Abstract
The Chimera antigen receptor (CAR)-T cell therapy has gained great success in the clinic. However, there are still major challenges for its wider applications in a variety of cancer types including lack of effectiveness due to the highly complex tumor microenvironment, and the forbiddingly high cost due to the personalized manufacturing procedures. In order to overcome these hurdles, numerous efforts have been spent focusing on optimizing Chimera antigen receptors, engineering and improving T cell capacity, exploiting features of subsets of T cell or NK cells, or making off-the-shelf universal cells. Here, we developed induced pluripotent stem cells (iPSCs)-derived, CAR-expressing macrophage cells (CAR-iMac). CAR expression confers antigen-dependent macrophage functions such as expression and secretion of cytokines, polarization toward the pro-inflammatory/anti-tumor state, enhanced phagocytosis of tumor cells, and in vivo anticancer cell activity. This technology platform for the first time provides an unlimited source of iPSC-derived engineered CAR-macrophage cells which could be utilized to eliminate cancer cells.
Keywords: Chimera antigen receptor (CAR), Induced pluripotent stem cells (iPSC)-derived macrophage cells (iMac), Antigen-dependent activation, Anti-cancer cell functions
To the Editor,
Recently, CAR-iPSC-differentiated CAR-expressing T cells and NK cells have been reported to have potent cytotoxic activity against cancer cells, and they represent a new family of engineered stem cell-derived immune cells for CAR therapies [1, 2]. Myeloid cells such as macrophages have been utilized as a type of effector cells to combat cancer cells by means of their phagocytosis function [3, 4]. However, immortalized macrophage cell lines are not applicable to clinical settings, and bone marrow or PBMC-derived primary macrophages are not efficiently engineered, thus leaving iPSC-derived macrophage cells as a great source for myeloid cell-based immunotherapy. Upon challenge with antigen-expressing cancer cells, CAR-expressing iPSC-induced macrophage (CAR-iMac) cells showed antigen-dependent macrophage functions. Expression of a CAR targeting tumor-associated antigen conferred CAR-iMac cells in vitro and in vivo anti-tumor effects.
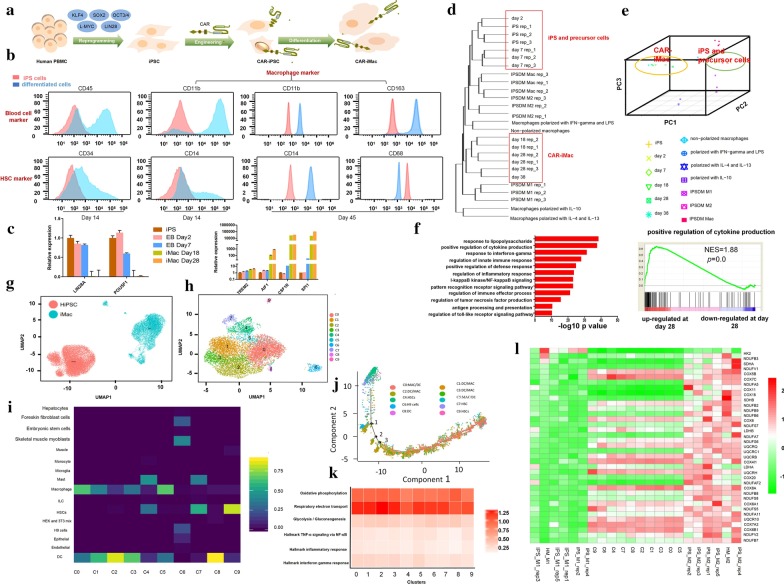
First, we started from deriving iPSCs from peripheral blood mononuclear cells (PBMC) of a healthy donor with non-integrating episomal vectors encoding reprogramming factors (Fig. 1a), and isolated single iPSC clones (Additional file 1: Fig. S1). The materials and methods are shown in detail in the Additional file 2. Then, we compared different CD19 CARs and chose the conventional CAR to introduce into the iPSCs by lentiviral transduction (Fig. 1a and Additional file 1: Fig. S2a–g). Then, we established a protocol of myeloid/macrophage differentiation to induce CAR-iPSCs toward myeloid cell lineages (Additional file 1: Fig. S2h). Differentiated cells showed typical macrophage marker gene expression (Fig. 1b, Additional file 1: Fig. S4b–i). The products can be collected from 20 to 30 day for multiple times to allow further expansion to have a final yield of above 50-fold of the starting iPSCs (Additional file 1: Fig. S3a), with high purity indicated by ~ 100% of CD11b and CD14 expression at later days (Fig. 1b). Key macrophage marker genes were induced, whereas pluripotent marker genes disappeared (Fig. 1c; Additional file 3: Table S1). RNA-sequencing using differentiated cells showed that iPSCs clustered with precursor cells, and late-day differentiated cells clustered with primary macrophage cells, or untransduced iPSC-differentiated macrophage cells from previous studies [5, 6](Fig. 1d, e). GO analyses showed strong enrichment of innate immunity-related functions in 28-day differentiated cells (Fig. 1f; Additional file 4: Table S2).
