Abstract
The microbiota profile of children changes with age. To investigate the differences in the gut microbiota profile of 1- and 4-year-old children, we collected fecal samples and sequenced the V3–V4 hypervariable region of the 16S rRNA gene via high-throughput DNA sequencing. From phylum to species level, the microbiota underwent significant changes with age. The abundance of phyla Proteobacteria and Actinobacteria declined with age, whereas phyla Firmicutes and Bacteroidetes increased with age and dominated the gut microbiota of 4-year-olds. The intestinal environment of children at age four is closer to maturity. Hence, the abundance of Bifidobacterium significantly decreased in the gut of 4-year-olds, whereas Akkermansia muciniphila increased from 0.14% in 1-year-olds to 4.25% in 4-year-olds. The functional change in gut microbiota is consistent with changes in infant food, as microbiota participating in amino acid and vitamin metabolism were enriched in 1-year-olds, whereas microbiota involved in lipid metabolism increased with age.
Subject terms: RNA sequencing, Microbiome
Introduction
Intestinal microbiota play an important role in human health, and dysbiosis (an imbalance in microbiota) could cause immunologic dysregulation1 and various chronic diseases, including diabetes2, obesity3, inflammatory bowel disease4, rheumatoid arthritis5, autism spectrum disorders6, and cancers7. The gut–brain axis8, gut–liver axis9 and gut–lung axis10 imply a bidirectional interaction between microbiota and tissues of human body, emphasizing the role of microbiota in both health and disease.
Strikingly, intestinal microbiota imbalance early in life may lead to disease conditions at a later age11,12. The gut microbiota taxa of 2-year-old infants showed an increasingly strong association with the BMI of 12-year-old children13, and reduced bacterial diversity in both 1- and 12-month-old infants’ intestinal microbiota was associated with an increased risk of allergic sensitization in the first 6 years of life14. These findings drew more attention on the development of pediatric intestinal microbiota15.
The formation of infant gastrointestinal (GI) microbiota is affected by many factors, including mode of delivery16,17, type of feeding18–20, race and cord blood vitamin D levels21. Most studies support the opinion that vaginal delivery and breast-feeding contribute to healthy gut microbiota22,23, but growing infants displayed increased alpha-diversity and reduced beta-diversity in the gut microbiota22. Analyses have shown that infants possess individually distinct microbial profiles by the end of the first year of life, but begin converging towards the characteristic microbiota of adults24, and by age three, the infant microbiota gradually changed towards an adult-like structure25. New opinion suggested that infant microbiota development may take longer than 3 years, but the gut microbiota composition in childhood beyond age three is often overlooked26.
To fill the knowledge gap about gut microbiota beyond age three, we collected the stool samples of 1- and 4-year-old healthy Chinese children and investigated whether they displayed a distinct microbial profile at age one and adult-like mature profile at age four.
Materials and methods
Sample collection
Study subjects were selected from the Shanghai-Minhang Birth Cohort Study (S-MBCS), which was reviewed and approved by the ethics committee board of the Shanghai Institute of Planned Parenthood Research (IRB00008297). Written informed consents were obtained from the parents of all participants involved in this study. All methods were performed in accordance with the Declaration of Helsinki. Fecal samples were collected from 1-year-old infants and 4-year-olds who were healthy, not overweight, had not taken antibiotics in one month, and had not developed eczema. All samples were stored at − 80 °C before DNA extraction.
DNA extraction, PCR amplification and 16S rRNA gene sequencing
DNA extraction and PCR amplification were performed as described previously27, with some modifications. In brief, the genomic DNA was extracted from 300 mg of feces using a QIAamp DNA stool mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The integrity of extracted genomic DNA was checked by 1.5% agarose gel electrophoresis. To generate 16S rRNA gene amplicons, the V3-4 hypervariable region of the 16S rRNA genes was amplified using the primers 338F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GACTACHVGGGTATCTAATCC-3′) with a TransStart Fastpfu DNA Polymerase (TransGen, Beijing, China) in 20 cycles. All amplicons were purified using the QIAquick PCR Purification Kit (Qiagen), quantified on Qubit (Life Technologies), then pooled into equal concentrations. The pooled amplicons were ligated with adaptors using TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme,China), then 2 × 300 bp paired-end sequencing was performed on an Illumina MiSeq instrument with MiSeq Reagent Kit v3.
