Highlights
-
•
COVID-19 cases are rising in the US, Brazil and India with no apparent end in sight.
-
•
The validated modified CDC assay performs well but its testing capacity is limited.
-
•
Laboratories need to acquire multiple assays to meet SARS-CoV-2 testing demand.
-
•
NeuMoDx has the highest throughput and shortest TAT than the other assays tested.
Keywords: COVID-19, SARS-CoV-2, Pandemic
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), has spread rapidly around the globe since it was first identified in December of 2019 in Wuhan, China. In a race to contain the infection, researchers and healthcare officials have developed several assays to help diagnose individuals with COVID-19. To help laboratories decide what assay to bring into testing lines, factors such as assay availability, cost, throughput, and TAT should be considered. Here we validated a modified version of the CDC assay and used it as a reference to evaluate the performance of the NeuMoDxTM SARS-CoV-2 and DiaSorin SimplexaTM Covid-19 Direct assays. In silico analysis and clinical sample testing showed that the primers/probes designed by the CDC were specific to the SARS-CoV-2 as they accurately detected all reactive samples with an assay LoD of 200 copies/mL. The performance of the three assays were analyzed using 159 nasopharyngeal swabs specimen tested within 1–5 days after routine testing. A 100 % agreement was observed between the commercial assays and the modified CDC SARS-CoV-2 assay. A deeper look at the Ct values showed no significant difference between NeuMoDx and the modified CDC SARS-CoV-2 assay, whereas DiaSorin had lower overall Ct values than the modified CDC SARS-CoV-2 assay. NeuMoDx and DiaSorin workflows were much easier to perform. NeuMoDx has the highest throughput and shortest TAT, whereas although the modified CDC SARS-CoV-2 assay has comparable throughput to DiaSorin, it has the longest hands-on time and highest TAT.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which started in Wuhan City in China at the end of December 2019, has spread to over 200 countries and was declared a global pandemic by the World Health Organization (WHO) on March 11, 2020 [1]. As of July 12, there were more than 12,740,000 confirmed cases of COVID-19 and 565,000 deaths (https://coronavirus.jhu.edu/map.html). The United States (US) led the world with over 3,280,000 cases of COVID-19 and over 135,000 deaths as of July 12, 2020. The rate of new cases continues to rise in the US, Brazil and India with no apparent end in sight.
To meet diagnostic needs as the pandemic grows, the U.S. Food and Drug Administration (FDA) granted several commercial SARS-CoV-2 tests Emergency Use Authorization (EUA). Since that, there has been a race against time to develop tests for SARS-CoV-2 detection so individuals with COVID-19 could be identified and isolated to slow the spread of the disease. In January 2020, the CDC developed a TaqMan probe-based molecular test [2]. In the following months, several commercial assays became available and have been used in the laboratory under the FDA’s EUA [[3], [4], [5], [6]]. With the rapid development of those tests, came the challenge of assay sensitivity and specificity. Our laboratory at Tampa General Hospital validated a modified version of the CDC assay following the FDA EUA guidelines and brought in commercial assays to help respond to the testing demand. The modified CDC SARS-CoV-2 assay involves off-instrument cell lysis and nucleic acid extraction steps, and an amplification step on a different instrument. The assay includes a panel of primer/probe sets targeting the viral N gene and the human RNase P gene. The NeuMoDx™ SARS-CoV-2 and DiaSorin Simplexa® Covid-19 Direct assays are fully automated sample-to-answer multiplex assays targeting two different regions of the viral genome. NeuMoDx assay targets the Nsp2 and the N genes while DiaSorin test targets the ORF1ab and S gene. The NeuMoDx™ SARS-CoV-2 assay is performed on a random-access, high throughput instrument allowing for with high priority and high volumes testing which is crucial in an outbreak. In this study, we sought to describe a modified CDC SARS-CoV-2 assay validation and compare its performance and workflow to that of the NeuMoDx SARS-CoV-2 and DiaSorin Simplexa Covid-19 Direct assays using respiratory specimens.
