Abstract
Background
There is a paucity of data on the prognostic value of programmed cell death protein 1 (PD-1) protein and gene expression in early breast cancer (BC) and the present study’s aim was to comprehensively investigate it.
Methods
The study consisted of three parts: a correlative analysis of PD-1 protein and gene expression from an original patient cohort of 564 patients with early BC; a systematic review and trial-level meta-analysis on the association between PD-1 protein expression and disease-free survival/overall survival (OS) in early BC; and a pooled gene expression analysis from publicly available transcriptomic datasets regarding PDCD1 expression.
Results
In the study cohort, PD-1 protein, but not gene expression, was associated with improved OS (HRadj=0.73, 95% CI 0.55 to 0.97, p=0.027 and HRadj=0.88, 95% CI 0.68 to 1.13, p=0.312, respectively). In the trial-level meta-analysis, PD-1 protein expression was not found to be statistically significantly associated with outcomes in the overall population. Finally, in the pooled gene expression analysis, higher PDCD1 expression was associated with better OS in multivariable analysis in the entire population (HRadj=0.89, 95% CI 0.80 to 0.99, p=0.025) and in basal-like tumours.
Conclusions
PD-1 protein and gene expression seem to be promising prognostic factors in early BC. Standardisation of detection and assessment methods is of utmost importance.
Keywords: breast cancer, gene expression, PD-1, prognosis, triple-negative
Key questions.
What is already known about this subject?
The prognostic role of programmed cell death protein 1 (PD-1) protein and gene expression in breast cancer is not well studied. Even fewer studies report on their association which remains unclear owing to analytical difficulties and the complex regulation of the PD-1/programmed cell death ligand 1 axis.
What does this study add?
PD-1 protein expression predicted better survival in an original patient cohort, though this finding was not confirmed in a trial-level meta-analysis of 15 studies. In addition, PD-1 gene expression predicted better survival in a pooled gene expression analysis. The prognostic effect was mainly driven by triple negative/basal-like breast cancer. PD-1 protein and gene expression were weakly, but significantly, correlated.
How might this impact on clinical practice?
Emerging data show that PD-1 gene expression might be a predictor of benefit from immune checkpoint inhibition. This study confirms its prognostic role and supports its future inclusion to management algorithms as a marker that selects a patient population, mainly with triple negative disease, with inherently better prognosis that may additionally benefit from immunotherapy.
What is already known about this subject?
What is already known about this subject?
The prognostic role of programmed cell death protein 1 (PD-1) protein and gene expression in breast cancer is not well studied. Even fewer studies report on their association which remains unclear owing to analytical difficulties and the complex regulation of the PD-1/programmed cell death ligand 1 axis.
What does this study add?
What does this study add?
PD-1 protein expression predicted better survival in an original patient cohort, though this finding was not confirmed in a trial-level meta-analysis of 15 studies. In addition, PD-1 gene expression predicted better survival in a pooled gene expression analysis. The prognostic effect was mainly driven by triple negative/basal-like breast cancer. PD-1 protein and gene expression were weakly, but significantly, correlated.
How might this impact on clinical practice?
How might this impact on clinical practice?
Emerging data show that PD-1 gene expression might be a predictor of benefit from immune checkpoint inhibition. This study confirms its prognostic role and supports its future inclusion to management algorithms as a marker that selects a patient population, mainly with triple negative disease, with inherently better prognosis that may additionally benefit from immunotherapy.
Introduction
While the era of immunotherapy of breast cancer (BC) through checkpoint inhibition1 has begun with the IMPassion130 trial,2 the quest to identify clinically useful biomarkers is far from over. The most widely used is protein expression of programmed cell death ligand 1 (PD-L1) as assessed by immunohistochemistry (IHC). Analytical difficulties and the demonstration that other biomarkers may more accurately predict benefit from immunotherapy are causes for concern.3 4 In addition, the improvement in pathological complete remission rates regardless of PD-L1 status in the KEYNOTE-522 trial5 further supports these concerns. At the same time, while the prognostic value of PD-L1 protein expression in BC remains unclear, PD-L1 gene expression is a marker clearly associated with good prognosis, whose potential clinical validity we have previously demonstrated.6 7 These observations pose the question whether results from IHC evaluation of PD-L1 are assay driven, rather than representative of the underlying biology.
Due to the complexity of programmed cell death protein 1 (PD-1)/PD-L1 axis regulation,8 other predictive biomarkers have been explored. Of those, high PD-1 mRNA expression, thought to represent a stable pre-existing adaptive immune response,4 is particularly promising and may outperform other markers, including PD-L1 IHC, as a predictor of benefit from PD-1/PD-L1 inhibition.4 The analytical validity and reproducibility of PD-1 mRNA evaluation have been previously demonstrated,9 underscoring the potential of this marker.
To our knowledge, few studies have reported on the prognostic value of PD-1 protein expression in BC, while available data on PD-1 gene expression are even more sparse. Herein, we aimed to investigate the prognostic value of PD-1 protein and gene expression in a cohort of patients treated in Stockholm, Sweden. In addition, in order to interpret our findings in the context of current evidence, we performed a trial-level meta-analysis of available data on PD-1 IHC in BC and a separate pooled analysis of transcriptomic data from publicly available datasets.
