Abstract
Here we describe the LifeTime Initiative, which aims to track, understand and target human cells during the onset and progression of complex diseases, and to analyse their response to therapy at single-cell resolution. This mission will be implemented through the development, integration and application of single-cell multi-omics and imaging, artificial intelligence and patient-derived experimental disease models during the progression from health to disease. The analysis of large molecular and clinical datasets will identify molecular mechanisms, create predictive computational models of disease progression, and reveal new drug targets and therapies. The timely detection and interception of disease embedded in an ethical and patient-centred vision will be achieved through interactions across academia, hospitals, patient associations, health data management systems and industry. The application of this strategy to key medical challenges in cancer, neurological and neuropsychiatric disorders, and infectious, chronic inflammatory and cardiovascular diseases at the single-cell level will usher in cell-based interceptive medicine in Europe over the next decade.
Subject terms: Systems biology, Biomarkers, Diseases
The LifeTime initiative is an ambitious, multidisciplinary programme that aims to improve healthcare by tracking individual human cells during disease processes and responses to treatment in order to develop and implement cell-based interceptive medicine in Europe.
Main
Although advances in medicine have led to remarkable progress in certain disease areas, most chronic disorders still cannot be completely cured. This is mainly because most such diseases are detected only late in their progression, once gross physiological symptoms manifest themselves, at which point tissues and organs have often undergone extensive or irreversible damage. At this stage, the choice of interventions is typically quite limited. It is difficult to predict whether a patient will respond to a particular treatment (often invasive or aggressive therapies that can be of modest benefit), or whether therapy resistance will emerge and lead to a relapse. Despite technology-driven revolutions that enable a patient’s physiology to be investigated at the level of molecules1,2 and placed in the context of tissues3,4, in most cases our ability to detect and predict diseases at an early stage is limited by our incomplete mechanistic understanding of disease at the cellular level.
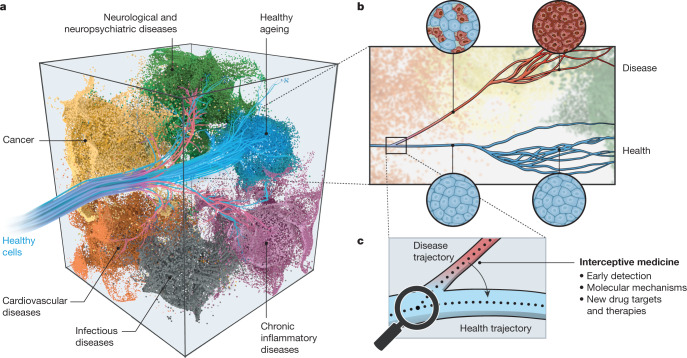
Cells develop and differentiate along specific lineage trajectories to form functionally distinct cell types and states5, which, together with their neighbouring cells, underlie and control normal physiology (Fig. 1). However, we have not been able to systematically detect and understand the molecular changes that propel an individual cell along these trajectories during normal development or ageing, or the molecular causes that trigger deviations from healthy trajectories and drive cells and tissues towards disease (Fig. 1). Timely detection and successful treatment of disease will depend crucially on our ability to understand and identify when, why, and how cells deviate from their normal trajectories. More accurate cellular and molecular diagnostics will enable us to intercept disease sufficiently early to prevent irreparable damage. To achieve this interceptive medicine (Fig. 1), we need to invest in approaches that provide a detailed molecular understanding of the basis of disease-related heterogeneity in tissues, with sufficient molecular, cellular and temporal resolution.
Fig. 1. Early disease detection and interception by understanding and targeting cellular trajectories through time.
a, Cells are programmed to develop and differentiate along many different specific lineage trajectories (blue trajectories) to reach their functional state. When these normal lineage processes go awry, it can cause a cell to deviate from a healthy state and move towards a complex disease space (coloured manifolds defined by multi-dimensional molecular space—including gene expression, protein modifications and metabolism), as shown by red trajectories. b, Many diseases are detected only at a relatively late stage with the onset of symptoms (red trajectory) and when pathophysiological changes can be at an advanced stage (red cells). At this point, cells, tissues and organs have undergone extensive and often irreversible molecular and physiological changes since the initial events that caused them to deviate from a healthy state. Hence, the choice of interventions may be limited and often involves harsh or invasive procedures. c, Understanding the early molecular mechanisms that cause cells to deviate from a healthy to a disease trajectory will provide biomarkers for the early detection of disease, and new drug targets and innovative therapies to intercept diseases before the onset of pathophysiology and the manifestation of symptoms.
Several challenges need to be overcome in order to understand complex disease landscapes, which comprise of vast numbers of potential cellular states (Fig. 1). First, we need to resolve normal cellular heterogeneity across space and time to begin to define the cell types, states and cell–cell interactions that normally exist in the body. This is a main goal of the Human Cell Atlas consortium6. However, to discover the cellular bases of diseases requires that we track cellular heterogeneity and the molecular composition of cell trajectories in health and during disease progression longitudinally—throughout an individual’s lifetime. Second, we need to understand the molecular mechanisms and complex networks that define a cell’s state, and control its function, fate and trajectory over time, to be able to reconstruct a cell’s history and predict its future. This is essential for selecting the optimal intervention for an individual patient. Thus, systematic and longitudinal profiling of samples from many individuals is required. Third, we have yet to develop the computational frameworks required for integrating temporal data and patient profiles, with large cohorts to identify regulatory changes and to dissect the causes and manifestations of disease. Current attempts to model human disease have not succeeded in integrating the thousands of molecular phenotypes that are acquired from patients. Finally, we are limited by our lack of knowledge of the underlying causes of disease. To predict any given patient’s response to a specific therapy may require testing or modifying cells from the patient in an experimental system, a challenge that has yet to be routinely implemented.
To address these challenges experts from different disciplines came together in 2018 to form the LifeTime Initiative (https://lifetime-initiative.eu). It has since grown to be a pan-European community consisting of more than 90 research institutions with support from 80 companies and several funding agencies and national science academies. In 2019 the initiative was awarded a Coordination and Support Action by the European Commission to develop a Strategic Research Agenda (SRA)7 for a large-scale, long-term initiative with a roadmap for implementing cell-based interceptive medicine in Europe in the next decade. The ambitious goal is the early detection and interception of complex diseases, as well as the ability to select the most effective therapeutic strategy for a patient. Between March 2019 and June 2020 the initiative established several multi-disciplinary working groups (listed in the Supplementary Information), organized numerous workshops, meetings and surveys (and thereby engaged the wider community) and commissioned stakeholder interviews and an impact study. The European Commission will use LifeTime’s SRA during the planning of the next research and innovation framework programme: Horizon Europe. Here, we outline LifeTime’s vision and key aspects of the SRA towards establishing cell-based interceptive medicine.
