Graphical abstract
List of Abbreviations: CHMs, Chinese herb medicines; TCM, Traditional Chinese medicine; ACE2, Angiotensin-converting enzyme II; RBD, Receptor-binding domain; SPR, Surface plasmon resonance; PPI, Protein-protein interaction; KD, Equilibrium dissociation constants
Keywords: COVID-19, SARS-CoV-2, Chinese herb medicine (CHM), ACE2, Network pharmacology, Surface plasmon resonance (SPR)
Highlights
-
•
Three CHM components with the potential against COVID-19 are identified using TCMSP.
-
•
SPR assay result shows good binding affinity of puerarin and quercetin to ACE2.
-
•
Puerarin and quercetin impairs the binding of viral S-protein to ACE2 receptor.
-
•
Quercetin can also directly bind to S-protein to exert an viral neutralizing effect.
-
•
Results from this study propose a prompt application of puerarin on COVID-19 patients.
Abstract
The outbreak of COVID-19 raises an urgent need for the therapeutics to contain the emerging pandemic. However, no effective treatment has been found for SARS-CoV-2 infection to date. Here, we identified puerarin (PubChem CID: 5281807), quercetin (PubChem CID: 5280343) and kaempferol (PubChem CID: 5280863) as potential compounds with binding activity to ACE2 by using Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP). Molecular docking analysis showed that puerarin and quercetin exhibit good binding affinity to ACE2, which was validated by surface plasmon resonance (SPR) assay. Furthermore, SPR-based competition assay revealed that puerarin and quercetin could significantly affect the binding of viral S-protein to ACE2 receptor. Notably, quercetin could also bind to the RBD domain of S-protein, suggesting not only a receptor blocking, but also a virus neutralizing effect of quercetin on SARS-CoV-2. The results from network pharmacology and bioinformatics analysis support a view that quercetin is involved in host immunomodulation, which further renders it a promising candidate against COVID-19. Moreover, given that puerarin is already an existing drug, results from this study not only provide insight into its action mechanism, but also propose a prompt application of it on COVID-19 patients for assessing its clinical feasibility.
1. Introduction
The emergence of a novel and highly pathogenic coronavirus SARS-CoV-2 caused an outbreak of acute infectious pneumonia in Wuhan, China, in late 2019. At the early stage of infection, patients generally present with flu-like clinical manifestations, such as fatigue, fever and dry cough, with a characteristic ground-glass opacity lesion of lung in CT findings. Thereafter, as the disease progresses, in severe cases, patients may show dyspnea, respiratory distress syndrome, septic shock and even death [1], [2]. Recent phylogenetic analysis result showed that SARS-CoV-2 virus belongs to Beta coronavirus, which is an enveloped, single-stranded RNA virus with the ability to infect animals and humans, causing sporadic zoonotic outbreaks and occasional epidemics [3]. SARS-CoV-2 shares a common bat coronavirus ancestor with the human-infecting SARS-CoV, and its S-protein shows high structural homology to that of SARS-CoV in the receptor-binding domain (RBD) that mediates the interaction with the host receptor [3], [4]. So far, the known human receptor of beta coronaviruses includes angiotensin-converting enzyme 2 (ACE2) for SARS-CoV and dipeptidyl peptidase-4 (DPP4) for MERS-CoV [5], [6]. Recent studies have demonstrated that ACE2 is the human receptor for SARS-CoV-2, and the cell entry of SARS-CoV-2 depends on ACE2, suggesting that ACE2-targeting strategy holds great promise for the drug discovery against COVID-19 [7], [8], [9], [10].
Traditional Chinese medicine (TCM) has been applied on protection against plagues in China since ancient times. As an integral part of TCM, Chinese herb medicine (CHM) shows a unique therapeutic effect on various infectious diseases, including the SARS epidemic of 2003, owing to its holistic treatment concept and multi-component, multi-target pharmacological characteristics [11]. More recently, the combined use of TCM and modern Western medicine has benefited the COVID-19 patients with shorter hospitalization and improved symptoms [12]. However, due to the vast diversity of components in CHM and the complexity of the interaction between those components and the disease, it is still a considerable challenge to uncover the mysteries of CHMs at the molecular level, which to a great extent hampers the general acceptance of TCM worldwide. Recently, a series of computational methods under the umbrella of TCM network pharmacology have been devised, such as the network platform-based prediction of the active compounds from CHMs, which opens up a new path for unraveling the action mechanism of CHM and thereby greatly accelerate the process of CHM-based new drug discovery [13], [14], [15], [16], [17].
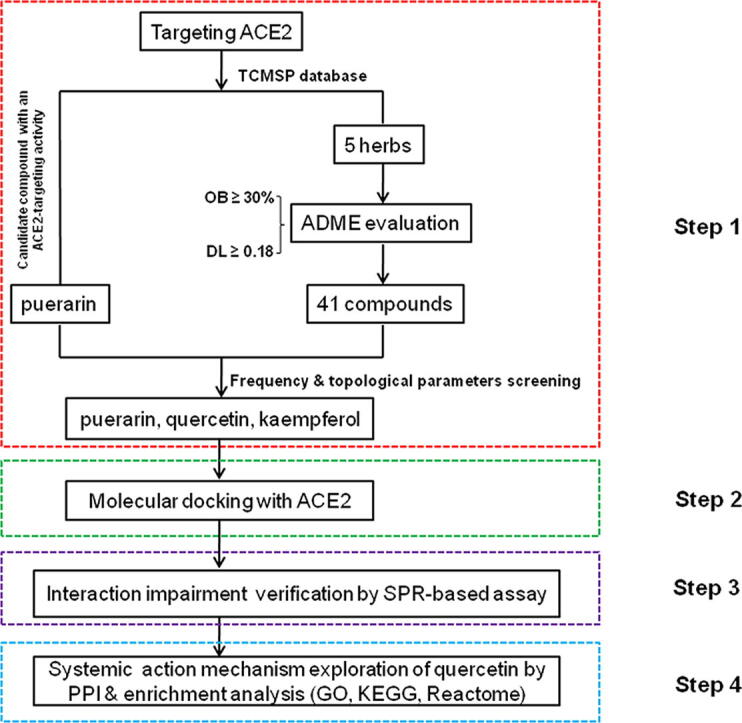
In this study, we first screened out puerarin as the candidate compound targeting human ACE2 from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database, and identified the potential effective herbs containing puerarin through TCMSP analysis platform. Next, according to the TCM theory that the compounds existing in the same CHM may have related and synergistic pharmacological activities [18], we screened all the active compounds contained in the above-obtained herbs and identified two other compounds with great anti-SARS-CoV-2 potential. The possible interaction of these compounds with ACE2 was further investigated by molecular docking and surface plasmon resonance assays, and the results support the view that puerarin and quercetin could significantly impair the binding of viral S-protein to its human ACE2 receptor, shedding light on CHM-based new drug discovery against COVID-19 (See workflow scheme in Fig. 1).
