Abstract
Background
Atrial fibrillation (AF) is related to structural and electrical atria remodeling. Atrial fibrosis development and progression is characteristic of structural remodeling and is taken as the AF perpetuation substrate. Increasing evidence has confirmed that microRNAs (miRNAs) are associated with AF, including cardiac fibrosis.
Methods
Pericardial fluid (PF) samples were collected from nine adult patients who had congenital heart disease with persistent AF or sinus rhythm (SR) undergoing surgery. Abnormally expressed miRNAs were acquired, and P<0.05 and fold change >2 were taken as the thresholds of differentially expressed miRNAs (DE-miRNAs). The predicted target genes were obtained by miRTarBase. The Database for Annotation, Visualization and Integrated Discovery was used to annotate functions and analyze pathway abundance for latent targets of DE-miRNAs. STRING database was applied to construct a protein–protein interplay (PPI) network, and Cytoscape software was used to visualize the miRNA-hub gene-Kyoto Encyclopedia of Genes and Genomes (KEGG) network. DE-miRNA expressions were evaluated by quantitative polymerase chain reaction (qPCR).
Results
Fifty-five exosomal DE-miRNAs were found between the AF and SR samples; these included 24 miRNAs that were upregulated and 31 that were downregulated. For the top 3 downregulated miRNAs (miR-382-3p, miR-3126-5p, and miR-450a-2-3p) 283 predicted target genes were identified, and were implicated in cardiac fibrosis-related pathways, including the hypoxia-inducible factor-1 (HIF1), mitogen-activated protein kinase (MAPK), and adrenergic and insulin pathways. The top 10 hub genes in the PPI network, including mitogen-activated protein kinase 1 (MAPK1) and AKT serine/threonine kinase 1 (AKT1), were identified as hub genes. By establishing the miRNA-hub gene-KEGG network, we observed that these hub genes, which were regulated by miR-382-3p, miR-3126-5p, and miR-450a-2-3p, were involved in many KEGG pathways associated with cardiac fibrosis, such as the AKT1/glycogen synthase kinsase-3β (GSK-3β) and transforming growth factor-β (TGF-β)/MAPK1 pathways.
Conclusions
The findings of the present study suggest that miR-382-3p, miR-450a-2-3p, and miR-3126-5p contained in exosomes in human PF are pivotal in the progression of AF. The results of qPCR showed that miR-382-3p was consistent with our sequencing data, which indicates its potential value as a therapeutic target for AF.
Keywords: Exosome, microRNA (miRNA), pericardial fluid (PF), atrial fibrillation (AF)
Introduction
Atrial fibrillation (AF), the most prevalent sustained supraventricular arrhythmia, increases the risk of stroke and heart failure. AF affects approximately 1% of the global population, with an increasing incidence among elderly people (1,2). The related mechanism is complicated and is considered to be associated with structural and electrical atrial remodeling (3,4).
Pericardial fluid (PF) is a plasma ultrafiltrate inside the double-walled pericardial sac (pericardium). PF surrounds the heart and the great vessel roots, which transport blood to the heart. Possibly derived from the myocardium, PF contains bioactive factors and brain and atrial natriuretic peptides, including endothelin-1 (5). The components of PF might be reflective of the myocardium expression profile.
MicroRNAs (MiRNAs) are regulators of gene expression. After transcription, they suppress gene expression by degrading or functionally suppressing target mRNAs (6). MiRNAs can be produced by parent cells in bioactive forms and are not destroyed in biologic fluids, thereby exhibiting their potential as novel biomarkers (7). The resilience of extracellular miRNA comes as a result of different partnerships, including extracellular vesicles (EVs), protein complexes, and lipoprotein complexes (6).
Exosomes, which are the smallest known endogenous EVs, exist inside multivesicles due to endosome compartmentalization. They are produced when multivesicles fuse with cell membranes (8). Non-parent cells take up exosomal miRNAs, which can affect their gene expression, and can function as messengers between cells in local and remote micro-communication mechanisms (6). Previous research has found that miRNAs can produce their effects via exosomes in human PF (9). However, their function in cardiac remodeling in AF remains unknown.
The rapid development of high-throughput sequencing (HTS) has facilitated the study of the transcription patterns of pathophysiological processes. Numerous studies have described ncRNAs associated with AF (10,11). However, miRNAs contained in exosomes in human PF are not well characterized. The exosomal miRNA expression change in human PF is speculated to be related to AF pathogenesis. In the present study, differential expression profiles of exosomal miRNAs in PF were obtained through integrating HTS, quantitative polymerase chain reaction (qPCR) validation, and related bioinformatic analyses. These findings may help to determine the underlying mechanism of AF and could facilitate the development of more promising treatment targets. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2066).