Fig. 1.
CAR-expressing iPSCs can differentiate into CAR-macrophage cells. a Overview of deriving CAR-iMacs from CAR-iPSCs. b Flow cytometry analysis of iPSC-derived cells at different stages of differentiation with stage-specific markers. c qRT-PCR showing pluripotent marker gene and key macrophage marker gene expression at different stages of CAR-iPSC differentiation. n = 3, error bar: standard error of the mean. d Hierarchical clustering of transcriptomes of CAR-iPSCs, their differentiated cells, and primary and untransduced iPSC-differentiated macrophages in different states. e Principal component analysis (PCA) of the same samples as in d. f Top GO terms enriched in genes up-regulated on day 28 differentiated CAR-iMac cells compared with CAR-iPSCs, and the right panel is an example of GSEA analysis of the GO terms. NES normalized enrichment score. P = 0: P value is a very small number. g UMAP plot showing separation between human iPSCs and CAR-iMac cells. h UMAP plot showing subpopulation clustering of CAR-iMac cells. Ten clustered C0–C9 were identified and labeled as 0–9 with different colors. i Heatmap showing blasting the C0-C9 clusters of cells illustrated in g against a human single-cell atlas database containing single-cell RNA-seq data of hundreds of cell types including macrophages (https://scibet.cancer-pku.cn). j Trajectory analysis of differentiated cells along a pseudotime axis. k Heatmap showing averaged expression of M1 or M2 signature pathway genes in different clusters of cells illustrated in i. l Heatmaps to compare (benchmark) the 10 clusters (C0-C9) of CAR-iMac cells against previously published M1 or M2 polarized macrophages using metabolism genes. Human iPS cells differentiated macrophages polarized by IFN-γ and LPS; IPS_M2: Human iPS cells differentiated macrophages polarized by IL-4; HM_M1: Human PBMC-derived macrophages polarized by IFN-γ and LPS; HM_M2: Human PBMC-derived macrophages polarized by IL-4
Next, we further dissected their subpopulations by performing single-cell RNA-sequencing analysis. These cells clustered away from undifferentiated CAR-iPSCs (Fig. 1g), and they appeared to be largely homogenous with only a small number of cells not clustered with the main population (Fig. 1h). Blasting the differentiated single cells in a database of human cell atlas containing single-cell RNA-sequencing data revealed that these iMac cells mainly clustered with macrophages (Fig. 1i and Additional file 1: Fig. S6a). Trajectory analysis revealed that CAR-iMac cells went through a path from HSC to macrophage and DC cells without major branches (Fig. 1j, Additional file 1: Fig. S5c).