Fig. 1.
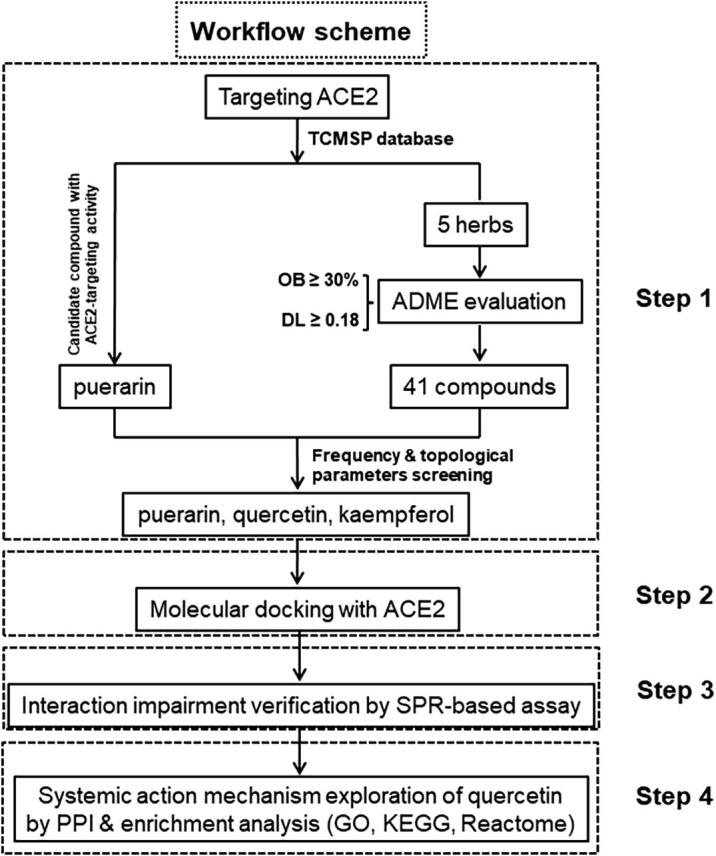
Workflow scheme. This work was composed of four main parts, including natural compound selection, molecular docking, SPR verification, and PPI network construction & enrichment analysis.
2. Material and methods
2.1. Screening of potential herbs and their active compounds targeting ACE2
To screen the potential CHMs and their compounds targeting ACE2, we input “angiotensin-converting enzyme 2″ to the search window of ”target name“ in TCMSP online platform. The parameters for selection of active compounds were set as oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 as standard.
2.2. Selection of the key active compounds against COVID-19
Statistical analysis of the overlapped compounds existing in the five individual herbs was presented in a Venn diagram. Prediction of potential targets for the selected compounds was performed using TCMSP database. Based on the selected candidate compounds and putative targets, a compound-target network was constructed by using Cytoscape. Then, the topological parameters of each candidate compounds in the network, including Degree, Betweenness Centrality and Clossness Centrality, were calculated by using a Cytoscape plugin CytoNCA.
2.3. Molecular docking analysis
Flexible docking process between chemical compounds and target proteins were conducted by the software AutoDock 4.2. A total of 30 docking conformations were extracted and ranked according to the docking energy value. The detailed docking process was performed as follows: (a) amino acids within 14 Å distance of the binding region on the receptor ACE2 were placed into the grid box for docking, and the different types of atoms were then used as probes to scan and calculate the grid energy, which was performed by AutoGrid program; (b) conformational searching for ligands within the box was performed by using Autodock program. The ultimately rankings were resulted according to the scorings based on the conformation, orientation, position and energy of the ligands. The top 1 conformation with the lowest binding energy was selected for further binding modes analysis.
2.4. Surface plasmon resonance assay
Surface plasmon resonance (SPR) analysis was conducted with Open SPR instrument (Nicoyalife, Canada). The COOH sensor chip was firstly installed on the OpenSPR instrument in accordance with the standard procedure. 1) Run the buffer at the maximum flow rate and exhaust the bubble after reaching the signal baseline. 2) Inject HCl (10 mM) to clean the chip surface and run for 1 min. 3) Slow down the flow rate of buffer solution (PBS) to 20 µL/min, then load 200 µL EDC (400 mM)/NHS (100 mM) (1:1) solution to activate COOH sensor chips and run for 4 min. 4) The ACE2 (40 µg/ml) and the S-protein (100 nM) were diluted with activation buffer (total 200 µL). 5) The injection port was rinsed with buffer solution and emptied with air. 6) Fill with 200 µL blocking solution (20 µL/min, 4 min), wash the sample ring with buffer solution and empty it with air. 7) Observe baseline for 5 min to ensure stability. Next, the selected compounds were diluted into a series of solutions with different concentration, which were then injected into the chip with the concentration from low to high. In each cycle, the sample (200 µL) was flowed through the chip for 7 mins at a constant flow rate of 20 µL/min (The binding time of the compounds and ACE2 was 240 s, and naturally dissociate for 180 s). After detection, 0.05% SDS was added as the regeneration buffer to dissociate the compounds from the target protein. The kinetic parameters of the binding reactions were calculated and analyzed by using Trace Drawer software (Ridgeview Instruments AB, The Kingdom of Sweden).
2.5. PPI network and module analysis
The PPI systematic network was constructed and visualized using Search Tool of Retrieval of Interacting Genes (STRING; version 11.0; https://stringdb.org) database and Cytoscape software 3.2.1, respectively. The proteins with a combined score > 0.7 were selected for PPI analysis. Molecular Complex Detection (MCODE) was used to screen the modules of the PPI network. The top modules were defined as having Degree cutoff > 5 and K-core > 5. The core subnetwork extraction from each of the parent PPI network was performed using the MCODE plugin with the same parameter settings as the above.
2.6. Screening of disease targets
The GeneCards database (http://www.genecards.org/) was used to acquire COVID-19 disease targeted genes. “Novel Coronavirus” was input as the keywords for searching, and the result was exported to an excel document. All the obtained targets were further confirmed by using Uniprot database.