Methods
PF specimen collection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethic Committee of Xiangya Hospital Central South University (201803209) and informed consent was taken from all individual participants. PF samples were collected from nine adult patients with congenital heart disease accompanied by sinus rhythm (SR) or persistent AF undergoing surgery. All participants provided signed informed consent before surgery. The specimens were instantly quickly frozen in liquid N2 and preserved at −80˚C for RNA isolation. Nine specimens were used for miRNA HTS using an Illumina/Solexa G1 sequencer (Shanghai Biotech). Additionally, for real-time PCR validation, total RNA was extracted from the exosomes in the PF of patients in the SR or AF groups (n=60 each).
Target gene prediction
The possible target genes of the top 3 most downregulated miRNAs were predicted with miRTarBase, an experimentally verified database for microRNA-target interplays (12).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
The Database for Annotation, Visualization and Integrated Discovery 6.8 was used to annotate functions and analyze pathway abundance for the possible targets of the three chosen differentially expressed miRNAs (DE-miRNAs) involved in GO and KEGG pathways (13,14). P<0.05 indicated statistical significance.
Protein–protein interplay (PPI) and miRNA-hub-KEGG pathway networks
The miRNA-hub KEGG and PPI networks were consecutively established. The target genes were first introduced into the STRING database to evaluate correlations in functions between them (15). A combined score >0.4 for interplays indicated significance. To obtain hub genes, the connectivity extent in the PPI network was investigated through Cytoscape 3.7.2, and the miRNA-hub-KEGG pathway network was then constructed through ClueGO + CluePedia.
Exosome precipitation from PF
Exosomes were enriched from PF using the ExoQuick kit (System Biosciences). The PF was collected and centrifuged at 3,000 ×g for 15 minutes to remove cells and cell debris. The supernatant was transferred to a sterile vessel and the appropriate volume of ExoQuick exosome precipitation solution was added to the PF. The ExoQuick/PF mixture was centrifuged at 1,500 ×g for 30 minutes. The residual ExoQuick solution was centrifuged at 1,500 ×g for 5 minutes to remove all traces of fluid by aspiration. The exosome pellet was resuspended in 100–500 µL phosphate-buffered saline.
Transmission electron microscopy (TEM) and Western blotting of exosomes
For TEM processing, 100 µL isolated exosomes were fixed in 2.5% glutaraldehyde for 2 hours. Next, the fixed exosomes were dropped onto a copper net and stained with 2% phosphotungstic acid oxalate for 2 minutes. Images were acquired by a transmission electron microscope (HT-7800; Hitachi).
To test specific proteins, the proteins were obtained from exosomes by using RIPA lysis buffer (Beyotime Biotechnology). The proteins were loaded and separated on 10% sodium dodecylsulfate–polyacrylamide gel electrophoresis (Dalian Meilun Biotechnology), transferred to a nitrocellulose membrane, blocked in 5% bovine serum albumin for 90 minutes at room temperature, and incubated in primary antibody 4°C overnight. The membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (HRP goat anti-mouse immunoglobulin G 1:5,000) at room temperature for 90 minutes and observed using the enhanced chemiluminescence system (ECL) reagent (Advansta).
The antibodies used were as follows: antibody against CD9 (Abgent category no. AP7251E, RRID:AB_2140114), antibody against CD63 (Abcam category no. S2377, RRID:AB_10641812), antibody against TSG101 (Abcam category no. ab70120, RRID:AB_1271357), antibody against GAPDH (Abcam category no. ab70700, RRID:AB_1209515), and antibody against immunoglobulin G (Abcam category no. ab47055, RRID:AB_955359).
RNA extraction and real-time PCR validation
Total RNA was extracted according to the manufacturer's instructions; 200 µL of sample was used with 1 mL of TRIzol (Thermo Fisher Scientific). Reverse transcription of miRNAs was conducted using a reverse transcription kit (Cowin), and qPCR primers were obtained from Sangon; qPCR was performed in triplicate using the UltraSYBR Mixture (Cowin). miRNA expression was normalized with U6 small nuclear RNA. The 2−ΔΔCT method was used to identify fold change in the RNA level of the sample relative to the reference sample.
Data analysis
Data were presented as mean ± standard deviation. Intergroup differences were determined through unpaired t-test. Two-tailed P<0.05 indicated data significance.