2. Materials and methods
2.1. Validation of the modified CDC SARS-CoV-2 assay
The primer/probe sets used in this validation were selected from regions of the SARS-CoV-2 virus nucleocapsid (N) gene and were described in the CDC EUA protocol for COVID-19 diagnostic testing [7]. The CDC original panel was designed for both universal detection of SARS-like coronaviruses (N3 primer/probe set) and specific detection of the SARS-CoV-2 (N1 and N2 primer/probe sets). Moreover, the original CDC protocol included two sets of controls: HSC (human specimen extraction control) and RP (human RNase P gene to assess specimen quality). Our modified protocol included the N1 and N2 primer/probe sets and the RP control. Primer and probe sets used in this validation were purchased from Integrated DNA Technologies (Coralville, IA).
Total nucleic acid extraction was carried out on the bioMérieux NucliSens® easyMAG® automated system (bioMerieux, France) from a 200 μL of the sample and eluted in 50 μl of EasyMag elution buffer. A separate reaction mix containing 5 μL of the eluate, 5 μL of TaqPath™ 1-Step RT-qPCR Master Mix (4x), 1.5 μL of combined primer/probe Mix (500 nM and 125 nM final concentration of primers and probes, respectively) and 13.5 μL of Nuclease-free Water was made for each of the assay target (N1, N2, and RP). A no-template water control (NTC) and nCoVPC plasmid were used as template for each of the primer/probe set; Hs_RPP30_Positive Control plasmid was used as template for RP primer/probe. The RT-PCR cycling conditions were set up on the Rotor-Gene 3000 thermocycler (Corbett Research, Australia) as followed: 25 °C for 2 min, 50 °C for 15 min, 95 °C for 2 min, followed by 45 cycles of 95 °C for 3 s and 55 °C for 30 s with fluorescence (FAM) detection during the 55 °C incubation step. A sample was positive for SARS-CoV-2 if at least one of the two targets (N1, N2) was detected regardless whether RP was amplified, negative if none of the targets was detected and RP was detected, and invalid if RP and the two targets were not detected.
2.2. Analytical evaluation of the modified CDC SARS-CoV-2 assay
For the analytical evaluation, we used SARS-CoV-2 RNA (strain USA_WA1/2020) kindly provided by the University of Texas Medical Branch (UTMB) in Galveston. A series of two-fold dilutions of the RNA were spiked in pooled sputum at concentrations of 800 copies/mL to 0.05 copies/mL to determine the limit of detection (LoD) of the assay. All samples were processed and tested in triplicate as described above. The LoD was confirmed by further testing in 20 replicates. The analytical specificity was determined by testing 22 samples previously positive for different respiratory species, including 15 patient samples, 4 ATCC strains, and 3 commercially available nucleic acid controls. The clinical performance was established by testing 30 contrived NP swabs and sputum specimens and 30 non-reactive specimens. Of the 30 contrived specimens, 20 were spiked with SARS-CoV-2 strain USA_WA1/2020 RNA at 1x-2x LoD concentrations and 10 were spiked at concentrations spanning the assay’s testing range (60,000 copies/mL to 234 copies/mL).
2.3. Comparison between the modified CDC SARS-CoV-2, the NeuMoDx SARS-CoV-2, and the DiaSorin Simplexa covid 19 Direct assays
A total of 159 NP swabs were used to compare clinical performance of the three SARS-CoV-2 assays. Samples were processed as described above for the modified CDC SARS-CoV-2 assay; for the NeuMoDx SARS-CoV-2 assay and the DiaSorin Simplexa Covid-19 Direct assay samples were processed according the manufacturer’s procedures. For NeuMoDx, 400 μL of sample was mixed with 400 μL Viral Lysis Buffer in a secondary tube before loading it onto the NeuMoDx 96 Molecular System (Ann Arbor, MI). A sample was positive if either N or Nsp2 genes were detected, negative if both targets were not amplified and the sample processing control (SPC2) was amplified, and indeterminate or unresolved if there was an instrument error or it failed to produce a valid result. For the DiaSorin, 50 μL of sample and 50 μL of the Reaction Mix were loaded to each well of an amplification disc and processed according to the manufacturer’s protocol on the LIASON® MDX instrument (Saluggia, Italy). A sample was positive for SARS-CoV-2 if either ORF1ab or S gene was detected, negative if both targets were not amplified and the internal control was amplified, and invalid if there was an internal control failure. The results obtained from each assay were compared with those obtained using the modified CDC SARS-CoV-2 assay. EP Evaluator was used to calculate positive percent agreement (PPA), negative percent agreement (NPA), and Cohen’s kappa (k) with 95 % confidence intervals.
