Abstract
High‐resolution matrix‐assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) is an emerging application for lipid research that provides a comprehensive and detailed spatial distribution of ionized molecules. Recent lipidomic approach has identified several phospholipids and phosphatidylinositols (PIs) are accumulated in breast cancer tissues and are therefore novel biomarker candidates. Because their distribution and significance remain unclear, we investigated the precise spatial distribution of PIs in human breast cancer tissues using high‐resolution MALDI IMS. We evaluated tissues from nine human breast cancers and one normal mammary gland by negative ion MALDI IMS at a resolution of 10 μm. We detected 10 PIs with different fatty acid compositions, and their proportions were remarkably variable in the malignant epithelial regions. High‐resolution imaging enabled us to discriminate cancer cell clusters from the adjacent stromal tissue within epithelial regions; moreover, this technique revealed that several PIs were specifically localized to cancer cell clusters. These PIs were heterogeneously distributed within cancer cell clusters, allowing us to identify two different populations of cancer cells that predominantly expressed either PI(18:0/18:1) or PI(18:0/20:3). Tracing the expression level of PIs during cancer cell progression suggested that the latter population is associated with the invasion. Our study documents a novel model for phospholipid analysis of breast cancer tissues by using high‐resolution MALDI IMS and identifies candidate PIs that can describe a specific phenotype of cancer cells.
Since Warburg's report on the upregulation of glycolysis and the increase in lactate production in cancer cells,1 cancer‐related metabolic processes have been studied intensively.2 In terms of lipid metabolism, de novo fatty acid synthesis is frequently upregulated in cancer cells, even in the early stages of cancer progression.2, 3 This “reprogramming” of lipid metabolism could be associated with the proliferation, membrane fluidity, and apoptosis of cancer cells by affecting membrane lipid remodeling.3 Recent studies demonstrated the accumulation of several phospholipids in breast cancer tissues and their association with sex hormone receptor expression, tumor grade, and prognosis.4, 5
Mass spectrometry (MS) is often used to investigate phospholipid profiles in living materials, and MALDI is the common ionization technique used for MS analysis. Matrix‐assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) is a new modality that facilitates the acquisition of mass spectra directly from tissue specimens and provides reconstructed density maps of detected ions.6 In contrast to conventional MS analysis, which requires lipid extraction, MALDI IMS does not destroy the histological structures of living materials, and the acquired mapping images can be compared with the corresponding histological images.7 Consequently, MALDI IMS is now used to investigate the localization of lipids in cancer tissue specimens.8 This technique also yields improved spatial resolution (less than 10 μm) and facilitates detailed two‐dimensional analysis of phospholipids, even in very small samples.9, 10, 11 However, the best method of analysis and interpretation of the vast amount of data produced by high‐resolution IMS has not been established.
Among the major classes of phospholipids, PIs are known for their involvement in intracellular signal transduction.12, 13 In particular, the PI3‐kinase pathway, which regulates many cellular functions, including lipid metabolism, is frequently mutated or activated in breast cancer tissues.14, 15 Unlike other phospholipids, the fatty acid composition of PIs is limited, and the PI profile shows a characteristic pattern in mammalian cells.16, 17 Specific stimulation can induce alterations in the composition of PIs, which can modulate PI3K signaling.18, 19 Moreover, PIs are suitable for analysis by IMS because they can be ionized by negative mode MALDI without exhibiting multiple ion peaks of adduct ions for the same molecule.20, 21 Based on this knowledge, we focused on the fatty acid composition of PIs and tried to examine their detailed spatial distribution in breast cancer tissues using high‐resolution IMS. High‐resolution imaging facilitated cancer cell‐oriented analysis and identified the PIs showing a possible association with specific cancer cell phenotypes.
Materials and Methods
Human tissue samples
Malignant breast cancer tissues from primary tumor sites and benign mammary gland tissues from a patient with fibrocystic mastopathy were collected by needle biopsy or surgical resection in the Department of Breast Surgery, Kyoto University Hospital. Prior written informed consents were obtained from all patients. All study protocols were approved by the Ethics Committee for Clinical Research, Kyoto University Hospital (authorization numbers G350 and G424). Each sample was quickly frozen in liquid nitrogen and stored at −80°C until analysis.
Sample preparation
Serial 10‐μm sections were prepared from each sample and mounted on indium‐tin‐oxide‐coated glass slides (Sigma‐Aldrich, St Louis, MO, USA). Each section was coated with 9‐aminoacridine hemihydrates (9‐AA) (Acros Organics, Geel, Belgium), which served as the matrix for MALDI MS. Each slide was anchored in vacuum deposition equipment (SVC‐700TM/700‐2; Sanyu Electron, Tokyo, Japan) and mechanically coated with 9‐AA evaporated at 220°C. The time required for this vapor deposition process was 8 min. For H&E staining, 9‐AA was removed from the slides by dipping them in methanol for 30 s after the MALDI MS analysis. The H&E‐stained sections were examined under an optical microscope (BZ‐9000; Keyence, Osaka, Japan).