Bioinformatics and statistical analysis
Paired-end 16S rRNA sequences were assembled using Mothur (version 1.41.1)28. DNA sequences were discarded using the following criteria: containing ambiguous bases, or containing chimeric or contaminant sequences, or homopolymers of > 8 nucleotides, or with lengths shorter than 350 bp. The chimeric sequences were identified by VSEARCH algorithm, and non-16S contaminants sequences were filtered based on the RDP database. Using SILVA reference databases (V132)29, the DNA sequences were clustered into OTUs at 97% similarity with reads number normalizing to 17,436. Community richness, evenness, and diversity (Shannon, Shannoneven, ACE, Chao, and Good’s coverage) were also assessed using Mothur. The online software, Ribosomal Database Project (RDP) classifier30 was used for taxonomy assignment for each Operational Taxonomic Units (OTU)31 using default parameters. Differences in bacterial diversity were assessed using analysis of similarities (ANOSIM), based on the unweighted UniFrac distance metrics using Mothur. The differences in features (taxonomy and OTU) were determined using STAMP tool via the Benjamini–Hochberg FDR test32. The prediction of microbiome functions were analyzed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software33, based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways34.
Representative OTUs were identified as species against the SILVA SSU database (132) and the NCBI online database with > 99% identity and highest total score29,35.
Results
Gut bacterial populations in 1- and 4-year-old children
A total of 10,210,871 (17,436–395,495) high-quality reads were obtained by high-throughput sequencing of 16S rRNA genes from 153 fecal samples (40 and 113 from 1- and 4-year-olds, respectively). To normalize data and avoid statistical bias, 17,436 16S rRNA genes of each sample were selected to calculate bacterial species richness, evenness, and diversity at 97% similarity. A total of 7195 OTUs (371 OTUs per sample on average) were obtained, and the Good’s coverage was over 99.8% for both groups (1- and 4-year-olds, Table 1), indicating that the sequencing depth was sufficient for gut microbiota investigation in children of two different ages.
Table 1.
The gut microbiota diversity evaluation of 1-year and 4-years old children.
| Group | Sample | OTUs | Coverage | Richness | Evenness | Diversity | |
|---|---|---|---|---|---|---|---|
| ACE | Chao | Shannoneven | Shannon | ||||
| 1-year | 40 | 1159 | 0.998975 | 1580.63 | 1584.31 | 0.566114 | 3.99411 |
| 4-years | 113 | 7089 | 0.999931 | 7103.50 | 7106.58 | 0.56353 | 4.99643 |
Bacterial composition changes with age
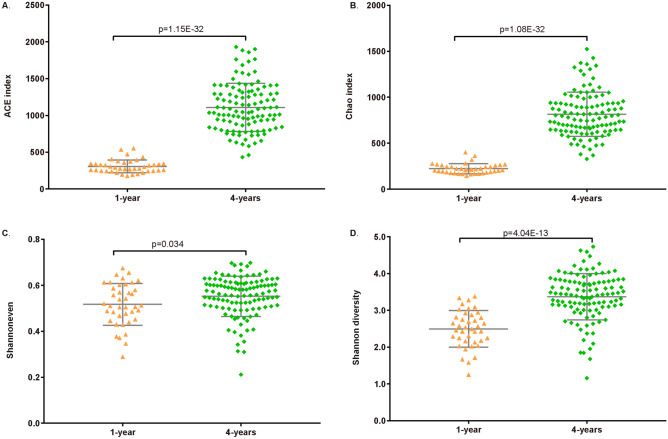
Based on the unweighted UniFrac distance metrics, principal coordinate analyses (PCoA) showed two significant parts divided by two different point of age (Fig. 1). ANOSIM analysis suggested that the microbial composition was significantly different (p < 0.001, R = 0.731) between 1- and 4-year-olds. Bacterial population evaluation showed that the species richness (ACE and Chao), species evenness, and diversity were significantly lower in the gut of 1-year-olds than in that of 4-year-olds (Fig. 2, p < 0.05). These results indicate that microbiota composition and diversity increased with the age of children.
Figure 1.