Central to LifeTime’s vision and approach is the development and integration of new technologies, such as single-cell multi-omics, high-content imaging, artificial intelligence (AI) and patient-derived experimental disease models. The application of these integrated approaches to medical challenges and their incorporation into both experimental and clinical workflows are expected to directly benefit patients. For example, appropriate single-cell based biomarkers will give physicians early warning that a cell or tissue is entering a disease trajectory. Understanding disease heterogeneity at the cellular level and knowing the molecular aetiology of a disease will allow researchers to systematically identify drug targets and resistance mechanisms and to define therapeutic approaches, based on a given disease’s molecular or cellular vulnerability. This strategy differs markedly from classical approaches to drug discovery8. The stratification of patients on the basis of underlying disease mechanisms, assessed in situ within single cells, will help physicians to select the most appropriate treatment(s) or to use combination therapies that are tailored to the individual. These will be used first to identify cells that are deviating from the healthy trajectory, to steer them away from disease, and later to reduce the threat of relapse (Fig. 1). This transformative single-cell data-driven approach has the potential to increase the success rates of clinical trials and the efficacy of novel therapeutic interventions in clinics over the next decade. Overall, the LifeTime strategy is likely to affect both diagnosis and treatment, to greatly improve health and quality of life, and to reduce the societal burden of diseases such as cancer, neurological and neuropsychiatric disorders, infectious diseases, and chronic inflammatory and cardiovascular diseases.
Below, we outline the development and implementation of technology at the heart of LifeTime’s approach, describe LifeTime’s mechanism for identifying medical priorities, discuss the required infrastructures in Europe, interactions with industry and innovation, ethical and legal issues, describe LifeTime’s education and training vision, and estimate the expected impact of the LifeTime approach on medicine and healthcare. LifeTime builds on and will collaborate with related international initiatives that are paving the way by producing reference maps of healthy tissues in the body, such as the Human Cell Atlas (HCA)6 and the NIH Human Biomolecular Atlas Program (HuBMAP)9.
Technology development and integration
Single-cell technologies—particularly transcriptomics—are generating the first reference cell atlases of healthy tissues and organs, and are revealing a previously hidden diversity of cell subtypes and functionally distinct cell states6. Single-cell analyses of patient samples are beginning to provide snap shots of changes in cell composition and pathways that are associated with diseases such as cancer10–15, chronic inflammatory diseases16,17, Alzheimer’s disease18–20, heart failure21, and sepsis22. Because pathophysiological processes within individual cells involve different molecular levels, understanding the underlying mechanisms requires the integration of current single-cell approaches. LifeTime proposes the integration of several approaches7. This includes combining transcriptomics (Fig. 2) with methodologies that provide additional information on chromatin accessibility, DNA methylation, histone modifications, 3D genome organization, and genomic mutations23–25. Future developments will enable the incorporation of single-cell proteomes, lipidomes, and metabolomes, which will add key insights into different cellular states and their roles in health and disease. In addition to specific cell subtypes and the role of cellular heterogeneity, it is crucial to investigate the surrounding tissue context and organ environment. New spatial ‘-omic’ approaches, particularly spatial transcriptomics, include information on the locations of diseased cells, their molecular makeup and aberrant cell–cell communication within the tissue26–32. Advanced imaging approaches also now enable the systematic spatial mapping of molecular components, in situ, within cells and of cells within tissues28,33–37. The cellular context, with respect to different immune and stromal cell types, extracellular components and signalling molecules that contribute to disease progression, will help to identify the roles of specific cell types and interactions in diseases32,38–40. The implementation of cell lineage tracing approaches41, which link cellular genealogies with phenotypic information about the same cells, may help us to understand how populations of cells develop dynamically to form the specific architecture of a healthy or a diseased tissue.
Fig. 2. Hallmarks of the LifeTime approach to disease interception and treatment.
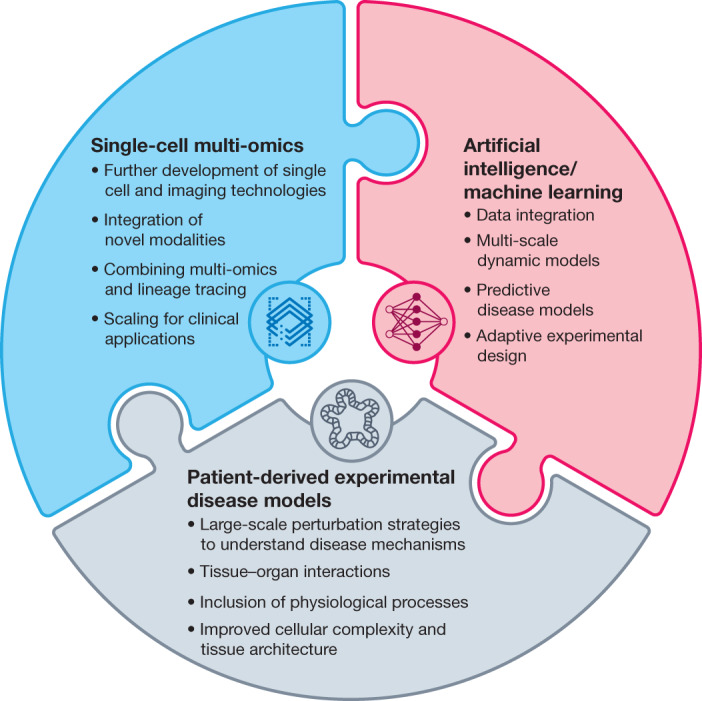
The schematic represents the development and integration of key technologies for investigating human diseases, as envisioned by the LifeTime Initiative. Single-cell multi-omics and imaging technologies will be developed for high-throughput applications. Different modalities will be combined to provide insight into underlying mechanisms, based on coordinated changes between different regulatory molecular layers. Insight into cellular genealogies and cellular dynamics will require the integration of lineage tracing tools. Technologies will also need to be scaled for clinical deployment. The integration and analysis of large, longitudinal multi-omics and imaging datasets will require the development of new pipelines and machine learning tools. These include the development of causal inference and interpretative machine learning approaches to create molecular networks for predictive and multiscale disease models. Patient-derived disease models such as organoids will be further developed to improve tissue architecture and the incorporation of physiological processes such as vasculature, nerve innervation and the immune system, to provide models that more faithfully recapitulate disease processes. Improved knowledge of disease mechanisms will require the application of large-scale perturbation tools to organoids. Tissue–tissue and organ–organ interactions will be recreated using microfluidics and organ-on-a-chip technologies to study key systemic interactions in diseases.
