Abstract
Background
Microsatellite instability (MSI) represents the first pan-cancer biomarker approved to guide immune checkpoint blockade (ICB) treatment. However its widespread testing, especially outside of gastrointestinal cancer, is hampered by tissue availability.
Methods
An algorithm for detecting MSI from peripheral blood was established and validated using clinical plasma samples. Its value for predicting ICB efficacy was evaluated among 60 patients with advanced gastrointestinal cancer. The landscape of MSI in blood was also explored among 5138 advanced solid tumors.
Results
The algorithm included 100 microsatellite markers with high capture efficiency, sensitivity, and specificity. In comparison with orthogonal tissue PCR results, the method displayed a sensitivity of 82.5% (33/40) and a specificity of 96.2% (201/209), for an overall accuracy of 94.0% (234/249). When the clinical validation cohort was dichotomized by pretreatment blood MSI (bMSI), bMSI-high (bMSI-H) predicted both improved progression-free survival and overall survival than the blood microsatellite stable (bMSS) patients (HRs: 0.431 and 0.489, p=0.005 and 0.034, respectively). Four patients with bMSS were identified to have high blood tumor mutational burden (bTMB-H) and trended towards a better survival than the bMSS-bTMB-low (bTMB-L) subset (HR 0.026, 95% CI 0 to 2.635, p=0.011). These four patients with bMSS-bTMB-H plus the bMSI-H group collectively displayed significantly improved survival over the bMSS-bTMB-L patients (HR 0.317, 95% CI 0.157 to 0.640, p<0.001). Pan-cancer prevalence of bMSI-H was largely consistent with that shown for tissue except for much lower rates in endometrial and gastrointestinal cancers, and a remarkably higher prevalence in prostate cancer relative to other cancer types.
Conclusions
We have developed a reliable and robust next generation sequencing-based bMSI detection strategy which, in combination with a panel enabling concurrent profiling of bTMB from a single blood draw, may better inform ICB treatment.
Keywords: biomarkers, tumor, immunotherapy, gastrointestinal neoplasms, genome instability
Background
Microsatellite instability (MSI) has leaped to the forefront of cancer molecular diagnosis ever since the Food and Drug Administration (FDA) approved pembrolizumab for treating mismatch repair-deficient (dMMR)/MSI-high (MSI-H) advanced solid tumors.1 2 Most frequently observed in colorectal and endometrial carcinomas, MSI has long been utilized to screen for Lynch syndrome and was also demonstrated to have prognostic relevance since patients with stage II colorectal cancer (CRC) with an MSI-H phenotype have a favorable prognosis and may not benefit from 5-fluorouracil-based adjuvant therapy.3–6 Given its clinical implications, MSI/MMR testing is nowadays recommended by the National Comprehensive Cancer Network (NCCN) guidelines for multiple cancer types.7–9
At present, MSI/MMR status is most commonly determined using immunohistochemistry (IHC) or polymerase chain reaction (PCR). IHC involves staining for the MMR proteins (MLH1, MSH2, MSH6, and PMS2) while PCR analyzes five to seven highly conserved microsatellite markers.10 11 However, IHC and PCR only correlate in approximately 90% of cases since an isolated loss of MMR protein expression may not lead to elevated microsatellite alterations and conversely, positive staining may result from retained antigenicity of non-functional MMR proteins and thus does not necessarily imply a microsatellite stable (MSS) status.12–14 Moreover, there exists significant inter-laboratory and intra-laboratory variability since the precision of IHC relies heavily on the antibodies used and the pathologists’ expertise.15 PCR-based detection may also have limited sensitivity in certain cancer types because most panels only cover a handful of MSI loci and were primarily developed for CRC.16
Over the last decade, a number of next generation sequencing (NGS)-based algorithms have been developed to characterize MSI from tissue samples.17–21 Nevertheless, their widespread application face the challenge of tissue availability, especially for patients with metastatic disease, where invasive tissue procurement is strongly contraindicated. In this regard, blood-based testing represents a viable alternative. To date, several blood-based MSI (bMSI) detection methods have been proposed, but each had some caveats.22–24 Primarily developed in early stage CRC, the bMSISEA (blood MSI signature enrichment analysis) algorithm only incorporated eight microsatellite markers and was never evaluated for its ability to guide immunotherapy. Georgiadis et al23 also only included five markers from the pentaplex panel and they arbitrarily used the tissue-derived MSI-H cut-off value to define bMSI-H. Willis et al22 used simulated data or contrived samples for both model construction and analytical validation, rendering the clinical applicability of their model questionable. Neither did they include patients with blood microsatellite stable (bMSS) in the clinical validation cohort for between-arm comparison.
