Abstract
Background
Immunogenic cell death (ICD) is a tumor cell death involving both innate and adaptive immune responses. Given published findings that oxaliplatin, but not irinotecan, drives ICD, we investigated whether single nucleotide polymorphisms (SNPs) in the ICD pathway are associated with the efficacy of oxaliplatin-based chemotherapy in metastatic colorectal cancer (mCRC).
Methods
Two randomized clinical trials data were analyzed: discovery cohort, FOLFOX/bevacizumab arm (MAVERICC); validation cohort, FOLFOXIRI/bevacizumab arm (TRIBE); and two control cohorts, FOLFIRI/bevacizumab arms (both trials). Genomic DNA extracted from blood samples was genotyped. Ten SNPs in the ICD pathway were tested for associations with clinical outcomes.
Results
In total, 648 patients were included. In the discovery cohort, three SNPs were significantly associated with clinical outcomes in univariate analysis: CALR rs1010222 with progression-free survival (G/G vs any A, HR=0.61, 95% CI 0.43–0.88), ANXA1 rs1050305 with overall survival (OS) (A/A vs any G, HR=1.87, 95% CI 1.04–3.35), and LRP1 rs1799986 with OS (C/C vs any T, HR=1.69, 95% CI 1.07–2.70). Multivariate analysis confirmed the trend, but statistical significance was not reached. In the validation cohort, ANXA1 rs1050305, and LRP1 rs1799986 were validated to have the significant associations with clinical outcome. No significant associations of these SNPs were observed in the two control cohorts. Treatment-by-SNP interaction test confirmed the predictive values.
Conclusions
The predictive utility of ICD-related SNPs for the efficacy of oxaliplatin-based chemotherapy was demonstrated, warranting further validation studies to be translated into personalized treatment strategies using conventional cytotoxic agents in mCRC.
Keywords: gastrointestinal neoplasms, adaptive immunity, immunity, innate, translational medical research
Background
The current standard of care for patients with microsatellite-stable metastatic colorectal cancer (mCRC) consists of the combination of a cytotoxic chemotherapy backbone and a monoclonal antibody based on RAS/BRAF testing.1 2 In clinical practice, there are two choices for cytotoxic agent, namely oxaliplatin or irinotecan in combination with 5-fluorouracil, and two for monoclonal antibodies targeting vascular endothelial growth factor (VEGF) or epidermal growth factor receptor (EGFR). Most research has focused on the efficacy of monoclonal antibodies, leading to the successful identification of predictive and prognostic biomarkers for EGFR inhibitors such as RAS and BRAF mutation status and primary tumor sidedness.3–6 However, there are no predictive biomarkers for the selection of oxaliplatin or irinotecan which have distinct mechanisms of action: oxaliplatin is a platinum compound forming intra-strand and inter-strand cross-links; irinotecan is a topoisomerase I (TOP1) inhibitor trapping TOP1 cleavage complex with DNA replication and transcription. The choice of these two drugs remains an important clinical question for optimizing treatment in individual patients.
Immunogenic cell death (ICD) is a novel concept of tumor cell death that engages both innate and adaptive immune responses, thereby conferring an additional immunogenic antitumor effect to cytotoxic agents.7–9 However, most chemotherapeutics kill tumor cells in a non-immunogenic manner, and only a few cytotoxic agents can prime antitumor immunity by inducing ICD.10 11 The mechanisms of ICD induction are characterized by the ability to induce endoplasmic reticulum stress, which in turn leads to the release of damage-associated molecular patterns (DAMPs) from dying tumor cells and the subsequent activation of pattern recognition receptors (PRRs) of host innate immune cells such as dendritic cells.12 The engagement of innate immune responses cross-primes adaptive responses controlled by cytotoxic T lymphocytes through proinflammatory cytokine release, potentially leading to the continuous elimination of tumor cells.12 13 Oxaliplatin has been recently well-recognized as an ICD inducer, similarly as other drugs such as anthracyclines and bortezomib.7 In a preclinical study, CRC cells treated with oxaliplatin exhibited increased pre-apoptotic exposure of calreticulin (CALR) and post-apoptotic release of high-mobility group box 1 (HMGB1), which are the signals of DAMPs required for ICD induction.14 By contrast, irinotecan lacks the mechanism of action that drives ICD, and thus, its cytotoxic effects depend on non-immunogenic mechanisms.10 15 These findings suggested that genetic variants in the ICD pathway are related to the efficacy of oxaliplatin but not irinotecan, prompting us to test whether these novel biomarkers will be useful for patient selection. Therefore, we investigated the predictive utility of single nucleotide polymorphisms (SNPs) in ICD-related genes encoding essential DAMPs and PRRs using genetic and clinical data from two first-line randomized clinical trials comparing oxaliplatin- (or oxaliplatin-containing) and irinotecan-based treatments in patients with mCRC.