2.7. Enrichment analysis
The functional and pathway enrichment analyses of the obtained putative and core targets were performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/). The analyses were mainly divided into two types, GO biological functional and KEGG signaling pathways. A p < 0.05 was considered as statistically significant. Meanwhile, WebGestalt (http://www.webgestalt.org) was also used as the enrichment method for quercetin and COVID-19 co-targeted over-representation analysis (ORA).
3. Results
3.1. Screening of potential CHMs and their active compounds targeting ACE2
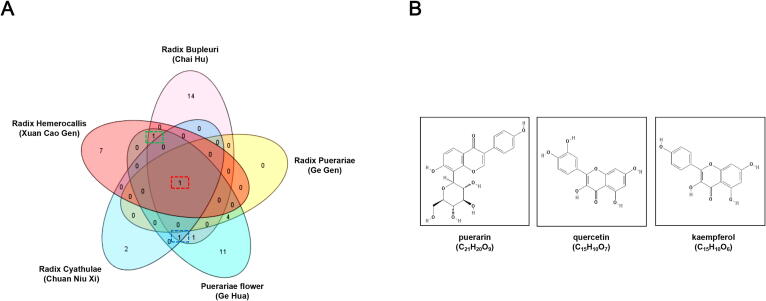
To screen the potential CHM compounds targeting human ACE2 receptor, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) was employed. From the above database, puerarin (PubChem CID: 5281807) was identified as the compound with potential binding activity to ACE2. Next, we screened out a total of five CHMs containing puerarin from the TCMSP database, namely Radix Bupleuri (Chinese name: Chaihu), Radix Puerariae (Chinese name: Gegen), Puerariae flower (Chinese name: Gehua), Radix Cyathulae (Chinese name: Chuanniuxi), and Radix Hemerocallis (Chinese name: Xuancaogen). According to the TCM theory, the compounds present in the same CHM generally have related and synergic pharmacological activities. We therefore expanded the screening range to all compounds contained in the five CHMs, with a screening criteria of oral bioavailability (OB) ≥ 30% and drug likeness (DL) ≥ 0.18 (Table 1). As such, a total of 41 compounds were obtained after removal of overlapped ones (Supplementary Table 1).
Table 1.
Specific information on the five chosen herbs and their potential active ingredients obtained from a virtual screening (TCMSP database).
| Name of herbs | MOL Number | Name of potential active ingredients | OB/% | DL |
|---|---|---|---|---|
| Radix Bupleuri(Chaihu) |
MOL004718 | α-spinasterol | 42.98 | 0.76 |
| MOL000354 | isorhamnetin | 49.60 | 0.31 | |
| MOL004644 | sainfuran | 79.91 | 0.23 | |
| MOL000490 | petunidin | 30.05 | 0.31 | |
| MOL000449 | stigmasterol | 43.83 | 0.76 | |
| MOL001645 | linoleyl acetate | 42.10 | 0.20 | |
| MOL013187 | cubebin | 57.13 | 0.64 | |
| MOL004624 | longikaurin A | 47.72 | 0.53 | |
| MOL000098 | quercetin | 46.43 | 0.28 | |
| MOL000422 | kaempferol | 41.88 | 0.24 | |
| MOL004628 | octalupine | 47.82 | 0.28 | |
| MOL004598 | 3, 5, 6,7-tetramethoxy-2-(3, 4,5-trimethoxyphenyl) chromone | 31.97 | 0.59 | |
| MOL004648 | troxerutin | 31.60 | 0.28 | |
| MOL004653 | (+)-anomalin | 46.06 | 0.66 | |
| MOL004609 | areapillin | 48.96 | 0.41 | |
| MOL002776 | baicalin | 40.12 | 0.75 | |
| MOL004702 | saikosaponin c_qt | 30.50 | 0.63 | |
| MOL012297 | puerarin | 24.03 | 0.69 | |
| Radix Puerariae (Gegen) |
MOL000392 | formononetin | 69.67 | 0.21 |
| MOL000358 | beta-sitosterol | 36.91 | 0.75 | |
| MOL003629 | daidzein-4,7-diglucoside | 47.27 | 0.67 | |
| MOL002959 | 3′-methoxydaidzein | 48.57 | 0.24 | |
| MOL012297 | puerarin | 24.03 | 0.69 | |
| Puerariae flower (Gehua) |
MOL011791 | kakkalide | 46.91 | 0.67 |
| MOL011793 | kakkatin | 55.25 | 0.24 | |
| MOL001749 | ZINC03860434 | 43.59 | 0.35 | |
| MOL000392 | formononetin | 69.67 | 0.21 | |
| MOL001792 | DFV | 32.76 | 0.18 | |
| MOL000449 | stigmasterol | 43.83 | 0.76 | |
| MOL003629 | daidzein-4,7-diglucoside | 47.27 | 0.67 | |
| MOL002959 | 3′-methoxydaidzein | 48.57 | 0.24 | |
| MOL005916 | irisolidone | 37.78 | 0.3 | |
| MOL000422 | kaempferol | 41.88 | 0.24 | |
| MOL000098 | quercetin | 46.43 | 0.28 | |
| MOL000468 | 8-o-methylreyusi | 70.32 | 0.27 | |
| MOL012976 | coumestrol | 32.49 | 0.34 | |
| MOL004957 | HMO | 38.37 | 0.21 | |
| MOL000358 | beta-sitosterol | 36.91 | 0.75 | |
| MOL000359 | sitosterol | 36.91 | 0.75 | |
| MOL008400 | glycitein | 50.48 | 0.24 | |
| MOL013305 | garbanzol | 83.67 | 0.21 | |
| MOL012297 | puerarin | 24.03 | 0.69 | |
| Radix Cyathulae (Chuanniuxi) |
MOL000098 | quercetin | 46.43 | 0.28 |
| MOL000358 | beta-sitosterol | 36.91 | 0.75 | |
| MOL012286 | betavulgarin | 68.75 | 0.39 | |
| MOL012298 | rubrosterone | 32.69 | 0.47 | |
| MOL012297 | puerarin | 24.03 | 0.69 | |
| Radix Hemerocallis (Xuancaogen) | MOL001255 | boswellic acid | 39.55 | 0.75 |
| MOL000422 | kaempferol | 41.88 | 0.24 | |
| MOL001243 | 3alpha-hydroxy-olean-12-en-24-oic-acid | 39.32 | 0.75 | |
| MOL002268 | rhein | 47.07 | 0.28 | |
| MOL013343 | hemerocallone | 63.01 | 0.54 | |
| MOL000471 | aloe-emodin | 83.38 | 0.24 | |
| MOL001771 | poriferast-5-en-3beta-ol | 36.91 | 0.75 | |
| MOL013345 | picraquassioside C | 53.99 | 0.69 | |
| MOL012297 | puerarin | 24.03 | 0.69 | |
3.2. Puerarin, quercetin and kaempferol were selected as the key compounds with anti-SARS-CoV-2 potential
Of the selected compounds from the five herbs, quercetin or kaempferol ranked the second in frequency (3 times), while puerarin ranked the highest, existing in all of the five herbs. Statistical analysis of the overlapped compounds existing in each of the five herbs was shown in a Venn diagram (Fig. 2A). Then the potential drug targets of the selected compounds from the five herbs were predicted using the online TCMSP analysis platform, which led to identification of 240 putative targets and subsequent construction of a compound-target network. Next, the topological parameters of each selected compound in the constructed network above, including Degree, Betweenness Centrality and Clossness Centrality, were calculated and scored by using a Cytoscape plugin CytoNCA. The higher the score, the higher the core degree and importance of a compound in the network. The top 10 compounds with the scores from high to low were listed in Table 2 (the rest compounds were listed in Supplementary Table 2). Among these, the top 2 compounds were quercetin and kaempferol. According to the above results, in addition to puerarin, quercetin and kaempferol were also selected as the key compounds for further analysis. The molecular formulas and chemical structures of puerarin, quercetin and kaempferol are shown in Fig. 2B.