3. Results
3.1. Primer/probe analysis and PCR efficiency
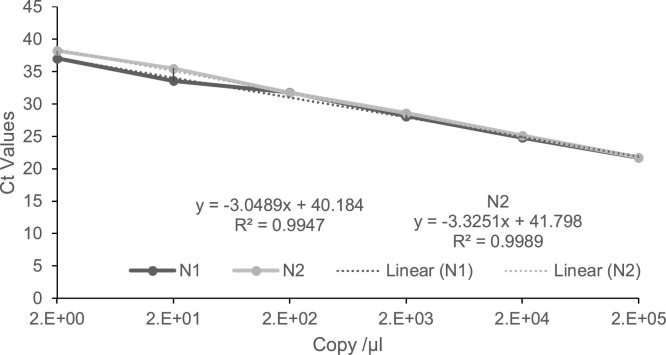
Since the CDC primer/probe panel became available, many SARS-CoV-2 isolates have been sequenced. Therefore, we ascertained that the primer/probe sets were specific to all available sequences for SARS-CoV-2 in the NCBI’s GenBank. Multiple sequence alignment of the nucleocapsid gene of SARS-CoV-2 and the two closely related coronaviruses, SARS-CoV and MERS-CoV, showed the positions of the primer/probe sets (Fig. 1 A). The N1 set amplified a 72 bp fragment of the nucleocapsid gene, N2 amplified a 67 bp of the same gene, and the primer/probe set for human RP gene internal control produced a 57 bp amplicon (Fig. 1B). The PCR efficiency was determined by testing a series of 10-fold dilutions of the 200,000 copies/μl concentration of the nCoVPC plasmid. The data showed that the PCR was linear over 6 orders of magnitude with great PCR efficiency (N1 = 111 % and N2 = 100) and R [2] of 0.99 for each of the primer/probe set (Fig. 2 ).
Fig. 1.
A) Multiple sequence alignment of partial sequence of the nucleocapsid gene of MERS-CoV (NC 038294.1), SARS-CoV-2 (NC_045512.2), SARS-CoV (NC_004718.3) showing target regions of N1 and N2 primer/probe sets. Forward primers sequences are bolded, probes are faded and itilicized, and reverse primers are underlined and bolded. Note: 780 bases between the two primer sets were omitted to shorted the length of the sequence. B) Capilary gel electrophoresis picture generated using the Agilent DNA 7500 kit on the Agilent 2100 bioanalyzer instrument. Image showed single band for each primer set. From left to right: L (ladder:100 – 7000bp), N1, N2, and RP of aproximately 72 bp, 67 bp, and 65 bp, respectively.
Fig. 2.
Real time PCR determining amplification efficiency of the primer/probe sets. Ten-fold serial dilution of 200,000 copies/μl of nCoVPC plasmid was tested. PCR linearity over 6 orders of magnitude with a limit of detection of 2 copies/μl; N1 slope of -3.05 with a correlation coefficient R2 = 0.99; N2 slope =-3.33 and R2 = 0.99.