Atmospheric pressure MALDI IMS and MS/MS analysis
Matrix‐assisted laser desorption/ionization IMS analysis was performed on a high‐resolution microscopic imaging mass spectrometer (RK27‐4050C; Shimadzu, Kyoto, Japan; the prototype model of iMScope) equipped with a 355‐nm Nd:YAG laser.22 Mass spectrometry data were acquired in negative mode in the 500.0–1200.0 m/z range by using an external calibration method. The region of interest was determined from the microscopic view of the slides; mass spectra were obtained at a spatial resolution of 10 μm. We used the same instrument for MS/MS analysis; the lipid class and fatty acid composition of the observed peaks were determined based on the spectrum pattern of the ion peaks of the product. Using SIMtool software (Shimadzu), we normalized the MS data to the total ion current to eliminate variations in ionization efficiency.
Acquisition of separate mass spectra from epithelial regions and stromal regions
The averaged mass spectra in epithelial regions and the surrounding stromal regions were extracted separately with BioMap software (Novartis, Basel, Switzerland). The accurate location of each region was determined by H&E staining. The regions for the acquisition of mass spectra were selected manually. The stromal regions were defined as the areas within approximately 100 μm of the edge of the epithelial region. To estimate the patterns of each mass spectrum, the proportion of each PI ion peak was calculated as a ratio to the sum of mass intensities of all detected PIs. Statistical differences were analyzed using the Welch's t‐test. Differences of P < 0.05 were considered statistically significant.
Hierarchical clustering
To examine the inter‐individual similarities of the spatial distribution of PIs, we digitalized their localization as the ratio of the intensity of each PI in epithelial regions to the intensity of each PI in stromal regions. The proportion of each PI was substituted as its relative intensity. The ratio was transformed to log2 scale, and hierarchical clustering was performed to evaluate correlations using MultiExperiment Viewer software (version 4.8.1; Dana‐Farber Cancer Institute, Boston, MA, USA).23 Similarities were assessed with the Pearson correlation coefficient.
Generating ion density maps and overlaying the maps on H&E staining images
The averaged ion density map of each PI was generated using BioMap software. The size and transparency of the ion density maps were adjusted, and they were overlaid on the corresponding H&E images with gimp 2.6.11 software (Free Software Foundation, Boston, MA, USA).
Histological diagnosis for H&E staining and immunohistochemistry
The histological diagnosis was performed by examination of H&E‐stained tissues by senior pathologists in the Department of Diagnostic Pathology, Kyoto University Hospital. Intrinsic breast cancer subtypes were determined by immunohistochemistry for estrogen and progesterone receptors, human epidermal growth factor receptor 2, and basal markers.
Comparison of PI expression between different cell clusters
The averaged mass spectra were obtained from each cell cluster using BioMap software. The number of cells in each cluster was counted manually in the H&E staining images. For the comparison of PI expression, each signal intensity was divided by the number of cells to eliminate the effect of cellular density. The results are presented as the mean of intensities from three independent cell clusters ± SD. Statistical differences were analyzed using the two‐tailed Student's t‐test. Differences of P < 0.05 were considered statistically significant.
Results
Patient backgrounds and clinicopathological features
Table 1 shows the clinicopathological features of the 10 patients recruited for this study. Breast cancer tissues were obtained from patients with luminal (Lu1‐3, n = 3), triple‐negative (TN1‐5, n = 5), and Her2‐positive (one patient) breast cancers. A benign mammary gland was obtained from a patient with fibrocystic mastopathy (MP). All cancer patients had invasive ductal carcinoma with T1 or T2 tumors. Lymph node metastasis was diagnosed in two triple‐negative (TN3 and 4) patients and a premenopausal patient with luminal disease (Lu3). The patient recorded as TN4 presented with distant metastasis. Based on immunohistochemical results for basal markers, all five triple‐negative cancers were of the basal‐like phenotype (Table S1).
Table 1.