Methods
Patient population and study design
This study included patients with mCRC enrolled in two first-line randomized trials: MAVERICC (NCT01374425)16 and TRIBE (NCT00719797).17 In the MAVERICC trial, patients were randomized to treatment with either FOLFOX plus bevacizumab or FOLFIRI plus bevacizumab. In the TRIBE trial, patients were randomized to the FOLFOXIRI plus bevacizumab or FOLFIRI plus bevacizumab arm. Patients without sufficient peripheral whole blood samples for analyzes were excluded from our study. To assess whether the ICD pathway is specifically related to the efficacy of oxaliplatin, we set the cohorts as follows: (1) discovery cohort, FOLFOX plus bevacizumab arm in MAVERICC; (2) validation cohort, FOLFOXIRI plus bevacizumab arm in TRIBE; (3) control cohort 1, FOLFIRI plus bevacizumab arm in MAVERICC; and (4) control cohort 2, FOLFIRI plus bevacizumab arm in TRIBE (figure 1). All patients provided informed consent for molecular research prior to study enrollment.
Figure 1.
Consort diagram. BEV, bevacizumab; SNP, single nucleotide polymorphism.
Genotyping and selecting polymorphisms
Genomic DNA was extracted from peripheral whole blood collected before treatment initiation using a QIAmp Kit (Qiagen, Valencia, California, USA) in accordance with the manufacturer’s protocol (www.qiagen.com). The OncoArray of 530 K SNPs was used for genotyping (Illumina, San Diego, California, USA).18 After genotyping, we planned this exploratory analysis focusing on five genes encoding DAMPs or PRRs related to chemotherapy-induced ICD,12 including three DAMP-encoding genes (CALR, HMGB1, and ANXA1) and two PRR-encoding genes (LRP1 and P2R×7) (online supplementary table S1). Although FPR1, TLR3, and TLR4 are also key PRR-encoding genes, these genes were not included in this study because they were not predictive of oxaliplatin efficacy in patients with CRC in another large biomarker study.19 The candidate SNPs for this study were arbitrarily selected according to the following criteria: (1) minor allele frequency in Caucasians of at least 10% in the Ensemble Genome Browser (https://www.ensembl.org), (2) having potential biological functions based on public databases (https://snpinfo.niehs.nih.gov; https://www.ncbi.nlm.nih.gov), and (3) tag SNPs chosen from HapMap genotype data with an r2 threshold of 0.8 (https://snpinfo.niehs.nih.gov). In total, 10 SNPs were selected, as presented in online supplementary table S2.
jitc-2020-001714supp001.pdf (145.4KB, pdf)
Statistical analysis
Selected SNPs were evaluated for their associations with tumor response, progression-free survival (PFS), and overall survival (OS). The overall response rate (ORR) was calculated as the percentage of patients with either a complete or partial response using the Response Evaluation Criteria in Solid Tumors version 1.1. PFS was defined as the time from randomization to disease progression or death from any cause. OS was defined as the time from randomization to death from any cause. Patients who experienced no events were censored at the last follow-up date. The correlation between each SNP and ORR was examined using the χ2 test. To test the association between each SNP and PFS or OS, univariate and multivariate analyzes using the Cox proportional hazards regression model and the log-rank test were performed. In the multivariate analyzes, the following study-specific adjusted covariates were used: the TRIBE trial included sex, age, Eastern Cooperative Oncology Group (ECOG) performance status (PS), primary tumor site, liver-limited disease, adjuvant chemotherapy, BRAF status, and RAS status; and the MAVERICC trial included ethnicity, sex, age, ECOG PS, primary tumor site, primary tumor resected, number of metastases, and KRAS status. SNPs were coded using a dominant genetic model for the number of variant alleles. To formally assess the predictive value, the treatment-by-SNP interaction was tested within each trial using the HR calculated in the multivariate analyzes. All analyzes were two-sided at a significance level of 0.05 and were performed using SAS V.9.4 software.
Results
Patient characteristics
In total, 648 patients were included in this study (figure 1). Some characteristics were unbalanced between the trial cohorts. In particular ECOG PS 0, left-sided primary tumor, metastasis in ≤2 organs, and primary tumor resection were more prevalent in the TRIBE trial arms than in the MAVERICC trial arms (online supplementary table S3). The median follow-up and survival time in each cohort are summarized in online supplementary table S4.