In this study, we sought to develop a bMSI detection strategy where both panel design and validation were performed using clinical blood samples. The predictive value of bMSI in response to immune checkpoint blockade (ICB) therapy was evaluated among 60 patients with advanced gastrointestinal cancer. Blood tumor mutational burden (bTMB) was also assessed as a complementary approach to inform ICB treatment. The pan-cancer landscape of bMSI was interrogated across 18 cancer types comprizing 5138 solid tumors to gain more insight into the population that may potentially benefit from bMSI testing and hence ICB treatment.
Methods
Samples and patients
Microsatellite loci were initially selected according to the whole exome sequencing (WES) data of 10 advanced CRC blood samples. Algorithm development was conducted using blood samples from 20 MSI-H and 100 MSS patients with gastrointestinal cancer (figure 1). Technical validation was performed in an independent cohort comprizing blood samples from 40 MSI-H and 209 MSS patients with advanced gastrointestinal cancer. For both algorithm development and technical validation, samples were obtained from Peking University Cancer Hospital Biobank. They were eligible if they had a confirmed pathological diagnosis of gastrointestinal cancer and available orthogonal tissue MSI (tMSI) results as characterized by PCR of the pentaplex panel.25 The MSI-H cell line SW48 and the MSS cell line HT55 were used to determine the limit of detection (LOD).26 Patients with advanced gastrointestinal cancer who received ICB treatment at Peking University Cancer Hospital from February 2016 until January 2020 were retrospectively enrolled for clinical validation. Patients were eligible if baseline blood samples were collected for circulating tumor DNA (ctDNA) sequencing within 2 weeks prior to ICB treatment using a panel targeting the 100 selected MSI loci and the exons of 150 cancer-related genes.27 The pan-cancer prevalence of bMSI-H was assessed among 5138 advanced tumors of 18 cancer types that were subjected to bMSI characterization using the same panel as part of routine clinical care (online supplemental table S1).
Figure 1.
A diagram illustrating the study design and samples used for each step. bMSI-H, blood-based MSI high; MS, microsatellite; MSI-H, microsatellite instability high; MSS, microsatellite stable; PD-1, programmed cell death protein-1; PD-L1, programmed death ligand 1.
jitc-2020-001297supp001.pdf (344.3KB, pdf)
Cell-free DNA preparation and NGS
DNA isolation, library preparation, targeted hybrid capture and sequencing were performed as previously described.27 Briefly, blood sample was centrifuged at 1600 g for 20 min at room temperature. Plasma was transferred to a new microcentrifuge tube, followed by centrifugation for 10 min at 16 000 g at 4°C to remove the residual cells and debris. DNA was isolated using the QIAamp Circulating Nucleic Acid Kit (Qiagen) and quantified using the Qubit dsDNA HS Assay Kit (Life Technologies). Library was prepared with 30 to 60 ng of cell-free DNA using the Accel-NGS 2S Plus DNA Library Kit (SWIFT) and the DNA fragments were tagged with unique molecular identifiers to reduce background noise. Hybrid capture was conducted using the xGen Exome Research Panel V.2 (Integrated DNA Technologies) with a custom panel covering the whole exon regions of 150 selected cancer-related genes and 100 selected MSI loci.27 Hybrid-captured libraries were loaded onto the NextSeq 500 (Illumina) for 75 bp paired-end sequencing.
bMSI detection by NGS
Sequencing reads were mapped against the human reference genome (hg19/GRCh37) with BWA V.0.7.12 and SAMtools V.1.3. Duplicate reads were removed using Picard V.1.130. Reads that were successfully mapped to each of the 100 loci were extracted from the de-duplicated BAM file. An in-house developed R script was employed to evaluate the distribution of read counts among various repeat length for each microsatellite locus of each sample. The model for determining the stability of each locus is described in detail in the Results section. A bMSI score was defined as the percentage of unstable loci. Any sample with a bMSI score of ≥0.2 was classified as bMSI-H, and otherwise bMSS.
bTMB determination by NGS
Sequence alignment and indel calling were carried out as described above. Blood TMB was defined as the total number of somatic single nucleotide variants and indels in the coding regions examined, including missense, silent, stop gain, stop loss, in-frame and frameshift mutations. Any variant with an allele frequency of more than 30% was suspected to be germline variant and thus was excluded from bTMB calculation. Any tumor with a bTMB of >13 was classified as bTMB-high (bTMB-H). The cut-off value was trained using the overall survival (OS) data of the clinical validation cohort (online supplemental figure S1).
ICB efficacy evaluation
Patients’ demographics and clinical outcome data were extracted from their medical records in a de-identified manner by two independent physicians and was reviewed by a third physician in case of discrepancy. Objective response rate (ORR) was assessed as per the Response Evaluation Criteria in Solid Tumors (RECIST), V.1.1. Progression-free survival (PFS) was defined as the time from the onset of ICB treatment to disease progression or death by any cause and OS was defined as the time from the onset of ICB treatment to death by any cause.
