Abstract
Nicotinic acetylcholine receptors are involved in a plethora of fundamental biological processes ranging from muscle contraction to formation of memories. The receptors are pentameric proteins whose subunits are encoded by distinct genes. Subunit composition of a mature nicotinic receptor is governed in part by the transcriptional regulation of each subunit gene. Here, using chromatin immunoprecipitation assays, we report the interaction of the transcription factors Sp1, Sp3, c-Jun and Sox10 with the β4 subunit gene promoter in neuronal-like cell lines and rodent brain tissue. Our results corroborate previous in-vitro data demonstrating that these transcription factors interact with the β4 promoter. Taken together, these data suggest that Sp1, Sp3, c-Jun and Sox10 regulate expression of the β4 subunit gene in the mammalian brain.
Keywords: c-Jun, chromatin immunoprecipitation, nicotinic acetylcholine receptor β4 subunit, Sox10, Sp factors
Introduction
Neuronal nicotinic acetylcholine receptors (nAChRs) are pentameric transmembrane ligand-gated ion channels that play an essential role in several physiological processes including learning, memory and attention [1], Functional channels are made up of homomeric or heteromeric combinations of α and β subunits, which to date include α2-α10 and β2-β4 [1]. Each combination of subunits produces a receptor that is electrophysiologically and pharmacologically distinct [1–4]. The subunit composition of a particular receptor is governed, at least in part, by the transcriptional regulation of individual subunit genes. Our laboratory has had a long-standing interest in the transcriptional mechanisms underlying expression of the α3, α5 and β4 subunit genes. The three nAChR subunits encoded by these genes make up the predominant nicotinic receptor subtype expressed in the peripheral nervous system [1]. The work described herein focuses on the transcriptional regulation of the β4 subunit gene.
The spatially restricted expression of the β4 gene in the mammalian nervous system [5] suggests that mature receptors including this subunit display distinct pharmacological properties critical for mediating neuronal processes that occur in and around these brain regions [2,6]. Studies done on β4 knock out animals have demonstrated that the β4 subunit plays a critical role in the molecular processes mediating nicotine-induced withdrawal [7] and the anxiety-related response [8]. These findings are of particular importance owing to the fact that nicotine is the addictive component of cigarettes, and tobacco use is the leading cause of preventable death in the United States [9].
Gaining a better understanding of the transcriptional regulation of the β4 subunit gene will lead to a better understanding of not only cholinergic signaling in the brain, but also nicotine addiction and withdrawal. Advancing research in these areas will likely contribute to the discovery of new drugs for the treatment of nicotine withdrawal and ultimately smoking cessation.
Much of our effort has been directed toward elucidating the transcriptional mechanisms underlying β4 subunit gene expression. Previous work from our laboratory identified a 226-base pair segment of DNA in the 5′-flanking DNA of the β4 gene capable of driving luciferase expression in a cell type-specific manner [10,11]. Deletional analysis and site-directed mutagenesis led to the identification of a unique regulatory element, located upstream of the β4 initiator methionine, that is, critical for luciferase expression from the 226-base pair β4 promoter region in neuronal-like cell lines [12]. This regulatory element is referred to as a CA box, reflecting its nucleotide composition.
Electrophoretic mobility shift assays demonstrated that the transcription factors Sp1 [13,14], Sp3 [14] and Sox10 [10] bind to the CA box in vitro. Importantly, transient transfection studies have shown that these transcription factors can transactivate expression of a luciferase reporter gene driven by the β4 promoter [10,14]. Furthermore, coimmunoprecipitation studies have demonstrated that Sp1 and Sp3 [14], as well as Sox10 and the Sp factors [15,16] can physically interact. In addition, these factors can transactivate the β4 promoter synergistically when cotransfected into neuronal-like cell lines [14,15]. Thus, it is our hypothesis that Sp1, Sp3, Sox10 and c-Jun functionally interact with the β4 promoter in vivo, the result of which is the transcriptional activation of the β4 subunit gene. To test whether the four regulatory factors interact with the β4 gene promoter in the context of native chromatin, we carried out an extensive chromatin immunoprecipitation (ChIP) analysis using two neuronal-like cell lines as well as chromatin isolated from rodent brain tissue.
