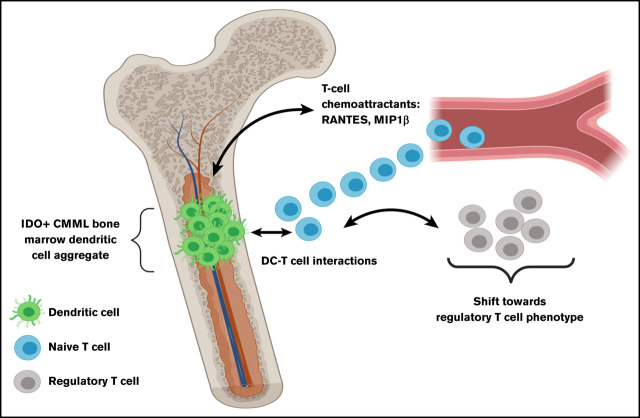
Key Points
Systemic immune microenvironment signatures in CMML indicate an altered T- and natural killer cell balance.
CMML bone marrow dendritic cell aggregates associate with disease progression and systemic regulatory T-cell phenotypic switch.
Visual Abstract
Introduction
Chronic myelomonocytic leukemia (CMML) is a myelodysplastic/myeloproliferative neoplasm with an inflammatory mileu1 and a clinical association with autoimmune disorders/inflammatory syndromes,2-4 suggesting pervasive immune deregulation. Upregulation of specific immune checkpoints such as programmed death ligand 1 in CD34+ cells and programmed cell death protein 1 in stromal cells from patients with CMML (and myelodysplastic syndromes/acute myeloid leukemia [AML]) has been shown previously5; however, specific mechanisms of immune tolerance remain to be elucidated.
Numerous studies have reported the presence of plasmacytoid dendritic cell (DC; pDC) aggregates in CMML bone marrow (BM),6,7 with a recent study concluding that these are clonal and associate with RAS-pathway mutations.7 Previous studies using melanoma mouse models have established a role for indoleamine 2,3-dioxygenase 1/2 (IDO1/2) in regulating pDC8-10 and myeloid DC (mDC) plasticity,11,12 inducing tolerogenic phenotypes in both contexts. IDO is an immune-checkpoint enzyme that induces systemic immune tolerance through multiple mechanisms, including regulatory T-cell (Treg) expansion and tryptophan catabolism.13,14 In AML and myelodysplastic syndromes, IDO expression has been identified as an independent adverse prognostic factor,15-18 and shown to impair immune response by Treg induction.19 In CMML, the role of IDO in DC (both CD123+ pDCs and CD11c+ mDCs) and T-cell interactions, and systemic immune microenvironment remains to be defined.
Methods
CMML patient samples (BM tissue blocks, peripheral blood mononuclear cell [PBMC], and plasma) from our clinically and molecularly annotated biobank were obtained after regulatory approval. Approval was obtained through the Mayo Clinic Institutional Review Board.
Immunohistochemistry (IHC; hematoxylin and eosin, IDO, CD123, and CD11c), multiplex immunostaining (IDO, CD123 and CD11c), liquid chromatography–mass spectrometry (LC-MS), time-of-flight mass cytometry (CyTOF), cytokine profiling (Luminex array), and gene-expression analysis (bulk RNA sequencing) were performed in-house. IHC slides were independently reviewed by 2 hematopathologists. Detailed protocols and methods are available in supplemental Methods.