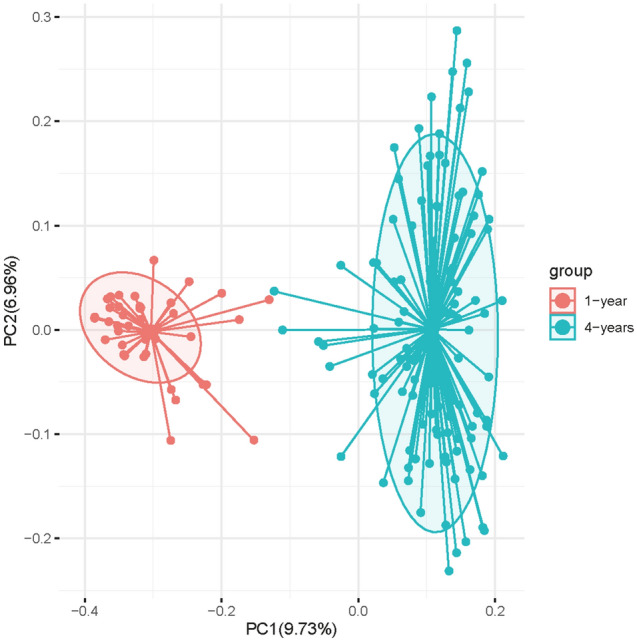
Principal coordinates analysis (PCoA) plots based on unweighted UniFrac distance metrics. Red points and blue points represent 1-year-old children and 4-year-old children, respectively.
Figure 2.
Richness, evenness and diversity of children gut microbiota. (A, B) were richness index, (C) was evenness index, (D) was diversity index. The p-value of top illustration was calculated by T-test, which indicated significant difference of gut microbiota between 1-year-old and 4-year-old children.
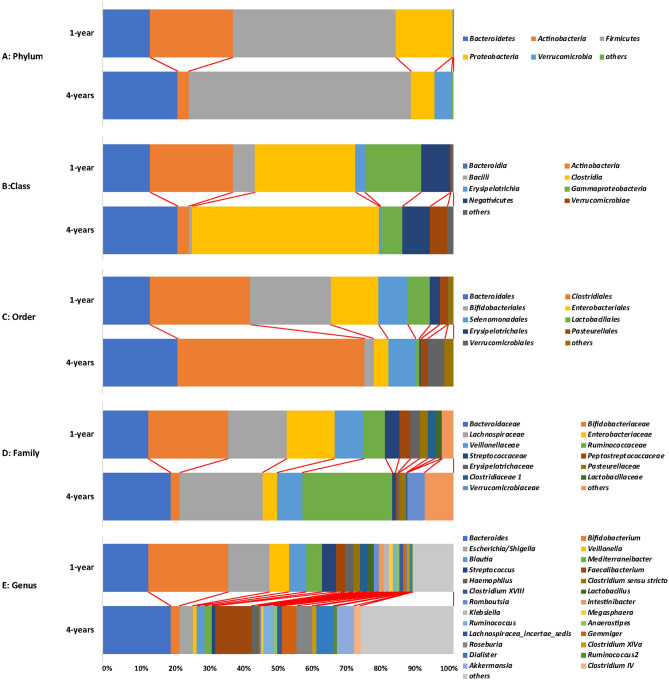
From phylum to species level, the microbiota differed significantly between 1- and 4-year-olds. At the phylum level, a total of 12 phyla were confirmed, with five major phyla (Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia) composing > 99% of gut microbiota (Fig. 3), and three phyla unique to 4-year-olds. Phyla Actinobacteria and Proteobacteria showed significant enrichment in 1-year-olds (q < 0.05), while phyla Firmicutes, Synergistetes, and Verrucomicrobia were significantly enriched in 4-year-olds (Fig. 4A).
Figure 3.
The gut microbiota composition of 1-year-old and 4-years-old children on different taxa level. The main bacteria phylum, class, order, family and genus in children gut were illustrated in different colors.
Figure 4.
Gut microbiota comparison between 1-year and 4-yearold children at phylum level (A), family level (B), genus level (C) and species level (D). The taxa with significant difference (q-value < 0.05, computed by STAMP) between the two groups are shown.
At the class level, a total of 20 classes were revealed; 16 classes were identified in both 1- and 4-year-olds and nine classes were significantly different between both groups (Figure S1), with Actinobacteria, Bacilli, Erysipelotrichia and Gammaproteobacteria significantly enriched in 1-year-olds, and Betaproteobacteria, Clostridia, Deltaproteobacteria, Synergistia and Verrucomicrobiae significantly increased in 4-year-olds.
At the order level, 38 orders were identified in total; 28 orders were identified in both groups, and nine orders were significantly different between the groups (Figure S2), including Bifidobacteriales, Enterobacteriales, Erysipelotrichales and Lactobacillales enriched in 1-year-olds and Burkholderiales, Clostridiales, Desulfovibrionales, Synergistales, and Verrucomicrobiales enriched in 4-year-olds.