Predictive value of ICD-related SNPs
All tested associations between each SNP and clinical outcomes are presented in online supplemental tables S5–8. Although several significant associations were observed in the discovery or validation cohort, few associations were confirmed in control cohort 1 or 2.
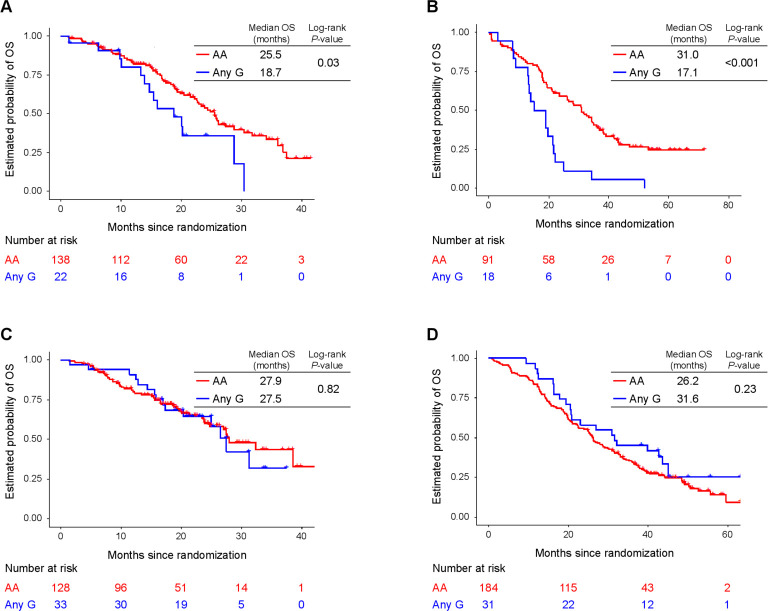
In particular, three SNPs displayed significant associations with outcome in the univariate analysis in the discovery cohort (table 1, figure 2). First, patients carrying any A allele of CALR rs1010222 had a better PFS than those with a G/G genotype (HR=0.61, 95% CI 0.43–0.88, p=0.008). Second, patients carrying any G allele of ANXA1 rs1050305 had worse OS than those with an A/A genotype (HR=1.87, 95% CI 1.04–3.35, p=0.03). Finally, patients carrying any T allele of LRP1 rs1799986 had worse OS than those with a C/C genotype (HR=1.69, 95% CI 1.07–2.70, p=0.03). Multivariate analysis confirmed these trends, but none of the associations reached statistical significance. In the validation cohort, two SNPs were confirmed to have significant associations with clinical outcome (table 1, figure 2). Namely, patients carrying any G allele of ANXA1 rs1050305 had worse ORR, PFS (revealed in univariate analysis), and OS (revealed in both univariate and multivariate analyzes) than those with an A/A genotype; and patients carrying any T allele of LRP1 rs1799986 had worse PFS (revealed in multivariate analysis) than those with a C/C genotype, even though this clinical endpoint was not same as that in the discovery cohort. Of note, in the two control cohorts, no significant associations were observed between these SNPs (CALR rs1010222, ANXA1 rs1050305, and LRP1 rs1799986) and clinical outcomes (table 1, figure 2).
Table 1.