Results and discussion
IDO+ DC aggregates in the BM microenvironment are associated with disease progression in CMML
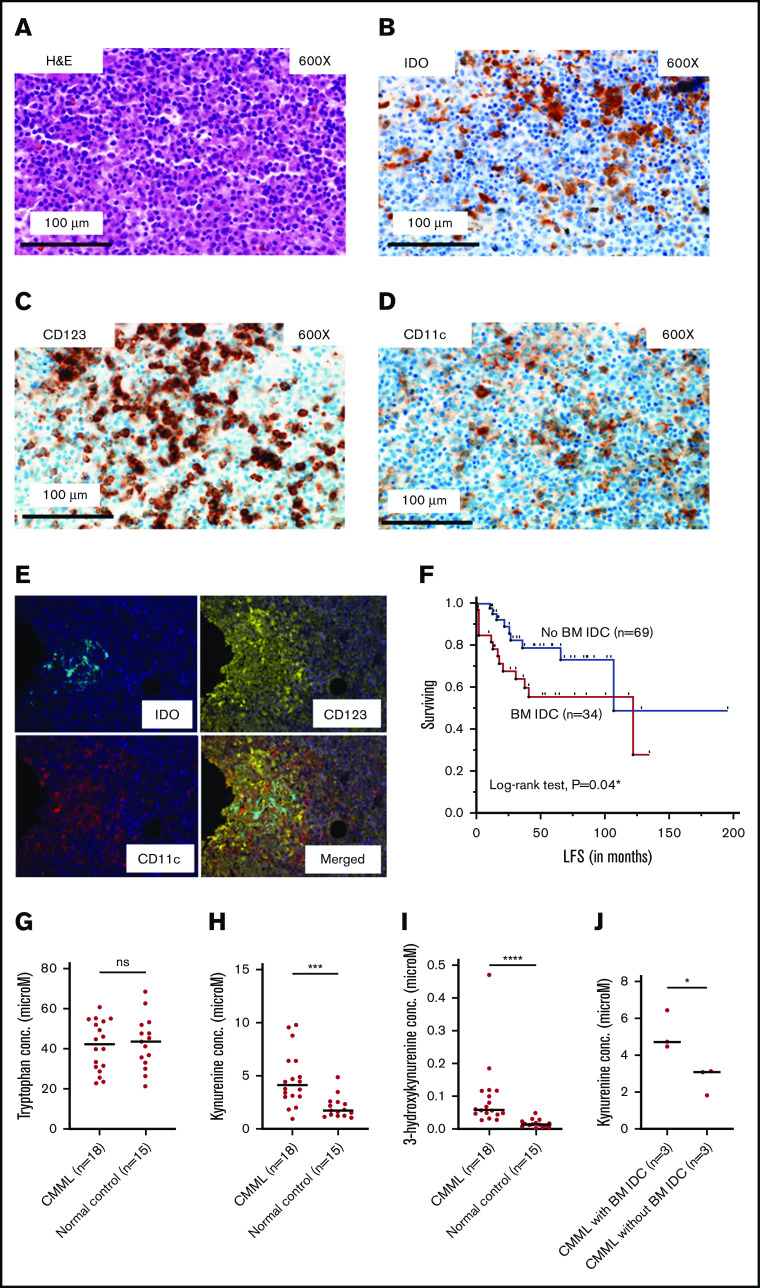
IHC with H&E and IDO stains were performed on BM biopsies of 103 CMML patients (80 at diagnosis). At least 1 morphologically defined BM IDO+ DC aggregate (IDC; morphologically defined as a cluster of ≥10 cells with DC morphology, consensus agreement by 2 hematopathologists with 90% concordance) was identified in 34 patients (33%; 25% at diagnosis). IDO expression was validated by 2 different commercially available antibodies (rabbit polyclonal [Sigma-Aldrich] and rabbit monoclonal [Cell Signaling Technology]; details in supplemental Methods). DC-phenotypic characterization was done by CD123 and CD11c staining (Figure 1A-D), with 18 patients (56%) being positive for both, 7 (22%) for CD123, and 5 (16%) for CD11c only (2 were negative for both, 2 were status unknown), suggesting that IDO expression is not limited to the previously described CD123-marked pDC aggregates in CMML.7 IDO expression in the DC vicinity was further confirmed by multiplex (IDO, CD123, and CD11c) staining in 2 biological replicates (Figure 1E). Although there was variability both within each IDC in a CMML BM and between each case, ∼30% of CD123+ and CD11c+ cells coexpressed IDO (as assessed by visual inspection). This again suggests that IDO expression is not limited to pDCs. To conclusively ascertain cell-type–specific IDO expression in CMML, we developed an 8-color flow cytometry panel to isolate DCs (panel details and gating strategy are available in supplemental Figure 1 and supplemental Methods). Flow results from CMML (n = 2 biological replicates) peripheral blood confirmed IDO coexpression with CD45+/CD3−/CD19−/CD56−/CD14−/CD16−/HLA-DR+ DCs, predominantly on CD11c+/CD123dim mDCs rather than CD11c−/CD123+ pDCs with interferon-γ stimulation (supplemental Figure 2). However, expression was also seen on monocytes, but not lymphocytes (similar results were seen with a normal sample; supplemental Figure 2). Collectively, the IHC and flow cytometry results establish that an IDO-expressing DC aggregate in CMML is not solely populated by CD123+ pDCs, but likely also includes other cell types such as mDCs, and CD123+/CD11c+ monocytes (although other known IDO-expressing populations such as myeloid-derived suppressor cells cannot be excluded) reflecting complex intercellular interactions.