Patient backgrounds and clinicopathological features
| ID | TN1 | TN2 | TN3 | TN4 | TN5 | Her2 | Lu1 | Lu2 | Lu3 | MP |
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 74 | 79 | 72 | 62 | 86 | 71 | 50 | 67 | 42 | 66 |
| Menopausal status | Post | Post | Post | Post | Post | Post | Post | Post | Pre | Post |
| Pathology | IDC | IDC | IDC | IDC | IDC | IDC | IDC | IDC | IDC | FC |
| Pathological T | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | |
| Nodal status | − | − | + | + | − | − | − | − | + | |
| Clinical M | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| Stage | IIA | IIA | IIA | IV | II | IIA | IIA | IIB | IIIC | |
| Intrinsic subtype | Basal | Basal | Basal | Basal | Basal | Her2 | Luminal | Luminal | Luminal | |
| Histological Grade | 3 | 2 | 3 | 2 | 3 | 3 | 2 | 3 | 2 | |
| ER (%) | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | |
| PgR (%) | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 80 | |
| Her2 status | − | − | − | − | − | + | − | − | − | |
| Sampling method | SR | SR | SR | SR | SR | SR | SR | NB | SR | NB |
ER, estrogen receptor; FC, fibrocystic mastopathy; IDC, invasive ductal carcinoma; NB, needle biopsy; PgR, progesteron receptor; SR, surgical resection.
Identification of PI molecular species with different fatty acid compositions by negative ion MALDI IMS and MS/MS analysis
Fifty ion peaks that exhibited the highest intensities were screened in all cases, and we identified 10 ion peaks that represented PIs at m/z 807.5, 809.5, 833.5, 835.5, 837.5, 861.5, 863.5, 885.5, 887.5, and 889.5. MS/MS analysis confirmed their lipid classes and fatty acid compositions (Table 2, Fig. S1). All identified PIs had one saturated and one unsaturated fatty acid. The observed saturated fatty acids were C16:0 and C18:0, and the unsaturated fatty acids were C16:1, C18:1, C18:2, C20:2, C20:3, and C20:4. The degree of unsaturation of each PI was defined by the degree of unsaturation of the fatty acids.
Table 2.
Lipid class and fatty acid composition identified by MS/MS analysis
| Precursor ion (m/z) | Lipid class | Fatty acid composition |
|---|---|---|
| 807.5 | PI | 16:0/16:1 |
| 809.5 | PI | 16:0/16:0 |
| 833.5 | PI | 16:0/18:2 |
| 835.5 | PI | 16:0/18:1 |
| 837.5 | PI | 16:0/18:0 |
| 861.5 | PI | 18:0/18:2 |
| 863.5 | PI | 18:0/18:1 |
| 885.5 | PI | 18:0/20:4 |
| 887.5 | PI | 18:0/20:3 |
| 889.5 | PI | 18:0/20:2 |
PI, phosphatidylinositol.
Cancer epithelia exhibit different mass spectra patterns of PIs than non‐malignant duct epithelium and surrounding stromal tissues
In mammalian cells, PIs carrying stearic (C18:0) and arachidonic acid (C20:4) in the acyl chain are the most abundant PIs.16, 17 We found that the averaged mass spectra of the benign epithelial region showed the largest ion peak at m/z 885.5, corresponding to PI(18:0/20:4) (n = 1) (Fig. 1a,b). The mass spectra acquired from the stromal region showed the same pattern; the ion peak at m/z 885.5 was also the largest among the observed peaks (Fig. 1a,c).
Figure 1.
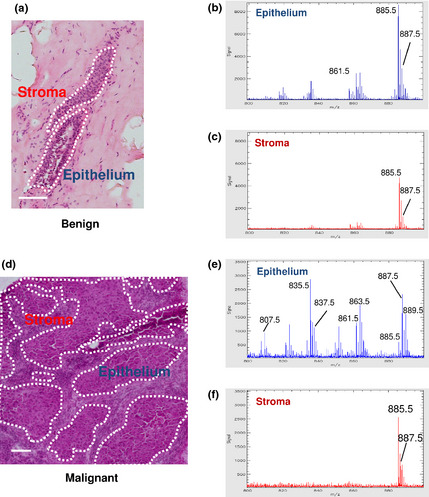
Averaged mass spectra obtained from benign and malignant mammary glands. (a–c) Representative image of a benign mammary gland. H&E staining (a), averaged mass spectrum obtained from the epithelial regions (b), and the stromal regions (c) are shown. (d–f) Representative image of malignant mammary glands. H&E staining (d), averaged mass spectrum obtained from the epithelial regions (e), and the stromal regions (f) are shown. Epithelial regions are encircled by dotted lines. The x‐ and y‐ axes represent m/z and the signal intensity, respectively. Scale bar, 100 μm.
The averaged mass spectra of malignant epithelial regions showed different patterns from those of the non‐malignant epithelial region (n = 9) (Fig. 1d,e). Phosphatidylinositols other than PI(18:0/20:4) exhibited large ion peaks, some of which were larger than that of PI(18:0/20:4) (Fig. 1e, Fig. S2). The averaged mass spectra obtained from the stromal area retained a pattern nearly the same as the typical pattern found in the non‐malignant condition; the ion peak of PI(18:0/20:4) was predominant (Fig. 1f, Fig. S3). These findings suggest that the proportion of PIs with different fatty acid compositions was changed in malignant epithelial regions in a manner different from their surrounding stromal cells.