Moreover, all 10 clusters of differentiated cells showed strong signatures of M2 state [7–10] (Fig. 1k, Additional file 1: Fig. S6b). We further compared the single-cell data in the 10 clusters with bulk RNA-seq data from the LPS/IFN-γ-polarized M1 cells or IL-4/IL-10-polarized M2 cells, by examining M1/M2-associated genes (Fig. 1l, Additional file 1: Fig. S5d), and found that most clusters were more similar to the M2 state particularly when using metabolism genes as markers [5, 6, 11–15].
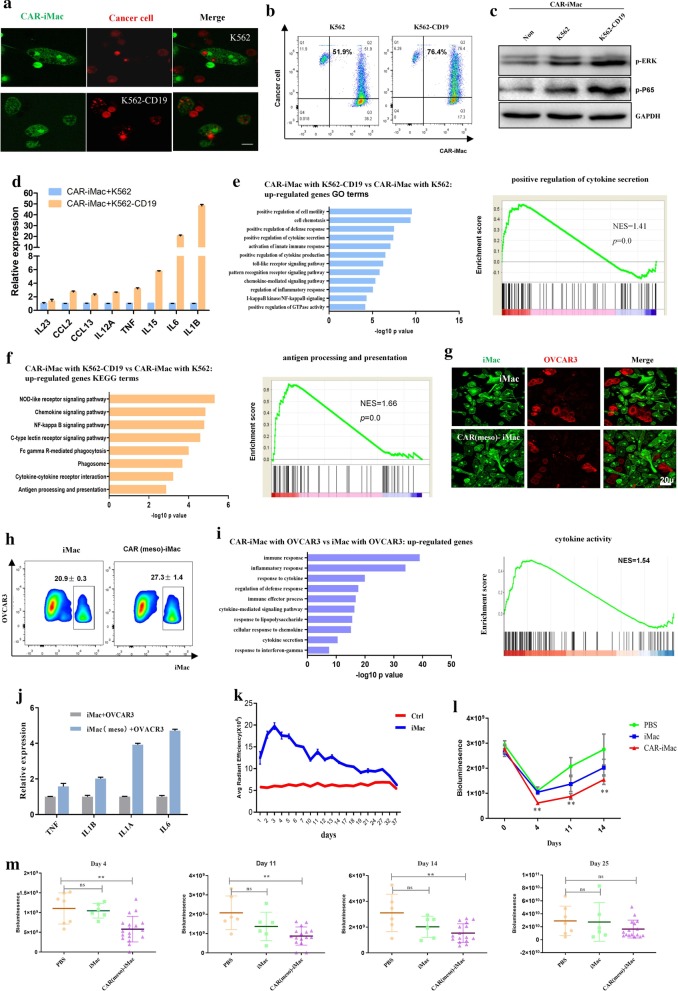
Next, we incubated the CAR (CD19)-iMac cells, CAR (meso)-iMac cells, or control iMac cells with CD19-expressing K562 leukemia cells or mesothelin-expressing OVCAR3/ASPC1 ovarian/pancreatic cancer cells. Compared with K562 alone, K562-CD19 cells were more likely to be phagocytosed by CAR (CD19)-iMacs (Fig. 2a, b), and compared with control cells, CAR (meso)-iMac showed increased phagocytosis activity against OVCAR3 and ASPC1 cells (Fig. 2g, h and Additional file 1: Fig. S7g). Intracellular signaling such as phosphorylation of ERK and NF-κB(P65) proteins were increased in CAR-iMacs co-cultured with CD19-expressing K562 cells compared to K562 cells, or to CAR-iMac cells cultured alone (Fig. 2c). We also examined cytokine gene expression in CAR(CD19)-iMac and CAR (meso)-iMac cells when they were incubated with tumor cells and found antigen-dependent increase in M1 pro-inflammatory cytokine expression(Fig. 2d, j and Additional file 1: Fig. S7h). Moreover, transcriptional analysis showed that CAR(CD19)-iMac cells and CAR(meso)-iMac cells showed strong enrichment of up-regulated genes in GO or KEGG terms of “positive regulation of cytokine secretion,” “antigen processing and presentation,” and “Toll-like receptor signaling pathway,” indicating these cells are more wired toward the pro-inflammatory state, when they encounter the antigen (Fig. 2e, f, i and Additional file 1: Fig. S7i).