Figure 1.
IDC populations are present in the CMML BM microenvironment, associate with adverse AML-free survival, and associate with metabolic signatures of systemic IDO activity. (A) Hematoxylin-and-eosin staining of an area of CMML BM with IDC. (B) IDO staining. (C) CD123 staining. (D) CD11c staining. (A-D) Original magnification ×600; scale bar, 100 μm. (E) Multiplex immunostaining showing clustering of IDO expression with CD123 and CD11c stains in a CMML BM, confirming expression in a DC population. Original magnification ×20. (F) Kaplan-Meier estimate of AML-free survival (LFS) in CMML patients with IDC at any disease time point (n = 34) vs CMML patients without IDC (n = 69; median, 122 vs 107 months; *log-rank P = .04). Although the median LFS is paradoxically higher in the IDC group due to late events that make the group reach the median, this is a statistical aberration. The survival curves clearly separate early and stay separated for the majority of follow-up (with the IDC group showing inferior survival; lower curve in red). Consequently, the corresponding hazard ratio from a Cox proportional hazard model is significantly greater than 1.00 (hazard ratio, 2.3 [95% CI, 1, 5.4; *P = .049], panel F, supplemental Table 1). (G-I) Similar tryptophan (42.4 vs 43.6 μM; P = .9), higher kynurenine (4.1 vs 1.8 μM; ***P = .0006), higher 3-hydroxykynurenine (0.06 vs 0.01 μM; ****P < .0001) concentrations in CMML patients (n = 18) and normal controls (n = 15), respectively, as evaluated by LC-MS in plasma samples. (J) Higher kynurenine concentration in diagnostic plasma samples (n = 6) of CMML patients with vs without BM IDC (4.7 vs 3 μM; *P = .049); data included for samples in which IHC and LC-MS assessment was done at the same time point. ns, not significant.
In patients without IDC at diagnosis (n = 60), 1 of 7 patients (14%) who progressed after a hypomethylating agent, and 2 of 7 patients (29%) who underwent AML transformation acquired IDC at time of progression and AML transformation, respectively, indicating a potential association of BM IDC in CMML with disease progression. This possibility is further strengthened by the observation of a higher frequency of AML transformation (41% vs 13%; **P = .002) and an adverse Kaplan-Meier estimate of AML-free survival (hazard ratio, 2.3 [95% confidence interval [CI], 1, 5.4] vs 0.4 [95% CI, 0.2, 0.9]; *P = .049; median, 122 vs 107 months; Figure 1F; supplemental Table 1) in CMML patients with, vs without, BM IDC aggregates at any time point (although the median AML-free survival is paradoxically higher in the IDC group due to late events that make the group reach the median, which is a statistical aberration; the survival curves clearly separate early and stay separated for the majority of follow-up with the IDC group, showing an inferior estimate of AML-free survival [lower curve in red; Figure 1F]). Additionally, CMML patients with and without IDC at any disease time point were evenly matched for clinical, genetic, and prognostic characteristics with only minimal differences as indicated in supplemental Table 1. Unlike in AML in which IDO expression is seen in leukemic blasts,15-17 in CMML, IDO is expressed in DC aggregates in the BM microenvironment and, similar to CD123+ infiltrating pDCs,7 are associated with an increased frequency of AML transformation.