Methods
Three parts comprised the present study: a correlative analysis of PD-1 protein and gene expression from an original patient cohort; a trial-level meta-analysis on PD-1 protein expression in early BC; and a pooled gene expression analysis from publicly available datasets regarding PDCD1 expression. The corresponding methods of the three parts are presented hereunder in succession.
Description of the study cohort
The study cohort consisted of patients treated for primary BC in Stockholm, Sweden during 1997–2005 and has been previously described in detail7 10 (online supplemental figure S1). The median follow-up for overall survival (OS), defined as time from date of diagnosis to death by any cause, was 15.7 years and the median follow-up for distant recurrence-free interval (DRFI), defined as time from date of diagnosis to distant metastasis or death from BC,11 was 12.6 years. The used methods and strategies in the present study are covered by the ethics application including amendments, approved by the ethics committee at Karolinska Institutet, Stockholm, Sweden.
esmoopen-2020-001032supp001.pdf (390.1KB, pdf)
Tissue microarray construction, staining and scoring for PD-1 protein expression
Tissue microarray (TMA) construction was performed as previously described.7 Immunostaining with the primary monoclonal antibody targeting PD-1 (clone NAT105, Roche, Basel, Switzerland) was performed using the Ventana (Roche, Basel, Switzerland) autostainer according to the manufacturer’s protocol. Reactive lymphoid node of the tonsil was used as a positive control. PD-1 was evaluated by two investigators (IZ, GR), while TMA cores with poor staining quality, folded, limited or missing tissue were excluded from the analysis. The total number of PD-1 positive (PD-1+) immune cells—defined as the presence of any membranous staining—was manually counted in each TMA core and then calculated between duplicate cores; thus, the average number of PD-1+ immune cells per sample was derived. Any staining of PD-1 +immune cells was used as cut-off for positivity.
Gene expression profiling
RNA was extracted from the obtained biopsies or surgical specimens and profiled on Rosetta/Merck Human RSTA Custom Affymetrix V.2.0 microarray as previously reported and are available at the Gene Expression Omnibus database under accession number GSE48091.7 Assignment of the intrinsic subtype in each tumour according to the PAM50 classification12–14 has been described previously in detail.10
Statistical methods for the PD-1 protein and gene expression analyses in the study cohort
In this exploratory study, association between PD-1 protein and mRNA expression levels and DRFI and OS was analysed. Kaplan-Meier curves were used to estimate the survival outcomes and dichotomised PD-1- protein levels (positive, negative) were compared with the non-parametric log-rank statistic. Univariate and multivariable Cox regression models with dichotomised PD-1 protein level as categorical variable were fitted and unadjusted and adjusted HRs and CIs were calculated. In the multivariable Cox regression models, adjustment was made for age (<50, 50+years), tumour size (≤2, 2–5,>5 cm), lymph node involvement (yes, no), histological grade (I, II, III), proliferation status (Ki67 ≤20%,>20%) and IHC-based subtype (ER+/HER2−, HER2+, ER−/HER2−). Missing values of adjustment variables were treated with separate categories in the models. Association between PD-1 mRNA expression levels and outcome was analysed with corresponding Cox regression models with PD-1 mRNA expression value as continuous variable after robust linear scaling (such that expression quantiles 2.5% and 97.5% were set to −one and +1, respectively). All statistical analysis was done in R (V.3.6.2).
Search strategy, study selection, data extraction and quality assessment in the meta-analysis of PD-1 protein expression
Comprehensive systematic electronic search was conducted by a librarian at the Karolinska Institutet University Library in November 2018 and updated in May 2019 in the following databases: Medline (Ovid), Embase, Cochrane Library (Wiley) and Web of Science Core Collection. The MeSH terms identified for searching Medline were adapted in accordance to corresponding vocabularies in Embase. Each search concept was also complemented with relevant free-text terms and these were, if appropriate, truncated and/or combined with proximity operators. Language restriction was made to English. Databases were searched from inception. The search strategy has been described in detail6 and provided in the online supplemental material.
The data collected from each study were: first author’s last name, year of publication, country where the study was conducted, type of study (retrospective/prospective); method of assessment, tissue used for analysis, threshold for PD-1 expression, antibody used; positivity rate of PD-1 in immune cells; characteristics of study cohort; follow-up time; outcome (time-to-event variables) within all patients and whenever possible within different BC subtypes including both univariate and multivariate results.
Only studies investigating the prognostic role (measured as time-to-event outcome) of PD-1 expression in patients with early-stage BC were included in our meta-analysis. Study selection was performed independently by two investigators (AM, IZ) and consensus was reached in all eligible studies. Data were independently extracted in a predefined form. A third investigator (AV) compared the databases and resolved any discrepancies. The concordance rate between the two investigators was 97.8%.