Associations between ICD-related SNPs and clinical outcomes
| Genotype | N | TR | PFS | OS | |||||||
| ORR (%) | P value* | Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | ||||||
| HR (95% CI) | P value† |
HR (95% CI) | P value‡ | HR (95% CI) | P value† | HR (95% CI) | P value‡ | ||||
| CALR rs1010222 | |||||||||||
| Discovery cohort | |||||||||||
| G/G | 72 | 66.2 | 0.48 | 1 | 0.008 | 1 | 0.10 | 1 | 0.07 | 1 | 0.38 |
| Any A | 87 | 60.7 | 0.61 (0.43 to 0.88) | 0.67 (0.42 to 1.07) | 0.67 (0.44 to 1.04) | 0.78 (0.46 to 1.35) | |||||
| Validation cohort | |||||||||||
| G/G | 44 | 71.4 | 0.33 | 1 | 0.63 | 1 | 0.61 | 1 | 0.76 | 1 | 0.35 |
| Any A | 65 | 62.3 | 0.90 (0.57 to 1.41) | 0.87 (0.52 to 1.46) | 0.93 (0.60 to 1.44) | 0.79 (0.49 to 1.29) | |||||
| Control cohort 1 | |||||||||||
| G/G | 81 | 67.5 | 1.00 | 1 | 0.98 | 1 | 0.88 | 1 | 0.41 | 1 | 0.94 |
| Any A | 82 | 67.5 | 0.99 (0.69 to 1.44) | 0.96 (0.58 to 1.60) | 0.81 (0.49 to 1.34) | 0.97 (0.50 to 1.89) | |||||
| Control cohort 2 | |||||||||||
| G/G | 90 | 63.6 | 0.16 | 1 | 0.37 | 1 | 0.56 | 1 | 0.75 | 1 | 0.79 |
| Any A | 119 | 53.9 | 1.16 (0.84 to 1.58) | 1.11 (0.79 to 1.56) | 1.05 (0.77 to 1.44) | 0.96 (0.69 to 1.33) | |||||
| ANXA1 rs1050305 | |||||||||||
| Discovery cohort | |||||||||||
| A/A | 138 | 63.4 | 0.99 | 1 | 0.26 | 1 | 0.47 | 1 | 0.03 | 1 | 0.10 |
| Any G | 22 | 63.6 | 1.33 (0.81 to 2.21) | 1.29 (0.66 to 2.51) | 1.87 (1.04 to 3.35) | 1.96 (0.93 to 4.12) | |||||
| Validation cohort | |||||||||||
| A/A | 91 | 70.6 | 0.03 | 1 | 0.009 | 1 | 0.17 | 1 | <0.001 | 1 | 0.04 |
| Any G | 18 | 44.4 | 2.17 (1.20 to 3.92) | 1.62 (0.83 to 3.16) | 2.69 (1.56 to 4.61) | 2.00 (1.07 to 3.72) | |||||
| Control cohort 1 | |||||||||||
| A/A | 128 | 66.9 | 0.67 | 1 | 0.72 | 1 | 0.85 | 1 | 0.82 | 1 | 0.63 |
| Any G | 33 | 71.0 | 1.08 (0.70 to 1.67) | 1.07 (0.54 to 2.11) | 1.07 (0.60 to 1.92) | 0.81 (0.33 to 1.98) | |||||
| Control cohort 2 | |||||||||||
| A/A | 184 | 57.6 | 0.70 | 1 | 0.92 | 1 | 0.72 | 1 | 0.23 | 1 | 0.09 |
| Any G | 31 | 61.3 | 1.02 (0.66 to 1.58) | 1.09 (0.68 to 1.74) | 0.76 (0.48 to 1.19) | 0.67 (0.42 to 1.09) | |||||
| LRP1 rs1799986 | |||||||||||
| Discovery cohort | |||||||||||
| C/C | 111 | 64.2 | 0.99 | 1 | 0.22 | 1 | 0.53 | 1 | 0.03 | 1 | 0.14 |
| Any T | 44 | 64.3 | 1.30 (0.85 to 1.97) | 1.18 (0.71 to 1.94) | 1.69 (1.07 to 2.70) | 1.55 (0.87 to 2.74) | |||||
| Validation cohort | |||||||||||
| C/C | 74 | 63.9 | 0.52 | 1 | 0.35 | 1 | 0.005 | 1 | 0.98 | 1 | 0.42 |
| Any T | 24 | 71.4 | 1.30 (0.75 to 2.25) | 2.71 (1.37 to 5.37) | 0.99 (0.58 to 1.71) | 1.30 (0.70 to 2.40) | |||||
| Control cohort 1 | |||||||||||
| C/C | 126 | 65.9 | 0.28 | 1 | 0.35 | 1 | 0.79 | 1 | 0.68 | 1 | 0.38 |
| Any T | 33 | 75.8 | 1.23 (0.79 to 1.92) | 0.93 (0.52 to 1.66) | 1.13 (0.63 to 2.02) | 0.71 (0.33 to 1.53) | |||||
| Control cohort 2 | |||||||||||
| C/C | 162 | 58.9 | 0.45 | 1 | 0.34 | 1 | 0.59 | 1 | 0.57 | 1 | 0.66 |
| Any T | 44 | 52.4 | 0.83 (0.57 to 1.22) | 0.90 (0.60 to 1.34) | 0.89 (0.61 to 1.31) | 1.10 (0.73 to 1.66) | |||||
Significant values are indicated in bold characters.
*P values were based on χ2 test.
†P values were based on log-rank test for PFS and OS in the univariate analysis.
‡P values were bases on Wald test in the multivariate Cox proportional hazards regression model.
ICD, immunogenic cell death; ORR, overall response rate; OS, overall survival; PFS, progression-free survival; SNP, single nucleotide polymorphism; TR, tumor response.
Figure 2.