Fig. 2.
Selection of the key compounds from CHMs. (A) The Venn diagram of 3 candidate core compounds from 5 herbs. Red box: puerarin; Blue box: quercetin; Green: kaempferol. (B) The 2D-chemical structure of puerarin, quercetin and kaempferol downloaded from the TCMSP database. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
The topological parameters of core candidate compounds (Top 10).
| MOL Number | Name | Degree | Betweenness Centrality | Closeness Centrality |
|---|---|---|---|---|
| MOL000098 | quercetin | 148 | 0.61714426 | 0.50391236 |
| MOL000422 | kaempferol | 61 | 0.17003082 | 0.39606396 |
| MOL012297 | puerarin | 55 | 0.19113992 | 0.38470729 |
| MOL000392 | formononetin | 38 | 0.05717885 | 0.37054085 |
| MOL000358 | beta-sitosterol | 37 | 0.08285532 | 0.37311703 |
| MOL000354 | isorhamnetin | 36 | 0.05139244 | 0.37225434 |
| MOL000449 | stigmasterol | 31 | 0.06871123 | 0.36884307 |
| MOL005916 | irisolidone | 31 | 0.03081572 | 0.36800000 |
| MOL000468 | 8-o-methylreyusi | 30 | 0.02290257 | 0.36716078 |
| MOL013343 | hemerocallone | 29 | 0.02611440 | 0.36800000 |
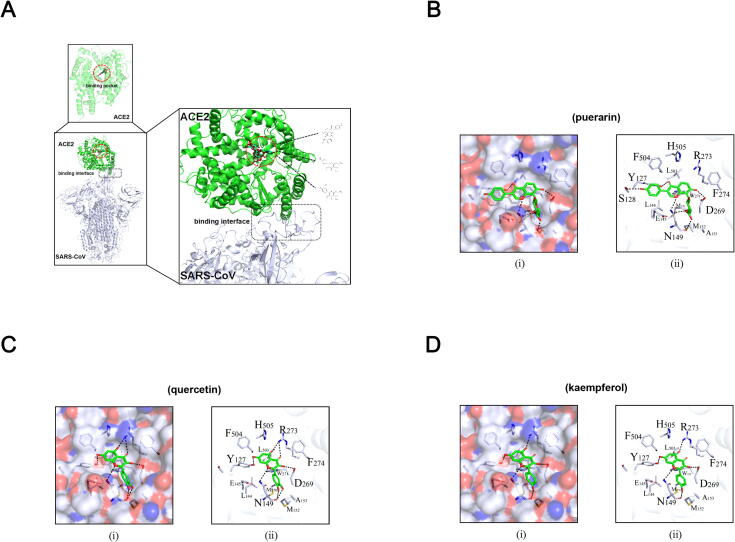
3.3. Molecular docking analysis showed the binding affinities between the key compounds and ACE2
Molecular docking was performed to determine the potential binding sites and binding affinity between the key compounds and ACE2 (PDB ID: 6ACG). The results showed that all of the three compounds could fit into a common binding pocket on ACE2 protein, which located at some distance from the binding interface of ACE2-SARS-CoV (Fig. 3A). Further docking analysis based on the recently resolved structure of ACE2 (PDB ID: 6M18) evaluated the binding affinity of ACE2 with all three compounds. Among these, quercetin displayed the highest binding affinity to ACE2 with an estimated binding free energy of −7.92 kcal/mol. It could form strong hydrogen bonds with the surrounding polar amino acids residues within the pocket, such as R273, D269 and N149, and form a weak hydrogen bond with the hydroxyl group of Y127. Meanwhile, the dihydroxyl group at the top of quercetin could forma double hydrogen bond with the –C O group on the backbone ofN149. The hydrophobic interactions formed between the planar ring and ACE2 residues M270 and W271 play also an important role in stabilization of the complex (Fig. 3B).
Fig. 3.
Molecular docking model. (A) The binding pattern between ACE2 and SARS-CoV (PDB: 6ACG). Red circle: the candidate protein binding pocket in ACE2; Black box: the protein binding interface between ACE2 and SARS-CoV. (B) Hydrophilic-hydrophobic interaction between (i) puerarin and ACE2 in protein binding pocket, and (ii) puerarin and relative amino acids residues. (C) Hydrophilic-hydrophobic interaction between (i) quercetin and ACE2 in protein binding pocket, and (ii) quercetin and relative amino acids residues. (D) Hydrophilic-hydrophobic interaction between (i) kaempferol and ACE2 in protein binding pocket, and (ii) kaempferol and relative amino acids residues. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Puerarin interacted with the residues D269 and N149 of ACE2 to form strong hydrogen bonds. Also, its top phenol group could form a hydrophobic interaction with L144, while the hydroxyl group forming a hydrogen bond with S128. However, compared to quercetin, puerarin had much less hydrophobic interactions with the surrounding residues within the pocket, due to a position shift caused by its hexose ring. Thus, the overall binding affinity of puerarin to ACE2 was lower than that of quercetin, with an estimated binding free energy of −7.46 kcal/mol (Fig. 3C).