Metabolic signatures of systemic IDO activity are evident in CMML
We then assessed for evidence of systemic IDO activity in CMML by measuring tryptophan and its metabolites (kynurenine and 3-hydroxykynurenine). Through LC-MS, although median tryptophan concentrations (42.4 vs 43.6 μM; P = .9; Figure 1G) were not significantly different between CMML (n = 18) and normal control plasma samples (n = 15), kynurenine (4.1 vs 1.8 μM; ***P = .0006; Figure 1H), and 3-hydroxykynurenine (0.06 vs 0.01 μM; ****P < .0001; Figure 1I), concentrations were higher in CMML (supplemental Table 2). These findings suggest accentuated tryptophan catabolism in CMML, a signature of systemic IDO activity.20 Furthermore, median kynurenine concentration was higher in CMML patients with vs without BM IDC at diagnosis (4.7 vs 3 μM; *P = .049; Figure 1J), suggesting increased systemic IDO activity in the former group.
CyTOF assessment highlights an altered T- and NK-cell balance in CMML
To characterize the immune microenvironment in CMML, we selected untreated (except hydroxyurea) CMML PBMC (n = 10; supplemental Table 1) samples and age-matched normal PBMC controls (n = 10, median age, 54 years [range, 50-65 years], voluntary blood bank donors) and performed CyTOF analysis (details in supplemental Methods). Quantitative comparisons (percentage of parent cell type) indicated significant differences between CMML and controls including decreased naive T cells (1.1 vs 12.3; *P = .02), CD8 central memory cells (2 vs 8.5; **P = .007), type 1 helper T cells (Th1; median, 4.04 vs 27.2; **P = .003), Th1/Th2 balance (ratio, 0.3 vs 9.6; **P = .003), CD4 terminal effector cells (11.6 vs 38.7; *P = .02), and γδ T cells (0.6 vs 1.6; **P = .006), with an expanded CD4 central memory (29.7 vs 13.4; **P = .004) and natural killer (NK)-cell population (2.6 vs 1.1; *P = .02) (supplemental Table 2). Altered NK- and T-cell balance has been shown in AML,21,22 and CML,23 suggesting similar patterns of immune dysregulation in CMML.
IDO+ BM DC aggregates are associated with expanded systemic Treg compartment in CMML
When untreated CMML patients with (n = 5) and without BM IDC (n = 5) were compared, the former group showed a higher percentage (of parent cell type) of Tregs (14.5% vs 3.6%; *P = .03; supplemental Table 3; Figure 2F-H). This suggests that IDC in the CMML BM microenvironment is likely associated with Treg differentiation in the systemic T-cell compartment. As shown in the context of other maligancies,8 this is likely due to DC–T-cell interactions driven by T-cell recruitment to these locations, as evidenced by cytokine profiling, which showed higher median levels (picograms per milliliter) of known T-cell chemoattractants,24,25 RANTES (CCL5, 8650 vs 349.7; *P = .04) and macrophage inflammatory protein-1β (MIP-1β; 28.7 vs 10.8; *P = .049), in diagnostic plasma samples of CMML patients with vs without BM IDO+ DC aggregates (supplemental Table 4). Furthermore, kynurenine levels showed a significant strong positive correlation with the percentage of Treg populations (Spearman ρ = 0.9; *P = .04; supplemental Figure 3) in diagnostic samples (plasma and PBMC, respectively, n = 5) collected at the same time point. Due to constraints of limited sample size in the above-mentioned analyses, additional validation was conducted using an alternative methodology: gene-set enrichment analysis (GSEA) was performed in a subset of PBMC RNA obtained from untreated patients at the same time as their BM IHC assessments. GSEA results confirmed Treg-associated gene upregulation (normalized enrichment score = 3.97; ***P < .0001) in untreated CMML patients with vs without BM IDC (n = 4 each; Figure 2I).
Figure 2.