At the family level, 81 families were identified in total; 58 families were identified in both groups, 19 families were significantly different between the groups (Fig. 4B). Eight families, Actinomycetaceae, Bifidobacteriaceae, Clostridiaceae 1, Enterobacteriaceae, Enterococcaceae, Erysipelotrichaceae, Peptostreptococcaceae and Streptococcaceae were significantly enriched in 1-year-olds, while the other 11 families, like Lachnospiraceae, Ruminococcaceae and Verrucomicrobiaceae were significantly enriched in 4-year-olds.
At the genus level, 203 genera were identified in total; 133 genera were identified in both groups, 18 of which were major genera in both groups (> 1% per group Table 2). The fecal microbiome of 1-year-olds was generally dominated by Bifidobacterium, Escherichia/Shigella, and Bacteroides, each genus composing > 12% (total 47.4%) of the microbiome. While in 4-year-olds, the dominant genera were Bacteroides (19.3%) and Faecalibacterium (10.2%). A total of 46 genera (including 11 major genera) were significantly different between both groups (Fig. 4C). Noteworthily, 12 genera including Actinomyces, Blautia, Clostridium sensu stricto, Clostridium XVIII, Eggerthella, Intestinibacter, Klebsiella, Streptococcus, Bifidobacterium, Escherichia/Shigella and Veillonella) were significantly enriched in 1-year-olds. The other 34 genera, including Akkermasia, Dialister, Faecalibacterium, Gemmiger, Roseburia, etc., were significantly enriched in 4-year-olds. The dominant genus, Bacteroides, maintained a stable population with age (12.9% and 19.3% in 1- and 4-year-olds, respectively).
Table 2.
Dominant genera and significant difference between 1-year and 4-years old children.
| Genus | Feature | 1-year: mean rel. freq. (%) | 1-year: std. dev. (%) | 4-years: mean rel. freq. (%) | 4-years: std. dev. (%) | q-values | Enriched in |
|---|---|---|---|---|---|---|---|
| Actinomyces | Difference | 0.1017 | 0.1187 | 0.0239 | 0.0296 | 0.0021 | 1-year |
| Akkermansia | Major and difference | 0.1368 | 0.7098 | 4.6110 | 13.4165 | 0.0052 | 4-years |
| Alistipes | Difference | 0.0612 | 0.2742 | 0.8799 | 1.8020 | 0.0002 | 4-years |
| Anaerofustis | Difference | 0.0006 | 0.0035 | 0.0034 | 0.0091 | 0.0268 | 4-years |
| Anaerotruncus | Difference | 0.0194 | 0.0529 | 0.0750 | 0.1206 | 0.0015 | 4-years |
| Barnesiella | Difference | 0.0000 | 0.0000 | 0.0307 | 0.1228 | 0.0376 | 4-years |
| Bifidobacterium | Major and difference | 22.4657 | 21.6951 | 2.4992 | 4.4353 | 0.0001 | 1-year |
| Bilophila | Difference | 0.0016 | 0.0061 | 0.0827 | 0.1719 | 0.0001 | 4-years |
| Blautia | Major and difference | 4.8976 | 5.9816 | 2.2828 | 2.9867 | 0.0458 | 1-year |
| Butyricicoccus | Difference | 0.0978 | 0.2197 | 0.3386 | 0.5161 | 0.0011 | 4-years |
| Butyricimonas | Difference | 0.0003 | 0.0017 | 0.0185 | 0.0657 | 0.0202 | 4-years |
| Christensenella | Difference | 0.0000 | 0.0000 | 0.0070 | 0.0177 | 0.0009 | 4-years |
| Cloacibacillus | Difference | 0.0000 | 0.0000 | 0.0017 | 0.0063 | 0.0293 | 4-years |
| Clostridium IV | Major and difference | 0.0136 | 0.0587 | 1.8773 | 3.7870 | 0.0001 | 4-years |
| Clostridium sensu stricto | Major and difference | 1.9968 | 3.8008 | 0.2728 | 0.6705 | 0.0324 | 4-years |
| Clostridium XlVa | Major and difference | 0.5308 | 0.7931 | 1.1052 | 1.8900 | 0.0381 | 4-years |
| Clostridium XlVb | Difference | 0.0067 | 0.0195 | 0.4134 | 0.9732 | 0.0004 | 4-years |
| Clostridium XVIII | Major and difference | 1.9694 | 3.7634 | 0.0981 | 0.1562 | 0.0186 | 1-year |