3.2. Analytical evaluation of the modified CDC SARS-CoV-2 assay
The analytical sensitivity of the modified CDC SARS-CoV-2 assay was determined by testing pooled sputum samples spiked with a series of two-fold dilutions at concentrations of 800 copies/mL to 50 copies/mL of SARS-CoV-2 strain USA_WA1/2020. The LoD was determined to be 200 copies/mL (Table 1 A). It was confirmed by further testing 20 replicates that were inoculated with 200 copies/mL; all 20 replicates were tested positive (Table 1B). The analytical specificity of the assay was determined by testing 22 previously positive samples, which included 15 patient samples, 4 ATCC strains, and 3 commercially available nucleic acid controls. The result showed that none of the targets was detected (Table 2 ). The performance of the assay was evaluated in 60 sputum samples to mimic the extent of viral colonization in respiratory specimens. The assay detected SARS-CoV-2 in all 30 contrived samples; no amplification was seen in the 30 non-reactive samples (Table 3 ).
Table 1.
Limit of detection of the modified CDC SARS-CoV-2 assay.
| A | |||
|---|---|---|---|
| SARS-CoV-2 RNA Concentration | Modified CDC SARS CoV-2 Ct Values |
Interpretation | |
| Target N1 | Target N2 | ||
| 800 copies/mL | 31.66 | 31.6 | Detected |
| 32.57 | 32.05 | Detected | |
| 32.25 | 32.76 | Detected | |
| 400 copies/mL | 34.02 | 33.65 | Detected |
| 34.72 | 34.97 | Detected | |
| 34.82 | 35.42 | Detected | |
| 200 copies/mL | 34.99 | 36.88 | Detected |
| 35.22 | 34.21 | Detected | |
| 34.76 | 35.19 | Detected | |
| 100 copies/mL | ND | 35.05 | Inconclusive |
| ND | 36.31 | Inconclusive | |
| 35.88 | 37.02 | Inconclusive | |
| 50 copies/mL | ND | ND | Not-Detected |
| 35.96 | 35.94 | Detected | |
| ND | 36.1 | Inconclusive | |
| B | ||
|---|---|---|
| Samples | Target N1 | Target N2 |
| 1 | Ct = 35.70 | Ct = 33.01 |
| 2 | Ct = 33.66 | Ct=32.52 |
| 3 | Ct = 32.99 | Ct=31.99 |
| 4 | Ct = 34.01 | Ct=35.53 |
| 5 | Ct = 32.57 | Ct=34.43 |
| 6 | Ct = 36.01 | Ct=34.61 |
| 7 | Ct = 33.87 | Ct=34.46 |
| 8 | Ct = 35.10 | Ct=37.87 |
| 9 | Ct = 34.77 | Ct=35.68 |
| 10 | Ct = 34.10 | Ct=34.83 |
| 11 | Ct = 35.70 | Ct=34.97 |
| 12 | Ct = 34.19 | Ct=36.04 |
| 13 | Ct = 38.77 | Ct=34.79 |
| 14 | Ct = 34.26 | Ct=34.97 |
| 15 | Ct = 38.79 | Ct=35.44 |
| 16 | Ct = 38.94 | Ct=35.15 |
| 17 | Ct = 34.85 | Ct=33.71 |
| 18 | Ct = 35.82 | Ct=33.52 |
| 19 | Ct = 33.34 | Ct=32.82 |
| 20 | Ct = 33.14 | Ct = 33.01 |
Table 2.
Proficiency panel of viral and bacterial samples tested using the modified CDC SARS-CoV-2 assay.
| Organisms | Source | Method of Identification | Interpretation |
|---|---|---|---|
| MERS CoV | Control Suspension | IDT# 10006623 | Not-Detected |
| SARS CoV | Control Suspension | IDT# 10006624 | Not Detected |
| Adenovirus | Control Suspension | Exact DX# ADVH102 | Not-Detected |
| Coronavirus HKU1 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Coronavirus NL63 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Coronavirus 229E | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Coronavirus OC43 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Influenza A | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Influenza B | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Respiratory Syncytial Virus | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Human Metapneumovirus | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Human Rhinovirus | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Chlamydia pneumoniae | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Mycoplasma pneumoniae | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Parainfluenza Virus 1 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Parainfluenza Virus 2 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Parainfluenza Virus 3 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Parainfluenza Virus 4 | NP Patient Sample | BioFire RVP2 | Not-Detected |
| Staphylococcus epidermidis | 0.5 McFarland Suspension | ATCC Strain 14990 | Not-Detected |
| Streptococcus pyogenes | 0.5 McFarland Suspension | ATCC Strain 19615 | Not-Detected |
| Pseudomonas aeruginosa | 0.5 McFarland Suspension | ATCC Strain 27853 | Not-Detected |
| Mycobacterium tuberculosis | 0.5 McFarland Suspension | ATCC Strain 25177 | Not-Detected |
Table 3.