Screening of PIs that demonstrate the characteristic localization in breast cancer tissues
As shown in Figure 2(a), the averaged proportion of each PI among all detected PIs was significantly altered in malignant epithelial regions as compared to the single sample of a benign epithelial region. The averaged proportion of PI(18:0/20:4), which was the predominant PI in the benign epithelial region, significantly decreased 33.3 ± 3.3% in the malignant epithelial regions (Table S2). As a result, PIs other than PI(18:0/20:4) accounted for a large percentage in these regions. In malignant stromal regions, the differences from the non‐malignant conditions were small, and few of them altered significantly (Table S2).
Figure 2.
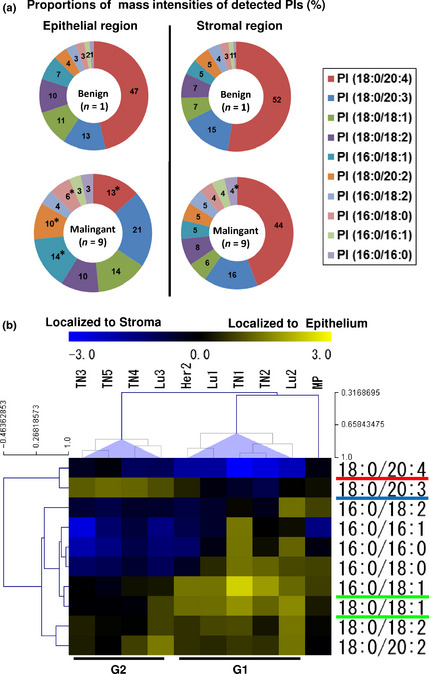
The distribution of PIs exhibited two distinctive features in breast cancer tissues. (a) Comparison of mass spectrum patterns between benign and malignant mammary glands. Circle graph shows the averaged proportion of each PI among all examined cases (%). The P‐values indicate significant differences from the benign condition. *P < 0.05. (b) Hierarchical clustering of PI distributions. The heat map shows the localization pattern of each PI. The two breast cancer tissue subgroups are identified as G1 and G2.
Hierarchical clustering identified the distinctive features of PIs in breast cancer cases by organizing their spatial information and inter‐individual differences. The PIs tended to cluster according to their degree of unsaturation (Fig. 2b). The breast cancer tissues showed two distinct clusters labeled G1 and G2. The benign mammary gland (MP) branched separately from these clusters. The classified PIs had different localization patterns in the G1‐ and G2‐subgroups. In the G1 subgroup, PI(16:0/18:1) and PI(18:0/18:1), which were classified in the single cluster, showed the highest specificity to epithelial regions. In contrast, PI(18:0/20:4) showed the highest specificity to stromal regions in the G1‐subgroup. PI(18:0/20:3), which was classified with PI(18:0/20:4), showed the highest specificity to the epithelial regions in the G2‐subgroup. The results of the cluster analysis suggested that the distributions of PI(16:0/18:1), PI(18:0/18:1), PI(18:0/20:3), and PI(18:0/20:4) effectively characterized the inter‐individual differences among breast cancer tissues and divided them into two distinct subgroups.
Ion density maps of IMS confirmed the characteristic localization of PIs
Based on our hierarchical clustering results, we evaluated the ion density maps of PI(18:0/18:1), PI(18:0/20:3), and PI(18:0/20:4), which commonly have C18:0 as their saturated fatty acyl chain.
In the benign mammary gland, the ion signals of PI(18:0/18:1) were preferentially localized in the epithelial area, whereas PI(18:0/20:3) and PI(18:0/20:4) were uniformly distributed both in the epithelium and in the surrounding stromal area (Fig. 3).
Figure 3.
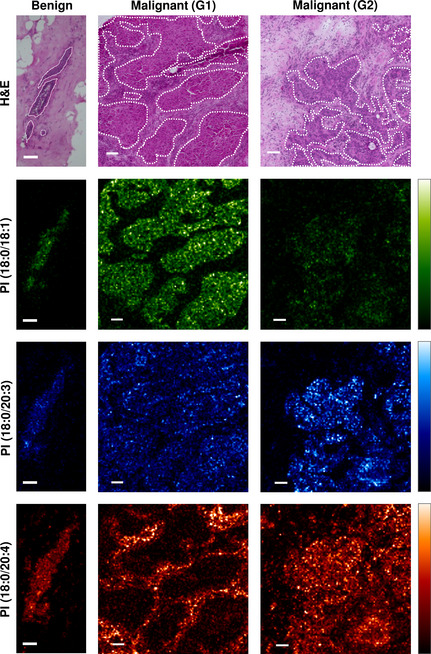
Mass spectrometry imaging showing the distributions of phosphatidylinositols (PIs) that characterize the two breast cancer tissue subgroups. Mass spectrometry (MS) images showing the distribution of PI(18:0/18:1) (green), PI(18:0/20:3) (blue), and PI(18:0/20:4) (red). Representative images of breast cancer tissues were selected from the G1‐ and G2‐subgroups. The corresponding H&E staining image is also shown. The epithelial regions are encircled by white dotted lines. Scale bar, 100 μm.