Results
Identification of DE-miRNAs and their target genes
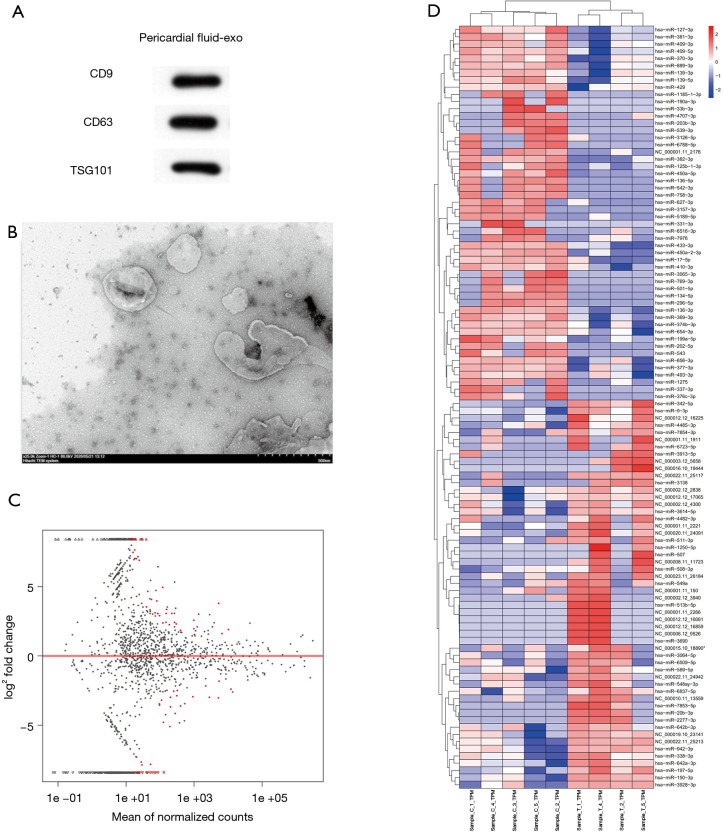
The exosomes from PF were validated by TEM and Western blot (Figure 1A,B). Fold change >2 and P<0.05 were taken as the DE-miRNA identification thresholds. Volcano plots and hierarchical clustering heatmap confirmed that the miRNA expressions differed significantly (Figure 1C,D). The miRNA profiling between the AF (n=4) and SR (n=5) groups was investigated. Fifty-five miRNAs in total were found to have significantly different expressions between the AF and SR groups; 24 had upregulated miRNAs and 31 had downregulated miRNAs, indicating a twofold change in the AF group relative to the SR group. The top 3 most differentially expressed miRNAs are presented in Table 1. Based on fold change, miR-382-3p, miR-3126-5p, and miR-450a-2-3p, were the top 3 most downregulated miRNAs; 283 possible target genes of the 3 miRNAs were predicted by using miRTarBase.
Figure 1.
Identification of exosomal microRNAs (miRNAs) and their target genes. (A,B) Exosomes from pericardial fluid (PF) were identified through transmission electron microscopy and Western blot. (C) Differentially expressed miRNA volcano plot. Black dots represent miRNAs that had no different expressions between 4 human PF specimens with atrial fibrillation (AF) and 5 without AF; red dots represent the differential expression of miRNAs. (D) Hierarchical clustering analysis (heatmap) indicating differential miRNA expressions between specimens from HTS. Expression levels are described in the color scale; red and blue indicate high and low relative expressions, respectively. Each miRNA is presented by a single row of colored boxes, and each specimen is represented by one column. Specimen T represents the AF group and specimen C represents the sinus rhythm group.
Table 1. Top 3 decreased miRNAs with different expressions between AF and SR groups.
| miRNA | Log2FC | P | Adj. P | Expression |
|---|---|---|---|---|
| hsa-miR-382-3p | −9.28675505 | 0.003279341 | 0.276672832 | Down |
| hsa-miR-3126-5p | −9.213282215 | 0.000141042 | 0.090964988 | Down |
| hsa-miR-450a-2-3p | −8.657240598 | 0.000522965 | 0.135646115 | Down |
AF, atrial fibrillation; SR, sinus rhythm.