Materials and methods
Cell culture and differentiation
The rat pheochromocytoma cell line PC12 [17] was maintained and differentiated for 72 h with nerve growth factor (NGF) as described previously [12]. OBL21 cells [18] were maintained at 37°C in 8% CO2 in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum.
Chromatin immunoprecipitation
ChIP experiments using cells grown in culture were carried out using a ChIP assay kit (Upstate Biotechnology, Lake Placid, New York, USA). At the end of the ChIP procedure, DNA was isolated with phenol chloroform extraction followed by ethanol precipitation. ChIP-derived DNA was resuspended in 10 mM Tris-HCl, pH 8.0. ChIP experiments performed using rat tissue samples were carried out using a slightly modified Upstate ChIP protocol. In short, frozen brain tissue was ground to a powder and subsequently triturated in phosphate-buffered saline with 1% formaldehyde. Cross-linking was carried out at 37°C for 10 min. Cells were then collected by centrifugation, washed twice with phosphate-buffered saline containing protease inhibitors, then collected again by centrifugation. The resulting pellet was resuspended in lysis buffer with protease inhibitors. After this step, the experiment was completed as described in the Upstate ChIP protocol. All experimental procedures were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals as well as the University of Massachusetts Medical School Institutional Animal Care and Use Committee guidelines.
Polymerase chain reactions
Each ChIP-derived DNA sample (5 μl) was used in 25-μl PCR reactions using the following primers designed to amplify a segment of the rat β4 promoter, 5′-TAAGCTGCCTCGGGTGAACTAAGA-3′, 5′-TGTCTGGGGGAACCTGTGGCTAT-3′. OBL21 ChIP-derived DNA was amplified using the following primers designed to amplify a segment of the mouse β4 promoter, 5′-TTGGGTAAGCCAGGCTAAGA-3′, 5′-GGTCCCGAGACTTTCTCACA-3′. After amplification, loading dye (5 μl) was added to each PCR reaction and PCR products were electrophoresed through a 2% agarose gel and visualized with ethidium bromide.
Northern blot analysis
Expression of Sox10 mRNA in the OBL21 cell line was determined by northern blot analysis as previously described [19] using a radioactively labeled Sox10 cDNA.
Western blot analysis
Expression of Sox10 protein in OBL21 cells was determined by western blot analysis as described previously [14]. Anti-Sox10 antibody (Santa Cruz Biotechnology, Santa Cruz, California, USA) was used at a dilution of 1: 200.
Results
The interaction of transcription factors Sp1, Sp3 and c-Jun with the β4 promoter in nerve growth factor-treated PC12 cells
Previously, we reported that Sp1, Sp3 and Sox10 are capable of interacting with a segment of DNA from the β4 promoter in vitro [10,14]. As an initial attempt to investigate whether these factors interact with the β4 promoter in a chromatin environment, ChIP assays were performed using NGF-differentiated PC12 cells. PCR reactions using ChIP-derived DNA from Sp1, Sp3 and c-Jun immunoprecipitations resulted in the amplification of the expected 207-base pair fragment of the β4 promoter (Fig. 1, lanes 4–6). The accuracy of PCR amplification was confirmed by sequence analysis (not shown).
Fig. 1.
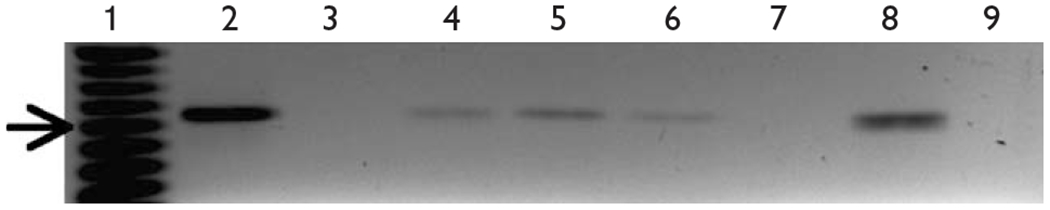
Differentiated PCI2 chromatin immunoprecipitation (ChIP)-derived DNA was used as template for PCR reactions designed to amplify a fragment of the rat β4 promoter. Amplification was observed for both histone H4 (lane 2) and input (lane 8) ChIP positive controls. The expected PCR product was also produced in Sp1, Sp3 and c-Jun reactions (lanes 4–6). No amplification was observed when using DNA immunoprecipitated by normal mouse IgG (lane 3), no antibody (lane 7) or in no-template PCR reactions (lane 9). The arrow indicates the 200-base pair marker (lane 1). Each ChIP experiment was carried out a minimum of three times.