LifeTime proposes to develop the necessary single-cell methodologies and end-to-end pipelines (Fig. 2), which will be integrated into robust, standardized multi-omics and imaging approaches, and scaled to profile hundreds of thousands of patients’ cells7. This will require an in-depth analysis of longitudinal human samples obtained from patients and cohorts, including European and national clinical trial groups as well as initiatives collecting longitudinal biological material connected to well-annotated clinical information (Fig. 3). Linking these data to clinical outcomes will identify the cellular parameters that are permissive to a therapeutic response, for example, during checkpoint blockade immunotherapy12,42,43 or treatment of multiple myeloma11. By detecting rare drug-resistant cells that are present before11,44 or that emerge during treatment45, therapeutic regimens and combinatorial treatments can be adapted to improve outcomes.
Fig. 3. Exploiting the LifeTime dimension to empower disease targeting.
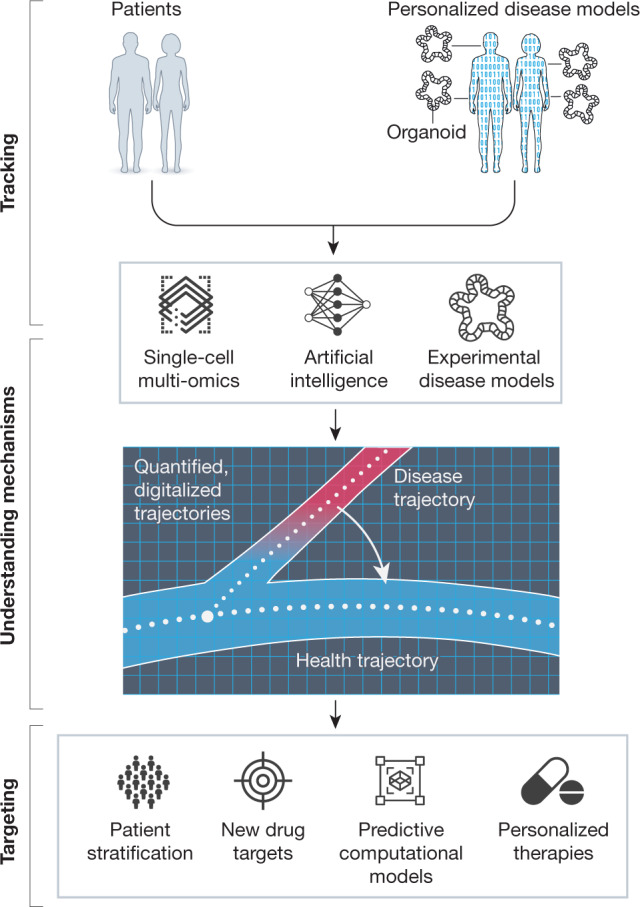
Single-cell multi-omics analysis of patient-derived samples (such as blood or tissue) or personalized disease models (for example, organoids and experimental disease models) will be profiled longitudinally to cover the different disease stages. Large-scale multidimensional datasets will provide quantitative, digitalized information that will provide information about the decision-making processes of cells. These will be analysed using AI and machine learning to arrive at predictive models for disease trajectories, providing cellular and molecular mechanisms of disease onset and progression. Models will be validated using large-scale perturbation analysis and targeted functional studies in disease models, which will be used in an iterative process to improve both computational and disease models.
Handling these large molecular datasets will require sophisticated and distributed computational and bioinformatics infrastructures (see ‘Implementation and infrastructure’), as well as the development of tools to integrate and ensure the interoperability of different data types, including single-cell multi-omics, medical information and electronic health records. LifeTime will work with ongoing European and national efforts to integrate molecular data into electronic health records and to establish standards and interoperable formats to address specific disease challenges. This process will promote the development of advanced personalized models of disease. To be able to implement routine longitudinal sampling of patients, we need to develop approaches for sampling small biopsies, including liquid biopsies, that will detect individual cells or cell-free DNA released from pathological cells before and during therapy46. Multi-dimensional descriptors of cell states from patients taken from different stages of disease or therapy will be used to derive new biomarker sets or to enhance current panels. Collaboration with ongoing atlas projects, industrial partners and regulatory authorities will be key for benchmarking and deriving the new standards that will enable us to deploy these new methods in the clinic. We hope that this will achieve earlier disease detection and guide the appropriate selection of drug targets and therapies (Fig. 3).
Unlocking the potential of unprecedented amounts of integrated digital information (including molecular data describing how individual cells make decisions) requires AI, in particular machine learning approaches that can identify meaningful molecular patterns and dependencies in the datasets47,48. Although such approaches have proven very useful when applied to medical imaging data and have enabled the identification of subtle disease-associated changes49, medical imaging cannot capture the full complexity of human physiology nor the status of a disease at the single-cell level. High-content imaging, together with information about gene expression, chromatin states, and protein and metabolic parameters, will contribute to the stratification of disease phenotypes. Machine learning and advanced modelling approaches will be used to integrate and analyse the different layers of cellular activity, and can generate multi-scale and potentially even causal models that will allow us to infer regulatory networks and to predict present and future disease phenotypes at the cellular level47,50–52 (Fig. 2).
The deep integration of machine learning technologies with spatial multi-omics and imaging technologies and data has the potential to usher in a new age of digital pathology to aid in decision-making by physicians (Fig. 3). By considering not only anatomical, physiological and morphological aspects, but also multidimensional molecular and cellular data, it will be possible to provide a more granular representation of a patient’s disease state to complement the pathologist’s slides and bulk measurements in tissues (for example, of mRNA or metabolites). We envision as the final goal the incorporation of new AI-based decision-aiding systems that will integrate and interpret available molecular, cellular, individual disease trajectory and imaging information. Interpretable and accountable AI systems will also provide the basis for clinical recommendations. The integration of cellular information should lead to a more precise description of a patient’s molecular and physiological history, and will guide early detection, allow predictive prognosis, and guide recommendations for therapeutic interventions to deliver more precise and effective treatments (Fig. 3).
Understanding the cellular origin and aetiology of disease from a patient-centred perspective requires systems that faithfully recapitulate key aspects of a patient’s pathophysiology, and render them experimentally tractable to test mechanistic hypotheses and predictions. Organoids are an emerging experimental system that allow aspects of organ development, regeneration and pathophysiology to be modelled3,4,53 (Fig. 2). Derived from adult or pluripotent human stem cells, organoids can capture individual features that are unique to each patient and can be interrogated molecularly in space and time. Importantly, by comparing organoid models from diseased and healthy individuals, unique disease features can be extracted even if the specific genetic cause of a disease is unknown. Therefore, organoid models offer a valuable tool for achieving some of the main goals of LifeTime, especially in cases in which repeated access to patient tissues is limited or impossible (for example, in neurological and neuropsychiatric disorders).