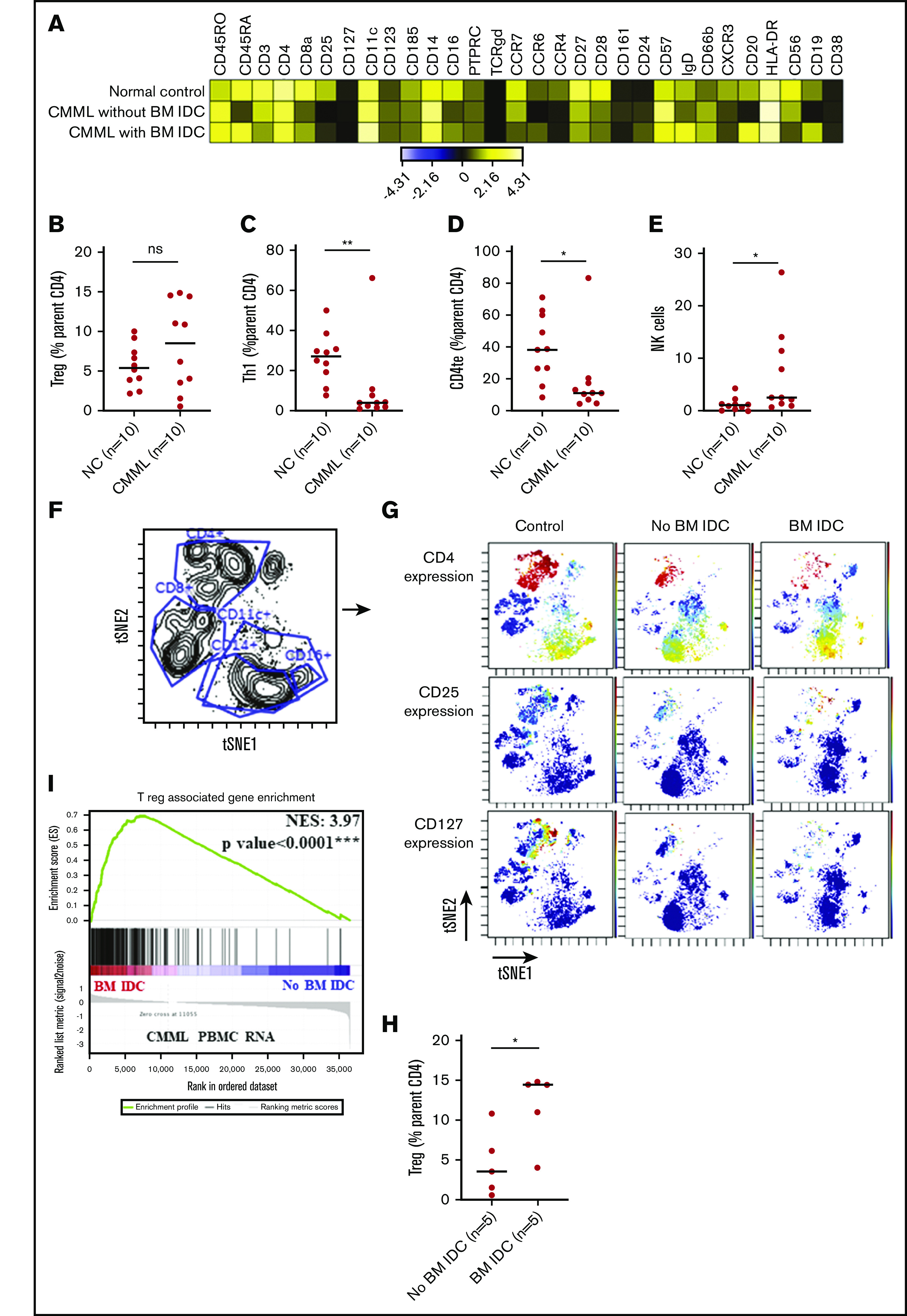
Mass cytometry (CyTOF) analysis of PBMC samples from untreated patients with CMML and age-matched normal PBMC controls. (A) Heatmap of all the markers used in the CyTOF panel (details in supplemental Table 3) and stratified by a representative sample in each group (normal control [NC], CMML without BM IDC aggregates, and CMML with BM IDC). Expression is indicated as transformed ratio of means. (B) Tregs, calculated as median value of percentage (%) of parent cell type (CD4 T), did not significantly (ns) differ between CMML cases and controls (8.5 vs 5.5; P = .4). (C-D) Significantly suppressed median percentage (%) of parent cell aggregates of Th1 (median, 4.04 vs 27.2; **P = .003) and CD4 terminal effector (te) cells (11.6 vs 38.7; *P = .02). (E) An expanded NK (2.6 vs 1.1; *P = .02) cell population in CMML vs normal controls. (F) Visualization of dimensionality reduction analysis plot with gates delineating CD4+, CD8+, CD11c+, CD14+, and CD16+ aggregates. (G) Expression of specific markers such as CD4, CD25, and CD127 (colors represent the median intensity of specified marker with red representing the brightest intensity and blue indicating no or minimal intensity). This figure provides a visualization of an expanded Treg (CD4+/CD25+/CD127dim) population in a CMML patient with vs without BM IDC. (H) Quantitative analysis confirming an expanded Treg (% parentCD4 T cell) population in untreated CMML patients with BM IDC (n = 5) vs untreated CMML patients without BM IDC (n = 5; median, 14.5 vs 3.6; *P = .03). (I) GSEA showing significant enrichment of Treg-associated genes (normalized enrichment score [NES] = 3.97; ***P < .0001) through RNA-sequencing analysis of untreated PBMC samples derived from CMML patients (n = 4, indicated in red) with vs without (n = 4, indicated in blue) BM IDC (samples collected at the same time point as IHC assessment).