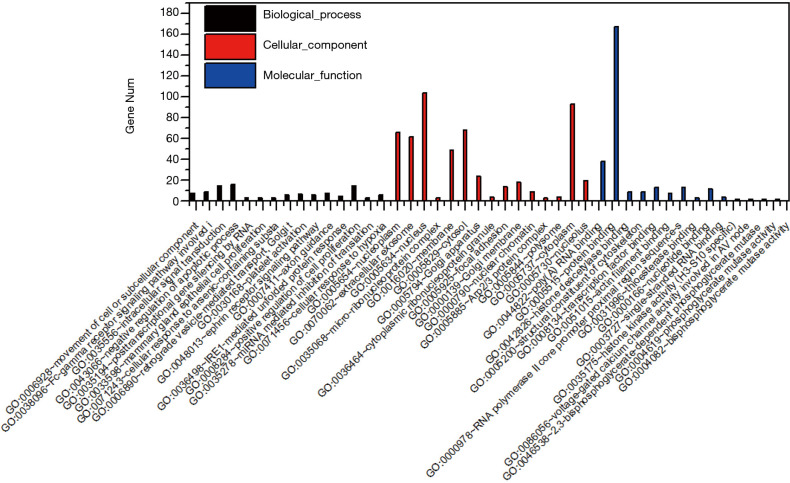
GO functional enrichment
Three types of annotations for GO functions were conducted on 3 possible target genes: biologic process (BP), cell composition (CC), and molecular function (MF). The various GO functions are shown in Figure 2, and include intracellular signal transduction, negative regulation of apoptotic process, platelet activation, cellular response to hypoxia, and positive regulation of cAMP-response element binding protein (CREB) transcription factor activity in the BP category; nucleus, micro-ribonucleoprotein complex, membrane, and focal adhesion in the CC category; and protein binding, transcription factor binding, and nucleotide binding in the MF category.
Figure 2.
Gene ontology functions for the target genes of the top 3 downregulated microRNAs.
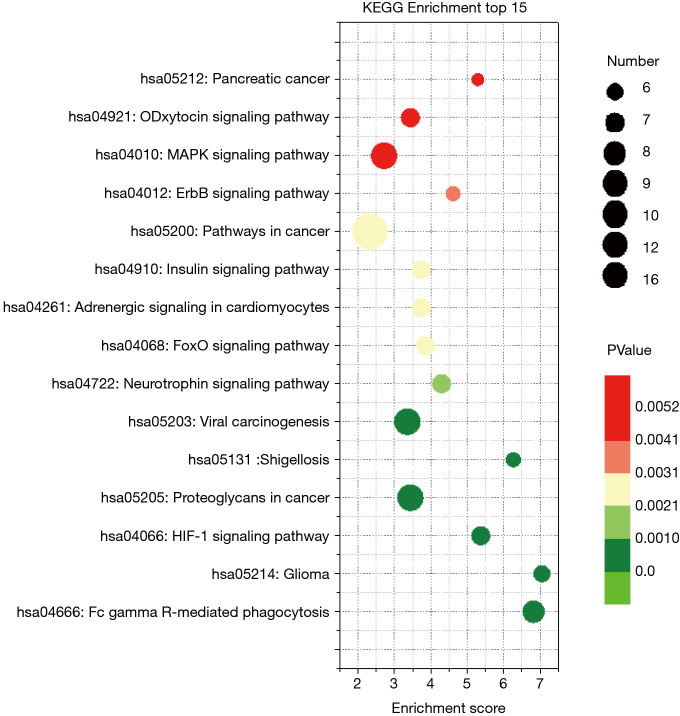
KEGG pathway enrichment
KEGG pathway enrichment was analyzed to investigate the pathways of these target genes, including the hypoxia-inducible factor-1 (HIF-1), mitogen-activated protein kinase (MAPK), and adrenergic and insulin pathways (Figure 3).
Figure 3.
Kyoto Encyclopedia of Genes and Genomes pathway abundance analysis of target genes of 3 regulated differentially expressed microRNAs. Only enriched pathways with P<0.05 are shown. Size of nodes represents gene count, and the colors represent P values.
PPI and miRNA-hub KEGG network establishment and analysis
The PPI network of target genes of the top 3 downregulated miRNAs was established through the STRING database. Data indicated that a number of target genes were able to interplay with each other. The top 10 hub genes were selected based on the node degree (Table 2). For the downregulated miRNAs, the hub genes were MAPK1, AKT1, ACTB, CCND1, VEGFA, GRB2, ACTR2, RAC1, YWHAZ, and RHOC; MAPK1 and AKT1 demonstrated the highest node degree (52 and 34, respectively), indicating that they were potentially important target genes related to cardiac fibrosis in AF.
Table 2. Hub genes of three decreased miRNAs performed in the PPI interplay.
| Gene | Degree |
|---|---|
| AKT1 | 52 |
| MAPK1 | 34 |
| ACTB | 34 |
| CCND1 | 22 |
| VEGFA | 21 |
| GRB2 | 20 |
| ACTR2 | 18 |
| RAC1 | 18 |
| YWHAZ | 18 |
| RHOC | 18 |
PPI, protein–protein interplay.