These PIs altered their localization patterns in malignant cancer tissues. In breast cancer tissues categorized in the G1‐subgroup, the ion signals of PI(18:0/18:1) were preferentially localized to the epithelial regions and exhibited strong intensities; however, in the G2‐subgroup, they showed neither specific localization nor signal upregulation (Fig. 3; green). In contrast, the ion signals of PI(18:0/20:3) were preferentially localized to epithelial regions and exhibited strong intensities in the G2‐subgroup but did not show specific localization in the G1‐subgroup (Fig. 3; blue). The strong ion signals of PI(18:0/20:4) were detected in the stromal regions in both the G1‐ and G2‐subgroups. They were almost undetectable in the epithelial regions in the G1‐subgroup but retained strong signals in the epithelial region in the G2‐subgroup. These findings suggested that the accumulation of PI(18:0/18:1) occurred in the epithelial regions of the G1‐subgroup, whereas the upregulation of PI(18:0/20:3) and PI(18:0/20:4) occurred in the epithelial regions of the G2‐subgroup. Because the accumulation of PI(18:0/20:4) was observed uniformly in the stromal regions in breast cancer tissues, the upregulation of PI(18:0/18:1) and PI(18:0/20:3) was identified as a malignant epithelial region‐specific process, and the expression patterns of PI(18:0/18:1) and PI(18:0/20:3) indicated the particular phenotype of the cancer cells.
The distribution of PI(18:0/18:1) and PI(18:0/20:3) in the epithelial region describes the heterogeneity of cancer cell populations
Because PI(18:0/18:1) and PI(18:0/20:3) seemed to exhibit a reciprocal pattern in their distribution, we attempted to evaluate their distributions within epithelial regions in a more detailed fashion. The average relative intensities of PI(18:0/20:3) in malignant epithelial regions was negatively associated with that of PI(18:0/18:1) (r = –0.72) (Fig. 4a). The merged image of ion density maps shows the heterogeneous distribution of these two molecules (Fig. 4b). The ion signals of PI(18:0/20:3) tended to be localized adjacent to the stromal regions and rarely overlapped with those of PI(18:0/18:1), which tend to be localized in the center of the cancer cell clusters. The overlaying image on the corresponding H&E staining confirmed that the ion signals of PI(18:0/20:3) in the epithelial edge matched with those in the cancer cells in contact with the stroma and not with the stromal cells (Fig. 4b). These findings suggested that the ratio of PI(18:0/20:3) and PI(18:0/18:1) in the averaged mass spectra in the epithelial region represents the ratio of the two different cellular populations: cancer cells expressing PI(18:0/20:3) and cancer cells expressing PI(18:0/18:1). Their localization also indicated that the phenotype of the former population is associated with their contact with the stroma.
Figure 4.
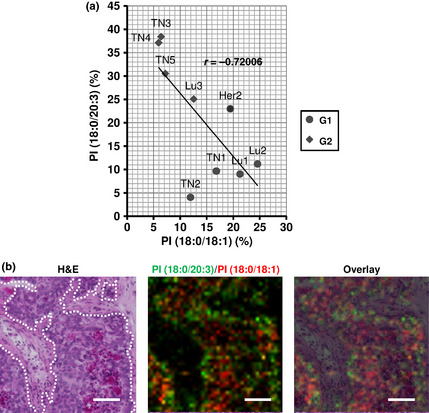
The distribution of PI(18:0/20:3) and PI(18:0/18:1) defines the two different populations of cancer cells. (a) Correlation of the relative intensities of PI(18:0/20:3) and PI(18:0/18:1). The circle dots and the crystal dots indicate breast cancer cases classified in the G1‐ and G2‐subgroup, respectively. r; cor‐relation coefficient. (b) Heterogeneous distribution of PI(18:0/20:3) and PI(18:0/18:1) in cancer cell clusters. The dotted lines in the H&E staining image indicate the border between cancer cell clusters and adjacent stroma. The green and red signals indicate PI(18:0/20:3) and PI(18:0/18:1), respectively. The ion density map is overlaid on H&E staining. Scale bar, 100 μm.