In summary, our constellation of IHC, flow cytometry, LC-MS, CyTOF, cytokine profiling, and gene-set enrichment analysis results provide evidence of systemic immune dysregulation in CMML, and highlight the association of BM IDC aggregates with a T-cell compartment shift toward a Treg phenotype. Future research is needed to clarify the genetic/epigenetic events responsible for CMML IDC formation to pinpoint the biology of DC–T-cell interactions, recapitulate the findings in in vivo models, and explore therapeutic vulnerabilities with IDO and/or CD123-directed therapies.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors acknowledge the services and expertise of the Pathology Research Core, the Immune Monitoring Core, and the Metabolomics Resource Core of Mayo Clinic (Rochester, MN). The authors also acknowledge the technical input of Hyo Jin Kim for the flow cytometry experiments.
This work was supported, in part, by the American Society of Clinical Oncology Conquer Cancer Foundation–sponsored Young Investigator Award, the Eagles 5th District Cancer Telethon Pilot Investigator Award, and the Mayo Clinic Hematology Small Grants Program Grant (A.A.M.), as well as grant UL1TR002377 from the National Institutes of Health, National Center for Advancing Translational Sciences.
Results and views do not necessarily reflect the views of the funding agencies.
Footnotes
For original data, please contact patnaik.mrinal@mayo.edu
Authorship
Contribution: A.A.M. designed the study with M.M.P., performed key experiments and data analysis, and wrote the first draft of the manuscript; G.C., T.L.L., R.M.C., and M.B., performed key experiments and provided critical feedback; A.C., V.N., and K.K.R. reviewed hematopathology slides and provided critical feedback; T.A., J.C.V., M.H., and S.M.A. provided feedback on mass cytometry experiments and data interpretation; M.M.T. and J.W.H. provided input and performed flow cytometry experiments; W.I.G. aided mass spectrometry experiments and data analysis; N.G., A.A.-K., M.A.E, A.P., A.T., W.I.G., M.R.L., K.H.B., H.B.A., M.S., A.P.W.-S., and W.J.H. contributed patient samples and edited the manuscript; M.M.P. supervised the study and edited all manuscript drafts; and all authors participated in writing and editing the manuscript.
Conflict-of-interest disclosure: M.M.P. is on the advisory board of Stemline Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Mrinal M. Patnaik, Division of Hematology, Department of Medicine, Mayo Clinic, 200 First St SW, Rochester, MN 55901; e-mail: patnaik.mrinal@mayo.edu.