Two investigators (AM, IZ) independently assessed each eligible study for methodological quality using the 20-item REMARK (Reporting recommendations for tumour marker prognostic studies) checklist, as previously described.15 Discrepancies were resolved by a third investigator (AV). The concordance rate between the two investigators considering quality assessment was 70.0%.
Statistical analysis for study-level meta-analysis of PD-1 protein expression
Each case was defined as positive when PD-1 expression exceeded the positivity threshold used in the specific study. For the time-to-event outcomes, we performed a meta-analysis first by transforming the HRs and their errors into their log counterparts, and then using the inverse variance method and transformed back into the HR scale. If time-to-event data were unavailable for direct extraction from the original publication, we extracted data according to the method described by Tierney et al.16 Whenever possible, we performed separate analyses for univariate and multivariate HRs in each outcome of interest. Subgroup analyses were performed if there were at least three studies in each subgroup.
We assessed the presence of statistical heterogeneity among the studies using the Q statistics and the magnitude of heterogeneity using the I2 statistic. We considered a p value<0.10 or an I2 value of greater than 50% as indicative of substantial heterogeneity. If substantial heterogeneity was detected, we used the random-effect model; otherwise, the fixed-effect model was used.
The presence of publication bias was evaluated qualitatively using a funnel plot. All reported p values are two sided, with significance set at p<0.05. Statistical analyses were performed with RevMan V.5.3 (The Cochrane Collaboration, 2014).
Gene expression data and statistical methods for pooled gene expression analysis
Thirty-nine datasets of gene expression profiles of nearly 9500 primary BC were used in the pooled transcriptomic analysis, as previously described.6 To ensure comparability of expression values across multiple datasets, a robust linear scaling was applied to each gene such that expression quantiles 2.5% and 97.5% were set to −1 and +1, respectively. Each tumour was assigned to a molecular intrinsic subtype according to the PAM50 classifier. Tumours of the normal-like subtype were not included in the subgroup analysis. ESR1 and ERBB2 status were classified based on the bimodal distribution of the expression values of these two genes, since not every dataset had information on ER and HER2 status.
Association between PD-1 mRNA expression levels and disease-free survival (DFS, DMFS [distant metastasis-free survival], RFS [relapse-free survival] or progression-free interval) and OS were analysed. Univariate and multivariable Cox regression models with scaled expression value as continuous variable and stratification by cohort were fitted, and unadjusted and adjusted HR and CI were calculated. In the multivariate Cox regression model, adjustment was made for age, tumour size, lymph node involvement, histological grade and ESR1 and ERBB2 expression status. No adjustment for ESR1 and ERBB2 status was made in models by molecular subtype. Each METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) study site (n=5) was treated as separate cohort. All data analysis was done in R/Bioconductor (V.3.6.2).
Results
Patient characteristics in the study cohort
The patients’ clinicopathological characteristics in the entire study cohort and according to PD-1 protein (positive vs negative; online supplemental figure S2) are summarised in table 1. In total, 220 (46.7%) had PD-1 protein positive expression and 251 (53.3%) negative. PD-1 protein expression was associated with biologically aggressive characteristics, such as oestrogen receptor negativity (p<0.001) and high proliferation according to Ki67 (p<0.001).
Table 1.
Patient characteristics for all patients in study cohort and according to programmed cell death protein 1 (PD-1) protein expression (positive vs negative)
| Clinicopathological characteristic | All | Patients with PD-1 IHC data N (%) |
PD-1 positive N (%)* |
PD-1 negative N (%)* |
P value† |
| Number of patients | 564 | 471 (100.0) | 220 (46.7) | 251 (53.3) | |
| Age | 0.795 | ||||
| Median in years (IQR) | 54.5 (23–76) | 54 (23–76) | 55 (23–75) | 53 (23–76) | |
| Tumour size | 0.799 | ||||
| Median in cm (IQR) | 2.20 (0.20–12.30) | 2.20 (0.20–12.30) | 2.20 (0.70–9.50) | 2.20 (0.20–12.30) | |
| Lymph node status | 0.760 | ||||
| Positive | 313 (55.5) | 271 (57.5) | 125 (56.8) | 146 (58.2) | |
