymorphisms that were statistically associated with host HLA class I alleles with a pre-defined threshold of p < 0.05, q < 0.2, as previously described21,27,62,63. The PDN model is a phylogenetically corrected logistic regression model that corrects the potential confounders, which include HLA linkage disequilibrium between host HLA class I alleles, evolutional relationship between the viral sequences, and viral codon covariation21,27,62,63. The HLA-associated amino acid residues in Nef protein were classified as adapted and non-adapted associations when amino acid enriched or depleted in the presence of a particular HLA21,28.
Association between viral polymorphisms and clinical parameters
The association of the number of HLA-adapted Nef amino acid variants with log10-transformed pVL or CD4 count was assessed by regression analysis. A two-tailed p-value < 0.05 was considered to be statistically significant.
Accession numbers
GenBank accession numbers for clonal nef sequences of subtypes B in this study are LC547123 to LC54720.
Supplementary information
Acknowledgements
This study was supported by a grant from Japan Agency for Medical Research and Development, AMED (Research Program on HIV/AIDS), and JSPS KAKENHI Grant-in-Aid for Scientific Research. MT received funding from JSPS KAKENHI Grant-in-Aid for Scientific Research. DK and TST are supported by the scholarship for The International Priority Graduate Programs; Advanced Graduate Courses for International Students (Doctoral Course) from the Ministry of Education, Science, Sports, and Culture (MEXT) of Japan. The funders played no role in determining the content of the manuscript or the authors’ decision to publish.
Author contributions
M.T, D.K. and T.U. designed the study; M.T., D.K., T.S.T., and K.G. performed the experiments; A.K-T., H.G., and S.O. provided access to patient samples and analyzed clinical data; M.P. provided important reagents; M.T., D.K., T.S.T., J.O. J.C. and T.U. analyzed data; and M.T, D.K. and T.U. wrote the paper. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mako Toyoda and Doreen Kamori.
Supplementary information
is available for this paper at 10.1038/s41598-020-76375-w.
References
- 1.Kestler HW, 3rd, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 2.Deacon NJ, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 3.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: Requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 4.Michel N, Allespach I, Venzke S, Fackler OT, Keppler OT. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. 2005;15:714–723. doi: 10.1016/j.cub.2005.02.058. [DOI] [PubMed] [Google Scholar]
- 5.Venzke S, Michel N, Allespach I, Fackler OT, Keppler OT. Expression of Nef downregulates CXCR4, the major coreceptor of human immunodeficiency virus, from the surfaces of target cells and thereby enhances resistance to superinfection. J. Virol. 2006;80:11141–11152. doi: 10.1128/jvi.01556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindler M, et al. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 2003;77:10548–10556. doi: 10.1128/jvi.77.19.10548-10556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. The human immunodeficiency virus-1 nef gene product: A positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz O, Marechal V, LeGall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 1996;2:338–342. doi: 10.1038/Nm0396-338. [DOI] [PubMed] [Google Scholar]
- 9.Chowers MY, et al. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 1994;68:2906–2914. doi: 10.1128/JVI.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 1995;69:5048–5056. doi: 10.1128/JVI.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MD, Warmerdam MT, Page KA, Feinberg MB, Greene WC. Expression of the human immunodeficiency virus type 1 (HIV-1) nef gene during HIV-1 production increases progeny particle infectivity independently of gp160 or viral entry. J. Virol. 1995;69:579–584. doi: 10.1128/JVI.69.1.579-584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosa A, et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usami Y, Wu Y, Gottlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J. Biol. Chem. 2017;292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akari H, et al. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J. Virol. 2000;74:2907–2912. doi: 10.1128/jvi.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg ME, et al. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. J. Article. 1997;16:6964–6976. doi: 10.1093/emboj/16.23.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piguet V, et al. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2000;2:163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren X, Park SY, Bonifacino JS, Hurley JH. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. Elife. 2014;3:e01754. doi: 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada T, et al. Proline 78 is crucial for human immunodeficiency virus type 1 Nef to down-regulate class I human leukocyte antigen. J. Virol. 2003;77:1589–1594. doi: 10.1128/jvi.77.2.1589-1594.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez PW, et al. Plasma membrane-associated restriction factors and their counteraction by HIV-1 accessory proteins. Cells. 2019 doi: 10.3390/cells8091020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brumme ZL, et al. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueno T, et al. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J. Immunol. 2008;180:1107–1116. doi: 10.4049/jimmunol.180.2.1107. [DOI] [PubMed] [Google Scholar]
- 23.Jin SW, et al. Natural HIV-1 Nef polymorphisms impair SERINC5 downregulation activity. Cell Rep. 2019;29:1449–1457. doi: 10.1016/j.celrep.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasan Z, et al. Minor contribution of HLA class I-associated selective pressure to the variability of HIV-1 accessory protein Vpu. Biochem. Biophys. Res. Commun. 2012;421:291–295. doi: 10.1016/j.bbrc.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Itoh Y, et al. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 2005;57:717–729. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 26.Chikata T, et al. HLA-associated viral polymorphism in chronically HIV-1-infected Japanese cohort: Analysis of four-digit HLA allele level. Retrovirology. 2012;9:P269. doi: 10.1186/1742-4690-9-s2-p269. [DOI] [Google Scholar]
- 27.Carlson JM, et al. Phylogenetic dependency networks: Inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput. Biol. 2008;4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson JM, et al. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J. Virol. 2012;86:13202–13216. doi: 10.1128/JVI.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Multilayered defense in HLA-B51-associated HIV viral control. J. Immunol. 2011;187:684–691. doi: 10.4049/jimmunol.1100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geels MJ, et al. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J. Virol. 2003;77:12430–12440. doi: 10.1128/jvi.77.23.12430-12440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams M, Roeth JF, Kasper MR, Filzen TM, Collins KL. Human immunodeficiency virus type 1 Nef domains required for disruption of major histocompatibility complex class I trafficking are also necessary for coprecipitation of Nef with HLA-A2. J. Virol. 2005;79:632–636. doi: 10.1128/Jvi.79.1.632-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu LX, et al. Mutation of a conserved residue (D123) required for oligomerization of human immunodeficiency virus type 1 Nef protein abolishes interaction with human thioesterase and results in impairment of Nef biological functions. J. Virol. 2000;74:5310–5319. doi: 10.1128/jvi.74.11.5310-5319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizzato M, et al. Dynamin 2 is required for the enhancement of HIV-1 infectivity by Nef. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6812–6817. doi: 10.1073/pnas.0607622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen GB, Rangan VS, Chen BK, Smith S, Baltimore D. The human thioesterase II protein binds to a site on HIV-1 Nef critical for CD4 down-regulation. J. Biol. Chem. 2000;275:23097–23105. doi: 10.1074/jbc.M000536200. [DOI] [PubMed] [Google Scholar]
- 35.Münch J, et al. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 2007;81:13852–13864. doi: 10.1128/jvi.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mwimanzi F, et al. An HIV-1 Nef genotype that diminishes immune control mediated by protective human leucocyte antigen alleles. Aids. 2020;34:1325–1330. doi: 10.1097/QAD.0000000000002559. [DOI] [PubMed] [Google Scholar]
- 37.Sudderuddin H, et al. Longitudinal within-host evolution of HIV Nef-mediated CD4, HLA and SERINC5 downregulation activity: A case study. Retrovirology. 2020;17:3. doi: 10.1186/s12977-019-0510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casartelli N, et al. The Pro78 residue regulates the capacity of the human immunodeficiency virus type 1 Nef protein to inhibit recycling of major histocompatibility complex class I molecules in an SH3-independent manner. J. Gen. Virol. 2006;87:2291–2296. doi: 10.1099/vir.0.81775-0. [DOI] [PubMed] [Google Scholar]