Despite their promise, organoids still require substantial development to harness their full potential for disease modelling (Fig. 2). LifeTime proposes to advance the models to capture the full degree of cellular heterogeneity and tissue-specific structural and metabolic conditions54, and to incorporate key physiological aspects, such as immune responses, vascularization or innervation. Because complex interactions between multiple tissues and organs are involved in many diseases, it will be necessary to develop tissue engineering principles that combine multiple organoids in pathophysiologically relevant crosstalk (‘organoids-on-a-chip’). To optimize translational potential, LifeTime will engage in standardizing, automating and scaling organoid approaches, to allow systematic derivation, propagation and banking of organoids. Such industrialization is also needed for large-scale chemical or genetic perturbations (for example, CRISPR–Cas screens), and for elucidating the genetic bases of disease variability and drug response at population-relevant scales, in both the preclinical and clinical contexts (Fig. 3). The resulting mechanistic dissection, enabled by large-scale perturbations, will be used to validate corresponding AI models of disease interception and progression.
In addition to organoids, in vivo model systems are necessary to translate the science from the bench to humans. A complex biological system is required to study the myriad of host–disease and host–pathogen interactions associated with complex diseases, such as infectious diseases, cancer or Alzheimer’s disease. The use of animal models is important for understanding the complex temporal relationships that occur in diseases, such as those involving the vasculature, immune system and pathogens as well as neuronal networks in the brain. LifeTime will therefore improve the clinical relevance of animal models and make use of approaches in which patient-derived tissues can be integrated into in vivo models55–59 to study the dynamics of cellular heterogeneity in space and time.
LifeTime, as a community, has the capacity to develop and integrate these technologies, which often require expertise and specialized instrumentation that are located in distinct laboratories. A coordinated effort can achieve the required benchmarking and standardization of technologies, workflows and pipelines. This will also ensure that the data, software and models generated adhere to FAIR (findable, accessible, interoperable, and reusable) principles60 (see ‘Implementation and infrastructure’), are available across national borders, and are in full compliance with international legislations such as the European General Data Protection Regulation. Moreover, LifeTime will ensure that technologies, including AI and organoids, will be developed in an ethically responsible way in collaboration with patients, putting the patient at the centre (see ‘Ethical and legal issues’).
Identification of medical priorities
LifeTime has initiated a mechanism, called Launchpad, to systematically identify medical challenges that can be addressed through LifeTime’s approach and have a direct effect on patient care. Initially, the focus has been on five disease areas that are a substantial burden to society: cancer, neurological and neuropsychiatric disorders, infectious diseases, chronic inflammatory diseases and cardiovascular diseases. Other disease areas will be continuously monitored (for example, rare Mendelian diseases and metabolic diseases), and research programmes initiated as technologies and infrastructures develop. The LifeTime Launchpad has defined several criteria to identify the medical challenges. These include: societal impact (including incidence and prevalence, disease severity, economic impact and the pressing need for new and more efficient clinical treatments and early detection), evidence for cellular heterogeneity that limits current clinical avenues, availability of samples from biobanks, relevant preclinical models, existence of patient cohorts including those enabling longitudinal studies, clinical feasibility and ethical considerations, as well as alignment with national and EU funding priorities. Subsequently, multidisciplinary working groups, including clinicians, in each disease area have used these criteria to define the following disease challenges and to develop ten-year roadmaps to address them in the LifeTime SRA7.
Despite cancer broadly covering hundreds of individual tumour types, there are critical knowledge gaps that are common to all cancer entities, including the mechanisms of early dissemination and therapy resistance. Metastatic dissemination of a subpopulation of cancer cells is a leading cause of death in almost all cancer types. Successful treatment of advanced and metastasized forms of cancer remains difficult, despite the development of targeted therapies and immunotherapies, owing to the emergence of drug or therapy resistance. To address these medical priorities, LifeTime recommends focusing on understanding the cell types and states—malignant cells and their microenvironment—that are involved in early stages of cancer dissemination, and the reprogramming of cellular states during disease and their effect on resistance to therapies.
For neurological disorders, a major challenge is a lack of understanding of the early events in disease onset to enable the development of disease-modifying therapies. The lack of access to longitudinal samples from patients necessitates the establishment of cohorts of patient-derived disease models to understand the cellular heterogeneity associated with disease. The discovery of pathways and biomarkers that will allow the stratification of patients on the basis of the cellular mechanisms that drive a disease will make it possible to design new clinical trials to reevaluate drugs that were previously tested without such stratification, and to broaden the drug target portfolio.
As seen during the coronavirus disease 2019 (COVID-19) pandemic, it is important to be able to understand infection mechanisms and the host response in order to rapidly identify the most likely effective treatment for an infection. At the same time, the continuous rise of antimicrobial resistance requires the discovery of new therapeutic strategies. A key medical challenge for infectious diseases is to understand the cellular response to infections and to develop precision, immune-based therapeutic strategies to combat infections.
Chronic inflammatory diseases impose a high burden owing to their long-term debilitating consequences, which result from the structural destruction of affected organs or tissues. Current therapies treat the symptoms but do not cure or fully control the chronic inflammatory pathophysiology. While different targeted therapies exist, they are expensive and their success is limited by high rates of non-response to treatment. Consequently, there is an urgent need to explore and understand how cellular heterogeneity contributes to the pathology of inflammatory diseases61 and how this relates to the predicted course of disease and the response of a patient to one of the numerous available therapies.
Many cardiovascular and metabolic diseases lack effective therapies owing to a lack of knowledge of their underlying causes and the link between abnormal cardiac cell structure or function and pathophysiology. The identified medical priority is to understand the cellular and molecular mechanisms involved, in order to enable early diagnosis and the design of new mechanism-based therapies for precise clinical treatment.
The LifeTime disease roadmaps can be divided broadly into three phases7: first, immediate research into the identified medical challenges using established, scaled single-cell technologies, computational tools and disease models; second, the development of new technologies that are required to address specific medical challenges, including the development of spatial multi-omics and imaging approaches and advanced patient-derived model systems for longitudinal analyses; and finally, the application of these next-generation technologies to the longitudinal analyses of patient samples, or patient-derived models, combined with machine learning to generate patient trajectories and predictive models of disease. The resulting predictions and biomarkers will be validated in prospectively collected patient cohorts within clinical trials that will also include longitudinal liquid biopsies. The routine clinical use of predictors and biomarkers for risk stratification of patients and resulting interventions—where feasible—is the pre-final step. The final step is the extension of predictors and biomarkers to the analysis of large longitudinal patient cohorts, such as national cohorts, for developing secondary and tertiary prevention approaches based on the new biomarkers.