| Negative | 234 (41.5) | 186 (39.5) | 87 (39.5) | 99 (39.4) | |
| Unknown | 17 (3.0) | 14 (3.0) | 8 (3.6) | 6 (2.4) | |
| ER status | <0.001 | ||||
| Positive | 399 (70.7) | 329 (69.9) | 133 (60.5) | 196 (78.1) | |
| Negative | 152 (27.0) | 129 (27.4) | 77 (35.0) | 52 (20.7) | |
| Unknown | 13 (2.3) | 13 (2.8) | 10 (4.5) | 3 (1.2) | |
| PR status | 0.005 | ||||
| Positive | 270 (47.9) | 223 (47.3) | 87 (39.5) | 136 (54.2) | |
| Negative | 152 (27.0) | 131 (27.8) | 73 (33.2) | 58 (23.1) | |
| Unknown | 142 (25.2) | 117 (24.8) | 60 (27.3) | 57 (22.7) | |
| HER2 status | |||||
| Positive | 97 (17.2) | 88 (18.7) | 39 (17.7) | 49 (19.5) | 0.888 |
| Negative | 385 (68.3) | 338 (71.8) | 160 (72.7) | 178 (70.9) | |
| Unknown | 82 (14.5) | 45 (9.6) | 21 (9.5) | 24 (9.6) | |
| Tumour grade | 0.088 | ||||
| Grade I | 46 (8.2) | 34 (7.2) | 17 (7.7) | 17 (6.8) | |
| Grade II | 244 (43.3) | 210 (44.6) | 85 (38.6) | 125 (49.8) | |
| Grade III | 245 (43.4) | 221 (46.9) | 114 (51.8) | 107 (42.6) | |
| Unknown | 29 (5.1) | 6 (1.3) | 4 (1.8) | 2 (0.8) | |
| Ki67 | <0.001 | ||||
| ≤20% | 230 (40.8) | 212 (45.0) | 120 (54.5) | 92 (36.7) | |
| >20% | 289 (51.2) | 253 (53.7) | 96 (43.6) | 157 (62.5) | |
| Unknown | 45 (8.0) | 6 (1.3) | 4 (1.8) | 2 (0.8) | |
| IHC subtype | 0.008 | ||||
| ER+/HER2− | 283 (50.2) | 248 (52.7) | 103 (46.8) | 145 (57.8) | |
| HER2+ | 97 (17.2) | 88 (18.7) | 39 (17.7) | 49 (19.5) | |
| ER−/HER2− | 93 (16.5) | 81 (17.2) | 51 (23.2) | 30 (12.0) | |
| Unknown | 91 (16.1) | 54 (11.5) | 27 (12.3) | 27 (10.8) | |
| PAM50-based subtype | 0.001 | ||||
| Luminal A | 199 (35.3) | 163 (34.6) | 67 (30.5) | 96 (38.2) | |
| Luminal B | 112 (19.9) | 98 (20.8) | 37 (16.8) | 61 (24.3) | |
| HER2-enriched | 78 (13.8) | 69 (14.6) | 30 (13.6) | 39 (15.5) | |
| Basal-like | 135 (23.9) | 113 (24.0) | 71 (32.3) | 42 (16.7) | |
| Normal-like | 40 (7.1) | 28 (5.9) | 15 (6.8) | 13 (5.2) | |
*Percentages calculated based on the number of patients with available data.
†P values from statistical tests where unknown categories are excluded. Wilcoxon rank sum test is used for the continuous variables and Fisher’s exact test is used for categorical variables.
ER, oestrogen receptor; HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry; PR, progesterone receptor.
Prognostic value of PD-1 protein expression and its correlation with PD-1 mRNA expression in the study cohort
In the study cohort, high PD-1 protein but not mRNA expression was generally found to be prognostic for improved patient outcomes. In multivariable analysis, PD-1 protein expression was associated with better DRFI (adjusted HR (HRadj)=0.66, 95% CI 0.48 to 0.91, p=0.010) and OS (HRadj=0.73, 95% CI 0.55 to 0.97, p=0.027; figure 1 and online supplemental table S1). In subgroup analysis, this prognostic effect was mainly driven by ER-positive/HER2-negative disease (for DRFI, HRadj=0.65, 95% CI 0.41 to 0.1.01, p=0.050; for OS, HRadj=0.70, 95% CI 0.47 to 1.04, p=0.076), although there was no interaction between PD-1 expression and IHC-based subtype in the survival analyses (figure 1; pinteraction 0.326 for DRFI and pinteraction 0.443 for OS).
Figure 1.
Kaplan-Meier curves and univariate HRs for distant metastasis-free survival (A) and overall survival (B) and forest plots for distant recurrence-free interval (C) and overall survival (D) in the overall populations and per immunohistochemistry-based subtype (multivariable HR), according to programmed cell death protein 1 (PD-1) protein expression (positive vs negative). HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry.
In contrast, high PD-1 mRNA expression was not prognostic in multivariable analysis for neither DRFI (HRadj=0.88, 95% CI 0.66 to 1.18, p=0.399), nor OS (HRadj=0.88, 95% CI 0.68 to 1.13, p=0.312) (online supplemental table S1). PDCD1 transcript levels were statistically significantly but weakly correlated with PD-1 protein expression (Spearman’s rho=0.123, p=0.007).
Characteristics and quality of eligible studies and between-study heterogeneity in the meta-analysis of immunohistochemical PD-1 expression
As shown in figure 2, the initial search identified 4736 entries, or 3298 entries following deduplication. Through exclusion by reading the title and/or abstract, 39 possibly eligible studies were retrieved as full text; 15 studies fulfilled the inclusion criteria and were included in the meta-analysis.17–30 The characteristics of eligible studies are presented in online supplemental table S2. The results from the study cohort described in the Methods section and presented in the Results section were included in the meta-analysis as one of the 15 eligible studies.
Figure 2.