The specificity of antibody–transcription factor complexes was demonstrated by negative results in ChIP assays when either normal mouse immunoglobulin G (IgG) or no antibody was used for the immunoprecipitations (Fig. 1, lanes 3 and 7, respectively). An additional negative control was performed by substituting water for ChIP-derived DNA at the final PCR step (Fig. 1, lane 9). Immunoprecipitation with antihistone H4 was performed as a ChIP positive control, which resulted in the amplification of the expected fragment (Fig. 1, lane 2). In addition, a sample of fragmented chromatin before immunoprecipitation was used as another positive control (referred to as Input), which also amplified the expected 207-base pair product (Fig. 1, lane 8). Taken together, these data indicate that Sp1, Sp3 and c-Jun are present at the endogenous β4 promoter in NGF-treated PC12 cells.
Sp1, Sp3, c-Jun and Sox10 assemble on the β4 promoter in the mouse olfactory bulb cell line OBL21
Our laboratory has previously shown that Sox10 binds to and transactivates the β4 promoter in neuronal-like cell lines, but not in nonneuronal cell lines [10]. As PC12 cells do not express Sox10, we extended our ChIP analysis to another cell line, OBL21, a line derived from mouse olfactory bulb cells [18]. Northern and western blot analyses indicate that OBL21 cells express high levels of Sox10 mRNA and protein, respectively (Fig. 2). ChIP experiments with OBL21 cells were carried out as described above for PC12 cells with an additional Sox10 immunoprecipitation. PCR reactions with ChIP-derived DNA from Sp1, Sp3, Sox10 and c-Jun immunoprecipitations resulted in the amplification of the expected 243-base pair product of the β4 promoter (Fig. 3, lanes 4–7). Once again, the specificity of antibody–transcription factor complexes was confirmed by the failure of ChIP-derived DNA from mouse IgG and no antibody immunoprecipitations to produce the β4 promoter PCR product (Fig. 3, lanes 3 and 8). Finally, H4 and input ChIP positive controls showed strong bands (Fig. 3, lanes 2 and 9). These data indicate that Sp1, Sp3, c-Jun and Sox10 interact with the β4 promoter in OBL21 cells.
Fig. 2.
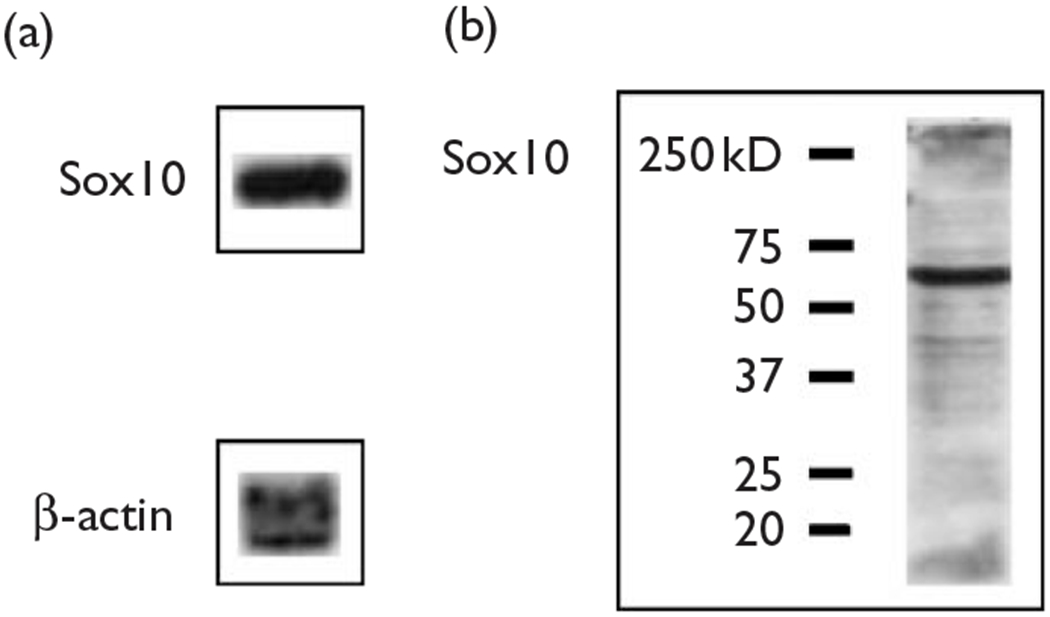
The mouse olfactory bulb cell line OBL21 expresses Sox10 mRNA and protein. (a) Northern blot analysis of OBL21 cells revealed relatively high levels of Sox10 mRNA. Northern analysis was also carried out for β-actin message as a loading control. (b) Western blot analysis was carried out on OBL21 cell lysates showed abundant levels of Sox10 protein (MW=68kDa). Lines on the left of the blot indicate the positions and weights of protein standards (Precision Plus Protein Standards, BioRad, Hercules, California, USA).