All samples were obtained with informed consent for research.
Statistical analyses
The difference in continuous variables between two groups were examined by the two-tailed unpaired t-test for normally distributed variables or Mann-Whitney U test for non-normally distributed variables. χ2 test or Fisher’s exact test was used to test the differences in categorical variables between two groups. The 95% CI for ORR was estimated using the Clopper and Pearson method. Kaplan-Meier curves of OS and PFS were compared by log-rank test. The HR was examined with a Cox proportional hazards regression model. All tests were two-sided. A p value of <0.05 was considered significant. Statistical analyzes were performed using GraphPad Prism V.7.01 (GraphPad Software Inc) and R software, V.3.6.1 (R Foundation for Statistical Computing).
Results
Algorithm development for bMSI detection
Five hundred loci with high capture efficiency were initially included based on the WES data of 10 CRC blood samples and were ranked by their susceptibility to instability (ie, sensitivity) in a cohort of blood samples from 20 tissue MSI-H (tMSI-H) and 100 tissue MSS (tMSS) gastrointestinal tumors while a high level of specificity was maintained. The top 100 loci were eventually selected for bMSI determination.
Locus-specific and sample-level stability thresholds were then determined using the same 20 tMSI-H and 100 tMSS blood samples mentioned above. The repeat lengths of each locus were first extracted using the microsatellite loci BED file, the BAM file and an in-house python script. The proportion of each repeat length among all reads for a specific locus was then calculated for all samples of the MSI-H subtype and the MSS subtype. The percentages for samples within each subtype were then averaged to obtain the cumulative percentage for that repeat length for each subtype. The repeat length (Ci) exhibiting the greatest difference in cumulative percentage between the MSI-H and the MSS subtypes was selected as the cut-off value to decide whether a sequencing read is unstable (if Lr≤Ci; Lr is the repeat length of a read) or stable (if Lr>Ci) for that locus. Subsequently, a locus’s stability could be assessed using a binomial probability model shown as follows,
where i is the locus being examined, pi stands for the cumulative percentage at the cut-point repeat length (Ci) of the MSS subtype, ni denotes the number of unstable reads, and Ni represents the total number of reads for that locus. A locus was considered unstable if the probability of P (X≥ni) was ≤0.001. For each sample, a bMSI score was calculated as the fraction of unstable loci in the 100 selected loci. To aim for a specificity of at least 95% according to pairwise comparison with tissue PCR results, the threshold for defining MSI-H was set to 0.2 (figure 2). Since variants with an allele frequency below 0.3% were undetectable according to our previous work, blood samples with a maximum somatic allele frequency (MSAF) of <0.3% were excluded from subsequent analyzes to reduce the risk of false negatives.28 Indeed, a preliminary analysis on blood samples from 24 tMSI-H patients revealed that those with an MSAF of <0.3% had a median bMSI score well below 0.2 (online supplemental figure S2).
Figure 2.

Cut-off value determination to define the blood microsatellite instability-high (bMSI-H) status. For each sample, a bMSI score was calculated as the fraction of unstable loci in the 100 selected loci. To aim for a specificity of at least 95% according to pairwise comparison with tissue PCR results, the threshold for defining MSI-H was determined to be 0.2 as shown by the dashed line. Red dots represent tissue PCR-defined MSI-H tumors, and blue dots represent tissue PCR-defined (microsatellite stable) MSS tumors.
Technical validation
In order to determine LOD of the bMSI assay, the genomic DNA of the MSI-H cell line SW48 was diluted in that of the MSS cell line HT55 to create a titration series consisting of 100%, 50%, 20%, 8%, 4%, 2%, 1%, 0.5%, and 0.25%, with triplicates for each titration point. Simulated bMSI score was plotted against titration gradient to generate a curve at a DNA input of 30 ng (online supplemental figure S3). The concentration of the MSI-H DNA had to be at least 0.5% in order for the sample’s bMSI score to reach 0.2, that is, to allow for MSI-H detection. Therefore, the LOD was determined to be 0.5% at 30 ng input.
To evaluate the performance of the assay, blood samples acquired from 40 tMSI-H and 209 tMSS patients with advanced gastrointestinal cancer were analyzed in reference with tissue PCR results. Our bMSI method detected 33 patients with tMSI-H and 201 patients with tMSS, yielding a sensitivity of 82.5% (33/40, 95% CI 70.2% to 94.8%), a specificity of 96.2% (201/209, 95% CI 93.5% to 98.8%) and an overall accuracy of 94.0% (234/249, 95% CI 91.0% to 97.0%) (table 1). By reviewing the patients’ records, one false positive and two false negative cases were discovered to have multiple lesions, and one false negative had blood sample collected over 1 year after tissue procurement, which could have contributed to tissue-blood discrepancy.
Table 1.