| Coprococcus | Difference | 0.0324 | 0.1297 | 0.2113 | 0.5193 | 0.0073 | 4-years |
| Dialister | Major and difference | 0.5084 | 2.8026 | 4.8636 | 10.4218 | 0.0011 | 4-years |
| Eggerthella | Difference | 0.1535 | 0.2235 | 0.0381 | 0.0516 | 0.0151 | 1-year |
| Eisenbergiella | Difference | 0.0000 | 0.0000 | 0.0294 | 0.0620 | 0.0001 | 4-years |
| Enterococcus | Difference | 0.2863 | 0.4723 | 0.0060 | 0.0254 | 0.0051 | 1-year |
| Escherichia/Shigella | Major and difference | 11.9535 | 14.2319 | 3.9337 | 10.4966 | 0.0126 | 1-year |
| Ezakiella | Difference | 0.0008 | 0.0038 | 0.0105 | 0.0362 | 0.0284 | 4-years |
| Faecalibacterium | Major and difference | 2.4513 | 4.7935 | 10.2261 | 13.3559 | 0.0000 | 4-years |
| Gemmiger | Major and difference | 0.7854 | 2.5049 | 4.2201 | 8.4445 | 0.0018 | 4-years |
| Holdemania | Difference | 0.0177 | 0.0735 | 0.0756 | 0.1338 | 0.0074 | 4-years |
| Hungatella | Difference | 0.0011 | 0.0040 | 0.0110 | 0.0356 | 0.0214 | 4-years |
| Intestinibacter | Major and difference | 1.4063 | 1.6740 | 0.1925 | 0.3323 | 0.0009 | 1-year |
| Intestinimonas | Difference | 0.0000 | 0.0000 | 0.0559 | 0.1416 | 0.0008 | 4-years |
| Klebsiella | Major and difference | 1.3742 | 2.7721 | 0.1372 | 0.4604 | 0.0355 | 1-year |
| Lactonifactor | Difference | 0.0008 | 0.0037 | 0.0051 | 0.0116 | 0.0050 | 4-years |
| Odoribacter | Difference | 0.0000 | 0.0000 | 0.0418 | 0.1476 | 0.0182 | 4-years |
| Oscillibacter | Difference | 0.0125 | 0.0326 | 0.8457 | 1.0766 | 0.0000 | 4-years |
| Parabacteroides | Difference | 0.2177 | 0.5702 | 0.7776 | 1.0870 | 0.0011 | 4-years |
| Parasutterella | Difference | 0.0118 | 0.0562 | 0.2985 | 0.9139 | 0.0085 | 4-years |
| Parvimonas | Difference | 0.0011 | 0.0032 | 0.0058 | 0.0148 | 0.0112 | 4-years |
| Phascolarctobacterium | Difference | 0.0089 | 0.0474 | 0.7095 | 1.9686 | 0.0023 | 4-years |
| Pseudoflavonifractor | Difference | 0.0000 | 0.0000 | 0.0011 | 0.0038 | 0.0111 | 4-years |
| Pyramidobacter | Difference | 0.0003 | 0.0017 | 0.0062 | 0.0245 | 0.0454 | 4-years |
| Roseburia | Major and difference | 0.5940 | 1.6508 | 4.4477 | 8.4708 | 0.0002 | 4-years |
| Ruminococcus2 | Major and difference | 0.3787 | 0.9090 | 1.0312 | 1.6200 | 0.0144 | 4-years |
| Scardovia | Difference | 0.0000 | 0.0000 | 0.0010 | 0.0040 | 0.0379 | 4-years |
| Streptococcus | Major and difference | 4.1816 | 6.7029 | 0.8335 | 1.3996 | 0.0182 | 1-year |
| Bacteroides | Major | 12.9731 | 19.2120 | 19.29113 | 19.1440 | 0.2184 | 4-year |
| Veillonella | Major and difference | 5.5698 | 6.5315 | 1.2074 | 3.7846 | 0.0023 | 1-year |
At the species (OTU) level, 23 of the top 30 OTUs were significantly different between 1- and 4-year-olds (Table S1), with 19 of them confirmed as known species via NCBI BLAST (Fig. 4D). Ten species were significantly enriched in 1-year-olds, including Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium pseudocatenulatum, Blautia wexlerae, Fusicatenibacter saccharivorans, Romboutsia timonensis, Ruminococcus gnavus, Streptococcus salivarius, Escherichia coli, and Veillonella dispar, which occupied 39.9% of gut microbiota in 1-year-olds. The abundance of the other nine species, including Akkermansia muciniphila, Bacteroides uniformis, Bacteroides xylanisolvens, Gemmiger formicilis, Roseburia faecis, Roseburia inulinivorans, Ruminococcus bromii, Faecalibacterium prausnitzii, and Dialister invisus, increased significantly in 4-year-olds compared to that in 1-year-olds.