Flow chart of search and study selection in the meta-analysis of studies evaluating protein programmed cell death protein 1 (PD-1) expression (A); and data availability in the pooled gene expression analysis (B). DFS, disease-free survival; OS, overall survival.
Fourteen studies were retrospective and one prospective. The median number of study quality score was 23.5 (range: 19–33) out of a maximum score of 40. Substantial between-study heterogeneity was noted among eligible studies regarding the threshold for PD-1 positivity rate used, the follow-up period, and the study population and BC subtypes.
Prognostic significance of pooled immunohistochemical PD-1 expression
PD-1 positive disease was not significantly associated with OS in the overall population (figure 3). The pooled (7 studies; 1975 patients) univariate HR was 1.65 (95% CI 0.94 to 2.90, p=0.08) and the pooled (four studies; 1395 patients) multivariate HR was 1.25 (95% CI 0.71 to 2.20, p=0.45). Significant heterogeneity was noted in both analyses (I2=88, pheterogeneity <0.001 and I2=78%, pheterogeneity=0.004, respectively). Similar results were observed for DFS (5 studies, n=633; pooled univariate HR=1.71, 95% CI 0.95 to 3.08, p=0.07; I2=57%, pheterogeneity=0.05).
Figure 3.
Forest plot for overall survival according to programmed cell death protein 1 (PD-1) protein expression in total population: pooled univariate HR (A) and pooled multivariable HR (B).
The association of PD-1 positivity and improved DFS in TNBC was statistically significant (7 studies, n=1245; pooled univariate HR=0.52, 95% CI 0.38 to 0.73, p<0.001; I2=52%, pheterogeneity=0.05 and 3 studies, n=417; pooled multivariate HR=0.57, 95% CI 0.29 to 0.90, p=0.02, I2=12%, pheterogeneity=0.32) but not with OS in pooled univariate (9 studies, n=1454; HR=0.76, 95% CI 0.44 to 1.31, p=0.32, I2=84%, pheterogeneity <0.001) and pooled multivariate analyses (3 studies, n=437; HR=0.74, 95% CI 0.47 to 1.17, p=0.20, I2=8%, pheterogeneity=0.34; online supplemental figure S3). Due to the limited number of studies, further analyses were not possible.
Pooled analysis of PD-1 gene expression as a prognostic factor
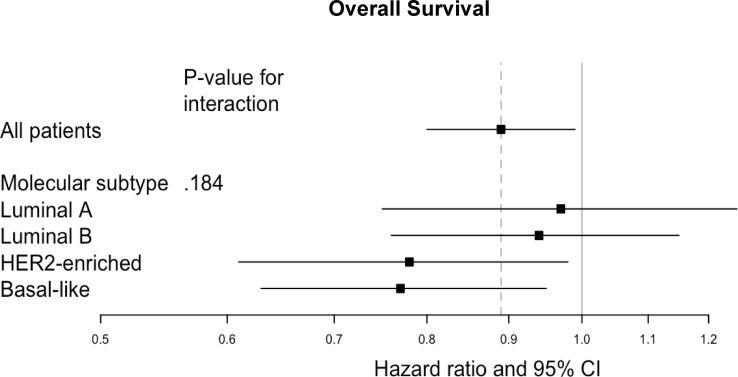
As previously described,6 39 datasets containing 9493 patients were included in the pooled gene expression analysis. The prognostic value of PD-1 gene expression in univariate and multivariable analysis is shown in figure 4 and in online supplemental tables S3 and S4). Higher PDCD1 expression was associated with better OS in multivariable analysis in the entire population (HRadj=0.89, 95% CI 0.80 to 0.99, p=0.025) and in basal-like (HRadj=0.77, 95% CI 0.63 to 0.95, p=0.014) as well as HER2-enriched tumours (HRadj=0.78, 95% CI 0.61 to 0.98, p=0.032), although no significant interaction between PD-1 expression and molecular subtype was detected (pinteraction=0.185). In contrast, PDCD1 expression was not predictive for DFS as shown in online supplemental table S3.
Figure 4.
Forest plot for overall survival according to programmed cell death protein 1 (PD-1) gene expression, in total population and per molecular intrinsic subtype. HER2, human epidermal growth factor receptor 2.