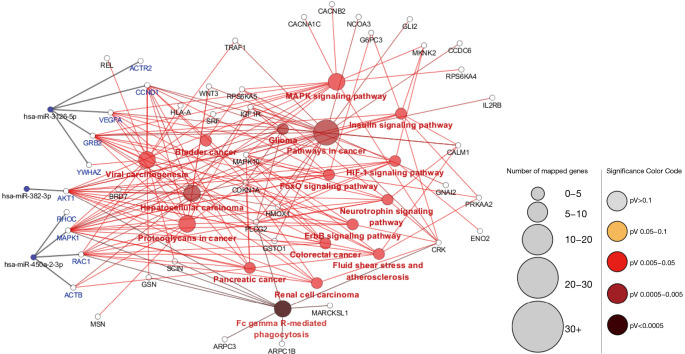
The miRNA-hub-KEGG network was then established through Cytoscape (Figure 4). We found that 5 genes (ACTR2, CCND1, VEGFA, GRB2, and YWHAZ) were likely to be regulated by miR-3126-5p, 4 hub genes (RHOC, MAPK1, RAC1, ACTB) were likely to be regulated by miR-450a-2-3p, and 1 hub gene (AKT1) were likely to be regulated by miR-382-3p. As known, an opposite correlation between miRNAs and target gene expressions exists. Therefore, these hub genes, regulated by downregulated miRNAs, might have higher expression in AF samples compared with SR samples. The data suggest that the 3 miRNAs might be potential modulators of cardiac fibrosis.
Figure 4.
Regulatory network of the three microRNAs, hub genes, and Kyoto Encyclopedia of Genes and Genomes pathways.
Quantitative reverse transcription PCR (qRT-PCR) verification of differentially expressed miRNAs
Three miRNAs were selected for qRT-PCR validation to confirm the confidence level of the sequencing results. As shown in Figure 5, miR-382-3p and miR-450a-2-3p were downregulated in the AF samples as compared to the SR samples. This result was in accordance with the HTS data, although the latter showed no statistical significance. However, miR-3126-5P was upregulated in the AF samples compared with the SR samples. To some extent, the functional roles of miR-3126-5P and miR-450a-2-3p in AF may be not as good as those of miR-382-3p, which plays a suppressive role in cardiac fibrosis.
Figure 5.
Expression of miR-382-3p, miR-3126-5p, and miR-450a-2-3p in atrial fibrillation. Data are expressed as mean ± standard deviation of three independent experiments. ****, P<0.0001.
Discussion
Structural and electrical remodeling, especially in atrial fibrosis, is identified as the AF onset substrate and progression and cardiac fibroblast (CF) expressing extracellular matrix (ECM) represents the most common cell type in the heart and plays a detrimental role in cardiac fibrosis (16). To date, effective therapeutic strategies against cardiac fibrosis are still rare, mainly because it is difficult to reverse the cardiac fibrosis process (17). The characteristics of atrial fibrosis include fibroblast migration, proliferation, and ECM protein precipitation. The ECM is an acellular component of the heart, comprising various fibers. Excessive ECM generation is usually related to the occurrence of cardiac remodeling. Fibroblast proliferation and matrix precipitation are related to collagen I and III upregulation, transforming growth factor-β1 (TGF-β1) activation, and downstream Smad pathways. The factors activating these pathways are various and include renin–angiotensin–aldosterone system activation, myocardial ischemia, and apoptosis. However, the underlying mechanism is not completely understood. Developing targets to suppress atrial fibrosis is beneficial to treating and preventing AF. Therefore, it is important to find a potential treatment target for atrial fibrosis prevention.
miRNAs are a series of endogenous small non-coding RNAs. They exhibit various biologic effects, such as regulating cardiac remodeling. Some miRNAs are associated with fibrogenesis. miR-133 and miR-30 are capable of modulating cardiac fibrosis through the inhibition of connective tissue growth factor (CTGF) expression. They have been observed to be downregulated in left ventricular hypertrophy related to upregulated CTGF expression (18). miR-29 is related to collagen I and III precipitation, and an increase in miR-29 results in a decrease in these proteins and vice versa (19).