The concordance between next generation sequencing (NGS) blood microsatelliteinstability (bMSI) with PCR tissue MSI (tMSI)
| PCR tMSI | Total | |||
| + | – | |||
| NGS bMSI | + | 33 | 8 | 41 |
| – | 7 | 201 | 208 | |
| Total | 40 | 209 | 249 | |
| Sensitivity=82.5% (33/40, 95% CI 70.2% to 94.8%) | ||||
| Specificity=96.2% (201/209, 95% CI 93.5% to 98.8%) | ||||
| PPV=80.5% (33/41, 95% CI 67.8% to 93.2%) | ||||
| NPV=96.6% (201/208, 95% CI 94.2% to 99.1%) | ||||
| Concordance=94.0% (234/249, 95% CI 91.0% to 97.0%) | ||||
NPV, negative predictive value; PPV, positive predictive value.
Clinical validation
Since tMSI may guide patient selection for ICB therapy, we set out to investigate whether pretreatment bMSI status has the same effect. Sixty patients with advanced gastrointestinal cancer who received ICB treatment between February 2016 and January 2020 were retrospectively enrolled (online supplemental figure S4). They were all subjected to ctDNA sequencing using the targeted 150-gene panel described above within 2 weeks before the onset of ICB treatment. The median duration of treatment was 3.53 months and the median duration of follow-up was 13.47 months. Of the 60 patients, 35 had CRC and 22 had gastric cancer (online supplemental table S2). The majority of the patients (56/60) had received at least one line of treatment before ICB. The ICB administered included both anti-programmed cell death protein-1 (PD-1) (55%) and anti-programmed death ligand 1 (PD-L1) (45%). According to our bMSI algorithm, 31 of the patients were classified as bMSI-H and 29 as bMSS. There were no statistically significant differences between the two groups in most of the baseline characteristics, except for a significantly higher lactate dehydrogenase (LDH) level among the bMSI-H tumors. Increased LDH level is known as a negative prognostic marker in many solid tumors and is also associated with resistance to ICB in advanced non-small cell lung cancer and metastatic melanoma.29–31 Despite the potential interference from an elevated LDH level, the bMSI-H patients exhibited a significantly higher ORR than the MSS patients (38.71% vs 6.90%, p=0.005) (figure 3). Notably, 13 patients of the bMSI-H subgroup experienced disease progression, consistent with the fact that 10 of them received ICB in the third line or above (figure 3A). Two of the patients with bMSS displayed partial response, one of which was a case of gastric cancer with a 40% PD-L1 expression which strongly predicts susceptibility to ICB (figure 3B). Overall, the bMSI-H subgroup had significantly prolonged PFS (5.57 months vs 2.03 months) (HR, 0.431; 95% CI 0.236 to 0.787, p=0.005) and OS (20.03 months vs 10.07 months) (HR, 0.489; 95% CI 0.249 to 0.961, p=0.034) compared with the patients with bMSS (figure 4).
Figure 3.
Best response of target lesions from the initiation of immune checkpoint blockade (ICB) treatment in the bMSI-H (A) and bMSS (B) subgroups. The upper and lower dashed lines mark progressive disease and partial response according to RECIST V.1.1, respectively. bMSI-H, blood-based microsatellite instability high; bMSS, blood microsatellite stable;RECIST, ResponseEvaluation Criteria in Solid Tumors.
Figure 4.
Kaplan-Meier analyzes of progression-free survival (PFS) (A) and overall survival (OS) (B) in the patients with bMSI-H versus bMSS. A cohort of 60 patients with advanced gastrointestinal cancer were stratified based on their pretreatment bMSI statuses and the Kaplan-Meier curves for PFS and OS were compared between the bMSI-H and the bMSS subgroups. The tick marks indicate censored data. bMSI-H, blood-based microsatelliteinstability high; bMSS, blood microsatellite stable.