The binding mode of kaempferol was similar to that of quercetin, except that only one hydrogen bond could be formed between the top mono-hydroxyl group of kaempferol and the N149 backbone -C = O group, which thereby changed the mutual position of kaempferol and the pocket. This structural shift resulted in an overall reduction of hydrogen bond formation. Thus, it formed a single hydrogen bond with R273 residue, instead of forming a double hydrogen bond with the same residue as quercetin did. As such, kaempferol displayed the lowest binding affinity to ACE2 among the three key compounds, with an estimated binding free energy of −7.21 kcal/mol (Fig. 3D).
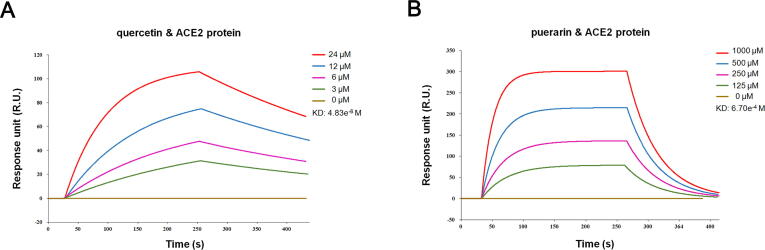
3.4. Interactions between the three compounds and ACE2 were validated by SPR assay
The interactions of the selected compounds and ACE2 were further evaluated by real-time biomolecular interaction analysis with SPR. The kinetics of the binding reaction was determined by injecting different concentrations of the compounds over a recombinant human ACE2 immobilized on the chip surface. The data were fitted to a monovalent binding model by non-linear regression, and the equilibrium dissociation constants (KD) for quercetin and puerarin were 4.83x10-6 M and 6.70x10-4 M, respectively, according to the obtained association and dissociation rates (Fig. 4A & 4B). Despite both compounds could bind to ACE2, quercetin exhibited a much higher affinity than puerarin with a KD value at the micromolar level. Unfortunately, attempts to dissolve kaempferol in 1% DMSO buffer to induce a significant response signal were unsuccessful, thus we failed to obtain the KD value for kaempferol (data not shown).
Fig. 4.
Interactions between the three compounds and ACE2 were determined by SPR assay. (A) The KD of the ACE2 protein with a series of concentrations of quercetin was calculated by SPR. (B) The KD of the ACE2 protein with a series of concentrations of puerarin was calculated by SPR.
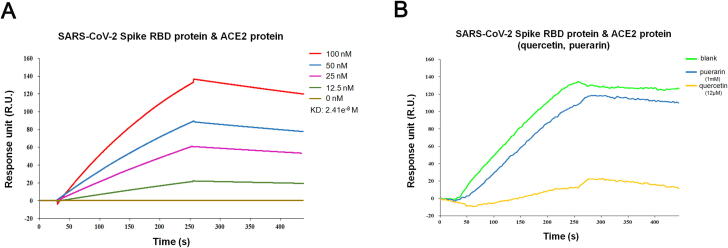
3.5. Puerarin and quercetin impaired the binding of S-protein to ACE2
An SPR-based competition assay was employed to determine whether puerarin and quercetin could affect the binding of S-protein to ACE2. We firstly determined the binding affinity of S-protein RBD to ACE2 by SPR assay, which unsurprisingly showed a very high KD value of 2.41x10-8 M (Fig. 5A). Having obtained the optimal concentration for each compound and the S-protein RBD according to their corresponding KD value, we injected either a mix of puerarin and S-protein RBD or a mix of quercetin and S-protein RBD over the immobilized human ACE2 for comparison. The result showed that addition of puerarin significantly attenuated the response signal by more than 20 units comparing to that of S-protein alone over the immobilized ACE2, indicating puerarin could indeed affect the binding of S-protein to ACE2. Notably, a substantial decrease in response signal was observed for quercetin, with a signal drop of over 100 units, indicating that the interaction between S-protein RBD and ACE2 was almost completely abrogated by quercetin (Fig. 5B).
Fig. 5.
SPR-based competition assay. (A) The KD of the ACE2 protein with a series of concentrations of SARS-CoV-2 Spike RBD protein was calculated by SPR. (B) quercetin (12 μM) and puerarin (1 mM) could block the combination of SARS-CoV-2 Spike RBD protein and ACE2 protein.
3.6. Quercetin could also bind to S-protein with a high binding affinity
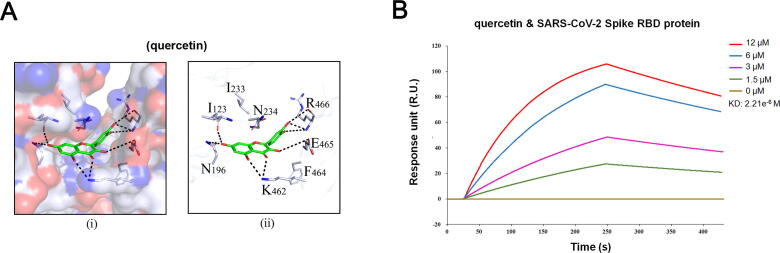
The striking disruptive effect of quercetin on the binding of S-protein with ACE2 inspired us to investigate the potential effect of quercetin on S-protein. The molecular docking analysis result showed that quercetin displayed a high binding affinity to S-protein, with an estimated binding free energy of −8.41 kcal/mol. Specifically, quercetin could form hydrogen bonds with the surrounding polar amino acids residues in the binding pocket of SARS-CoV-2 (PDB ID: 6VXX), such as K462, E465, R466 and N146, and its top dihydroxyl group could form a double hydrogen bond with the –C O group on the backbone of I123. Furthermore, the planar ring of quercetin could form hydrophobic interactions with I123 and F464 RBD residues (Fig. 6A). We subsequently carried out SPR assay again to determine the interaction between quercetin and S-protein. Consistent to the molecular docking result, quercetin exhibited a high binding affinity to SARS-CoV-2 RBD domain, with a KD value of 2.21x10-6 M (Fig. 6B). It should be noted that the KD value of quercetin on SARS-CoV-2 RBD domain was even lower than that of quercetin on ACE2, suggesting that quercetin has both receptor blocking and virus neutralizing effect on SARS-CoV-2.