In the present study, we profiled miRNA expression in human PF exosomes with AF, which, to the best of our knowledge, has not been done before, and discovered some predicted target genes of the top 3 regulated miRNAs after conducting a different expression analysis on an miRNA array from our HTS. Fifty-five miRNAs in total were identified, involving 31 downregulated miRNAs and 24 upregulated miRNAs. In total, 283 possible target genes were predicted for the 3 downregulated miRNAs by using miRTarBase. GO and KEGG enrichment analyses were performed to explore interactions among the target genes, which were mainly enriched in the BP terms associated with intracellular signal transduction, negative regulation of apoptotic process, platelet activation, cellular response to hypoxia, positive regulation of CREB transcription factor activity, and in the HIF-1 signaling pathway, adrenergic signaling in cardiomyocytes, insulin signaling pathway, and MAPK signaling pathway. These biologic processes and pathways are likely to be involved in the progression of cardiac fibrosis. Based on node degree, the top 10 hub genes of 3 downregulated miRNAs were selected, and the miRNA-hub-KEGG network was established. The hub genes that were potentially involved in cardiac remodeling were MAPK1, AKT1, CCND1, VEGFA, RAC1, and RHOC; MAPK1 and AKT1 exhibited the highest node degree (52 and 34, respectively). This result suggests that these genes, especially MAPK1 and AKT1, modulated by the three downregulated miRNAs, might play significant roles in the progression of AF.
qPCR indicated that miR-382-3p was significantly downregulated in exosomes from human PF. Recent studies have reported that miR-382 is involved in renal tubulointerstitial fibrosis through decreasing heat shock protein family D member 1 (HSPD1) (20), and is one of the 43 significantly upregulated miRNAs in idiopathic pulmonary fibrosis. However, to the best of our knowledge, the roles of miR-382-3p, miR-3126-5P, and miR-450a-2-3p in AF, especially in cardiac fibrosis, have not been investigated. It is therefore important to identify their effects in AF to determine the underlying mechanisms of cardiac remodeling modulation.
Among the various regulatory stimuli, TGF-β1 and angiotensin II (AngII) are the most effective factors of collagen preparation by cardiac fibrosis (21,22). TGF-β1 signals pass through no less than two isolated routes: one is the Smad-dependent canonical pathway, and the other is the SMAD-independent or non-canonical pathway. In the canonical pathway, the dimerized TGF-β1 receptor (TbRI and TbRII) is activated. Ligand-receptor binding causes cascade reactions of phosphorylation, inactivating Smad proteins 2, 3, and 4 of the Smad complex, which then translocates to the nucleus to regulate transcriptional responses (23). TGF-β1 is also able to signal through noncanonical SMAD-independent pathways, such as the AKT1/glycogen synthase kinsase-3β (GSK-3β), ras/Mitogen-Activated Protein Kinase Kinase 7 (MEK)/MAPK1(extracellular signal-regulated kinase [ERK]), TNF receptor-associated factor 4 (TRAF4), TRAF6, TGF-β-activated kinase 1, rhodopsin (RHO), nuclear factor-κB, and transient receptor potential cation channel subfamily C member 6 (TRPC6) (24,25).
AKT1, also known as AKT, is one of 3 important serine/threonine protein kinases (AKT1, AKT2, and AKT3). Its biologic processes regulate metabolism, proliferation, cell survival, and angiogenesis. AKT1 is a upstream effector of GSK-3β and inhibits its expression. Lal et al. reported that the AKT1/GSK-3β pathway induces cardiac fibrosis (26). They used 2 fibroblast-specific GSK-3β knockdown mouse models to show that GSK-3β loss in cardiac fibrosis causes left ventricle impairment, fibrogenesis, and excessive ischemic heart scarring. Recent research has found that TGF-β1 elicits an increase in INa, which is regulated by activating the phosphoinositide 3-kinase-AKT pathway through forkhead box protein O1 phosphorylation, contributing to the electrical remodeling related to myocardial injury (27).
MAPK1, also known as ERK, belongs to the MAPK family and is related to various cell processes, including proliferation, differentiation, transcription modulation, and development. Numerous studies have demonstrated that MAPK1 is an important molecule correlated to fibrogenesis (25,28-36); for example, miR-21 modulates fibroblast growth and survival factor secretion via the MAPK1 signaling pathway, controlling the interstitial fibrosis degree and cardiac hypertrophy (28). In addition, AngII induces endothelin 1 (ET-1) via MAPK1 and reactive oxygen species (ROS), thereby driving fibroblast activation and fibrosis (29). ET-1 induces ECM production and myofibroblast differentiation in fibroblasts (30). Some studies have suggested that ET-1 works together with TGF-β to promote myofibroblast differentiation (31). Furthermore, MAPK1, induced by TGF-β, is required for CTGF expression, which is one of the key markers of myofibroblast differentiation (32-34). Harada et al. found that TRPC3 modulates cardiac fibrosis proliferation by influencing Ca2+ influx via the MAPK1/miRNA-26/nuclear factor of activated T cells (NFAT) pathway to increase TRPC3 expression in an AF dog model (35). TRPC3 is related to cardiovascular diseases, including AF and cardiac fibrosis, directly eliciting migration and proliferation (36).