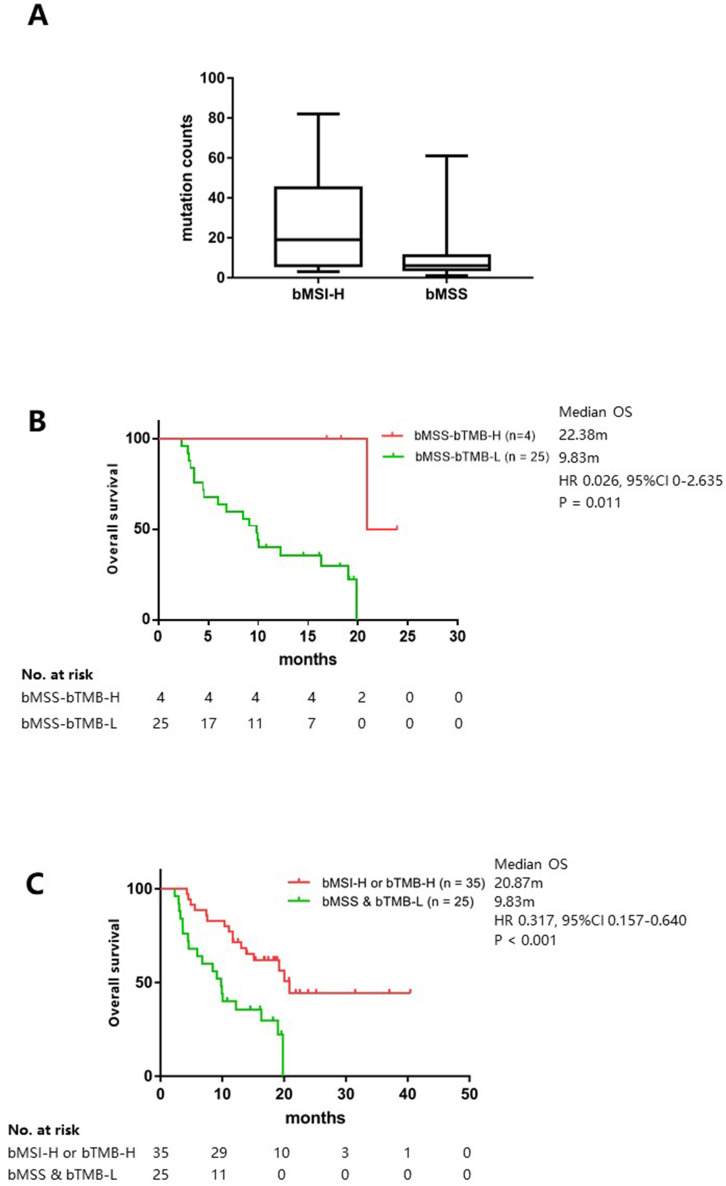
In view of recent publication showing that MSS CRC could be further dichotomized into TMB-H and TMB-L subsets in tissue, we were interested to see if it was the case with patients with bMSS.32 Using the same 150-gene panel for bTMB estimation, which was fully validated previously, the bMSI-H group presented a significantly higher bTMB load than the bMSS group (19 vs 6, p=0.001) (figure 5A).27 Intriguingly, 4 of the 29 bMSS tumors were identified to be bTMB-H according to a cut-off of 13 as trained using the OS data of the ICB treatment cohort and they trended towards a better survival than the bMSS-bTMB-L subset (22.38 months vs 9.83 months) (HR 0.026, 95% CI 0 to 2.635, p=0.011) (online supplemental table S2, figure 5B). These four patients combined with the bMSI-H group collectively displayed significantly improved survival (20.87 months vs 9.83 months) (HR 0.317, 95% CI 0.157 to 0.640, p<0.001) over the bMSS-bTMB-L patients, indicating that patients are able to benefit from ICB treatment as long as one of the two biomarkers predicts response (figure 5C).
Figure 5.
Blood tumor mutational burden (bTMB) as a complement to bMSI to inform ICB treatment. (A) The bTMB levels in the bMSI-H versus the bMSS groups; (B) Kaplan-Meier analysis of OS in the patients with bMSS-TMB-H versus bMSS-TMB-L; (C) Kaplan-Meier analysis of OS in the bMSI-H or bMSI-H versus the bMSS-bTMB-L patients. A cohort of 60 patients with advanced gastrointestinal cancer were stratified based on pretreatment bMSI and bTMB statuses, and those classified as bMSI-H-bTMB-L, bMSS-bTMB-H, or bMSI-H-bTMB-H were pooled as one group in (C). The tick marks indicate censored data. bMSI-H, blood-based microsatellite instability high; bMSS, blood microsatellite stable;H, high; ICB, immune checkpoint blockade; L, low; OS, overallsurvival.
Prevalence of bMSI-H in pan-cancer
Although pembrolizumab was approved for treating MSI-H solid tumors years ago, the rate of MSI testing outside of gastrointestinal cancer remained scarce. In order to better understand the clinical impact MSI testing could have on other malignancies, we investigated the bMSI landscape across 18 cancer types comprizing 5318 advanced tumors that had undergone ctDNA sequencing using the 150-gene panel. The majority of the patients had advanced stage disease (30.5% with stage III and 54.4% with stage IV). Eighty patients were identified to be bMSI-H, equivalent of a pan-cancer prevalence of 1.6% (figure 6). The relative prevalence of bMSI-H across cancer types was in general consistent with that shown for tumor tissue, with endometrial, colorectal, and gastric cancers exhibiting the highest prevalence and other tumors such as breast, kidney and urothelial cancers on the lower end. However, prostate cancer was distinguished from other urinary tumors and ranked second to endometrial cancer with a prevalence of 4%.21 It was also noteworthy that bMSI-H were detected at much lower rates in endometrial (5.7%), colorectal (3.3%) and gastric (3.1%) cancers than those estimated for tissue MSI.3 21 33–35
Figure 6.