During the implementation of these roadmaps, the initiative will establish an experimental design working group to develop systematic procedures to ensure that research samples are acquired from diverse cohorts (including age, sex, and ethnicity). This will require the development of strict criteria for the inclusion of samples and to ensure appropriate coverage of critical metadata. They will also define standardized procedures for the acquisition and processing of samples from different pathology sites (depending on the disease area). It is envisaged that during disease challenge pilot projects, an experimental design oversight body will determine, using early data, the number of diseases that should be studied as the initiative develops, with recommendations on the sample sizes required to obtain sufficient statistical power.
Implementation and infrastructure
The scale of the data that will be generated and analysed, the cross-disciplinary and international structure, and the ambition of LifeTime to pioneer novel analytics using AI, place LifeTime in an excellent position to shape the next generation of computational infrastructure for medical and biological data in Europe. This will require close interaction with and evolution of the established European infrastructure (Fig. 4), such as the European Open Science Cloud (EOSC) and high-performance computing infrastructures through the European High-Performance Computing (EuroHPC) initiative. LifeTime will also interact with related European Life Sciences Research Infrastructures62 to create added value and to avoid duplication of effort in strategies and tools for sharing and accessing data and the development and application of standards. As medicine is inherently decentralized, LifeTime will also help to connect EU medical systems and develop large federated European data infrastructures.
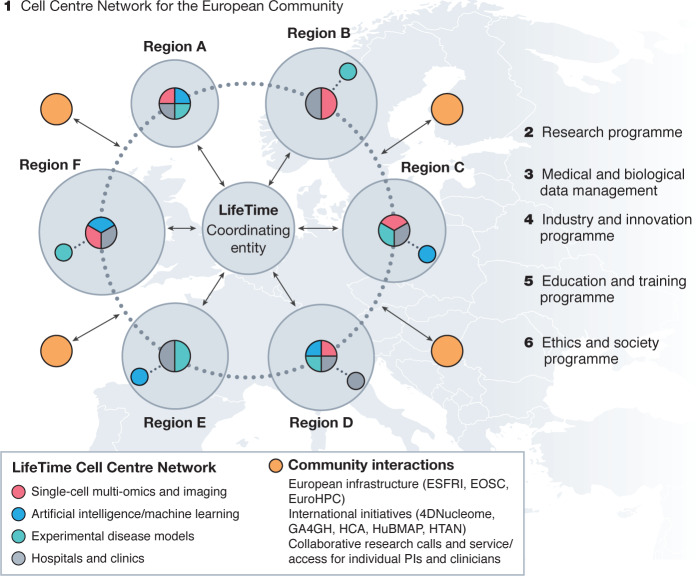
Fig. 4. Blueprint of the LifeTime Initiative.
LifeTime proposes a large-scale research initiative to coordinate national efforts, and to foster collaboration and knowledge exchange between the public and private sectors. LifeTime recommends the implementation of several programmes. (1) A network of Cell Centres to support the European Community. The interdisciplinary centres would complement each other’s strengths and expertise in the three LifeTime technology areas and operate in tight association with hospitals, integrating technology development with clinical practice. The connected but geographically distributed nodes would serve as both innovation hubs with strong links to industry and open education and training centres. Community coordination would avoid duplication of effort and increase effectiveness; this model requires funding instruments for a central coordination body. (2) The LifeTime research and technology integration programme includes both technology development and integration and the discovery of disease mechanisms and clinical applications. (3) Medical and biological data management platform. (4) Programmes fostering industry and innovation. (5) Education and training. (6) Ethics and societal engagement.
Fragmentation of research across borders, disciplines and timeframes needs to be overcome. The generation of data and development of technology by LifeTime will be harmonized across expert groups and centres, allowing the results to be quickly applied in clinics. Thus, a coordinated approach is required that integrates the multidisciplinary expertise of single-cell technologies, data science, and organoids as well as in vivo models across Europe. It must also engage clinicians and patients to achieve medical impact. To address these challenges, LifeTime proposes a multidisciplinary network of LifeTime Centres (Fig. 4) with different complementary thematic clusters across Europe, each working in close association with hospitals. These connected, flexible innovation nodes will share resources, gather the necessary critical mass for global competitiveness, and be open for collaboration with the entire scientific community. LifeTime Centres should deliver a number of key functions:
∙ Serve as platforms for the development and advancement of breakthrough technologies for single-cell research in -omics and imaging, AI (in particular machine learning), and experimental and computational disease models.
∙ Closely and actively collaborate with patients, clinicians, hospitals and healthcare systems, in some cases with a specific disease focus.
∙ Set standards in data generation, standardization and management, implementing FAIR principles.
∙ Set standards in ethical, legal and societal issues (ELSI) programmes by working together in multidisciplinary teams aimed at responsible research and innovation.
∙ Offer opportunities to collaborate, test and benchmark new methodologies and analysis methods; for example, in adaptive experimental design.
∙ Offer unique opportunities to industry to translate recent knowledge and novel technologies from the laboratory to the market.
∙ Provide an early access programme to new technologies developed by companies.
∙ Function as open, interconnected education hubs, delivering training in the new technologies to researchers, scientific personnel and clinicians, as well as providing engagement activities for patients and the public.
LifeTime aims to analyse data that are inherently distributed across different clinical centres in different countries, which is a substantial challenge. These data are usually not accessible outside a national, regional clinical care system or specified data ‘safe havens’; when they are accessible, accredited systems are often required for storing the data and information governance may be at the hospital, federal or international level. This means that a federated approach is the only way to access and integrate information from various European healthcare systems. Thus, the LifeTime data and computational network, building on cloud technologies, will provide the necessary capacities to enable federated analytics across the LifeTime centres and will provide a technical and legal framework for integrating core information structures, multi-omics assays, imaging, AI and machine learning technologies, and health records (Fig. 4). A joint Data Coordination Centre, following a multi-level approach, will ensure transparent data access control, compatibility and standardization. Within this framework, LifeTime will also coordinate and pioneer open data sharing and reuse and collaboration, including models of access before publication of data.