Discussion
By using multiple approaches (IHC, gene-expression profiling data, trial-level meta-analysis and pooled transcriptomic analysis), we demonstrate in this study that PD-1 expression at the protein and mRNA levels seem to be associated with better prognosis in early BC, especially—but not exclusively—in the triple-negative and basal-like subtypes. These findings are in line with our previously published study on the correlation of PD-L1 gene and immune cell protein expression with improved outcomes,6 further highlighting the prognostic significance of the PD-L1/PD-1 axis and the necessity for biomarker development.3 31
The favourable survival outcomes with higher PD-1 expression might seem paradoxical given that it is considered to be a coinhibitory molecule. However, it is not certain if PD-1 represents a T-cell exhaustion marker, thus leading to impaired antitumour immune response, or a marker indicating T-cell activation and receptor signalling.32 Indeed, an association of PD-1 expression with genes reflecting T-cell regulation and activation status has been described.33 Of note, additional immune checkpoint markers (ie, CD39, LAG-3, TIM-3) can also define T-cell exhaustion34 and are linked with better outcomes.35 Moreover, the improved patient outcomes among high expressors may also reflect the potential predictive value of PD-1 in patients receiving chemotherapy, and thus higher benefit from adjuvant therapy, in accordance with previous data showing that immune-related gene expression is a driver of chemosensitivity in BC.36 37
Even though improved OS in PD-1high vs PD-1low triple-negative/basal-like patients was observed in the pooled analyses with no benefit in DFS owing likely to the exploratory nature of this study, a discrepancy in the prognostic information between protein and mRNA levels was noted in our study cohort. Regarding protein expression, the lack of survival benefit in ER-positive/HER2-negative BC in the meta-analysis could be due to the short follow-up of these studies compared with our cohort masking a possible long-term effect of immune activation on tumour dormancy.38 The observed weak correlation between PD-1 protein and transcript levels raises concerns on how informative PD-1 IHC evaluation really is. Besides analytical difficulties, this discrepancy might be related to the complex regulation of PD-1 expression, especially at the post-transcriptional/translational level.8 39 Data on PDCD1 expression are scarce, with few published studies demonstrating an association with improved survival outcomes.29 33 These observations further underscore the need for validation of the genomic platforms for the detection of PD-1,9 while its clinical utility regarding both its prognostic and predictive4 role needs to be tested in a prospective manner.
This study suffers from certain limitations that need to be addressed. First, the meta-analysis was performed at a trial level, with relatively few retrospective studies identified and included, which did not allow for further subgroup analyses of potential interest and led to significant heterogeneity in some of the analyses. Second, variations in IHC scoring, antibodies used and cut-offs, mRNA detection methodology and patient selection, as well as variations in defining ER and HER2 positivity also contributed to the high heterogeneity among the studies. In addition, the smaller size, different patient population of our original study and non-randomised allocation to adjuvant chemotherapy, as well as PD-1 IHC evaluation with different cut-off and on TMA rather than in whole sections, which possibly underestimates the level of spatial heterogeneity,6 might explain the observed discrepant results as compared with the pooled analyses. The risk for bias and the limitations regarding analyses of publicly available transcriptomic datasets have been also described elsewhere.40 Finally, our study cannot provide information on the effect of systemic therapy in either the study cohort or the meta-analysis studies or the pooled gene expression analysis, since allocation to adjuvant therapy was not randomised.
Given the emerging impact of PD-L1/PD-1 signalling axis, its complex regulation37 39 and the early translational research data on immune checkpoint blockade in BC,4 it is of utmost importance to rethink and better understand its impact in the context of tumor-immune microenvironment. Multiplexed imaging and multiomics approaches31 can further delineate the expression patterns and specific immune cell composition as well as their prognostic and/or predictive value. Considering the possible predictive value of PD-1 mRNA for immunotherapy across tumour types and the inconsistent results of PD-L1 as a predictive marker in BC in both the neoadjuvant5 41 and metastatic settings,2 42 PD-1 mRNA may very well represent a potential candidate for integration into future treatment algorithms of both early and advanced BC as a fourth biomarker besides ER, PR and HER2.
In conclusion, this multilevel study reveals the promising prognostic capacity of PD-1 expression in patients with early BC, especially in the triple-negative and basal-like subtypes. However, the low concordance between protein and gene expression and analytical inconsistencies in reported methodologies of PD-1 IHC assessment underscore the need for prospective validation of our findings.
Acknowledgments
The authors would like to acknowledge the contribution of Magdalena Svanberg, librarian, Karolinska Institutet University Library during the preparation of this manuscript. Alexios Matikas was supported by the Stockholm Region (clinical postdoctorial appointment). Theodoros Foukakis is a recipient of the Senior Clinical Investigator Award from the Swedish Cancer Society. The authors would also like to acknowledge the contribution of Susanne Agartz and Georgia Kokaraki, lab engineers for their technical support with patient material (TMAs) and immunohistochemical stainings. An abstract of the study has been presented as a poster at the Annual Meeting of the American Society of Clinical Oncology (ASCO) 2020.
Footnotes
AM and IZ contributed equally.
Contributors: Study concept: AM, IZ and TF. Study design: AM, IZ, AV and TF. Data acquisition: AM, IZ, JL, FR, CS and GR. Data analysis and interpretation: AM, IZ, JL, ES, AV and TF. Manuscript preparation: AM, IZ and TF. All the authors edited, read and approved the final manuscript.
Funding: This study was supported by the Swedish Cancer Society (grant number CAN 2018/846 to TF), the Cancer Society in Stockholm (174113 to TF); the Swedish Breast Cancer Association (IZ, TF); Alexios Matikas was supported by the Stockholm Region (clinical postdoctorial appointment, dnr K 2017-4577); Theodoros Foukakis is recipient of the Senior Clinical Investigator Award from the Swedish Cancer Society (grant number CAN 2017/1043); Jonas Bergh’s research group receives funding from the Stockholm region, the Swedish Cancer Society, the funds at Radiumhemmet, the Swedish Research Council, the Knut and Alice Wallenberg fund.