Fig. 6.
Quercetin could bind to RBD domain of S-protein with a high binding affinity. (A) Hydrophilic-hydrophobic interaction between (i) quercetin and SARS-CoV-2 Spike in candidate protein binding pocket, and (ii) quercetin and relative amino acids. (B) The KD of the SARS-CoV-2 Spike RBD protein with a series of concentrations of quercetin was calculated by SPR.
3.7. Systemically exploring the potential action mechanism of quercetin through PPI target network and pathway enrichment analyses
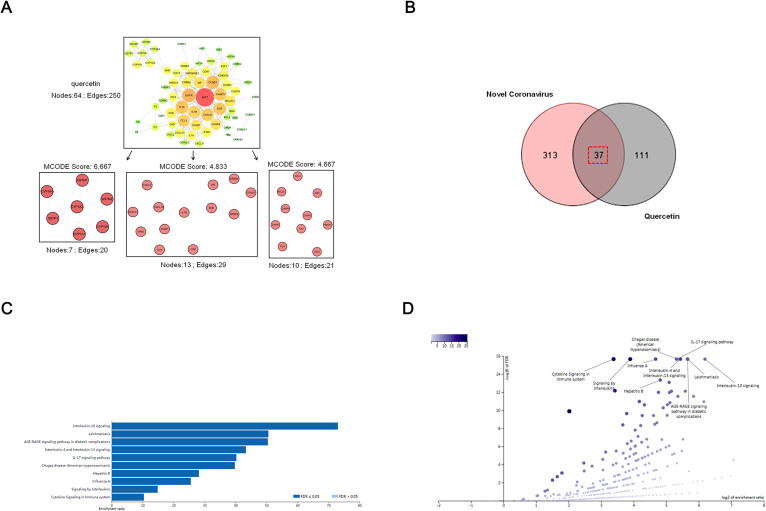
To explore the potential action mechanism of quercetin in a systemic view, we set about to construct a drug-target PPI network to comprehend the complex interaction among quercetin targets using the MCODE plugin (the detailed 148 putative targets of quercetin were listed in Supplementary Table 3). Based on a stringent parameter setting for target screening, the quercetin-target PPI network with 64 nodes and 250 edges was constructed. Next, we performed core subnetwork extraction from the PPI network, as the number of the putative targets (nodes) and the interaction (edges) were still high and complicated. The top 3 core subnetworks with the highest MCODE scores, containing a total of 30 nodes and 70 edges, were thus extracted from the parental PPI network (Fig. 7A). The hub targets consisting of subnetwork-1 were mainly the members of cytochrome P450 and glutathione S-transferase superfamilies, including CYP1A1, CYP1A2, CYP3A4, CYP1B1, GSTM1, GSTM2 and GSTP1. The hub targets in subnetwork-2 included various cytokines and molecules involved in immunoregulatory and inflammatory processes, such as CXCL2, CXCL10, IL1A, IL1B and ICAM1, whereas the hub targets in subnetwork-3 were mainly composed of apoptosis associated factors, such as BCL2, BAX, BCL2L1, CASP8 and CASP3. Consistently, GO functional enrichment analysis based on the 64 targets from the parental PPI network revealed that quercetin was closely related to gene transcription and expression, response to drugs, inflammatory response, signal transduction, apoptosis and oxidation–reduction process. Meanwhile, KEGG signaling pathway enrichment result showed that quercetin was involved in Pathways in cancer, Hepatitis B, PI3K-Akt signaling pathway, TNF signaling pathway, Influenza A, and MAPK signaling pathway (Table 3).
Fig. 7.
Systematic exploration of the action mechanism of quercetin against COVID-19. (A) PPI network construction and excavation of hub targets. Three core subnetworks and 30 hub targets was excavated by using the MCODE plugin. PPI: protein–protein interaction. (B) A Venn diagram of quercetin and COVID-19 co-targeted genes. (C) The ORA enrichment results in bar chart. (D) The ORA enrichment results in volcano plot.
Table 3.
The enrichment analysis of candidate targets of quercetin.
| Type | Term | Gene count | P-value |
|---|---|---|---|
| GO | GO-BP:positive regulation of transcription from RNA polymerase II promoter | 38 | 1.26E-16 |
| GO-BP:positive regulation of transcription, DNA-templated | 27 | 1.31E-14 | |
| GO-BP:response to drug | 25 | 6.40E-18 | |
| GO-BP:inflammatory response | 22 | 1.04E-12 | |
| GO-BP:signal transduction | 22 | 1.79E-04 | |
| GO-BP:positive regulation of gene expression | 20 | 1.17E-13 | |
| GO-BP:negative regulation of cell proliferation | 19 | 1.23E-09 | |
| GO-BP:apoptotic process | 18 | 1.44E-06 | |
| GO-BP:negative regulation of transcription from RNA polymerase II promoter | 18 | 3.32E-05 | |
| GO-BP:oxidation–reduction process | 17 | 1.12E-05 | |
| GO-MF:protein binding | 102 | 1.06E-10 | |
| GO-MF:identical protein binding | 29 | 1.31E-12 | |
| GO-MF:enzyme binding | 25 | 3.80E-17 | |
| GO-MF:transcription factor activity, sequence-specific DNA binding | 23 | 2.66E-06 | |
| GO-MF:DNA binding | 23 | 0.0066762 | |
| GO-MF:protein homodimerization activity | 22 | 1.16E-07 | |
| GO-MF:transcription factor binding | 19 | 4.28E-12 | |
| GO-MF:ATP binding | 19 | 0.0310054 | |
| GO-MF:sequence-specific DNA binding | 18 | 3.44E-07 | |
| GO-MF:protein heterodimerization activity | 16 | 2.20E-06 | |
| KEGG | KEGG:Pathways in cancer | 42 | 3.31E-23 |
| KEGG:Hepatitis B | 27 | 1.44E-20 | |
| KEGG:PI3K-Akt signaling pathway | 25 | 1.06E-09 | |
| KEGG:HTLV-I infection | 23 | 8.94E-11 | |
| KEGG:Proteoglycans in cancer | 22 | 6.43E-12 | |
| KEGG:MAPK signaling pathway | 21 | 3.60E-09 | |
| KEGG:MicroRNAs in cancer | 21 | 2.95E-08 | |
| KEGG:HIF-1 signaling pathway | 20 | 4.96E-16 | |
| KEGG:TNF signaling pathway | 20 | 4.34E-15 | |
| KEGG:Influenza A | 20 | 3.66E-11 |
To further investigate the potential mechanism of quercetin against COVID-19, we performed the ORA pathway enrichment analysis for quercetin and COVID-19 co-targeted genes. Firstly, COVID-19 disease-related genes were collected by using GeneCards online database, which resulted in a list of 350 obtained genes (the detailed information of these disease-targets were listed in Supplementary Table 4). Then, 37 overlapped targets among 148 quercetin drug-targets and 350 COVID-19 disease-related targets were obtained and shown in a Venn diagram (Fig. 7B and Supplementary Table 5). Finally, we performed an ORA systematic pathway enrichment analysis for these 37 co-targets using WebGestalt. The KEGG pathway and Reactome enrichment results showed that a total of 10 positively related categories were identified, which were mainly involved in immuno-modulation and virus infection activities, including IL-10 signaling, IL-4 and IL-13 signaling, IL-17 signaling, Influenza A, Signaling by Interleukins, Cytokine signaling in immune system (Fig. 7C & 7D). Together, the above results suggest that quercetin is a promising candidate against COVID-19, owing to its multifaceted pharmacological activities.