To start this cooperative LifeTime Centre network, the initiative can build on initial developments and programmes by LifeTime members in a number of European countries; for example, the VIB Single-cell Accelerator Programme in Belgium, the Berlin Cell Hospital/Clinical Single-cell Focus in Germany, the UK’s Sanger/EBI/Babraham Single Cell Genomics Centre, and the LifeTime Single-Cell Centre in Poland. To avoid duplication and lack of standardization, the LifeTime Cell Centre network should be coordinated through an entity or framework that optimizes coordination and support to achieve the LifeTime vision. Funding for specific research projects that involve one or more LifeTime Centres could come from a portfolio of private and public funding opportunities, on both the national and pan-European levels. The network will interact closely with key European efforts and will contribute to EU strategies and programmes.
Interaction with industry and innovation
Collaborations with the private sector will be key for the rapid translation and delivery of technologies, instrumentation, diagnostics and therapies (Fig. 4). Currently, more than 80 companies support LifeTime’s vision. These span multiple sectors as well as industrial associations and networks such as the European Federation of Pharmaceutical Industries (EFPIA) and the Euro-BioImaging Industry Board (EBIB).
The transformation of breakthrough discoveries into solutions to improve the health of European citizens will involve several crucial steps. These include the creation of a unifying framework that fosters and streamlines pre-competitive interactions between academia and industry at the interfaces of computer science, single-cell biology, -omics, imaging, patient-derived disease modelling and precision medicine. A large-scale collaboration platform across Europe should be developed that provides umbrella agreements, regular meetings, dual training of early-career scientists in academia and industry, and exchange programmes. This will enable joint projects between public and private sectors that span the entire biomedical innovation cycle from discovery research and technology development to implementation in hospitals and the healthcare industry.
Cross-sectoral collaborations between small, medium-size and large companies with different development timelines and distinct business models is crucial to stimulate innovation. To expedite the identification of, and investment in, emerging technologies developed in academic and industrial laboratories, successful local initiatives such as tech watch and accelerator programmes (for example, the VIB Single-cell Accelerator) should be scaled and coordinated at the EU level. LifeTime aims to create a networking and match-making platform for individuals and academic and industry organizations that share the goal of developing and integrating breakthrough technologies and applying them in the clinic to benefit patients. Further measures could foster innovation and entrepreneurship. For example, a pre-seed, pre-incubator funding scheme based on competitive calls to support start-up or technology transfer ideas.
The creation of a dedicated European ecosystem is also essential. This will require additional key measures, such as the development of enabling digital environments and the promotion of early disease interception with all necessary stakeholders (for example, patients, regulators, payers, and others), as described in the LifeTime call for action launched in December 2019 (https://lifetime-initiative.eu/make-eu-health-research-count/).
Ethical and legal issues
The implementation of LifeTime’s vision triggers relevant ethical questions from all societal groups that are directly affected by the project (patients, clinicians and scientists), and from society in general. LifeTime aims to pioneer a real-time or parallel ELSI programme that will predict, identify, monitor and manage the ethical impact of the biomedical innovations that arise from research and technology development, ensuring that implementation follows ethical guidelines. LifeTime’s ELSI programme can be used as a testing ground for other international interdisciplinary initiatives (Fig. 4). Ethical issues will be identified and managed as early as possible, and the programme will ensure that ethical and research integrity guidance is implemented throughout the entire research process to stimulate positive effects and mitigate negative ones63.
Specialists in bioethics, public engagement, ethics of technology and lawyers have identified LifeTime’s ethical and societal priority areas. These include questions related to the derivation, storage and use of organoids, the use of AI, data ownership and management, anonymization of data, equity of access to such revolutionary medical care, the definition of health and illness, and transparent science communication to society64. To initiate a relationship of trust with the public, we will include diverse modes of communication and engagement, for example through art, citizen science and public dialogue, contributing to scientific literacy, and promoting individual critical thinking and public participation in decision-making processes.
Education and training
The introduction of interceptive medicine into clinical practice in parallel with a multidisciplinary research programme will require capacity building in health and research systems, and substantial deployment of technology in clinics. This will lead to a collaborative, fast-developing and interdisciplinary environment in research and in hospitals, which will require new training inputs. To respond to these needs, LifeTime will create an Education and Training Programme, ensuring the sustainable application of new technologies and the implementation of new medical and scientific approaches (Fig. 4). Importantly, this will be done in an integrative scheme that intersects the multiple LifeTime disciplines and areas of action: disruptive technologies applied to medical challenges, technology transfer and innovation, research integrity, data management and stewardship, ethical and societal issues, communication and emotional skills, or management of medico-scientific and collaborative projects.
Each LifeTime training activity will be based on multi-lateral education: basic researchers will teach other researchers and clinicians about the potential of the technological solutions, while clinicians will teach researchers about the clinical needs and biological challenges of the diseases in focus. This will strictly follow the idea of bench to bedside and back. The programme will have an inclusive philosophy to ensure that it can provide training to the wider community, including researchers, clinicians, technical operators, managers and staff of technology platforms, as well as administrators, patients and the lay public.
LifeTime envisions the organization of cycles of colloquia and outreach activities to inform the public, the formulation of short-term courses compatible with a culture of lifelong learning and adaptability, and interdisciplinary Masters and PhD programmes. Through education and training, LifeTime will engage and inform society, will develop new professional curricula and will train a new generation of highly skilled medical scientists and support staff, in order to foster scientific and medical excellence in an ethical, responsible and inclusive framework.
Impact on medicine and healthcare
Medicine and healthcare are rapidly expanding pillars of our economy. EU countries collectively spend more than €1,400 billion per year on healthcare for their 500 million citizens. Given the dimensions and spiralling healthcare costs associated with an ageing population, these numbers will continue to increase unless we can mitigate the damaging effects of ageing. We expect that coupling current health monitoring with early detection and disease interception will have a major economic impact. In Europe, 20% of the population will soon be over 65 years old, with an age distribution that will continue to change until 12% are over 80 years old in 208065. Given the prevalence and cost of caring for people with degenerative conditions and the increase in chronic lifestyle-induced diseases, the knowledge and technologies developed by LifeTime are urgently needed to detect these diseases earlier, and to avoid their worst manifestations. LifeTime would also have an impact in the era of unexpected pandemics such as COVID-19 by rapidly determining the cellular and molecular basis of the disease. This would identify potential therapeutic strategies for patient subgroups as well as representing a starting point for the development of effective new therapies.
One of healthcare’s largest outstanding issues is that many patients do not respond to commonly prescribed treatments. Whereas well-controlled randomized clinical trials provide evidence for the statistical utility of a given therapy, in practice often many patients must be treated before a single patient will show a measurable benefit. Other patients may not benefit at all or even be harmed66, leading to an economic loss that is estimated to be in the hundreds of billions of Euros per year. The variable therapeutic responses that originate from the cellular and genetic heterogeneity that exists in cancer and other complex diseases, contributes not only to the failure of treatments, but also to the rising cost of drug development, which is currently estimated at around €1–2 billion per drug. In silico models for disease trajectories generated by LifeTime will enable the integration of personal genetic and lifestyle information into predictive models of disease course. This will allow physicians to determine and implement optimal therapeutic strategies that are tailored to the individual (precision medicine) with sophisticated timing of disease interception. The knowledge gained will also contribute to more appropriate selection of patients for clinical trials.