Competing interests: JL: employment in RaySearch laboratories. Christos Sotiriou: advisory board (a receipt of honoraria or consultation fees) from Astellas, Cepheid, Vertex, Seattle Genetics, Puma; participation in company-sponsored speaker’s bureau: Eisai, Prime Oncology, Teva, Foundation Medicine; travel and accommodation expenses from Roche, Genentech and Pfizer. JB receives research funding from Merck paid to Karolinska Institutet and from Amgen, Bayer, Pfizer, Roche and Sanofi-Aventis paid to Karolinska University Hospital. No personal payments. Payment from UpToDate for a chapter in breast cancer prediction paid to Asklepios Medicine HB. TF: institutional grants from Roche and Pfizer and personal fees from Novartis, Pfizer, Roche and UpToDate.
Patient consent for publication: Not required.
Ethics approval: The used methods and strategies in the present study are covered by the ethics application including amendments, approved by the ethics committee at Karolinska Institutet, Stockholm, Sweden (Dnr 2006/394-31/3, 2006/1183-31/2, and amendments 2016/1505-32, 2018/789-32, 2018/790-32), which decided that no additional written informed consent was needed for each patient.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. Microarray GEP data are available from the Gene Expression Omnibus depository (accession number GSE48091).
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Ishida Y, Agata Y, Shibahara K, et al. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. Embo J 1992;11:3887–95. 10.1002/j.1460-2075.1992.tb05481.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med 2018;379:2108–21. 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 3.Afkhami M, Schmolze D, Yost SE, et al. Mutation and immune profiling of metaplastic breast cancer: correlation with survival. PLoS One 2019;14:e0224726. 10.1371/journal.pone.0224726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paré L, Pascual T, Seguí E, et al. Association between PD1 mRNA and response to anti-PD1 monotherapy across multiple cancer types. Ann Oncol 2018;29:2121–8. 10.1093/annonc/mdy335 [DOI] [PubMed] [Google Scholar]
- 5.Schmid P, Cortes J, Pusztai L, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med 2020;382:810–21. 10.1056/NEJMoa1910549 [DOI] [PubMed] [Google Scholar]
- 6.Matikas A, Zerdes I, Lövrot J, et al. Prognostic implications of PD-L1 expression in breast cancer: systematic review and meta-analysis of immunohistochemistry and pooled analysis of transcriptomic data. Clin Cancer Res 2019;25:5717–26. 10.1158/1078-0432.CCR-19-1131 [DOI] [PubMed] [Google Scholar]
- 7.Zerdes I, Sifakis EG, Matikas A, et al. Programmed death-ligand 1 gene expression is a prognostic marker in early breast cancer and provides additional prognostic value to 21-gene and 70-gene signatures in estrogen receptor-positive disease. Mol Oncol 2020;14:951–63. 10.1002/1878-0261.12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zerdes I, Matikas A, Bergh J, et al. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 2018;37:4639–61. 10.1038/s41388-018-0303-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, Navarro A, Paré L, et al. Immune-Related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res 2017;77:3540–50. 10.1158/0008-5472.CAN-16-3556 [DOI] [PubMed] [Google Scholar]
- 10.Lundberg A, Lindström LS, Harrell JC, et al. Gene expression signatures and immunohistochemical subtypes add prognostic value to each other in breast cancer cohorts. Clin Cancer Res 2017;23:7512–20. 10.1158/1078-0432.CCR-17-1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudis CA, Barlow WE, Costantino JP, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the steep system. J Clin Oncol 2007;25:2127–32. 10.1200/JCO.2006.10.3523 [DOI] [PubMed] [Google Scholar]
- 12.Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. 10.1200/JCO.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis C, Shah SP, Chin S-F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altman DG, McShane LM, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med 2012;9:e1001216. 10.1371/journal.pmed.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ács B, Madaras L, Tőkés A-M, et al. PD-1, PD-L1 and CTLA-4 in pregnancy-related - and in early-onset breast cancer: A comparative study. Breast 2017;35:69–77. 10.1016/j.breast.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 18.Asano Y, Kashiwagi S, Goto W, et al. Prediction of treatment responses to neoadjuvant chemotherapy in triple-negative breast cancer by analysis of immune checkpoint protein expression. J Transl Med 2018;16:87. 10.1186/s12967-018-1458-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bottai G, Raschioni C, Losurdo A, et al. An immune stratification reveals a subset of PD-1/LAG-3 double-positive triple-negative breast cancers. Breast Cancer Res 2016;18:121. 10.1186/s13058-016-0783-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockhoff G, Seitz S, Weber F, et al. The presence of PD-1 positive tumor infiltrating lymphocytes in triple negative breast cancers is associated with a favorable outcome of disease. Oncotarget 2018;9:6201–12. 10.18632/oncotarget.23717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byun KD, Hwang HJ, Park KJ, et al. T-Cell immunoglobulin mucin 3 expression on tumor infiltrating lymphocytes as a positive prognosticator in triple-negative breast cancer. J Breast Cancer 2018;21:406–14. 