Predicted functional change of gut microbiota between 1-year-old and 4-year-old children
We used PICRUSt software to predict the potential functional changes with age. Sixty-nine predicted Metabolism Pathways, 17 pathways related to Genetic Information Processing, two pathways belonging to Cellular Processes, and three pathways participating Environmental Information Processing, were identified as having significant differences (q-value < 0.05) between 1- and 4-year-olds (Figure S3). Analysis revealed the relative abundance of genes involved in O-glycan biosynthesis, steroid biosynthesis, secondary bile acid biosynthesis, carotenoid biosynthesis, etc., were significantly increased in 4-year-olds. Genes participating in cell motility, membrane transport, galactose metabolism, folate biosynthesis, glutathione metabolism, etc., were significantly decreased in 4-year-olds.
Discussion
In this study, we compared the gut microbiota of 1- and 4-year-old Chinese children by investigating the V3–V4 hypervariable region of the 16S rRNA gene via high-throughput DNA sequencing. Our results revealed that the gut microbiota in children increased significantly from age 1 to 4. In terms of population, species richness, species evenness, and diversity were mainly dominated by five phyla (Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes and Verrucombicrobia). These findings are consistent with previous reports that the gut microbiome of children gradually mature as that of adults during the first three years of life25.
A lot of research in the past decades have focused on the development of the infant gut microbiota during the first 3 years of life; only, few reports have investigated the variation in gut microbiota in children above 3 years of age26, and further studies are needed. For example, Fiona et al. indicated that the gut microbiota of children was dynamic before age four due to the effects of perinatal factors36, and Ringel-Kulka et al. revealed that by age four the microbiota of children were still not as mature as those of adults, suggesting that the microbiota of children continue to progress after age 437.
Hence, our research paid attention to the development of children’s gut microbiota at age four for a deeper understanding of the intestinal microbiota of children. Through the unweighted UniFrac distance metrics, we demonstrated that there was a significant difference in gut microbiota compositions between 1- and 4-year-old children, suggesting that the gut microbiota of infants matures with age.
We analyzed specific differences from the phylum to the species level. At the phylum level, Actinobacteria and Proteobacteria were significantly reduced in the intestines of 4-year-olds. This result is consistent with previous reports that Actinobacteria, represented by Bifidobacterium, declined after weaning due to decreased protein requirements22,38,39. However, Firmicutes, Synergistetes, and Verrucomicrobia increased significantly in the intestines of 4-year-olds. It was recently noted that the abundance of Firmicutes is suppressed while children receive breast milk20. Once weaning begins, Firmicutes increase in abundance and dominate gut microbiota. It is supposed that the introduction of solid foods can increase bacterial load and short-chain fatty acid levels, which may be due to the ability of Firmicutes, such as Roseburia spp., to metabolize carbohydrates in the diet40,41. Bacteroides, which can breakdown complex plant polysaccharides42, maintain dominance of the gut microbiota with age, indicating that Bacteroides already attained stability at infancy. These results are consistent with previous reports that Firmicutes and Bacteroidetes are the most dominant phyla in healthy adult subjects43, suggesting a maturation of gut microbiota at age 4.
At the genus level, Bifidobacterium, Escherichia/Shigella, and Veillonella were enriched in the intestines of 1-year-olds. Bifidobacterium levels declined with age, in agreement with reports that Bifidobacterium is more abundant in children than in adults44. It is generally known that Bifidobacterium has several subspecies relating to infants. Bifidobacterium longum subsp. is a kind of archetypical bacteria capable of using human milk oligosaccharide (HMO) as substrates. B. longum subsp. infantis is an infant commensal that thrives in the presence of milk45. Our results showed that the abundance of Bifidobacterium decreased significantly in the gut microbiota of 4-year-olds, coinciding with the beginning of weaning and the introduction of table foods. In the gut microbiota of 4-year-olds, the abundance of Faecalibacterium, Dialister, Gemmiger, and Akkermansia increased significantly. Notably, Akkermansia is a probiotic, and may be more beneficial for growing children as they face a more complex diet and other environmental factors. Akkermansia helps regulate the thickness of intestinal mucus and maintain intestinal barrier integrity to reduce sugar absorption. It is beneficial for weight loss, blood sugar regulation and diabetes mellitus management46.