The findings of the present study demonstrate that miR-382-3p, miR-3126-5P, and miR-450a-2-3p are abnormally expressed in clinical specimens and are important modulators of cardiac fibrosis by inhibiting the expression of their target genes, such as MAPK1 and AKT1, which are regulated by miR-382-3p and miR-450a-2-3p, respectively, in AF. We described a new possible mechanism by which cardiac fibroblasts and/or myofibroblasts facilitate cardiac fibrosis via a previously unknown PF exosomal miR-382-3p→AKT1/GSK-3β and miR-450a-2-3p→TGF-β/MAPK1(ERK) regulatory pathway. However, the present study has several limitations as follows: (I) only the abundance of target genes of the top 3 most downregulated miRNAs were analyzed; (II) only three miRNAs were selected for experimental verification, possibly excluding several functional miRNAs; (III) studies on specific mechanisms underlying miR-382-3p, miR-3126-5P, and miR-450a-2-3p modulating AF remodeling were limited; and (IV) there was a lack of related in vitro and in vivo experiments in the study.
Conclusions
In the present study, we successfully identified three cardiac fibrosis-associated miRNAs secreted from exosomes in human PF (miR-382-3p, miR-450a-2-3p, and miR-3126-5P), according to the bioinformatic analysis. The results of the study indicate that these miRNAs play a pivotal role in AF atrial remodeling and can serve as potential targets for AF treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
The raw data of HTS have been submitted to SRA of NCBI. Accession Number were: SRR11862887-SRR11862895.
Funding: This work was supported by the National Natural Science Foundation of China (to Fanyan Luo; no. 81873494) and the Hunan Natural Science Foundation (to Fanyan Luo; no. 2018JJ2665).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethic Committee of Xiangya Hospital Central South University (201803209) and informed consent was taken from all individual participants.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2066
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2066
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2066). The authors have no conflicts of interest to declare.
References
- 1.Conen D. Epidemiology of atrial fibrillation. Eur Heart J 2018;39:1323-4. 10.1093/eurheartj/ehy171 [DOI] [PubMed] [Google Scholar]
- 2.Dzeshka MS, Lip GY, Snezhitskiy V, et al. Cardiac Fibrosis in Patients With Atrial Fibrillation: Mechanisms and Clinical Implications. J Am Coll Cardiol 2015;66:943-59. 10.1016/j.jacc.2015.06.1313 [DOI] [PubMed] [Google Scholar]
- 3.Burstein B, Nattel S. Atrial fibrosis: mechanisms and clinical relevance in atrial fibrillation. J Am Coll Cardiol 2008;51:802-9. 10.1016/j.jacc.2007.09.064 [DOI] [PubMed] [Google Scholar]
- 4.Corradi D. Atrial fibrillation from the pathologist's perspective. Cardiovasc Pathol 2014;23:71-84. 10.1016/j.carpath.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Kuosmanen SM, Hartikainen J, Hippeläinen M, et al. MicroRNA profiling of pericardial fluid samples from patients with heart failure. PLoS One 2015;10:e0119646. 10.1371/journal.pone.0119646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol 2014;63:2177-87. 10.1016/j.jacc.2014.01.050 [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 8.Emanueli C, Shearn AI, Angelini GD, et al. Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vascul Pharmacol 2015;71:24-30. 10.1016/j.vph.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beltrami C, Besnier M, Shantikumar S, et al. Human Pericardial Fluid Contains Exosomes Enriched with Cardiovascular-Expressed MicroRNAs and Promotes Therapeutic Angiogenesis. Mol Ther 2017;25:679-93. 10.1016/j.ymthe.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva AM, de Araújo JN, de Freitas RC, et al. Circulating MicroRNAs as Potential Biomarkers of Atrial Fibrillation. Biomed Res Int 2017;2017:7804763. 10.1155/2017/7804763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao H, Zhang M, Yang JJ, et al. MicroRNA-21 via Dysregulation of WW Domain-Containing Protein 1 Regulate Atrial Fibrosis in Atrial Fibrillation. Heart Lung Circ 2018;27:104-13. 10.1016/j.hlc.2016.01.022 [DOI] [PubMed] [Google Scholar]