References
- 1.Niyongere S, Lucas N, Zhou JM, et al. Heterogeneous expression of cytokines accounts for clinical diversity and refines prognostication in CMML. Leukemia. 2019;33(1):205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahid MF, Barraco D, Lasho TL, et al. Spectrum of autoimmune diseases and systemic inflammatory syndromes in patients with chronic myelomonocytic leukemia. Leuk Lymphoma. 2017;58(6):1488-1493. [DOI] [PubMed] [Google Scholar]
- 3.Grignano E, Mekinian A, Braun T, et al. ; GFM, SNFMI and CRI . Autoimmune and inflammatory diseases associated with chronic myelomonocytic leukemia: a series of 26 cases and literature review. Leuk Res. 2016;47:136-141. [DOI] [PubMed] [Google Scholar]
- 4.Loghavi S, Sui D, Wei P, et al. Validation of the 2017 revision of the WHO chronic myelomonocytic leukemia categories. Blood Adv. 2018;2(15):1807-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, Bueso-Ramos C, DiNardo C, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2014;28(6):1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermi W, Facchetti F, Rosati S, et al. Nodal and extranodal tumor-forming accumulation of plasmacytoid monocytes/interferon-producing cells associated with myeloid disorders. Am J Surg Pathol. 2004;28(5):585-595. [DOI] [PubMed] [Google Scholar]
- 7.Lucas N, Duchmann M, Rameau P, et al. Biology and prognostic impact of clonal plasmacytoid dendritic cells in chronic myelomonocytic leukemia. Leukemia. 2019;33(10):2466-2480. [DOI] [PubMed] [Google Scholar]
- 8.Munn DH, Sharma MD, Hou D, et al. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest. 2004;114(2):280-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pallotta MT, Orabona C, Volpi C, et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat Immunol. 2011;12(9):870-878. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, Liang X, Peterson AJ, Munn DH, Blazar BR. The indoleamine 2,3-dioxygenase pathway is essential for human plasmacytoid dendritic cell-induced adaptive T regulatory cell generation. J Immunol. 2008;181(8):5396-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munn DH, Sharma MD, Lee JR, et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297(5588):1867-1870. [DOI] [PubMed] [Google Scholar]
- 12.Théate I, van Baren N, Pilotte L, et al. Extensive profiling of the expression of the indoleamine 2,3-dioxygenase 1 protein in normal and tumoral human tissues [published correction appears in Cancer Immunol Res. 2020;8(6):842]. Cancer Immunol Res. 2015;3(2):161-172. [DOI] [PubMed] [Google Scholar]
- 13.Munn DH, Mellor AL. IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 2016;37(3):193-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munn DH, Zhou M, Attwood JT, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281(5380):1191-1193. [DOI] [PubMed] [Google Scholar]
- 15.Mangaonkar A, Mondal AK, Fulzule S, et al. A novel immunohistochemical score to predict early mortality in acute myeloid leukemia patients based on indoleamine 2,3 dioxygenase expression. Sci Rep. 2017;7(1):12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folgiero V, Goffredo BM, Filippini P, et al. Indoleamine 2,3-dioxygenase 1 (IDO1) activity in leukemia blasts correlates with poor outcome in childhood acute myeloid leukemia. Oncotarget. 2014;5(8):2052-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamuleau ME, van de Loosdrecht AA, Hess CJ, et al. High INDO (indoleamine 2,3-dioxygenase) mRNA level in blasts of acute myeloid leukemic patients predicts poor clinical outcome. Haematologica. 2008;93(12):1894-1898. [DOI] [PubMed] [Google Scholar]
- 18.Müller-Thomas C, Heider M, Piontek G, et al. Prognostic value of indoleamine 2,3 dioxygenase in patients with higher-risk myelodysplastic syndromes treated with azacytidine. Br J Haematol. 2020;190(3):361-370. [DOI] [PubMed] [Google Scholar]
- 19.Curti A, Trabanelli S, Onofri C, et al. Indoleamine 2,3-dioxygenase-expressing leukemic dendritic cells impair a leukemia-specific immune response by inducing potent T regulatory cells. Haematologica. 2010;95(12):2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34(3):137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizzolo G, Trentin L, Vinante F, et al. Natural killer cell function and lymphoid subpopulations in acute non-lymphoblastic leukaemia in complete remission. Br J Cancer. 1988;58(3):368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian T, Yu S, Liu L, et al. The profile of T helper subsets in bone marrow microenvironment is distinct for different stages of acute myeloid leukemia patients and chemotherapy partly ameliorates these variations. PLoS One. 2015;10(7):e0131761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiani A, Habermann I, Schäke K, Neubauer A, Rogge L, Ehninger G. Normal intrinsic Th1/Th2 balance in patients with chronic phase chronic myeloid leukemia not treated with interferon-alpha or imatinib. Haematologica. 2003;88(7):754-761. [PubMed] [Google Scholar]
- 24.Crawford A, Angelosanto JM, Nadwodny KL, Blackburn SD, Wherry EJ. A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog. 2011;7(7):e1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taub DD, Conlon K, Lloyd AR, Oppenheim JJ, Kelvin DJ. Preferential migration of activated CD4+ and CD8+ T cells in response to MIP-1 alpha and MIP-1 beta. Science. 1993;260(5106):355-358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.