At the species level, F. prausnitzii, D. invisus, A. muciniphila, R. bromii, B. xylanisolvens, G. formicilis, R. faecis and R. inulinivorans increased significantly with age, while E. coli, B. pseudocatenulatum, B. longum, B. wexlerae, F. saccharivorans, R. timonensis, R. gnavus, S. salivarius, V. dispar and B. breve were more abundant in the intestines of 1-year-olds than in those of 4-year-olds. F. prausnitzii is one of the most abundant microorganisms in the intestinal tract of healthy people; it can generate butyrate as an anti-inflammatory to help slow down inflammatory bowel disease46. A. muciniphila, a mucin-degrading probiotic, increased from 0.14% in 1-year-olds to 4.25% in 4-year-olds. A. muciniphila can regulate immune responses by promoting relevant gene expression47, and reverse high-fat diet-induced metabolic disorders by improving host metabolism46. R. inulinivorans belong to the genus Roseburia, which was reported to produce short-chain fatty acids and play a major role in maintaining gut health and immune defense48. The increased levels of these profitable species suggest that the intestinal environment of 4-year-olds is attaining adult-like maturity.
Bifidobacterium is able to thrive on HMOs and is dominant in infant gut microbiota before weaning24. The abundance of these milk-related Bifidobacterium, including B. pseudocatenulatum, B. longum, and B. breve, significantly decreased in 4-year-olds due to the change in their diet (from milk to solid food). The onset of weaning is usually associated with an increase in a diversity of intestinal microbiota, with Bifidobacteria-dominated intestinal microbiota gradually being replaced by more complex microbial communities capable of degrading carbohydrates from plant and animal sources.
In this study, PICRUSt software was used to predict the potential function of the gut microbiota of 1- and 4-year-olds. Bacteria involved in galactose metabolism, amino acid metabolism, cofactor and vitamin metabolism like folate biosynthesis, nicotinate and nicotinamide metabolism, vitamin B6 metabolism, etc., were significantly enriched in 1-year-olds. While the abundance of microbiota participating in lipid metabolism, metabolism of terpenoids and polyketides pathways increased with age. This functional change in microbiota is consistent with children’s diet changes.
Nevertheless, the current study has limitations as this study used fecal samples. As is known, the fecal microbiota does not fully represent the luminal or mucosal communities of the GI tract49,50. Although previous study revealed that fecal microbial community has a good potential to identify most taxa in the chicken gut51, non-invasively sampling at different gut locations would be preferred, and recently developed smart sampling capsule would achieve this goal52. Another limitation of our study is lacking of paying attention to the factors influencing the development of intestinal microbiota in children, such as breast milk feeding, dietary habits and antibiotic use.
In conclusion, the first 4 years of life is a crucial period for the formation of intestinal microbiota in young children, and has a profound impact on subsequent physical development and health. Our study demonstrates that the intestinal microbiota composition of infants changed from Bifidobacteria-dominated to a more complex microbiota, and attained adult-like intestinal microbiota maturation by age four. It is worthy of note that owing to various influencing factors, there are great differences in the composition and development of intestinal microbiota among different populations; our study is scientifically relevant among existing studies involving other ethnic groups.
Supplementary information
Acknowledgements
This study was supported by grants from the National Key Research and Development Program of China (Grant no: 2016YFC1000505); Shanghai Municipal Health Commission (Grant no: 20194Y0160); the Shanghai Sailing Program (Grant no: 17YF1416100); and Innovation-oriented Science and Technology Grant from NHC Key Laboratory of Reproduction Regulation (Grant no: CX2017-06).
Abbreviations
- GI
Gastrointestinal
- S-MBCS
Shanghai-Minhang Birth Cohort Study
- RDP
Ribosomal Database Project
- OTU
Operational Taxonomic Units
- ANOSIM
Analysis of similarities
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PCoA
Principal coordinate analyses
- HMO
Human milk oligosaccharide
Author contributions
W.Y. and H.Z. designed the project. Material preparation was performed by M.M., F.Y. and Y.C. DNA extraction and sequencing was performed by M.G. and M.D. Bioinformatics analysis was performed by M.G., M.M. and Y.W. The first draft of the manuscript was written by M.G., Y.W. and H.Z., and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The microbiota sequence data for the 1- and 4-year-old children have been deposited in the National Omics Data Encyclopedia (NODE, https://www.biosino.org/node/index) under the accession numbers OEX010570 and OEX010571, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Min Guo and Maohua Miao.