Outlook summary
Recent advances in key single-cell technologies, AI and patient-based experimental systems, such as induced pluripotent cells and organoids, have set the stage for their integration and deployment to improve mechanistic molecular understanding, prediction, and treatment of disease onset and progression. Patients will benefit from cell-based medicine through the earlier detection of diseases at a stage where they can be effectively intercepted. The integrated technologies will enable the selection, monitoring and, if necessary, modification of therapeutic strategies for an individual to improve clinical outcomes based on high-resolution cellular information. Within the next decade, the obtained molecular mechanistic information has the potential to revolutionize drug discovery processes and clinical trial design, and eventually to be incorporated into clinicians’ daily decision-making processes. As the LifeTime community continues to grow, new individuals, institutions and companies are encouraged to join and contribute to establishing a European platform to implement single-cell and data-driven medicine to address the growing burden of complex and chronic diseases.
Supplementary information
Author and affiliation list for members of the LifeTime Working Groups.
Acknowledgements
We acknowledge all participants that have attended and contributed to LifeTime meetings and workshops through many presentations and discussions. We thank J. Richers for artwork and A. Sonsala, A. Tschernycheff and C. Lozach for administrative support. LifeTime has received funding from the European Union’s Horizon 2020 research and innovation framework programme under grant agreement 820431.
Author contributions
All authors contributed to the writing of the article and provided comments and feedback. They all approved submission of the article for publication. The individuals listed at the end of the paper are members of Working Groups that contributed to the writing of the LifeTime Strategic Research Agenda (listed in full in the Supplementary Information). Please note that the complete LifeTime Community is much broader and includes many associates and supporters that are actively contributing to and advocating for LifeTime (further information can be found at https://lifetime-initiative.eu).
Competing interests
C.B. is an inventor on several patent applications in genome technology and cofounder of Aelian Biotechnology, a single-cell CRISPR screening company. H.C. is a non-executive board member of Roche Holding, Basel. A.P. holds European and US patents on ‘Genome Architecture Mapping’ (EP 3230465 B1, US 10526639 B2). W.R. is a consultant and shareholder of Cambridge Epigenetix. T.V. is co-inventor on licensed patents WO/2011/157846 (methods for haplotyping single cells), WO/2014/053664 (high-throughput genotyping by sequencing low amounts of genetic material), WO/2015/028576 (haplotyping and copy number typing using polymorphic variant allelic frequencies). All other authors declare no competing interests.
Footnotes
Peer review information Nature thanks Michael Snyder, Ali Torkamani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nikolaus Rajewsky, Geneviève Almouzni, Stanislaw A. Gorski
A list of affiliations appears at the end of the paper
Change history
3/17/2021
A Correction to this paper has been published: 10.1038/s41586-021-03287-8
Contributor Information
Nikolaus Rajewsky, Email: rajewsky@mdc-berlin.de.
Geneviève Almouzni, Email: genevieve.almouzni@curie.fr.
Stanislaw A. Gorski, Email: stan.gorski@mdc-berlin.de
LifeTime Community Working Groups:
Lavinia Alberi, Stephanie Alexander, Theodore Alexandrov, Ernest Arenas, Claudia Bagni, Robert Balderas, Andrea Bandelli, Burkhard Becher, Matthias Becker, Niko Beerenwinkel, Monsef Benkirame, Marc Beyer, Wendy Bickmore, Erik E. A. L. Biessen, Niklas Blomberg, Ingmar Blumcke, Bernd Bodenmiller, Barbara Borroni, Dimitrios T. Boumpas, Thomas Bourgeron, Sarion Bowers, Dries Braeken, Catherine Brooksbank, Nils Brose, Hilgo Bruining, Jo Bury, Nicolo Caporale, Giorgio Cattoretti, Nadia Chabane, Hervé Chneiweiss, Stuart A. Cook, Paolo Curatolo, Marien I. de Jonge, Bart Deplancke, Bart De Strooper, Peter de Witte, Stefanie Dimmeler, Bogdan Draganski, Anna Drews, Costica Dumbrava, Stefan Engelhardt, Thomas Gasser, Evangelos J. Giamarellos-Bourboulis, Caroline Graff, Dominic Grün, Ivo Gut, Oskar Hansson, David C. Henshall, Anna Herland, Peter Heutink, Stephane R. B. Heymans, Holger Heyn, Meritxell Huch, Inge Huitinga, Paulina Jackowiak, Karin R. Jongsma, Laurent Journot, Jan Philipp Junker, Shauna Katz, Jeanne Kehren, Stefan Kempa, Paulus Kirchhof, Christine Klein, Natalia Koralewska, Jan O. Korbel, Malte Kühnemund, Angus I. Lamond, Elsa Lauwers, Isabelle Le Ber, Ville Leinonen, Alejandro Lopez Tobon, Emma Lundberg, Astrid Lunkes, Henrike Maatz, Matthias Mann, Luca Marelli, Vera Matser, Paul M. Matthews, Fatima Mechta-Grigoriou, Radhika Menon, Anne F. Nielsen, Massimiliano Pagani, R. Jeroen Pasterkamp, Asla Pitkänen, Valentin Popescu, Cyril Pottier, Alain Puisieux, Rosa Rademakers, Dory Reiling, Orly Reiner, Daniel Remondini, Craig Ritchie, Jonathan D. Rohrer, Antoine-Emmanuel Saliba, Raquel Sanchez-Valle, Amedeo Santosuosso, Arnold Sauter, Richard A. Scheltema, Philip Scheltens, Herbert B. Schiller, Anja Schneider, Philip Seibler, Kelly Sheehan-Rooney, David Shields, Kristel Sleegers, August B. Smit, Kenneth G. C. Smith, Ilse Smolders, Matthis Synofzik, Wai Long Tam, Sarah Teichmann, Maria Thom, Margherita Y. Turco, Heleen M. M. van Beusekom, Rik Vandenberghe, Silvie Van den Hoecke, Ibo Van de Poel, Andre van der Ven, Julie van der Zee, Jan van Lunzen, Geert van Minnebruggen, Alexander van Oudenaarden, Wim Van Paesschen, John van Swieten, Remko van Vught, Matthijs Verhage, Patrik Verstreken, Carlo Emanuele Villa, Jörg Vogel, Christof von Kalle, Jörn Walter, Sarah Weckhuysen, Wilko Weichert, Louisa Wood, Anette-Gabriele Ziegler, and Frauke Zipp
Supplementary information
is available for this paper at 10.1038/s41586-020-2715-9.