OS of patients with ANXA1 rs1050305 variants. (A) Discovery cohort (FOLFOX plus bevacizumab arm in MAVERICC), (B) validation cohort (FOLFOXIRI plus bevacizumab arm in TRIBE), (C) control cohort 1 (FOLFIRI plus bevacizumab arm in MAVERICC), and (D) control cohort 2 (FOLFIRI plus bevacizumab arm in TRIBE). OS, overall survival.
The results of the treatment-by-SNP interaction test are presented in table 2. In total, six SNPs exhibited significant interactions with treatment: HMGB1 rs1360485 (PFS in TRIBE), ANXA1 rs1050305 (OS in TRIBE), LRP1 rs1799986 (PFS in TRIBE), LRP1 rs11172113 (both PFS and OS in TRIBE), P2R×7 rs208294 (OS in MAVERICC and OS in TRIBE), and P2R×7 rs1718119 (OS in MAVERICC).
Table 2.
Treatment-by-single nucleotide polymorphism interaction test
| MAVERICC | TRIBE | |||||||||||
| Hazard ratio on PFS | Hazard ratio on OS | Hazard ratio on PFS | Hazard ratio on OS | |||||||||
| FOLFOX+ BEV |
FOLFIRI+ BEV |
Interaction p value |
FOLFOX+ BEV |
FOLFIRI+ BEV |
Interaction p value |
FOLFOXIRI+ BEV |
FOLFIRI+ BEV |
Interaction p value |
FOLFIRI+ BEV |
FOLFIRI+ BEV |
Interaction p value |
|
| CALR rs1049481 | ||||||||||||
| T/T | 1 | 1 | 0.87 | 1 | 1 | 0.98 | 1 | 1 | 0.11 | 1 | 1 | 0.42 |
| Any G | 0.85 (0.54–1.36) | 0.91 (0.54–1.51) | 1.03 (0.60–1.76) | 0.92 (0.47–1.78) | 0.69 (0.39–1.21) | 1.14 (0.81–1.62) | 0.74 (0.45–1.24) | 0.97 (0.69–1.36) | ||||
| CALR rs1010222 | ||||||||||||
| G/G | 1 | 1 | 0.41 | 1 | 1 | 0.41 | 1 | 1 | 0.34 | 1 | 1 | 0.57 |
| Any A | 0.67 (0.42–1.07) | 0.96 (0.58–1.60) | 0.78 (0.46–1.35) | 0.97 (0.50–1.89) | 0.87 (0.52–1.46) | 1.11 (0.79–1.56) | 0.79 (0.49–1.29) | 0.96 (1.69–1.33) | ||||
| HMGB1 rs1045411 | ||||||||||||
| C/C | 1 | 1 | 0.35 | 1 | 1 | 0.58 | 1 | 1 | 0.07 | 1 | 1 | 0.50 |
| Any T | 1.43 (0.85–2.41) | 1.09 (0.63–1.89) | 1.46 (0.79–2.69) | 1.20 (0.58–2.48) | 1.93 (1.06–3.53) | 1.11 (0.74–1.65) | 1.03 (0.59–1.80) | 0.95 (0.65–1.38) | ||||
| HMGB1 rs1412125 | ||||||||||||
| T/T | 1 | 1 | 0.87 | 1 | 1 | 0.51 | 1 | 1 | 0.30 | 1 | 1 | 0.11 |
| Any C | 0.74 (0.43–1.28) | 0.78 (0.45–1.35) | 0.80 (0.43–1.48) | 1.06 (0.52–2.16) | 1.22 (0.64–2.30) | 0.98 (0.69–1.40) | 1.14 (0.64–2.01) | 0.83 (0.58–1.17) | ||||
| HMGB1 rs1360485 | ||||||||||||
| T/T | 1 | 1 | 0.28 | 1 | 1 | 0.73 | 1 | 1 | 0.04 | 1 | 1 | 0.42 |
| Any C | 1.45 (0.89–2.38) | 1.03 (0.62–1.69) | 1.38 (0.78–2.43) | 1.19 (0.62–2.25) | 1.70 (0.95–3.06) | 1.06 (0.74–1.50) | 0.93 (0.54–1.60) | 0.98 (0.70–1.37) | ||||
| ANXA1 rs1050305 | ||||||||||||
| A/A | 1 | 1 | 0.82 | 1 | 1 | 0.15 | 1 | 1 | 0.16 | 1 | 1 | 0.002 |
| Any G | 1.29 (0.66–2.51) | 1.07 (0.54–2.11) | 1.96 (0.93–4.12) | 0.81 (0.33–1.98) | 1.62 (0.83–3.16) | 1.09 (0.68–1.74) | 2.00 (1.07–3.72) | 0.67 (0.42–1.09) | ||||
| LRP1 rs1799986 | ||||||||||||
| C/C | 1 | 1 | 0.66 | 1 | 1 | 0.23 | 1 | 1 | 0.03 | 1 | 1 | 0.55 |
| Any T | 1.18 (0.71–1.94) | 0.93 (0.52–1.66) | 1.55 (0.87–2.74) | 0.71 (0.33–1.53) | 2.71 (1.37–5.37) | 0.90 (0.60–1.34) | 1.30 (0.70–2.40) | 1.10 (0.73–1.66) | ||||
| LRP1 rs11172113 | ||||||||||||
| T/T | 1 | 1 | 0.31 | 1 | 1 | 0.73 | 1 | 1 | 0.004 | 1 | 1 | 0.004 |
| Any C | 1.05 (0.63–1.74) | 1.22 (0.72–2.06) | 0.97 (0.55–1.69) | 0.66 (0.34–1.26) | 0.55 (0.31–0.98) | 1.35 (0.96–1.89) | 0.58 (0.34–0.96) | 1.20 (0.87–1.67) | ||||
| P2R×7 rs208294 | ||||||||||||
| C/C | 1 | 1 | 0.33 | 1 | 1 | 0.046 | 1 | 1 | 0.62 | 1 | 1 | 0.02 |