Fig. 2.
CAR-iMac cells showed antigen-dependent phagocytosis and anticancer cell functions in vitro and in vivo. a Confocal microscopy pictures showing phagocytosis of K562 or K562-CD19 cells (red) by CAR (CD19)-iMac cells (green). b Flow cytometry showing phagocytosis of K562 or K562-CD19 cells by CAR (CD19)-iMac cells. c Western blotting showing phosphorylation of ERK and NF-κB P65 in CAR (CD19)-iMac cells in the indicated conditions. d qRT-PCR showing cytokine gene mRNA expression when CAR (CD19)-iMac cells were incubated with K562 or K562-CD19 cancer cells for 24 h. n = 3, error bar: standard error of the mean. e Top GO terms enriched in genes up-regulated in CAR-iMac cells. Right panel is GSEA analysis of “positive regulation of cytokine production.” f Top KEGG pathways enriched in genes up-regulated in CAR-iMac cells. Right panel is GSEA analysis of “antigen processing and presentation.” g Confocal microscopic images showing phagocytosis of OVCAR3 cells (red) by iMac or CAR (meso)-iMac cells (green). h Flow cytometry showing phagocytosis of OVCAR3 ovarian cancer cells by iMac or CAR (meso)-iMac cells. i GO term analysis with RNA-seq data showing the up-regulated genes in CAR (meso)-iMac cells. Right panel is GSEA analysis of “cytokine activity gene.” j qRT-PCR showing cytokine gene mRNA expression when iMac or CAR (meso)-iMac cells were incubated with OVCAR3 cells for 24 h. n = 3. Error bar: standard error of the mean. k 3 × 106 DiR dye-labeled iMac cells were intraperitoneally injected into NSG mice. n = 3. Error bars represent standard error of the mean. l 4 × 105 of luciferase-expressing ovarian cancer cells (HO8910) were intraperitoneally injected into NSG mice. Mice were treated 4 h later with I.P. injection of PBS, 4 × 106 iMac or 4 × 106 CAR (meso)-iMac cells. Bioluminescence showing tumor development on the indicated days. Statistical analysis was calculated via one-way ANOVA with multiple comparisons between the PBS group and the CAR-iMac group. *P < 0.05; **P < 0.01. m Quantification of tumor burden (total flux) by bioluminescent imaging on day 4, 11, 14, and day 25 after CAR-iMac treatment was plotted. Data are presented as the median ± SD, with statistical significance calculated via one-way ANOVA with multiple comparisons. *P < 0.05; **P < 0.01, ns not significant
When injected into NSG mice, these CAR-iMac cells expanded in vivo till around day 3 for about two fold, and persisted till more than 20 days and gradually disappeared after around 30 days (Fig. 2k). To test their anti-tumor cell activity, we first intraperitoneally injected ovarian cancer cells HO8910 expressing a luciferase gene into NSG mice. In order to further polarize CAR-iMac cells toward M1, we treated them with IFN-γ and washed IFN-γ away before injection (Additional file 1: Fig. S8). CAR(meso)-iMac-treated mice showed reduced tumor burden compared to the control group on day 4, 11, and 14 (Fig. 2l, m). These data demonstrate that the CAR confers anti-cancer cell activities in iMacs in vivo. The efficacy and persistency need to be further improved by designing more effective CARs or further modifying the CAR-iMac cells to stay constitutively active.
Supplementary information
Additional file 1. The main results of differentiation of CAR-expressing iPSCs into CAR-macrophage cells with antigen-dependent phagocytosis and anti-cancer cell functions in vitro and in vivo.
Additional file 2. Materials and Methods.
Additional file 3: Table S1. Primer sequences for qRT-PCR.
Additional file 3: Table S2. RNA-sequencing data analysis of iMac during the differentiation process from iPSCs.