Prevalence of bMSI across 18 cancer types comprizing 5138 solid tumors. bMSI, blood-based microsatellite instability; GIST, gastrointestinal stromal tumor; NSCLC, non-small cell lung cancer.
Discussion
In the present study, we developed and validated an NGS-based bMSI detection strategy using clinical blood samples. The method exhibited high overall concordance with tissue PCR and proved effective in predicting response to ICB therapy among advanced gastrointestinal tumors. The panel also allowed for simultaneous determination of bTMB which, in combination with bMSI, may maximize patient access to ICB treatment. Furthermore, this study represents the first effort to uncover the pan-cancer MSI landscape in ctDNA among Asian patients.
While liquid biopsy becomes increasingly adopted to profile biomarkers in cancer diagnosis, some argue that it is still in early development. The main challenges associated with MSI detection from ctDNA include panel design and low tumor cell fraction in the blood.36 Loci that cannot be effectively captured or mapped or those substantially variable in MSS tumors (ie, less specific) are considered uninformative and therefore should be excluded from panel design.22 In our study, each locus was selected based on a comprehensive assessment of its capture efficiency and sensitivity/specificity. The thresholds for determining locus-specific and sample-level stability were also trained using clinical blood samples to ensure high reliability. It is true that tumor-derived DNA can sometimes be present at an extremely low fraction in cell-free DNA but this problem could be readily resolved by implementing quality control to exclude blood samples with an MSAF of <0.3%.
Apart from loci selection and tumor fraction, sample-related factors may also affect bMSI detection. Among the 15 tissue-blood discordant cases, three had multiple lesions while one had an extended interval between tissue and blood collection. Although there was not enough sample left for confirmatory tests, it was not irrational to speculate that intralesional and interlesional heterogeneity may have contributed to tissue-blood inconsistency, especially when the vast majority of the validation population were previously treated advanced patients. Previous literature showed that MSI-H and MSS cell populations may co-exist within the same MSI-H tumor and patients burdened with multiple tumors could also have discrete genotypes in different lesions, both of which are expected to be better captured by liquid biopsy.37–41 Temporal discordance may also have played a role since a previous study showed that tissue-blood concordance dropped from 100% to 60% as the time interval between tissue biopsy and blood draw increased from ≤2 weeks to >6 months.42
The feasibility of predicting ICB efficacy by NGS-determined bMSI has been explored previously. As Willis et al22 reported, 16 patients with pretreated MSI-H metastatic gastric cancer achieved an ORR of 63% after ICB monotherapy. In Georgiadis et al’s ICB-treated pan-cancer cohort, the bMSI-H subgroup displayed a median PFS of 16.2 months and a median OS of 16.3 months, which were significantly and numerically prolonged compared with those of the bMSS subset, respectively.23 In comparison, the bMSI-H patients in our validation cohort showed an ORR of 38.71% and a median PFS of 5.57 months, which we feel are in better agreement with the 39.6% ORR and the 4.1-month median PFS reported previously for patients with pretreated gastrointestinal cancer.43–45
Given the precedent of patients with tMSS being diagnosed as TMB-H and responding to ICB therapy, it would be desirable to examine bTMB in parallel instead of relying solely on bMSI for ICB efficacy prediction.32 Indeed, our study took these findings forward by demonstrating in blood that bMSS patients could be further dichotomized into bTMB-H and bTMB-L subsets. Although the survival benefit of the bTMB-H subset was not statistically significant due to the small number of bTMB-H patients, the trend was prominent. Moreover, the bMSS-TMB-H and the bMSI-H groups collectively predicted significantly improved outcome, indicating that bMSI combined with bTMB may maximize the scope of ICB therapy. Therefore, the information regarding bTMB as provided by our method will serve as a valuable complement to ICB efficacy prediction by bMSI.
Additionally, improving patient access to ICB therapy also involves identifying patients potentially benefiting from ICB in tumor types where MSI is rarely tested. Even though the 4% bMSI-H rate in prostate cancer was only marginally higher than the 3.1% to 3.7% seen in previous tissue and blood analysis, its relative prevalence compared with other cancer types was striking.46 47 This was not completely unprecedented as Willis et al observed a similar trend for prostate cancer in their pan-cancer analysis.22 A possible explanation could be that patients with castration-resistant prostate cancer (CRPC) were over-represented in our cohort, and MSI-H was reported to be found at a higher rate in CRPC (4.5%) than in hormone-sensitive prostate cancer (2.4%).46 The lower than expected prevalence of bMSI-H in endometrial, colorectal and gastric cancers could have resulted from a much larger proportion of advanced patients in our pan-cancer cohort than that included in tissue-based studies, since MSI-H is observed more frequently in early stage cancers.48
A limitation of our study is that both the development and validation were performed using gastrointestinal cancer samples, although the algorithm was intended to be used in a pan-cancer setting. Therefore, the applicability of the bMSI algorithm described herein needs to be further validated in other cancer types. In addition, the clinical validation was conducted retrospectively in a small population. Prospective trials with larger sample sizes will be warranted to confirm these findings. Orthogonal tMSI results would also be valuable for a concordance analysis between tMSI-predicted and bMSI-predicted ICB efficacy. Taken together, we have provided a reliable NGS-based bMSI detection strategy, which in combination with a panel that allows for concurrent profiling of bTMB may better inform ICB treatment.