| Any T | 0.93 (0.55–1.58) | 1.26 (0.72–2.22) | 0.62 (0.33–1.16) | 1.84 (0.84–4.06) | 0.92 (0.54–1.58) | 1.02 (0.71–1.47) | 0.54 (0.33–0.89) | 0.92 (0.64–1.32) | ||||
| P2R×7 rs1718119 | ||||||||||||
| G/G | 1 | 1 | 0.66 | 1 | 1 | 0.03 | 1 | 1 | 0.85 | 1 | 1 | 0.77 |
| Any A | 1.02 (0.62–1.70) | 1.26 (0.74–2.14) | 0.97 (0.53–1.78) | 2.57 (1.16–5.67) | 0.77 (0.45–1.31) | 0.87 (0.62–1.21) | 0.69 (0.42–1.15) | 0.83 (0.60–1.15) | ||||
Significant values are indicated in bold characters. Each HR is presented with a point estimate and a 95% CI (in parentheses) calculated in the multivariate analyzes.
BEV, bevacizumab; OS, overall survival; PFS, progression-free survival.
Discussion
This is the first study to identify genetic variants in ICD-related pathways that may predict the efficacy of oxaliplatin-based first-line chemotherapy in patients with mCRC. No predictive biomarkers for the personalized use of cytotoxic agents in mCRC have been identified so far. DNA damage response genes and the transcriptome-based consensus molecular subtype have been examined in biomarker research, but their predictive value has not been validated.16 20 Gray et al reported that genetic variants of three PRR genes (FPR1, TLR3, and TLR4) were not related to the efficacy of oxaliplatin in patients with CRC in the adjuvant setting (using data from the SCOT trials) and the first-line setting for patients with metastasis (using data from the COIN/COIN-B trials).19 However, DAMPs are also essential tumor-sided factors for ICD induction, whereas PRRs are host-side factors. Therefore, in our study, we extensively assessed the functional variants of both DAMPs and PRRs. According to our hypothesis, ICD-related SNPs were significantly associated with the efficacy of oxaliplatin-based combination. These findings imply the linkage between ICD pathway and oxaliplatin efficacy, which can be coupled with the potential synergy of oxaliplatin and immunotherapy.21 22
Our findings identified ANXA1 rs1050305 and LRP1 rs1799986 as SNPs exhibiting significant associations with the efficacy of oxaliplatin but not irinotecan as confirmed in the discovery and validation cohorts. ANXA1 is one of the DAMPs released from the cytoplasm of dying cells, and it interacts with FPR1, which is expressed on dendritic cells.23 The extracellular accumulation of ANXA1 constitutes a hallmark of ICD.12 ANXA1 rs1050305 is a synonymous SNP located at an exonic splicing enhancer/silencer site that disrupts splicing activity and causes alternative splicing (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html). LRP1 is a PRR activated via docking of cell surface-exposed CALR, which is another hallmark of chemotherapy-induced ICD.12 24 LRP1 rs1799986 is a missense SNP that may participate in splicing regulation. This SNP is reported to be associated with susceptibility to Alzheimer’s disease, supporting its functional significance.25 Of note, while the predictive value of ANXA1 rs1050305 was consistently validated on OS, that of LRP1 rs1799986 was observed on inconsistent clinical outcomes between the discovery and validation cohorts: the discovery cohort showed a significant association with OS, and the validation cohort showed that with PFS. This discrepancy might be due to some unidentified factors such as different contribution of second-line treatment between two cohorts, which might affect the associations between biomarker and clinical outcome.