- 12.Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46:D296-302. 10.1093/nar/gkx1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 14.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561-8. 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res 2009;105:1164-76. 10.1161/CIRCRESAHA.109.209809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Yu Y, Zhang P, et al. The crucial role of activin A/ALK4 pathway in the pathogenesis of Ang-II-induced atrial fibrosis and vulnerability to atrial fibrillation. Basic Res Cardiol 2017;112:47. 10.1007/s00395-017-0634-1 [DOI] [PubMed] [Google Scholar]
- 18.Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 2009;104:170-8, 6p following 178. [DOI] [PubMed]
- 19.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027-32. 10.1073/pnas.0805038105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, Xie T, Xue N, et al. miR-382 Contributes to Renal Tubulointerstitial Fibrosis by Downregulating HSPD1. Oxid Med Cell Longev 2017;2017:4708516. 10.1155/2017/4708516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan R, Sheppard R. Fibrosis in heart disease: understanding the role of transforming growth factor-beta in cardiomyopathy, valvular disease and arrhythmia. Immunology 2006;118:10-24. 10.1111/j.1365-2567.2006.02336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leask A. Potential therapeutic targets for cardiac fibrosis: TGFbeta, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ Res 2010;106:1675-80. 10.1161/CIRCRESAHA.110.217737 [DOI] [PubMed] [Google Scholar]
- 23.Greene RM, Nugent P, Mukhopadhyay P, et al. Intracellular dynamics of Smad-mediated TGFbeta signaling. J Cell Physiol 2003;197:261-71. 10.1002/jcp.10355 [DOI] [PubMed] [Google Scholar]
- 24.Hecker L, Vittal R, Jones T, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med 2009;15:1077-81. 10.1038/nm.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis J, Burr AR, Davis GF, et al. A TRPC6-dependent pathway for myofibroblast transdifferentiation and wound healing in vivo. Dev Cell 2012;23:705-15. 10.1016/j.devcel.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lal H, Ahmad F, Zhou J, et al. Cardiac fibroblast glycogen synthase kinase-3β regulates ventricular remodeling and dysfunction in ischemic heart. Circulation 2014;130:419-30. 10.1161/CIRCULATIONAHA.113.008364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur K, Zarzoso M, Ponce-Balbuena D, et al. TGF-β1, released by myofibroblasts, differentially regulates transcription and function of sodium and potassium channels in adult rat ventricular myocytes. PLoS One 2013;8:e55391. 10.1371/journal.pone.0055391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980-4. 10.1038/nature07511 [DOI] [PubMed] [Google Scholar]
- 29.Shephard P, Hinz B, Smola-Hess S, et al. Dissecting the roles of endothelin, TGF-beta and GM-CSF on myofibroblast differentiation by keratinocytes. Thromb Haemost 2004;92:262-74. 10.1160/TH03-11-0669 [DOI] [PubMed] [Google Scholar]
- 30.Leask A. Targeting the TGFbeta, endothelin-1 and CCN2 axis to combat fibrosis in scleroderma. Cell Signal 2008;20:1409-14. 10.1016/j.cellsig.2008.01.006 [DOI] [PubMed] [Google Scholar]
- 31.Shi-wen X, Kennedy L, Renzoni EA, et al. Endothelin is a downstream mediator of profibrotic responses to transforming growth factor beta in human lung fibroblasts. Arthritis Rheum 2007;56:4189-94. 10.1002/art.23134 [DOI] [PubMed] [Google Scholar]
- 32.Leask A, Holmes A, Black CM, et al. Connective tissue growth factor gene regulation. Requirements for its induction by transforming growth factor-beta 2 in fibroblasts. J Biol Chem 2003;278:13008-15. 10.1074/jbc.M210366200 [DOI] [PubMed] [Google Scholar]
- 33.Xie S, Sukkar MB, Issa R, et al. Regulation of TGF-beta 1-induced connective tissue growth factor expression in airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2005;288:L68-76. 10.1152/ajplung.00156.2004 [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Sun SQ, Hassid A, et al. cAMP inhibits transforming growth factor-beta-stimulated collagen synthesis via inhibition of extracellular signal-regulated kinase 1/2 and Smad signaling in cardiac fibroblasts. Mol Pharmacol 2006;70:1992-2003. 10.1124/mol.106.028951 [DOI] [PubMed] [Google Scholar]
- 35.Harada M, Luo X, Qi XY, et al. Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation 2012;126:2051-64. 10.1161/CIRCULATIONAHA.112.121830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han L, Li J. Canonical transient receptor potential 3 channels in atrial fibrillation. Eur J Pharmacol 2018;837:1-7. 10.1016/j.ejphar.2018.08.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as