Contributor Information
Wei Yuan, Email: yuanwei11@yahoo.com.
Huajun Zheng, Email: zhenghj@chgc.sh.cn.
Supplementary information
is available for this paper at 10.1038/s41598-020-76591-4.
References
- 1.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aw W, Fukuda S. Understanding the role of the gut ecosystem in diabetes mellitus. J. Diabetes Investig. 2018;9:5–12. doi: 10.1111/jdi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. ILAR J. 2015;56:192–204. doi: 10.1093/ilar/ilv030. [DOI] [PubMed] [Google Scholar]
- 5.Maeda Y, Takeda K. Role of gut microbiota in rheumatoid arthritis. J. Clin. Med. 2017;6:60. doi: 10.3390/jcm6060060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Han Y, Dy ABC, Hagerman RJ. The gut microbiota and autism spectrum disorders. Front. Cell Neurosci. 2017;11:120. doi: 10.3389/fncel.2017.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zitvogel L, et al. Cancer and the gut microbiota: An unexpected link. Sci. Transl. Med. 2015;7:271. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carabotti M, Scirocco A, Maselli MA, Severi C. The gut–brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. Quart. Publ. Hellenic Soc. Gastroenterol. 2015;28:203–209. [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathi A, et al. The gut–liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsland BJ, Trompette A, Gollwitzer ES. The gut–lung axis in respiratory disease. Ann. Am. Thorac. Soc. 2015;12(Suppl 2):S150–156. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez JM, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: Health and disease. Front. Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanislawski MA, et al. Gut microbiota in the first 2 years of life and the association with body mass index at age 12 in a norwegian birth cohort. MBio. 2018;9:e01751–e1818. doi: 10.1128/mBio.01751-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bisgaard H, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011;128(646–652):e641–645. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 15.Ihekweazu FD, Versalovic J. Development of the pediatric gut microbiome: Impact on health and disease. Am. J. Med. Sci. 2018;356:413–423. doi: 10.1016/j.amjms.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 17.Mandar R, Mikelsaar M. Transmission of mother's microflora to the newborn at birth. Biol. Neonate. 1996;69:30–35. doi: 10.1159/000244275. [DOI] [PubMed] [Google Scholar]
- 18.Balmer SE, Wharton BA. Diet and faecal flora in the newborn: Breast milk and infant formula. Arch. Dis. Child. 1989;64:1672–1677. doi: 10.1136/adc.64.12.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen HJ, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 20.Pannaraj PS, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sordillo JE, et al. Factors influencing the infant gut microbiome at age 3–6 months: Findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART) J. Allergy Clin. Immunol. 2017;139:482–491. doi: 10.1016/j.jaci.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Azad MB, et al. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derrien M, Alvarez AS, de Vos WM. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27:997–1010. doi: 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Zheng H, Liang H, Wang Y, Miao M, Shi T, Yang F, et al. Altered gut microbiota composition associated with eczema in infants. PLoS ONE. 2016;11:e0166026. doi: 10.1371/journal.pone.0166026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escapa IF, et al. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): A resource for the microbiome of the human aerodigestive tract. mSystems. 2018;3:e00187–e218. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouhy F, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat. Commun. 2019;10:1517. doi: 10.1038/s41467-019-09252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ringel-Kulka T, et al. Intestinal microbiota in healthy US young children and adults—A high throughput microarray analysis. PLoS ONE. 2013;8:e64315. doi: 10.1371/journal.pone.0064315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arboleya S, Watkins C, Stanton C, Ross RP. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan SH, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 43.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agans R, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol. Ecol. 2011;77:404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derrien M, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Jolivet-Gougeon A. Roseburia spp.: A marker of health? Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 49.Durban A, et al. Assessing gut microbial diversity from feces and rectal mucosa. Microb. Ecol. 2011;61:123–133. doi: 10.1007/s00248-010-9738-y. [DOI] [PubMed] [Google Scholar]
- 50.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan W, et al. Efficacy of fecal sampling as a gut proxy in the study of chicken gut microbiota. Front. Microbiol. 2019;10:2126. doi: 10.3389/fmicb.2019.02126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waimin JF, et al. Smart capsule for non-invasive sampling and studying of the gastrointestinal microbiome. RSC Adv. 2020;10:16313–16322. doi: 10.1039/C9RA10986B. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microbiota sequence data for the 1- and 4-year-old children have been deposited in the National Omics Data Encyclopedia (NODE, https://www.biosino.org/node/index) under the accession numbers OEX010570 and OEX010571, respectively.