Acknowledgments
The authors would like to acknowledge all study coordinators and operation staff at both Peking University Cancer Hospital and 3D Medicines Inc who supported this study.
Footnotes
ZW and XZ contributed equally.
Funding: This work was supported by the National Key Research and Development Program of China (NO. 2016YFC0905302).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The study protocol was approved by the institutional review board of Beijing Cancer Hospital (Approval ID: 2018KT04).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Le DT, Le DT, Diaz LA, Jr. Pd-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;373:1979. 10.1056/NEJMc1510353 [DOI] [PubMed] [Google Scholar]
- 2.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland CR, Thibodeau SN, Hamilton SR, et al. A national cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–57. [PubMed] [Google Scholar]
- 4.Warthin AS. Classics in oncology. heredity with reference to carcinoma as shown by the study of the cases examined in the pathological laboratory of the University of Michigan, 1895-1913. by Aldred Scott Warthin. 1913. CA Cancer J Clin 1985;35:348–59. 10.3322/canjclin.35.6.348 [DOI] [PubMed] [Google Scholar]
- 5.Lynch HT, Shaw MW, Magnuson CW, et al. Hereditary factors in cancer. study of two large midwestern kindreds. Arch Intern Med 1966;117:206–12. [PubMed] [Google Scholar]
- 6.Benson AB, Arnoletti JP, Bekaii-Saab T, et al. Colon cancer. J Natl Compr Canc Netw 2011;9:1238–90. 10.6004/jnccn.2011.0104 [DOI] [PubMed] [Google Scholar]
- 7.Benson AB, Venook AP, Al-Hawary MM, et al. Small bowel adenocarcinoma, version 1.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2019;17:1109–33. 10.6004/jnccn.2019.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koh W-J, Abu-Rustum NR, Bean S, et al. Cervical cancer, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2019;17:64–84. 10.6004/jnccn.2019.0001 [DOI] [PubMed] [Google Scholar]
- 9.Network NCC Gastric cancer, version 5.2017, NCCN clinical practice guidelines in oncology. Available: https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf
- 10.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004;96:261–8. 10.1093/jnci/djh034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goel A, Nagasaka T, Hamelin R, et al. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS One 2010;5:e9393. 10.1371/journal.pone.0009393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M-L, Chen J-Y, Hu J, et al. Comparison of microsatellite status detection methods in colorectal carcinoma. Int J Clin Exp Pathol 2018;11:1431–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley JC, Lin M-T, Le DT, et al. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res 2016;22:813–20. 10.1158/1078-0432.CCR-15-1678 [DOI] [PubMed] [Google Scholar]
- 14.Evrard C, Tachon G, Randrian V, et al. Microsatellite instability: diagnosis, heterogeneity, discordance, and clinical impact in colorectal cancer. Cancers 2019;11. 10.3390/cancers11101567. [Epub ahead of print: 15 10 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen R, Hain E, Buhard O, et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol 2019;5:551–5. 10.1001/jamaoncol.2018.4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Soong TD, Elemento O. A novel approach for characterizing microsatellite instability in cancer cells. PLoS One 2013;8:e63056. 10.1371/journal.pone.0063056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kautto EA, Bonneville R, Miya J, et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget 2017;8:7452–63. 10.18632/oncotarget.13918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6. 10.1093/bioinformatics/btt755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salipante SJ, Scroggins SM, Hampel HL, et al. Microsatellite instability detection by next generation sequencing. Clin Chem 2014;60:1192–9. 10.1373/clinchem.2014.223677 [DOI] [PubMed] [Google Scholar]
- 20.Zhu L, Huang Y, Fang X, et al. A novel and reliable method to detect microsatellite instability in colorectal cancer by next-generation sequencing. J Mol Diagn 2018;20:225–31. 10.1016/j.jmoldx.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 21.Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342–50. 10.1038/nm.4191 [DOI] [PubMed] [Google Scholar]
- 22.Willis J, Lefterova MI, Artyomenko A, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 2019;25:7035–45. 10.1158/1078-0432.CCR-19-1324 [DOI] [PubMed] [Google Scholar]
- 23.Georgiadis A, Durham JN, Keefer LA, et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin Cancer Res 2019;25:7024–34. 10.1158/1078-0432.CCR-19-1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Z, Wang Z, Liu C, et al. Detection of Microsatellite Instability from Circulating Tumor DNA by Targeted Deep Sequencing. J Mol Diagn 2020;22:860–70. 10.1016/j.jmoldx.2020.04.210 [DOI] [PubMed] [Google Scholar]