References
- 1.Claussnitzer M, et al. A brief history of human disease genetics. Nature. 2020;577:179–189. doi: 10.1038/s41586-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat. Rev. Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 4.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 5.Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regev A, et al. The human cell atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The LifeTime Initiative. LifeTime Strategic Research Agenda. https://lifetime-initiative.eu/wp-content/uploads/2020/08/LifeTime-Strategic-Research-Agenda.pdf (2020).
- 8.Yofe I, Dahan R, Amit I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat. Med. 2020;26:171–177. doi: 10.1038/s41591-019-0736-4. [DOI] [PubMed] [Google Scholar]
- 9.HuBMAP Consortium The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019;574:187–192. doi: 10.1038/s41586-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 11.Ledergor G, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 2018;24:1867–1876. doi: 10.1038/s41591-018-0269-2. [DOI] [PubMed] [Google Scholar]
- 12.Li H, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176:775–789.e718. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Galen P, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–1281.e1224. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Der E, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 2019;20:915–927. doi: 10.1038/s41590-019-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubman A, et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019;22:2087–2097. doi: 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- 19.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e1217. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- 22.Reyes M, et al. An immune-cell signature of bacterial sepsis. Nat. Med. 2020;26:333–340. doi: 10.1038/s41591-020-0752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argelaguet R, et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019;576:487–491. doi: 10.1038/s41586-019-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark SJ, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 2018;9:781. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooijers K, et al. Simultaneous quantification of protein–DNA contacts and transcriptomes in single cells. Nat. Biotechnol. 2019;37:766–772. doi: 10.1038/s41587-019-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WT, et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell. 2020;182:976–991.e19. doi: 10.1016/j.cell.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 27.Giladi A, et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 2020;38:629–637. doi: 10.1038/s41587-020-0442-2. [DOI] [PubMed] [Google Scholar]
- 28.Moffitt JR, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362:eaau5324. doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nitzan M, Karaiskos N, Friedman N, Rajewsky N. Gene expression cartography. Nature. 2019;576:132–137. doi: 10.1038/s41586-019-1773-3. [DOI] [PubMed] [Google Scholar]
- 30.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 31.van den Brink SC, et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature. 2020;582:405–409. doi: 10.1038/s41586-020-2024-3. [DOI] [PubMed] [Google Scholar]
- 32.Vickovic S, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bintu B, et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362:eaau1783. doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardozo Gizzi AM, et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol. Cell. 2019;74:212–222.e215. doi: 10.1016/j.molcel.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo LJ, et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019;568:49–54. doi: 10.1038/s41586-019-1035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medaglia C, et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science. 2017;358:1622–1626. doi: 10.1126/science.aao4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson HW, et al. The single-cell pathology landscape of breast cancer. Nature. 2020;578:615–620. doi: 10.1038/s41586-019-1876-x. [DOI] [PubMed] [Google Scholar]
- 39.Keren L, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174:1373–1387.e1319. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniatis S, et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science. 2019;364:89–93. doi: 10.1126/science.aav9776. [DOI] [PubMed] [Google Scholar]
- 41.Baron CS, van Oudenaarden A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 2019;20:753–765. doi: 10.1038/s41580-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 42.Helmink BA, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krieg C, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 44.Kim C, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173:879–893.e813. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambow F, et al. Toward minimal residual disease-directed therapy in melanoma. Cell. 2018;174:843–855.e819. doi: 10.1016/j.cell.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 47.Eraslan G, Avsec Ž, Gagneur J, Theis FJ. Deep learning: new computational modelling techniques for genomics. Nat. Rev. Genet. 2019;20:389–403. doi: 10.1038/s41576-019-0122-6. [DOI] [PubMed] [Google Scholar]
- 48.Lähnemann D, et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 50.Argelaguet R, et al. Multi-Omics Factor Analysis—a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 2018;14:e8124. doi: 10.15252/msb.20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Efremova M, Teichmann SA. Computational methods for single-cell omics across modalities. Nat. Methods. 2020;17:14–17. doi: 10.1038/s41592-019-0692-4. [DOI] [PubMed] [Google Scholar]
- 52.Pearl, J. & Mackenzie, D. The Book of Why: The New Science of Cause and Effect (Penguin, 2019).
- 53.Amin ND, Paşca SP. Building models of brain disorders with three-dimensional organoids. Neuron. 2018;100:389–405. doi: 10.1016/j.neuron.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Knoblich JA. Lab-built brains. Sci. Am. 2016;316:26–31. doi: 10.1038/scientificamerican0117-26. [DOI] [PubMed] [Google Scholar]
- 55.Bleijs M, van de Wetering M, Clevers H, Drost J. Xenograft and organoid model systems in cancer research. EMBO J. 2019;38:e101654. doi: 10.15252/embj.2019101654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrne AT, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer. 2017;17:254–268. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 57.Espuny-Camacho I, et al. Hallmarks of Alzheimer’s disease in stem-cell-derived human neurons transplanted into mouse brain. Neuron. 2017;93:1066–1081.e1068. doi: 10.1016/j.neuron.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Hasselmann J, et al. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron. 2019;103:1016–1033.e1010. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mancuso R, et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 2019;22:2111–2116. doi: 10.1038/s41593-019-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkinson MD, et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schultze JL, The SYSCID Consortium & Rosenstiel, P. Systems medicine in chronic inflammatory diseases. Immunity. 2018;48:608–613. doi: 10.1016/j.immuni.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 62.Life Science RI European Life Science Research Infrastructureshttps://lifescience-ri.eu/home.html (2020).
- 63.Sugarman J, Bredenoord AL. Real-time ethics engagement in biomedical research: ethics from bench to bedside. EMBO Rep. 2020;21:e49919. doi: 10.15252/embr.201949919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres-Padilla ME, et al. Thinking ‘ethical’ when designing a new biomedical research consortium. EMBO J. 2020;39:e105725. doi: 10.15252/embj.2020105725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.European Commission. People in the EU: who are we and how do we live? https://ec.europa.eu/eurostat/documents/3217494/7089681/KS-04-15-567-EN-N.pdf/8b2459fe-0e4e-4bb7-bca7-7522999c3bfd (Eurostat, 2015).
- 66.What happened to personalized medicine? Nat. Biotechnol. 30, 1 (2012). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Author and affiliation list for members of the LifeTime Working Groups.