Ct values of contrived clinical specimen spiked with different concentrations of SARS-CoV-2 RNA.
| Sample ID (SARS-CoV-2 RNA Copies) | Modified CDC SARS-CoV-2 assay Ct Values |
Interpretation | ||
|---|---|---|---|---|
| Target N1 | Target N2 | Target RP | ||
| 001−020 (200−400) | 32.75 ± 2.14* | 32.12 ± 2.23* | 23.44 ± 3.24* | Detected |
| 021 (60,000) | 25.15 | 24.81 | 22.58 | Detected |
| 022 (30,000) | 28.17 | 26.17 | 21.61 | Detected |
| 023 (15,000) | 26.84 | 27.46 | 21.58 | Detected |
| 024 (7500) | 28.37 | 28.5 | 21.62 | Detected |
| 025 (3750) | 29.33 | 29.75 | 21.7 | Detected |
| 026 (1875) | 30.23 | 31.02 | 21.54 | Detected |
| 027 (937) | 31.13 | 31.97 | 21.38 | Detected |
| 028 (468) | 32.59 | 32.66 | 21.59 | Detected |
| 029 (300) | 32.55 | 32.66 | 21.59 | Detected |
| 030 (234) | 32.85 | 34.41 | 21.63 | Detected |
| 031- 060 (negative) | ND | ND | 20.9 ± 0.5 | Not Detected |
Average Ct values and standard deviation of the mean for samples 1–19 spiked with 200–400 copies of SARS-CoV-2 RNA and 1 known positive patient sample; ND: No Ct value detected.
3.3. Clinical performance and workflow comparison between the modified CDC SARS-CoV-2 assay, NeuMoDx SARS-CoV-2 assay, and DiaSorin Simplexa Covid-19 Direct assay
One hundred and fifty-nine NP samples with a wide range of Ct values were used to compare the performance of the three assays. Of the 43 samples used for comparison between modified CDC SARS-CoV-2 assay and Simplexa Covid 19 Direct assay, 37 samples were run within 2 days and 6 were run within 5 days of first testing. Of the 116 samples used for comparison between the modified CDC SARS-CoV-2 assay and NeuMoDx SARS-CoV-2 assay, 102 samples were run within a day and 14 were run within 5 days of first testing. All the samples tested by the modified CDC SARS-CoV-2 assay matched the results using the two commercial assays evaluated yielding a 100 % PPV and NPV for each assay (Table 4 A). NeuMoDx did not yield a Ct value for one of the two targets in 5 samples: 4 were negative for Nsp2 gene target and 1 was negative for the N gene target. However, they were still considered positive as only one detected target was needed for a positive result.
Table 4.
Clinical performance and workflow comparison between modified CDC SARS-CoV-2 assay and the commercial assays.
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Commercial assays | Modified CDC SARS-COV-2 assay |
Ct ranges | Kappa (%) | PPA/NPA(%) | 95 % CI | |||
| Positive | Negative | Total | ||||||
| NeuMoDx | Positive | 67 | 0 | 67 | 10.04−40.07 | 100 | 100/100 | 96.9−100 |
| Negative | 0 | 49 | 49 | |||||
| Total | 67 | 49 | 116 | |||||
| DiaSorin | Positive | 22 | 0 | 22 | 12.00−36.10 | 100 | 100/100 | 91.8−100 |
| Negative | 0 | 21 | 21 | |||||
| Total | 22 | 21 | 43 | |||||
| B | |||
|---|---|---|---|
| *Workflow | Assays |
||
| Modified CDC | NeuMoDx | DiaSorin | |
| Hands-on | ∼45 min | ∼5min | ∼15 min |
| Extraction | ∼35 min | ∼1h20min | N/A |
| PCR | ∼90 min | ∼1h25min | |
| Overall TAT | ∼2h50min | ∼1h 25min | ∼1h40min |
| Max samples/run | 10 | 24 | 8 |
| **Throughput/8-h shift | ∼40 | ∼96 | ∼40 |
PPA: Positive percent agreement; NPA negative percent agreement; CI: confidence interval.