- 25.Du C, Zhao J, Xue W, et al. Prognostic value of microsatellite instability in sporadic locally advanced rectal cancer following neoadjuvant radiotherapy. Histopathology 2013;62:723–30. 10.1111/his.12069 [DOI] [PubMed] [Google Scholar]
- 26.Gaffney EV, Tsai SC, Dell'Aquila ML, et al. Production of growth-inhibitory activity in serum-free medium by human monocytic leukemia cells. Cancer Res 1983;43:3668–73. [PubMed] [Google Scholar]
- 27.Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 2019;5:696–702. 10.1001/jamaoncol.2018.7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang N, Li Y, Liu Z, et al. The characteristics of ctDNA reveal the high complexity in matching the corresponding tumor tissues. BMC Cancer 2018;18:319. 10.1186/s12885-018-4199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mezquita L, Auclin E, Ferrara R, et al. Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non-small cell lung cancer. JAMA Oncol 2018;4:351–7. 10.1001/jamaoncol.2017.4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buder-Bakhaya K, Hassel JC. Biomarkers for clinical benefit of immune checkpoint inhibitor Treatment-A review from the melanoma perspective and beyond. Front Immunol 2018;9:1474. 10.3389/fimmu.2018.01474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrelli F, Cabiddu M, Coinu A, et al. Prognostic role of lactate dehydrogenase in solid tumors: a systematic review and meta-analysis of 76 studies. Acta Oncol 2015;54:961–70. 10.3109/0284186X.2015.1043026 [DOI] [PubMed] [Google Scholar]
- 32.Goodman AM, Sokol ES, Frampton GM, et al. Microsatellite-Stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol Res 2019;7:1570–3. 10.1158/2326-6066.CIR-19-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmerman S, Van Rompuy AS, Van Gorp T, et al. Analysis of 108 patients with endometrial carcinoma using the promise classification and additional genetic analyses for MMR-D. Gynecol Oncol 2020;157:245–51. 10.1016/j.ygyno.2020.01.019 [DOI] [PubMed] [Google Scholar]
- 34.Cancer genome atlas research N. comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361–75. 10.1038/s41575-019-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Ajani JA. Ushering in liquid biopsy for the microsatellite status: advantages and caveats. Clin Cancer Res 2019;25:6887–9. 10.1158/1078-0432.CCR-19-2585 [DOI] [PubMed] [Google Scholar]
- 37.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. 10.1038/s41591-018-0101-z [DOI] [PubMed] [Google Scholar]
- 38.Goyal L, Saha SK, Liu LY, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov 2017;7:252–63. 10.1158/2159-8290.CD-16-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pectasides E, Stachler MD, Derks S, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov 2018;8:37–48. 10.1158/2159-8290.CD-17-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tachon G, Frouin E, Karayan-Tapon L, et al. Heterogeneity of mismatch repair defect in colorectal cancer and its implications in clinical practice. Eur J Cancer 2018;95:112–6. 10.1016/j.ejca.2018.01.087 [DOI] [PubMed] [Google Scholar]
- 41.He W-Z, Hu W-M, Wang F, et al. Comparison of mismatch repair status between primary and matched metastatic sites in patients with colorectal cancer. J Natl Compr Canc Netw 2019;17:1174–83. 10.6004/jnccn.2019.7308 [DOI] [PubMed] [Google Scholar]
- 42.Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016;22:5772–82. 10.1158/1078-0432.CCR-16-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcus L, Lemery SJ, Keegan P, et al. FDA approval summary: pembrolizumab for the treatment of microsatellite Instability-High solid tumors. Clin Cancer Res 2019;25:3753–8. 10.1158/1078-0432.CCR-18-4070 [DOI] [PubMed] [Google Scholar]
- 44.Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with Noncolorectal high microsatellite Instability/Mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 2020;38:1–10. 10.1200/JCO.19.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le DT, Kim TW, Van Cutsem E, et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite Instability-High/Mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J Clin Oncol 2020;38:11–19. 10.1200/JCO.19.02107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abida W, Cheng ML, Armenia J, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol 2019;5:471–8. 10.1001/jamaoncol.2018.5801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraune C, Simon R, Höflmayer D, et al. High homogeneity of mismatch repair deficiency in advanced prostate cancer. Virchows Arch 2020;476:745–52. 10.1007/s00428-019-02701-x [DOI] [PubMed] [Google Scholar]
- 48.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003;349:247–57. 10.1056/NEJMoa022289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001297supp001.pdf (344.3KB, pdf)