4. Discussion
To date, the COVID-19 has been spreading around the world at an exponential rate, raising an urgent need for effective therapeutics to contain this emerging pandemic. The ultimate solution is obviously the targeted vaccination against SARS-CoV-2. However, to develop a vaccine may take months or even years, and the developed vaccine may not be effective against the newly emerging strains of the virus, making the efforts futile. Alternatively, CHM could be a promising approach to combat COVID-19, as the patients treated with CHM have benefited from shorter hospitalization and improved symptoms [19], [20]. Combined application of TCM and modern Western medicine on COVID-19 patients can effectively relieve symptoms such as fever, cough, sore throat, myalgia and fatigue, and shorten the course of disease [12]. Therefore, screening the active compounds from CHM provides a convenient path to the rapid drug discovery against COVID-19.
In this study, taking advantage of the recent advances in network pharmacology and computational methods, we screened out 3 compounds from CHMs that hold great potential against SARS-CoV-2 virus. Interestingly, despite identified through different approaches, these compounds are all present in a single herb, Radix Bupleuri, which not only confirms the aforementioned theory that the key compounds existing in the same CHM are likely to have related and synergic pharmacological activities, but also validates the appropriateness of our screening strategy. Furthermore, a recent molecular docking analysis showed that baicalin, another compound of Radix Bupleuri, could also interact with ACE2 receptor through ASN-149, ARG-273 and HIS-505 residues, identical to those recognized by puerarin, quercetin and kaempferol within the same binding pocket [21]. As a popular CHM in China, Radix Bupleuri has been widely used for treating various symptoms and diseases, such as fever, inflammations, malaria, hepatitis B and influenza A [22], [23], [24], [25], [26]. A recent study showed that Radix Bupleuri was among the 26 selected CHMs with great potential to directly inhibit SARS-CoV-2 infection, and was advised to use at the early course of infection by senior TCM practitioners [27]. In this connection, it is not surprising that the multiple compounds it contains may possess anti-SARS-CoV-2 activity through a similar action mechanism. Although the common binding pocket shared by these compounds locates at some distance from the binding interface between the S-protein and ACE2, the binding of these compounds significantly compromised the interaction of RBD domain of S-protein with ACE2, at least in the cases of puerarin and quercetin as shown by the SPR-based competition assays. Therefore, these compounds are likely adopting alternative mechanisms, such as causing conformational changes to impair the interaction, rather than binding to the interface to competitively block SARS-CoV-2 virus.
As an efficient drug discovery strategy, drug repurposing could greatly shorten the time and reduce the cost compared to de novo drug discovery. Now, in the view of an urgent need to contain the epidemic, drug repurposing appears to be the best option for timely drug discovery against COVID-19. Recently, several existing drugs have been tested for efficacy in inhibiting SARS-CoV-2 in vitro. For example, remdesivir, an experimental drug being developed against Ebola virus, and hydroxychloroquine, a front-line drug for treating malarial and autoimmune disease, has shown a promising inhibitory effect on SARS-CoV-2 [28]. As an existing drug approved by China Food and Drug Administration (CFDA), puerarin has been widely used in clinical practice for decades in China. As such, and with its good safety profile, puerarin could be an ideal repurposable drug for COVID-19. It is worth noting that the safe dose of puerarin is as high as 0.5 g (~hundreds of mM in concentration) per day, indicating that puerarin may achieve a desirable antiviral effect at a safe dose, although the binding ability of puerarin to ACE2 is relatively low compared with the other tested drugs. Furthermore, because puerarin has the well-known beneficial effects on fever, cardiovascular diseases, neurological dysfunction and liver injury [29], [30], it can be co-administrated as a adjuvant to help improve the symptoms and prognosis of COVID-19 patients. In this context, the “old drug new use” of puerarin on COVID-19 can minimize the translational gap between drug development and clinical outcomes, providing an optimal outlet for timely developing effective therapeutics against the emerging pandemics.
Compared with puerarin, quercetin showed a much higher binding affinity to ACE2 with a KD at 4.83x10-6 M, which is actually a very good value for an unmodified natural compound. Moreover, quercetin showed an even higher binding affinity to the RBD domain of S-protein with a KD value of 2.41x10-8 M, and more importantly, the viral binding to ACE2 receptor was almost abrogated in the presence of quercetin, as shown in the subsequent SPR-based competition assay. Molecular docking analysis revealed that quercetin could form hydrogen bonds with the K462, E465 and R466 residues of SARS-CoV-2 RBD domain to make a stable complex. Notably, according to the recently resolved cryo-EM structure of full-length ACE2-RBD-B0AT1 complex [9], the interaction between ACE2 and the virus is to a great extend dependent on the residues Tyr453 and Gln474 of RBD, which are only several residues away from the quercetin binding pocket on RBD. These results support the view that the conformational change of these RBD residues upon the binding of quercetin plays an important role in the disrupted binding of S-protein to ACE2. Thus, quercetin impairs the interaction between S-protein and ACE2 through simultaneous binding with both of them, and this dual-targeting approach may generate a synergistic disruptive effect, and therefore result in a better anti-SARS-CoV-2 activity.