Workflow and overall turnaround time (TAT) is based on 8 samples per run.
Number of samples that can be resulted in an 8-h shift.
Although 100 % agreement was observed among the assays, further analysis showed that there were some differences in the Ct values. The average Ct value difference in samples ran within 24 h between NeuMoDx SARS-CoV-2 and the modified CDC SARS-CoV-2 assay was -0.14, and -2.13 between samples ran within 5 days. The overall Ct value difference for all samples ran between the two assays was -0.346. On the other hand, the average Ct values difference between samples ran within 2 days between DiaSorin Simplexa Covid 19 Direct assay and the modified CDC SARS-CoV-2 assay was -2.42, and -6.0 between samples ran within 5 days. The overall Ct value difference for all samples between the two assays was -3.47.
We assessed the workflow and compared the throughput, time of sample-to-result, and cost per sample for each assay. Based on 8 samples/run, it was estimated to be 2 h and 50 min for modified CDC SARS-CoV-2 assay which included 35 min extraction time, 90 min PCR run, and 45 min of hands-on time, 1 h and 40 min for DiaSorin Simplexa Covid 19 Direct assay including 15 min hands-on time, and 1 h and 25 min for NeuMoDx SARS-CoV-2 assay which included a 5 min hands-on time (Table 4B). The NeuMoDx 96 Molecular System is a random-access platform that can process and result 24 samples in less than two hours. That enabled the NeuMoDx assay to have a throughput of 96 samples per 8-hr shift compared to 40 samples on the DiaSorin Simplexa Covid 19 Direct assay, and 40 samples on the modified CDC SARS-CoV-2 assay if sample processing and PCR runs were staggered. The most cost-effective test was the NeuMoDx. Reagents for this test is a little more expensive than the modified CDC SARS-CoV-2 assay, but less expensive than the DiaSorin. The labor involved on the modified CDC SARS-CoV-2 assay, however, is considerably greater than on the NeuMoDx and DiaSorin assays, which ends up increasing the cost of the test.
4. Discussion
The ongoing pandemic still poses great risks for many around the world, and with the easing of certain restrictions, the need for health care facilities to be equipped and accurately test for the virus to limit its spread is as crucial as it will ever be. To that extent, laboratories have brought in SARS-CoV-2 assays and molecular platforms to respond to the need of their communities. There have been a few publications on head-to-head comparisons of those assays, including a couple very recently as we were preparing this article, in order to shed light on their performance characteristics and help laboratories make informed decisions on acquiring those assays [4,[8], [9], [10]].
In this study, we validated a modified CDC SARS-CoV-2 assay and found that the assay is very sensitive and specific to SARS-CoV-2. The clinical performance comparison between NeuMoDx SARS-CoV-2 assay, Simplexa Covid-19 Direct assay, and the modified CDC SARS-CoV-2 assay showed an overall agreement of 100 %. The Ct value difference between the modified CDC assay and the NeuMoDx assay suggests that there is not a significant difference between the two assays; however, there seems to be a greater difference in Ct values between Simplexa SARS-CoV-2 Direct assay and the modified CDC SARS-CoV-2 assay, with the former having lower Ct values. The difference is greater in samples that were run 5 days after the routine testing on the modified CDC SARS-CoV-2 assay. This is in line with previously published data that showed Ct values on Simplexa SARS-CoV-2 Direct assay was much lower than those on an a modified CDC SARS-CoV-2 assay by an average Ct difference of -2.1 [9]. The overall data also suggest that depending on the viral burden NP samples can be refrigerated for at least 5 days and still maintain the RNA integrity for viral detection by the assays in this study.