Acknowledgements
We thank George Q. Daley and Thorsten M. Schlaeger for providing reprogramming plasmids. We thank the Core Facilities of Zhejiang University School of Medicine for technical support and Center for Drug Safety Evaluation at Zhejiang University for animal technical support. We thank Ming Chen (College of Life Sciences, Zhejiang University) for computational analyses support and Harvey Lodish (MIT) for helpful Discussion. We thank CellOrigin for supporting this project.
Abbreviations
- CAR
Chimera antigen receptor
- DC
Dendritic cells
- ERK
Extracellular-regulated protein kinase
- HSC
Hematopoietic stem cell
- ICT
Immune-checkpoint therapy
- IFN-γ
Interferon gamma
- IP
Intraperitoneal injection
- iPSC
Induced pluripotent stem cells
- LPS
Lipopolysaccharide
- MPS
Mononuclear phagocyte system
- PBMC
Peripheral blood mononuclear cells
- qRT-PCR
Quantitative reverse transcription PCR
Authors’ contributions
JZ, QW, HH, YX, and YH designed and supervised the study. LZ, LT, XD, JW, AL, MZ, JX, WZ, YZ, ZS, HZ, and YW performed the experiments. HY performed the bioinformatics analysis. HH, HL, and GC provided key reagents and advice for the manuscript. All authors read and approved the final manuscript.
Funding
J.Z. is supported by the National Key Research and Development Program of China (2018YFA0107100, 2018YFA0107103, 2018YFC1005002), the National Natural Science Foundation projects of China (31871453, 91857116), the Zhejiang Innovation Team grant (2019R01004) and the Zhejiang Natural Science Foundation projects of China (LR19C120001).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The animal study was performed in compliance with relevant regulatory standards. All animal studies are approved by ZJU IRB committee.
Consent for publication
Written informed consent for publication was obtained from all participants.
Competing interests
A patent has been filed related to the data presented. JZ and GC are founders and scientific advisors for CellOrigin.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Zhang, Lin Tian, Xiaoyang Dai, Hua Yu and Jiajia Wang have contributed equally to this work
Contributor Information
He Huang, Email: hehuangyu@126.com.
Qinjie Weng, Email: wengqinjie@zju.edu.cn.
Jin Zhang, Email: zhgene@zju.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13045-020-00983-2.
References
- 1.Themeli M, Kloss CC, Ciriello G, Fedorov VD, Perna F, Gonen M, et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013;31(10):928–933. doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Hermanson DL, Moriarity BS, Kaufman DS. Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell. 2018;23(2):181–92e5. doi: 10.1016/j.stem.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrissey MA, Williamson AP, Steinbach AM, Roberts EW, Kern N, Headley MB, et al. Chimeric antigen receptors that trigger phagocytosis. Elife. 2018;7:e36688. doi: 10.7554/eLife.36688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol. 2020;38(8):947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Xue C, Shah R, Bermingham K, Hinkle CC, Li W, et al. Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circ Res. 2015;117(1):17–28. doi: 10.1161/CIRCRESAHA.117.305860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep. 2017;8(6):1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Investig. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viola A, Munari F, Sanchez-Rodriguez R, Scolaro T, Castegna A. The metabolic signature of macrophage responses. Front Immunol. 2019;10:1462. doi: 10.3389/fimmu.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvan-Pena S, O'Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly B, O'Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25(7):771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40(2):274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 15.Lachmann N, Ackermann M, Frenzel E, Liebhaber S, Brennig S, Happle C, et al. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Rep. 2015;4(2):282–296. doi: 10.1016/j.stemcr.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The main results of differentiation of CAR-expressing iPSCs into CAR-macrophage cells with antigen-dependent phagocytosis and anti-cancer cell functions in vitro and in vivo.
Additional file 2. Materials and Methods.
Additional file 3: Table S1. Primer sequences for qRT-PCR.
Additional file 3: Table S2. RNA-sequencing data analysis of iMac during the differentiation process from iPSCs.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.