- 39.Fackler OT, et al. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology. 2006;351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 40.Lindwasser OW, et al. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J. Virol. 2008;82:1166–1174. doi: 10.1128/JVI.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray LR, et al. CD4 and MHC class 1 down-modulation activities of nef alleles from brain- and lymphoid tissue-derived primary HIV-1 isolates. J. Neurovirol. 2011;17:82–91. doi: 10.1007/s13365-010-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann JK, et al. Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology. 2013;10:100. doi: 10.1186/1742-4690-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mwimanzi P, et al. Dynamic range of Nef functions in chronic HIV-1 infection. Virology. 2013;439:74–80. doi: 10.1016/j.virol.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Kuang XT, et al. Impaired Nef function is associated with early control of HIV-1 viremia. J. Virol. 2014;88:10200–10213. doi: 10.1128/JVI.01334-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann JK, et al. Nef-mediated down-regulation of CD4 and HLA class I in HIV-1 subtype C infection: Association with disease progression and influence of immune pressure. Virology. 2014;468–470:214–225. doi: 10.1016/j.virol.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toyoda M, et al. Differential ability of primary HIV-1 Nef isolates to downregulate HIV-1 entry receptors. J. Virol. 2015;89:9639–9652. doi: 10.1128/JVI.01548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mwimanzi F, et al. Resistance of major histocompatibility complex class B (MHC-B) to Nef-mediated downregulation relative to that of MHC-A is conserved among primate lentiviruses and influences antiviral T cell responses in HIV-1-infected individuals. J. Virol. 2018 doi: 10.1128/JVI.01409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poe JA, Smithgall TE. HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication. J. Mol. Biol. 2009;394:329–342. doi: 10.1016/j.jmb.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jere A, Fujita M, Adachi A, Nomaguchi M. Role of HIV-1 Nef protein for virus replication in vitro. Microbes Infect. 2010;12:65–70. doi: 10.1016/j.micinf.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Moroco JA, et al. Remodeling of HIV-1 Nef structure by Src-family kinase binding. J. Mol. Biol. 2018;430:310–321. doi: 10.1016/j.jmb.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heigele A, et al. The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe. 2016;20:381–391. doi: 10.1016/j.chom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Sousa-Pereira P, et al. The antiviral activity of rodent and lagomorph SERINC3 and SERINC5 is counteracted by known viral antagonists. J. Gen. Virol. 2019;100:278–288. doi: 10.1099/jgv.0.001201. [DOI] [PubMed] [Google Scholar]
- 53.Meribe SC, et al. Association between a naturally arising polymorphism within a functional region of HIV-1 Nef and disease progression in chronic HIV-1 infection. Arch. Virol. 2015;160:2033–2041. doi: 10.1007/s00705-015-2480-5. [DOI] [PubMed] [Google Scholar]
- 54.Pond SL, Frost SD, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 55.Ueno T, Idegami Y, Motozono C, Oka S, Takiguchi M. Altering effects of antigenic variations in HIV-1 on antiviral effectiveness of HIV-specific CTLs. J. Immunol. 2007;178:5513–5523. doi: 10.4049/jimmunol.178.9.5513. [DOI] [PubMed] [Google Scholar]
- 56.Mahiti M, Brumme ZL, Jessen H, Brockman MA, Ueno T. Dynamic range of Nef-mediated evasion of HLA class II-restricted immune responses in early HIV-1 infection. Biochem. Biophys. Res. Commun. 2015;463:248–254. doi: 10.1016/j.bbrc.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 57.Mwimanzi P, et al. Attenuation of multiple Nef functions in HIV-1 elite controllers. Retrovirology. 2013;10:1. doi: 10.1186/1742-4690-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mwimanzi P, et al. Effects of naturally-arising HIV Nef mutations on cytotoxic T lymphocyte recognition and Nef's functionality in primary macrophages. Retrovirology. 2011;8:50. doi: 10.1186/1742-4690-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahiti M, et al. Relative resistance of HLA-B to downregulation by naturally occurring HIV-1 Nef sequences. mBio. 2016;7:e01516. doi: 10.1128/mBio.01516-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pizzato M, et al. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods. 2009;156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 61.Trautz B, et al. The antagonism of HIV-1 Nef to SERINC5 particle infectivity restriction involves the counteraction of virion-associated pools of the restriction factor. J. Virol. 2016;90:10915–10927. doi: 10.1128/JVI.01246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brumme ZL, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PloS ONE. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore CB, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.