A limitation of this study was that the same samples were not tested by the three different assays, so a head-to-head comparison of the three assays was not performed due to the limited kits available for routine patient care. However, a head-to-head comparison of the assay’s workflow was performed. NeuMoDx has the shortest time-to-result, the highest throughput, and is the most cost-effective of the three assays evaluated. Therefore, it allows the clinical laboratory to significantly increase throughput and reduce TAT in a time when more testing is needed, and faster result is crucial to speed up isolation measures. The TAT comparison between the three assays was based on 8 samples/run since the LIASON® MDX instrument can only accommodate 8 samples/run. The overall throughput of the modified SARS-CoV-2 CDC assay can be improved to 24 samples if the 72-PCR tube adaptor option for the Rotor-Gene 3000 is used. Laboratories that are equipped with 96-well nucleic acid extraction platforms and PCR instruments, as well as the personnel can further scale up testing and significantly increase the throughput of the CDC assay. Nevertheless, the increased demand for testing highlighted a well-known short resource of skilled laboratory personnel and having to train new staff usually takes some time. Acquiring two NeuMoDx96 instruments helped our laboratory to increase the testing capacity from 200 tests/day to about 700 tests/day while maintaining the same number of laboratory personnel. Therefore, although the three assays have the same accuracy, due to its random-access and high throughput capacity, NeuMoDx SARS-CoV-2 assay outperforms the other two assays.
Diagnostic laboratories around the world have faced with unprecedented challenges due to the SARS-CoV-2 pandemic. The testing requirement has not only forced laboratories to bring in new technologists to help with testing, but it has also led to shortage of reagents. Consequently, multiple assays and platforms are used to meet testing demand. As much as LDT assays, were instrumental at the onset of the pandemic, their overall testing capacity are very limited. Therefore, it is necessary for laboratories to acquire multiple high throughput automated instruments that can test high volume of samples quickly with almost the same number of qualified laboratory professionals.
CRediT authorship contribution statement
Amorce Lima: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft. Vicki Healer: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Supervision. Elaine Vendrone: Validation, Investigation. Suzane Silbert: Conceptualization, Methodology, Formal analysis, Writing - review & editing, Project administration.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We are grateful to University of Texas Medical Branch for providing SARS CoV-2 RNA Template for the validation of the modified CDC SARS-CoV-2 assay.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104688.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. Epub 2020 Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. Epub 2020 Jan 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D., Deconinck L., Lescure F.X., Lucet J.C., Bouadma L., Timsit J.F., Descamps D., Yazdanpanah Y., Casalino E., Houhou-Fidouh N. Evaluation of the QIAstat-Dx respiratory SARS-CoV-2 panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nörz D., Fischer N., Schultze A., Kluge S., Mayer-Runge U., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J. Clin. Virol. 2020;128:104390. doi: 10.1016/j.jcv.2020.104390. Epub 2020 Apr 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordi L., Piralla A., Lalle E., Giardina F., Colavita F., Tallarita M., Sberna G., Novazzi F., Meschi S., Castilletti C., Brisci A., Minnucci G., Tettamanzi V., Baldanti F., Capobianchi M.R. Rapid and sensitive detection of SARS-CoV-2 RNA using the Simplexa™ COVID-19 direct assay. J. Clin. Virol. 2020;128:104416. doi: 10.1016/j.jcv.2020.104416. Epub 2020 May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poljak M., Korva M., Knap Gašper N., Fujs Komloš K., Sagadin M., Uršič T., Avšič Županc T., Petrovec M. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(8) doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J. Clin. Microbiol. 2020;17 doi: 10.1128/JCM.00760-20. 00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman J.A., Pepper G., Naccache S.N., Huang M.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 2020;29 doi: 10.1128/JCM.00821-20. 00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-Answer platforms for the detection of SARS-CoV-2. J. Clin. Microbiol. 2020;24 doi: 10.1128/JCM.00783-20. 00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.