To further explore the action mechanism of quercetin, we constructed a quercetin-target PPI network and performed core subnetwork analysis. Interestingly, a cluster of hub targets from the extracted subnetwork were mainly composed of various cytokines and molecules involved in immunoregulatory and inflammatory processes, suggesting an important role of quercetin in immunomodulation. Consistently, the subsequent ORA enrichment analysis for quercetin and COVID-19 co-targeted genes led to a similar result, underlying a potential immunomodulation effect of quercetin on COVID-19 patients. Recent studies have shown that the levels of cytokines, such as IL-6, IL-10 and TNF, were increased dramatically in COVID-19 patients with severe symptoms compared to those with mild symptoms [1]. Given that the severe patients are inclined to form cytokine storm, leading to severe respiratory distress syndrome and multiple-organ failure, quercetin therefore may help improve the symptoms and outcomes of the COVID-19 patients through immunomodulation. Moreover, a recent single-cell RNA-Seq study found that multiple functional genes related to viral assembly and replication were highly co-expressed with ACE2 in type II alveolar cells [31], among which ICAM1 and EGFR were also present in the subnetwork as the hub targets. These results suggest that quercetin may have an adverse effect on infectiousness of SARS-CoV-2.
5. Conclusions
Taken together, three compounds from CHMs, namely puerarin, quercetin and kaempferol, exhibit high ACE2-targeting potential. Among these, puerarin and quercetin were demonstrated to impair the interaction between S-protein and ACE2 by SPR assays. Further results showed that puercetin has a dual binding ability to both ACE2 receptor and the viral S-protein, along with its potential immunomodulation effect, making it a good anti-SARS-CoV-2 drug candidate.
6. Funding sources
This study was supported by the National Natural Science Foundation of China (No.81572416), and the National Key Technologies R&D Program of China (No.2016YFC1303200).
7. Contributors
LL designed, supervised the study and wrote the manuscript. ML and BP engaged in study design. BP, SF, JZ and YP performed experiments and analyzed data. HL and YW assisted with performing experiments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.010.
Contributor Information
Min Li, Email: limin@mail.csu.edu.cn.
Liren Liu, Email: liuliren@tmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lake M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin Med. 2020;20(2):124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W.u., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virol. 2012;86(5):2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan R., Zhang Y., Li Y., Xia L.u., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science:abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q.i., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y.u., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z., Nakamura T. Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phytother Res. 2004;18(7):592–594. doi: 10.1002/ptr.1485. [DOI] [PubMed] [Google Scholar]
- 12.Xu X., Wu X., Jiang X., Xu K., Ying L., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S., Zhang B.o., Jiang D., Wei Y., Zhang N. Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinf. 2010;11(S11) doi: 10.1186/1471-2105-11-S11-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S., Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11:110–120. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- 15.Yang M., Chen J.-L., Xu L.-W., Ji G. Navigating traditional chinese medicine network pharmacology and computational tools. Evidence-Based Complementary and Alternative Medicine. 2013;2013:1–23. doi: 10.1155/2013/731969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan B., Ren Y., Liu L. Uncovering the action mechanism of polydatin via network pharmacological target prediction. RSC Adv. 2018;8(34):18851–18858. doi: 10.1039/C8RA03124J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan B., Shi X., Ding T., Liu L. Unraveling the action mechanism of polygonum cuspidatum by a network pharmacology approach. Am J Transl Res. 2019;11:6790–6811. [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y., Xu Q., Chen Q., Li R. People's Medical Publishing House; Beijing: 2004. Pharmacology of traditional Chinese medicine; p. 1099. [Google Scholar]
- 19.Runfeng L.i., Yunlong H., Jicheng H., Weiqi P., Qinhai M.a., Yongxia S., Chufang L.i., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L.i., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu K.e., Guan W.-J., Bi Y., Zhang W., Li L., Zhang B., Liu Q., Song Y., Li X., Duan Z., Zheng Q., Yang Z., Liang J., Han M., Ruan L., Wu C., Zhang Y., Jia Z.-H., Zhong N.-S. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. 2020:153242. doi: 10.1016/j.phymed.2020.153242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H., Du Q. Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection. Preprints. 2020;2020:2020010358. doi: 10.20944/preprints202001.0358.v3. [DOI] [Google Scholar]
- 22.Yang F., Dong X., Yin X., Wang W., You L., Ni J. Radix Bupleuri: a review of traditional uses, botany, phytochemistry, pharmacology, and toxicology. Biomed Res Int. 2017;2017:1–22. doi: 10.1155/2017/7597596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan B., Yang R., Ma Y., Zhou S., Zhang X., Liu Y. A systematic review of the active saikosaponins and extracts isolated from Radix Bupleuri and their applications. Pharm Biol. 2017;55(1):620–635. doi: 10.1080/13880209.2016.1262433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang L.C., Ng L.T., Liu L.T., Shieh D.E., Lin C.C. Cytotoxicity and anti-hepatitis B virus activities of saikosaponins from Bupleurum species. Planta Med. 2003;69:705–709. doi: 10.1055/s-2003-42797. [DOI] [PubMed] [Google Scholar]
- 25.Wen S.u., Huifu X.u., Hao H. In vitro anti-influenza A H1N1 effect of extract of Bupleuri Radix. Immunopharmacol Immunotoxicol. 2011;33(3):433–437. doi: 10.3109/08923973.2010.527985. [DOI] [PubMed] [Google Scholar]
- 26.Cao S.-L., Chen E.n., Zhang Q.-Z., Jiang X.-G. A novel nasal delivery system of a chinese traditional medicine,Radix Bupleuri, based on the concept of ion-activated in situ gel. Arch Pharm Res. 2007;30(8):1014–1019. doi: 10.1007/BF02993971. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D.-H., Wu K.-L., Zhang X., Deng S.-Q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integrative Med. 2020;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W.u., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y.-X., Zhang H., Peng C. Puerarin: a review of pharmacological effects: activity of puerarin. Phytother Res. 2014;28(7):961–975. doi: 10.1002/ptr.5083. [DOI] [PubMed] [Google Scholar]
- 30.WEI S.-Y., CHEN Y.i., XU X.-Y. Progress on the pharmacological research of puerarin: a review. Chinese J Natural Med. 2014;12(6):407–414. doi: 10.1016/S1875-5364(14)60064-9. [DOI] [PubMed] [Google Scholar]
- 31.Yu Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.