Fig. 3.
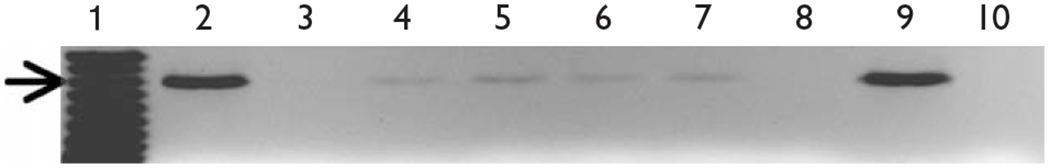
OBL21 chromatin immunoprecipitation (ChIP)-derived DNA was used as a template for PCR reactions designed to amplify a 243-base pair product of the mouse β4 promoter.The expected product was observed in PCR reactions for both histone H4 (lane 2) and input (lane 9) positive controls. Amplification was also observed in Sp1, Sp3, Sox10 and c-Jun immunoprecipitations (lanes 4–7). No amplification was seen in PCR reactions using DNA immunoprecipitated by normal mouse IgG (lane 2), no antibody (lane 8) or in no-template PCR reactions (lane 10). The arrow indicates the 250-base pair marker (lane 1). Each ChIP experiment was carried out a minimum of three times.
Sp1, Sp3, Sox10 and c-Jun interact with the β4 promoter in fetal rat brain tissue
To investigate the interaction of Sp1, Sp3, c-Jun and Sox10 with the β4 promoter in a more physiologically relevant setting, ChIP experiments were performed using cells isolated from rodent brain tissue. PCR reactions using fetal rat brain ChIP-derived DNA as template resulted in the amplification of the expected β4 promoter fragment for Sp1, Sp3, c-Jun and Sox10 immunoprecipitations (Fig. 4a, lanes 4–7). The specificity of antibody–transcription factor complexes in these ChIP experiments was demonstrated by failure of ChIP-derived DNA from mouse IgG and no antibody immunoprecipitations to produce the expected product (Fig. 4a, lanes 3 and 8). These fetal rat brain ChIP experiments indicate that Sp1, Sp3, c-Jun and Sox10 bind to the β4 promoter in vivo.
Fig. 4.