Tracking PI expression levels through cancer cell development and progression
A patient in the G2 subgroup (TN5) harbored a small cluster of invasive ductal carcinoma (IDC) within a ductal carcinoma in situ (DCIS). Because a single tissue section from this patient contained normal duct epithelium, hyperplastic ductal epithelium, DCIS, and IDC, we assessed the cellular expression of PI(18:0/20:3), PI(18:0/18:1), and PI(18:0/20:4) throughout cancer progression using a section from this patient.
Mass spectrometry imaging showed that PI(18:0/18:1) was preferentially localized to the hyperplastic duct epithelium. In contrast, PI(18:0/20:3) was preferentially localized both in DCIS and in IDC (Fig. 5a). The signals of PI(18:0/20:4) were distributed in DCIS and IDC lesions as well as in stromal cells (Fig. S4). Looking at the estimated amounts of these PIs, PI (18:0/18:1) was transiently accumulated in hyperplastic cells and decreased in DCIS and IDC (Fig. 5b). The amount of PI(18:0/20:3) gradually increased in accordance with disease progression, and it showed the largest increase from DCIS to IDC (38.1 ± 5.3, P < 0.05). Although the amount of PI(18:0/20:4) also increased in accordance with cancer progression, a significant difference was not observed during any steps of progression (Table S3). These findings suggested that the accumulation of PI(18:0/20:3) occurs in cancer cells during the process of invasion and contact with the stromal component. In addition, because the G2‐subgroup, in which PI(18:0/20:3)‐expressing cells were predominant, manifested a high incidence of nodal metastasis (Table 1 and Fig. 2b), we posited that cancer cells expressing PI(18:0/20:3) had a propensity to invade into stromal components, resulting in nodal metastasis.
Figure 5.
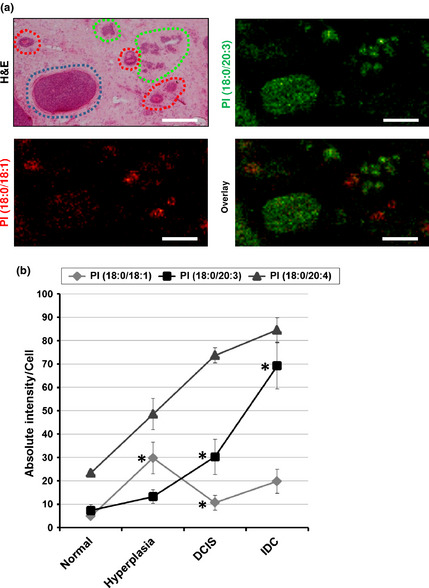
The association of PI(18:0/20:3) expression with cancer cell progression. (a) Mass spectrometry (MS) images of PI(18:0/18:1) and PI(18:0/20:3) and merged images of the two molecular species. The corresponding H&E staining image is also shown. The red, blue, and green circles indicate the hyperplastic epithelium, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC), respectively. Scale bar, 100 μm. (b) The stepwise change in the intensity of phosphatidylinositols (PIs) in normal epithelium, hyperplastic epithelium, DCIS, and IDC. The P‐values indicate significant differences from the previous stage of progression. *P < 0.05. Error bar, SD.
Discussion
In the present study, we report for the first time the unique spatial distribution of PIs with different fatty acid compositions in breast cancer tissues as determined by high‐resolution IMS. Moreover, we identified PI(18:0/20:3) as a possible marker for specific populations of breast cancer cells. Since the accumulation of PI(18:0/20:3) showed the association with the stromal contact and nodal status, we developed a plausible hypothesis that the accumulation of PI(18:0/20:3) in breast cancer cells may account for their invasion capacity. This hypothesis could be supported by the fact that the accumulation of PI(18:0/20:3) could affect not only cellular membrane fluidity but also the activity of PI3K signaling pathway.24, 25, 26, 27, 28 This is a preliminary study with a limited number of cases, and whether PI(18:0/20:3) expression is directly related to cellular invasion remains to be tested. However, high‐resolution IMS raised intriguing possibility that the alteration of lipid metabolites, as a consequence of the complicated physiological process, can directly reflect tumor invasiveness or sensitivity to PI3K inhibition, which cannot be satisfactorily predicted by the existing clinicopathological features.