Our study had great strengths in its design, featuring discovery, validation, and control cohorts obtained from two randomized clinical trials. The validated effects of the SNPs on clinical outcomes in the discovery and validation cohorts, lacking of that in the two control cohorts, strongly supports the predictive value of ANXA1 rs1050305 and LRP1 rs1799986 specifically for oxaliplatin efficacy. The multiple significant associations observed in the discovery and validation cohorts but not in the control cohorts further indicate that the ICD pathway plays an important role in the efficacy of oxaliplatin but not irinotecan. Another strength was that the treatment-by-SNP interaction test confirmed the predictive value of ANXA1 rs1050305 and LRP1 rs1799986 and highlighted the predictive utility of 6 out of 10 tested SNPs.26
This study had two key limitations. First, this was a prospective–retrospective study; thus, the results need to be validated in prospective clinical trials. According to published guidelines, one or more validation studies are required to reach a sufficient level of evidence for medical utility.27 28 Second, the validation cohort consisted of patients treated with FOLFOXIRI plus bevacizumab, which is both an oxaliplatin-containing and irinotecan-containing regimen. To validate the different effects of SNPs on oxaliplatin and irinotecan efficacy, it will be better to use a treatment arm containing only oxaliplatin-based regimens as the validation cohort. However, our study provides reliable evidence describing the specific connection of ICD-related SNPs with the efficacy of “oxaliplatin-containing” treatment based on the use of “oxaliplatin-non-containing” cohorts as controls.
In conclusion, our study provides the first evidence that genetic variants in the ICD pathway are significantly associated with the efficacy of oxaliplatin-based, but not irinotecan-based, first-line chemotherapy in patients with mCRC. Further translational studies are warranted to validate our novel findings and realize the personalized use of conventional cytotoxic chemotherapeutic agents.
Acknowledgments
We thank all patients who contributed to this study.
Footnotes
Contributors: HA and H-JL primarily planned, designed, and drafted the manuscript. YX and JM did statistical analyses. All authors made substantial contributions to data collection and drafting the manuscript. All authors read and approved the final manuscript.
Funding: This work was supported by the National Cancer Institute [P30CA 014089 to HJL], Gloria Borges WunderGlo Foundation, Dhont Family Foundation, Victoria and Philip Wilson Research Fund, San Pedro Peninsula Cancer Guild, and Daniel Butler Research Fund.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: All patients provided informed consent for molecular research prior to study enrollment. The study protocol was approved by the Institutional Review Board of each participating institution and was conducted in accordance with the tenets of the Declaration of Helsinki as well as the Good Clinical Practice and REMARK guidelines.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. De-identified datasets analyzed in the current study are available from the corresponding author on reasonable request.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.NCCN clinical practice guidelines in oncology: colorectal cancer. version 4, 2020. Available: https://www.ncn.org/professionals/physician_gls/pdf/colon.pdf [Accessed 11 July 2020].