10.4048/jbc.2018.21.e61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang R, Cui Y, Guo Y. Programmed cell death protein-1 predicts the recurrence of breast cancer in patients subjected to radiotherapy after breast-preserving surgery. Technol Cancer Res Treat 2018;17:153303381879342. 10.1177/1533033818793425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitano A, Ono M, Yoshida M, et al. Tumour-Infiltrating lymphocytes are correlated with higher expression levels of PD-1 and PD-L1 in early breast cancer. ESMO Open 2017;2:e000150. 10.1136/esmoopen-2016-000150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manson QF, Ter Hoeve ND, Buerger H, et al. Pd-1 and PD-L1 expression in male breast cancer in comparison with female breast cancer. Target Oncol 2018;13:769–77. 10.1007/s11523-018-0610-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muenst S, Soysal SD, Gao F, et al. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat 2013;139:667–76. 10.1007/s10549-013-2581-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noske A, Möbus V, Weber K, et al. Relevance of tumour-infiltrating lymphocytes, PD-1 and PD-L1 in patients with high-risk, nodal-metastasised breast cancer of the German adjuvant intergroup node-positive study. Eur J Cancer 2019;114:76–88. 10.1016/j.ejca.2019.04.010 [DOI] [PubMed] [Google Scholar]
- 27.Ren X, Wu H, Lu J, et al. Pd1 protein expression in tumor infiltrated lymphocytes rather than PDL1 in tumor cells predicts survival in triple-negative breast cancer. Cancer Biol Ther 2018;19:373–80. 10.1080/15384047.2018.1423919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Fei X, Mao Y, et al. PD-1(+) immune cell infiltration inversely correlates with survival of operable breast cancer patients. Cancer Immunol Immunother 2014;63:395–406. 10.1007/s00262-014-1519-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeong J, Lim JCT, Lee B, et al. Prognostic value of CD8 + PD-1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J Immunother Cancer 2019;7:34. 10.1186/s40425-019-0499-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T, Xu D, Tang B, et al. Expression of programmed death ligand-1 and programmed death-1 in samples of invasive ductal carcinoma of the breast and its correlation with prognosis. Anticancer Drugs 2018;29:904–10. 10.1097/CAD.0000000000000683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JS, Ruppin E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol 2019. 10.1001/jamaoncol.2019.2311. [Epub ahead of print: 22 Aug 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egelston CA, Avalos C, Tu TY, et al. Human breast tumor-infiltrating CD8+ T cells retain polyfunctionality despite PD-1 expression. Nat Commun 2018;9:4297. 10.1038/s41467-018-06653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang C, Cao S, Li N, et al. Pd-1 and PD-L1 correlated gene expression profiles and their association with clinical outcomes of breast cancer. Cancer Cell Int 2019;19:233. 10.1186/s12935-019-0955-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chrétien S, Zerdes I, Bergh J, et al. Beyond PD-1/PD-L1 inhibition: what the future holds for breast cancer immunotherapy. Cancers 2019;11. 10.3390/cancers11050628. [Epub ahead of print: 05 May 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burugu S, Gao D, Leung S, et al. LAG-3+ tumor infiltrating lymphocytes in breast cancer: clinical correlates and association with PD-1/PD-L1+ tumors. Ann Oncol 2017;28:2977–84. 10.1093/annonc/mdx557 [DOI] [PubMed] [Google Scholar]
- 36.Matikas A, Lövrot J, Ramberg A, et al. Dynamic evaluation of the immune infiltrate and immune function genes as predictive markers for neoadjuvant chemotherapy in hormone receptor positive, HER2 negative breast cancer. Oncoimmunology 2018;7:e1466017. 10.1080/2162402X.2018.1466017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foukakis T, Lövrot J, Matikas A, et al. Immune gene expression and response to chemotherapy in advanced breast cancer. Br J Cancer 2018;118:480–8. 10.1038/bjc.2017.446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H-F, Wang S-S, Huang M-C, et al. Targeting immune-mediated dormancy: a promising treatment of cancer. Front Oncol 2019;9:498. 10.3389/fonc.2019.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med 2016;375:1767–78. 10.1056/NEJMra1514296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramasamy A, Mondry A, Holmes CC, et al. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med 2008;5:e184. 10.1371/journal.pmed.0050184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittendorf EA, Zhang H, Barrios CH, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet 2020;396:1090–100. 10.1016/S0140-6736(20)31953-X [DOI] [PubMed] [Google Scholar]
- 42.Miles DW, Gligorov J, Andre F. Primary results from IMpassion131, a double-blind placebo-controlled randomised phase III trial of first-line paclitaxel (PAC) ± atezolizumab (atezo) for unresectable locally advanced/metastatic triple-negative breast cancer (mTNBC). European Society for Medical Oncology annual meeting, 2020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2020-001032supp001.pdf (390.1KB, pdf)