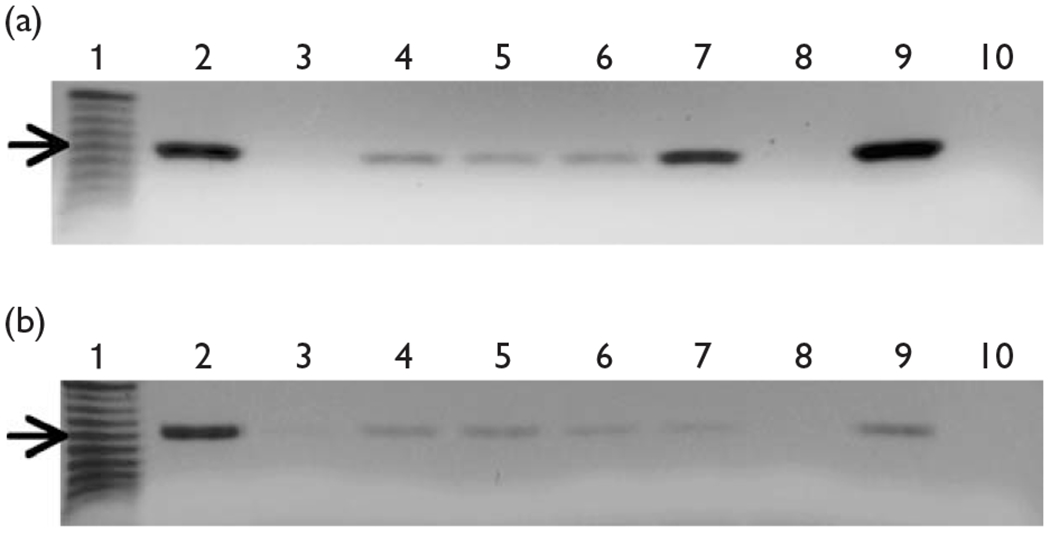
Rat brain chromatin immunoprecipitation (ChIP) experiments. (a) Fetal rat brain ChIP-derived DNA was used as template for PCR reactions designed to amplify the rat β4 promoter. Histone H4 (lane 2) and input (lane 9) positive controls produced the expected product. Amplification was also observed for Sp1, Sp3, Sox10 and c-Jun PCR reactions (lanes 4–7). ChIP negative controls, normal mouse IgG (lane 3), mock immunoprecipitation (lane 8) and no-template PCR control (lane 10) produced no amplified products. The arrow indicates the 200-base pair marker (lane 1). (b) Adult rat brain ChIP-derived DNA was used in PCR reactions designed to amplify the rat β4 promoter. Again, both the histone H4 (lane 2) and input (lane 9) positive controls produced the expected product. Amplification was also observed for Sp1, Sp3, Sox10 and c-Jun PCR reactions (lanes 4–7). Finally, ChIP negative controls, normal mouse IgG (lane 3), mock immunoprecipitation (lane 8) and no-template PCR control (lane 10) produced no amplified products. The arrow indicates the 200-base pair marker (lane 1). Each ChIP experiment was carried out a minimum of three times.
Chromatin immunoprecipitation of Sp1, Sp3, c-Jun and Sox10 from adult rat brain tissue
We hypothesized that as Sp1, Sp3, c-Jun and Sox10 were found to interact with the β4 promoter in two neuronal-like cell lines as well as fetal rat brain tissue and given that the β4 gene is expressed in adult brain, the transcription factors would also interact with the β4 promoter in adult rat brain tissue. To test this hypothesis, we performed an additional round of ChIP assays using adult rat brain tissue. PCR using ChIP DNA from Sp1, Sp3, c-Jun and Sox10 immunoprecipitations resulted in the amplification of the expected fragment of the β4 promoter (Fig. 4b, lanes 4–7), which was sequenced to confirm the specificity of the PCR reaction (not shown). No amplification occurred in PCR reactions using DNA derived from mouse IgG and mock immunoprecipitations (Fig. 4b, lanes 3 and 8), further suggesting that the results shown in Fig. 4b are specific to the antibodies used to precipitate the adult rat brain chromatin.
Discussion
nAChR subunit genes are expressed in a variety of locations in both the peripheral and central nervous systems. The molecular mechanisms underlying the temporally and spatially specific expression of nAChR subunit genes, however, remain to be completely elucidated. Here, we extended our previous in-vitro work on the transcriptional regulation of the β4 subunit gene to further define the DNA-protein interactions that occur on the β4 gene promoter. Using both cell lines and brain extracts in ChIP assays, we have shown that Sp1, Sp3, Sox10 and c-Jun are present at the β4 gene promoter in vivo. The positive ChIP results for the transcription factors Sp1 and Sp3 are in agreement with electrophoretic mobility shift experiments carried out previously in our laboratory that demonstrated direct interactions between these proteins and the β4 promoter in vitro [14]. In related studies performed by others, it was shown that there are functional Sp1-binding sites in the promoters of the α3, β4 and α5 nAChR subunit genes [20–22]. Interestingly, these three subunit genes are located in a tight cluster in the mammalian genome, implying that they are coordinately regulated [23,24]. This coordinate regulation is likely responsible for directing the overlapping expression patterns of the clustered subunits in the mammalian nervous system [1]. It is our hypothesis that the coordinate regulation of these clustered subunits takes place at least in part through the action of the transcription factor Sp1.