- 2.Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016;27:1386–422. 10.1093/annonc/mdw235 [DOI] [PubMed] [Google Scholar]
- 3.Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for clinical pathology, College of American pathologists, association for molecular pathology, and the American Society of clinical oncology. J Clin Oncol 2017;35:1453–86. 10.1200/JCO.2016.71.9807 [DOI] [PubMed] [Google Scholar]
- 4.Arnold D, Lueza B, Douillard J-Y, et al. Prognostic and predictive value of primary tumour side in patients with Ras wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol 2017;28:1713–29. 10.1093/annonc/mdx175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with Ras wild-type metastatic colorectal cancer: retrospective analyses of the crystal and FIRE-3 trials. JAMA Oncol 2017;3:194–201. 10.1001/jamaoncol.2016.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 2015;107 10.1093/jnci/dju427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroemer G, Galluzzi L, Kepp O, et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013;31:51–72. 10.1146/annurev-immunol-032712-100008 [DOI] [PubMed] [Google Scholar]
- 8.Galluzzi L, Buqué A, Kepp O, et al. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 2015;28:690–714. 10.1016/j.ccell.2015.10.012 [DOI] [PubMed] [Google Scholar]
- 9.Galluzzi L, Senovilla L, Zitvogel L, et al. The secret ally: immunostimulation by anticancer drugs. Nat Rev Drug Discov 2012;11:215–33. 10.1038/nrd3626 [DOI] [PubMed] [Google Scholar]
- 10.Obeid M, Tesniere A, Ghiringhelli F, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med 2007;13:54–61. 10.1038/nm1523 [DOI] [PubMed] [Google Scholar]
- 11.Garg AD, More S, Rufo N, et al. Trial Watch: immunogenic cell death induction by anticancer chemotherapeutics. Oncoimmunology 2017;6:e1386829. 10.1080/2162402X.2017.1386829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galluzzi L, Buqué A, Kepp O, et al. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97–111. 10.1038/nri.2016.107 [DOI] [PubMed] [Google Scholar]
- 13.Ruan H, Leibowitz BJ, Zhang L, et al. Immunogenic cell death in colon cancer prevention and therapy. Mol Carcinog 2020;59:783–93. 10.1002/mc.23183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 2010;29:482–91. 10.1038/onc.2009.356 [DOI] [PubMed] [Google Scholar]
- 15.Pozzi C, Cuomo A, Spadoni I, et al. The EGFR-specific antibody cetuximab combined with chemotherapy triggers immunogenic cell death. Nat Med 2016;22:624–31. 10.1038/nm.4078 [DOI] [PubMed] [Google Scholar]
- 16.Parikh AR, Lee F-C, Yau L, et al. MAVERICC, a randomized, Biomarker-stratified, phase II study of mFOLFOX6-Bevacizumab versus FOLFIRI-Bevacizumab as first-line chemotherapy in metastatic colorectal cancer. Clin Cancer Res 2019;25:2988–95. 10.1158/1078-0432.CCR-18-1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loupakis F, Cremolini C, Masi G, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med 2014;371:1609–18. 10.1056/NEJMoa1403108 [DOI] [PubMed] [Google Scholar]
- 18.Amos CI, Dennis J, Wang Z, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomarkers Prev 2017;26:126–35. 10.1158/1055-9965.EPI-16-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray V, Briggs S, Palles C, et al. Pattern recognition receptor polymorphisms as predictors of oxaliplatin benefit in colorectal cancer. J Natl Cancer Inst 2019;111:828–36. 10.1093/jnci/djy215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aderka D, Stintzing S, Heinemann V. Explaining the unexplainable: discrepancies in results from the CALGB/SWOG 80405 and FIRE-3 studies. Lancet Oncol 2019;20:e274–83. 10.1016/S1470-2045(19)30172-X [DOI] [PubMed] [Google Scholar]
- 21.Boku N, Ryu MH, Oh D-Y, et al. LBA7_PR nivolumab plus chemotherapy versus chemotherapy alone in patients with previously untreated advanced or recurrent gastric/gastroesophageal junction (G/GEJ) cancer: ATTRACTION-4 (ONO-4538-37) study. Annals of Oncology 2020;31:S1192 10.1016/j.annonc.2020.08.2297 [DOI] [Google Scholar]
- 22.Moehler M, Shitara K, Garrido M, et al. LBA6_PR Nivolumab (nivo) plus chemotherapy (chemo) versus chemo as first-line (1L) treatment for advanced gastric cancer/gastroesophageal junction cancer (GC/GEJC)/esophageal adenocarcinoma (EAC): First results of the CheckMate 649 study. Annals of Oncology 2020;31:S1191 10.1016/j.annonc.2020.08.2296 [DOI] [Google Scholar]
- 23.Baracco EE, Petrazzuolo A, Kroemer G. Assessment of annexin A1 release during immunogenic cell death. Methods Enzymol 2019;629:71–9. 10.1016/bs.mie.2019.06.010 [DOI] [PubMed] [Google Scholar]
- 24.Garg AD, Krysko DV, Verfaillie T, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. Embo J 2012;31:1062–79. 10.1038/emboj.2011.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Liu S, Wang J, et al. Association between LRP1 C766T polymorphism and Alzheimer's disease susceptibility: a meta-analysis. Sci Rep 2017;7:8435. 10.1038/s41598-017-08335-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol 2015;33:3968–71. 10.1200/JCO.2015.63.3651 [DOI] [PubMed] [Google Scholar]
- 27.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446–52. 10.1093/jnci/djp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polley M-YC, Freidlin B, Korn EL, et al. Statistical and practical considerations for clinical evaluation of predictive biomarkers. J Natl Cancer Inst 2013;105:1677–83. 10.1093/jnci/djt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001714supp001.pdf (145.4KB, pdf)