We demonstrated several methodological advantages of high‐resolution IMS compared with the conventional lipidomic studies on cancer tissues. Conventional IMS (resolution greater than 10 μm) can roughly distinguish histologically different regions, such as epithelial regions and stromal regions. However, this lower‐resolution imaging failed in discriminating the intricate histological structures assembled by cancer epithelial cells which were composed of heterogeneous populations.29, 30 High‐resolution IMS could exclude stromal cells from epithelial regions and identify cancer cell heterogeneity. Especially, the diversity of PIs has not been previously described in human breast cancer tissues by a conventional MS analysis.4, 31 Our results show for the first time the stromal component of breast cancer tissues is predominantly occupied with PI(18:0/20:4) without significant inter‐individual differences. Because breast cancer tumors often contain abundant stromal components, the PI(18:0/20:4) within stroma may disrupt the detection of other PIs.32
Our results also suggest that differences in cellular density seemed to distort the true picture of molecular distribution when interpreting the ion density map of IMS. High‐resolution IMS can acquire the mass profiles by targeting small clusters containing only epithelial cells (average of 35 cells in this study), which enables us to estimate the amount of each PIs adjusted by cellular density. As a result, it revealed the significant increase in PI(18:0/20:3) from DCIS‐ to IDC‐lesions, which was difficult to recognize from ion density images alone. The ion intensity produced by the MALDI technique could be affected by other factors, such as tissue conditions, the affinity for the matrix, and ion suppression by other ionized molecules.25 However, studies that compared the MALDI IMS ion intensity and the results of quantitative analyses of electrospray ionization MS showed that MALDI IMS ion intensities were representative of the relative amount of PIs when comparisons were carried out between the same lipid classes.33, 34, 35
In conclusion, high‐resolution MALDI IMS identified the detailed spatial distribution of PIs with different fatty acid compositions in breast cancer specimens. It facilitated a cancer cell‐oriented analysis and revealed the accumulation of PI(18:0/20:3) as a phenotype of cancer cells that might be associated with their invasion capacity. Moreover, our study demonstrated a useful model for lipid research in breast cancer tissues using a new modality.
Disclosure Statement
N. Iwamoto, T.A. Sato and K. Tanaka are full‐time employees of Shimadzu Corporation. K. Shimizu is an advisory board member of Toray Industry.
Supporting information
Table S1. Immunohistochemistry results for basal markers.
Table S2. Differences in averaged proportions of phosphatidylinositols (PIs) (Malignant‐ vs benign‐region).
Table S3. Differences in signal intensities during the progression.
Fig. S1. Mass spectrometry (MS)/MS spectra of phosphatidylinositols (PIs) observed in this study.
Fig. S2. The averaged mass spectra obtained from epithelial areas in breast cancer tissues.
Fig. S3. The averaged mass spectra obtained from stromal areas in breast cancer tissues.
Fig. S4. Mass spectrometry (MS) images of PI (18:0/20:4) in the same case of Fig. 5.
Acknowledgments
This research was supported by a grant from the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for World‐Leading Innovative R&D on Science and Technology (FIRST Program)”, initiated by the Council for Science and Technology Policy (CSTP). We thank the medical staff of the Department of Breast Surgery of Kyoto University Hospital for helping recruit patients and collect samples.
(Cancer Sci 2013; 104: 1372–1379
References
- 1. Warburg O. On the origin of cancer cells. Science 1956; 123: 309–14. [DOI] [PubMed] [Google Scholar]
- 2. Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012; 491: 364–73. [DOI] [PubMed] [Google Scholar]
- 3. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 2007; 7: 763–77. [DOI] [PubMed] [Google Scholar]
- 4. Hilvo M, Denkert C, Lehtinen L et al Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res 2011; 71: 3236–45. [DOI] [PubMed] [Google Scholar]
- 5. Kang HS, Lee SC, Park YS et al Protein and lipid MALDI profiles classify breast cancers according to the intrinsic subtype. BMC Cancer 2011; 11: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI‐TOF MS. Anal Chem 1997; 69: 4751–60. [DOI] [PubMed] [Google Scholar]
- 7. Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry – Looking beyond classical histology. Nat Rev Cancer 2010; 10: 639–46. [DOI] [PubMed] [Google Scholar]
- 8. Amstalden van Hove ER, Blackwell TR, Klinkert I, Eijkel GB, Heeren RM, Glunde K. Multimodal mass spectrometric imaging of small molecules reveals distinct spatio‐molecular signatures in differentially metastatic breast tumor models. Cancer Res 2010; 70: 9012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kubo A, Ohmura M, Wakui M et al Semi‐quantitative analyses of metabolic systems of human colon cancer metastatic xenografts in livers of superimmunodeficient NOG mice. Anal Bioanal Chem 2011; 400: 1895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schober Y, Guenther S, Spengler B, Römpp A. Single cell matrix‐assisted laser desorption/ionization mass spectrometry imaging. Anal Chem 2012; 84: 6293–7. [DOI] [PubMed] [Google Scholar]