We have also shown that Sox10 interacts with the endogenous β4 promoter in vivo, this finding is also corroborated by previous electrophoretic mobility shift results from our laboratory, which indicate that the Sox10 protein can interact with the β4 promoter in vitro [10]. The c-Jun protein was not found to interact with the β4 promoter using electrophoretic mobility shift assay (data not shown); however, c-Jun can in fact synergistically transactivate the β4 promoter with Sp1 [25]. This is in agreement with the data presented here, demonstrating that c-Jun is associated with the endogenous β4 promoter in neuronal-like cell lines and in brain tissue. Taken together, these results suggest that, in vivo, c-Jun either binds directly to the β4 promoter or interacts with other regulatory factors that bind to the promoter. As we suggested before [25], likely candidates for such factors are the Sp proteins.
In sum, we have demonstrated that four transcription factors, Sp1, Sp3, Sox10 and c-Jun, previously shown to interact functionally with the nAChR β4 gene promoter in vitro, also assemble on the β4 promoter in the context of native chromatin, strongly suggesting that they are critical players in the transcriptional regulation of the β4 subunit gene in vivo.
Acknowledgements
The authors express their sincere appreciation to Connie Cepko and Liz Ryder for their kind gift of OBL21 cells and Hsien-Sung Huang for useful advice regarding ChIP assays. The project was supported by Grant Number R01NS030243 from the National Institute of Neurological Disorders and Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institutes of Health.
References
- 1.Leonard S, Bertrand D. Neuronal nicotinic receptors: from structure to function. Nicotine Tob Res 2001; 3:203–223. [DOI] [PubMed] [Google Scholar]
- 2.Duvoisin RM, Deneris ES, Patrick J, Heinemann S. The functional diversity of the neuronal nicotinic acetylcholine receptors is increased by a novel subunit: β4. Neuron 1989; 3:487–496. [DOI] [PubMed] [Google Scholar]
- 3.Harvey SC, Luetje CW. Determinants of competitive antagonist sensitivity on neuronal nicotinic receptor β subunits. J Neurosci 1996; 16:3798–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker MJ, Beck A, Luetje CW. Neuronal nicotinic receptor β2 and β4 subunits confer large differences in agonist binding affinity. Mol Pharmacol 1998; 54:1132–1139. [PubMed] [Google Scholar]
- 5.Dineley-Miller K, Patrick J. Gene transcripts for the nicotinic acetylcholine receptor subunit, β4, are distributed in multiple areas of the rat central nervous system. Brain Res Mol Brain Res 1992; 16: 339–344. [DOI] [PubMed] [Google Scholar]
- 6.Gahring LC, Persiyanov K, Rogers SW. Neuronal and astrocyte expression of nicotinic receptor subunit β4 in the adult mouse brain. J Comp Neurol 2004; 468:322–333. [DOI] [PubMed] [Google Scholar]
- 7.Salas R, Pieri F, De Biasi M. Decreased signs of nicotine withdrawal in mice null for the β4 nicotinic acetylcholine receptor subunit. J Neurosci 2004; 24:10035–10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salas R, Cook KD, Bassetto L, De Biasi M. The α3 and β4 nicotinic receptor subunits are necessary for nicotine-induced seizures and hypolocomotion in mice. Neuropharmacology 2004; 47:401–407. [DOI] [PubMed] [Google Scholar]
- 9.McPhillips-Tangum C, Bocchino C, Carreon R, Erceg C, Rehm B. Addressing tobacco in managed Care: results of the 2002 survey. Prev Chronic Dis 2004; 1:1–11. [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Melnikova IN, Hu M, Gardner PD. Cell type-specific activation of neuronal nicotinic acetylcholine receptor subunit genes by Sox10. J Neurosci 1999; 19:9747–9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu M, Bigger CB, Gardner PD. A novel regulatory element of a nicotinic acetylcholine receptor gene interacts with a DNA binding activity enriched in rat brain. J Biol Chem 1995; 270:4497–4502. [DOI] [PubMed] [Google Scholar]
- 12.Hu M, Whiting Theobald NL, Gardner PD. Nerve growth factor increases the transcriptional activity of the rat neuronal nicotinic acetylcholine receptor β4 subunit promoter in transfected PC12 cells. J Neurochem 1994; 62:392–395. [DOI] [PubMed] [Google Scholar]
- 13.Bigger CB, Casanova EA, Gardner PD. Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. Functional interactions between Sp1 and the rat β4 subunit gene promoter. J Biol Chem 1996; 271:32842–32848. [DOI] [PubMed] [Google Scholar]
- 14.Bigger CB, Melnikova IN, Gardner PD. Sp1 and Sp3 regulate expression of the neuronal nicotinic acetylcholine receptor β4 subunit gene. J Biol Chem 1997; 272:25976–25982. [DOI] [PubMed] [Google Scholar]
- 15.Melnikova IN, Lin HR, Blanchette AR, Gardner PD. Synergistic transcriptional activation by Sox10 and Sp1 family members. Neuropharmacology 2000; 39:2615–2623. [DOI] [PubMed] [Google Scholar]
- 16.Melnikova IN, Yang Y, Gardner PD. Interactions between regulatory proteins that bind to the nicotinic receptor β4 subunit gene promoter. Eur J Pharmacol 2000; 393:75–83. [DOI] [PubMed] [Google Scholar]
- 17.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A 1976; 73:2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryder EF, Snyder EY, Cepko CL. Establishment and characterization of multipotent neural cell lines using retrovirus vector-mediated oncogene transfer. J Neurobiol 1990; 21:356–375. [DOI] [PubMed] [Google Scholar]
- 19.Fanger GR, Brennan C, Henderson LP, Gardner PD, Maue RA. Differential expression of sodium channels and nicotinic acetylcholine receptor channels in nnr variants of the PC12 pheochromocytoma cell line. J Membr Biol 1995; 144:71–80. [DOI] [PubMed] [Google Scholar]
- 20.Campos-Caro A, Carrasco-Serrano C, Valor LM, Viniegra S, Ballesta JJ, Criado M. Multiple functional Sp1 domains in the minimal promoter region of the neuronal nicotinic receptor α5 subunit gene. J Biol Chem 1999; 274:4693–4701. [DOI] [PubMed] [Google Scholar]
- 21.Deneris ES, Francis N, McDonough J, Fyodorov D, Miller T, Yang X. Transcriptional control of the neuronal nicotinic acetylcholine receptor gene cluster by the β43′ enhancer, Sp1, SCIP and ETS transcription factors. Eur J Pharmacol 2000; 393:69–74. [DOI] [PubMed] [Google Scholar]
- 22.Valor LM, Campos-Caro A, Carrasco-Serrano C, Ortiz JA, Ballesta JJ, Criado M. Transcription factors NF-Y and Sp1 are important determinants of the promoter activity of the bovine and human neuronal nicotinic receptor β4 subunit genes. J Biol Chem 2002; 277: 8866–8876. [DOI] [PubMed] [Google Scholar]
- 23.Boulter J, O’Shea-Greenfield A, Duvoisin RM, Connolly JG, Wada E, Jensen A, et al. Alpha 3, alpha 5, and beta 4: three members of the rat neuronal nicotinic acetylcholine receptor-related gene family form a gene cluster. J Biol Chem 1990; 265:4472–4482. [PubMed] [Google Scholar]
- 24.Yang X, Yang F, Fyodorov D, Wang F, McDonough J, Herrup K, et al. Elements between the protein-coding regions of the adjacent β4 and α3 acetylcholine receptor genes direct neuron-specific expression in the central nervous system. J Neurobiol 1997; 32:311–324. [PubMed] [Google Scholar]
- 25.Melnikova IN, Gardner PD. The signal transduction pathway underlying ion channel gene regulation by SP1-C-Jun interactions. J Biol Chem 2001; 276:19040–19045. [DOI] [PubMed] [Google Scholar]