- 11. Miura D, Fujimura Y, Yamato M et al Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Anal Chem 2010; 82: 9789–96. [DOI] [PubMed] [Google Scholar]
- 12. van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci 2011; 124: 5–8. [DOI] [PubMed] [Google Scholar]
- 13. Cantley LC. The phosphoinositide 3‐kinase pathway. Science 2002; 296: 1655–7. [DOI] [PubMed] [Google Scholar]
- 14. Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3‐kinase and antiestrogen resistance in breast cancer. J Clin Oncol 2011; 29: 4452–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holub BJ, Kuksis A. Structural and metabolic interrelationships among glycerophosphatides of rat liver in vivo . Can J Biochem 1971; 49: 1347–56. [DOI] [PubMed] [Google Scholar]
- 17. Baker RR, Thompson W. Positional distribution and turnover of fatty acids in phosphatidic acid, phosphinositides, phosphatidylcholine and phosphatidylethanolamine in rat brain in vivo . Biochim Biophys Acta 1972; 270: 489–503. [DOI] [PubMed] [Google Scholar]
- 18. Milne SB, Forrester JS, Ivanova PT, Armstrong MD, Brown HA. Multiplexed lipid arrays of antiimmunoglobulin M‐induced changes in the glycerophospholipid composition of WEHI‐231 cells In: Feng L, Prestwich GD, eds. Functional Lipidomics. Boca Raton, FL: CRC Press, 2006; 263–83. [Google Scholar]
- 19. Lee HC, Kubo T, Kono N et al Depletion of mboa‐7, an enzyme that incorporates polyunsaturated fatty acids into phosphatidylinositol (PI), impairs PI 3‐phosphate signaling in Caenorhabditis elegans . Genes Cells 2012; 17: 748–57. [DOI] [PubMed] [Google Scholar]
- 20. Cerruti CD, Benabdellah F, Laprévote O, Touboul D, Brunelle A. MALDI imaging and structural analysis of rat brain lipid negative ions with 9‐aminoacridine matrix. Anal Chem 2012; 84: 2164–71. [DOI] [PubMed] [Google Scholar]
- 21. Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev 2011; 111: 6491–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harada T, Yuba‐Kubo A, Sugiura Y et al Visualization of volatile substances in different organelles with an atmospheric‐pressure mass microscope. Anal Chem 2009; 81: 9153–7. [DOI] [PubMed] [Google Scholar]
- 23. Saeed AI, Sharov V, White J et al TM4: a free, open‐source system for microarray data management and analysis. Biotechniques 2003; 34: 374–8. [DOI] [PubMed] [Google Scholar]
- 24. Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J 2000; 79: 328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dirat B, Bochet L, Dabek M et al Cancer‐associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res 2011; 71: 2455–65. [DOI] [PubMed] [Google Scholar]
- 26. Nieman KM, Kenny HA, Penicka CV et al Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011; 17: 1498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoshino D, Jourquin J, Emmons SW et al Network analysis of the focal adhesion to invadopodia transition identifies a PI3K‐PKCα invasive signaling axis. Sci Signal 2012; 5: ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wander SA, Zhao D, Besser AH et al PI3K/mTOR inhibition can impair tumor invasion and metastasis in vivo despite a lack of antiproliferative action in vitro: implications for targeted therapy. Breast Cancer Res Treat 2013; 138: 369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. MALDI‐based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J Chromatogr B Analyt Technol Biomed Life Sci 2007; 855: 98–103. [DOI] [PubMed] [Google Scholar]
- 30. Metzger‐Filho O, Tutt A, de Azambuja E et al Dissecting the heterogeneity of triple‐negative breast cancer. J Clin Oncol 2012; 30: 1879–87. [DOI] [PubMed] [Google Scholar]
- 31. Dória ML, Cotrim CZ, Simões C et al Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J Cell Physiol 2013; 228: 457–68. [DOI] [PubMed] [Google Scholar]
- 32. Alowami S, Troup S, Al‐Haddad S, Kirkpatrick I, Watson PH. Mammographic density is related to stroma and stromal proteoglycan expression. Breast Cancer Res 2003; 5: 129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hankin JA, Murphy RC. Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Anal Chem 2010; 82: 8476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Löhmann C, Schachmann E, Dandekar T, Villmann C, Becker CM. Developmental profiling by mass spectrometry of phosphocholine containing phospholipids in the rat nervous system reveals temporo‐spatial gradients. J Neurochem 2010; 114: 1119–34. [DOI] [PubMed] [Google Scholar]
- 35. Burnum KE, Cornett DS, Puolitaival SM et al. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res 2009; 50: 2290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Immunohistochemistry results for basal markers.
Table S2. Differences in averaged proportions of phosphatidylinositols (PIs) (Malignant‐ vs benign‐region).
Table S3. Differences in signal intensities during the progression.
Fig. S1. Mass spectrometry (MS)/MS spectra of phosphatidylinositols (PIs) observed in this study.
Fig. S2. The averaged mass spectra obtained from epithelial areas in breast cancer tissues.
Fig. S3. The averaged mass spectra obtained from stromal areas in breast cancer tissues.
Fig. S4. Mass spectrometry (MS) images of PI (18:0/20:4) in the same case of Fig. 5.


