Abstract
Fungal infections in humans are increasing worldwide and are currently mostly treated with a relative limited set of antifungals. Resistance to antifungals is increasing, for example, in Aspergillus fumigatus and Candida auris, and expected to increase for many medically relevant fungal species in the near future. We have developed and patented a set of cathelicidin-inspired antimicrobial peptides termed ‘PepBiotics’. These peptides were initially selected for their bactericidal activity against clinically relevant Pseudomonas aeruginosa and Staphylococcus aureus isolates derived from patients with cystic fibrosis and are active against a wide range of bacteria (ESKAPE pathogens). We now report results from studies that were designed to investigate the antifungal activity of PepBiotics against a set of medically relevant species encompassing species of Aspergillus, Candida, Cryptococcus, Fusarium, Malassezia, and Talaromyces. We characterized a subset of PepBiotics and show that these peptides strongly affected metabolic activity and/or growth of a set of medically relevant fungal species, including azole-resistant A. fumigatus isolates. PepBiotics showed a strong inhibitory activity against a large variety of filamentous fungi and yeasts species at low concentrations (≤1 μM) and were fungicidal for at least a subset of these fungal species. Interestingly, the concentration of PepBiotics required to interfere with growth or metabolic activity varied between different fungal species or even between isolates of the same fungal species. This study shows that PepBiotics display strong potential for use as novel antifungal compounds to fight a large variety of clinically relevant fungal species.
Keywords: antifungal peptide, cathelicidin, fungal infections, antifungal resistance, antimicrobial peptide
Introduction
Fungal infections in humans and animals are currently treated with a relatively limited set of antifungals.1 However, resistance to antifungals is increasing with an unprecedented rate, among others due to their extensive use in the environment outside medical care facilities.2 Consequently, extended exposure of humans and animals to resistant pathogens is expected to lead to a rise in incurable infections. Resistance to antifungals is frequently reported for important fungal pathogens.1,3 In addition, the global increase in immunocompromised patients, for example, due to higher use of immunoblockers results in more fungal infections. Clearly, these developments urge for new therapeutic compounds to fight and prevent resistance and suboptimal treatments. Further, restricted or even exclusive use in medical settings will be necessary to prevent rapid resistance development in the environment.4
Antimicrobial peptides (AMPs, also known as host-defense peptides) exhibit strong potential to fight microbial infections. AMPs are peptides with a broad spectrum of antimicrobial activity, are produced by all eukaryotic organisms, including fungi,5 and are an important part of the innate immune system of vertebrates.6 They vary in length between 12 and 50 amino acids and are generally positively charged and amphipathic allowing strong interactions with hydrophobic surfaces and membranes. AMPs are very diverse with respect to sequence and structure and are generally classified based on their conformation: α-helical, β-sheet (disulfide bridges), cyclic or peptides with extended structure/random-coil.7,8 AMPs kill target cells through diverse mechanisms primarily by disrupting the membrane. It has also been found that AMPs target key cellular processes, including DNA and protein synthesis, protein folding, cell wall synthesis and enzymatic activity.9,10 Clearly, AMPs have the advantage of a less specific but efficient mechanism of action as compared to traditional antifungals, a feature expected to limit resistance development.
In this paper we focus on the AMPs belonging to the cathelicidin family that are synthesized as prepropeptide and share a highly conserved N-terminal cathelin domain.11 However, the antimicrobial mature peptide varies in amino acid sequence, structure, size and activity between species.6,11–13 Cathelicidins can be constitutively produced or induced in response to a variety of microbial infections and inflammation.6,11,14,15 The only human cathelicidin LL-37 is produced by neutrophils, macrophages, epithelial cells, and natural killer (NK) cells.6,14 Chicken genomes encode four cathelicidins, CATH-B1, CATH-1, CATH-2, and CATH-3,15 the latter three were shown to possess strong antibacterial activity without inducing notable resistance.6,16 Previously, CATH-2 and LL-37 were reported to have candidacidal activities.17 Here we compared the antifungal activity of LL-37 and L-CATH-2 with a set of cathelicidin-inspired variants called ‘PepBiotics’ against several medically relevant species encompassing species belonging to the fungal genera Aspergillus, Candida, Cryptococcus, Fusarium, Malassezia, and Talaromyces. These fungal pathogens are responsible for an annual high number of human infections globally and have been extensively described in numerous overviews.18,19 Just four fungal species causing invasive infections—A. fumigatus, Cryptococcus neoformans, Candida albicans, and Histoplasma capsulatum—kill 1.5 million people annually.20 The rise in resistance frequency to the azole antifungal compounds in the case of A. fumigatus is worrying, especially due to the development of multi-azole resistance and difficulties to treat patients infected with tri-azole resistant fungi.21,22 Azoles target the ergosterol biosynthesis pathway and inhibit the demethylation of precursor sterols of 14-α-demethylase encoded by the cyp51A gene23 and a variety of mutations, including TR34/L98H and TR46/Y121F/T289, are detected in a large number of azole-resistant A. fumigatus strains in the Netherlands.24A. fumigatus is globally responsible for over 14 million cases per annum causing invasive and non-invasive infections in immunocompromised and competent humans, respectively.25 Aspergillosis is a lung-associated infection and patients with for example COPD or cystic fibrosis are frequently infected. A recent survey in five Dutch CF centers revealed that A. fumigatus is isolated most frequently with a mean prevalence of 31.7% and 7.1% of these strains were azole-resistant strains.26 The PepBiotics used in this study vary in their primary sequence and were initially screened for their activity against gram-negative and -positive bacteria (manuscript in preparation) and a subset was now used to study antifungal activity. Here we show that PepBiotics interfere with metabolic activity, fungal growth, and/or viability of several clinically relevant fungal strains. These results illustrate the strong potential of PepBiotics as a promising novel class of antifungal drugs to treat infections caused by clinically relevant fungal species in humans.
Methods
Strains and culture conditions
Tables 1 and 2 summarize the fungal species and strains used. New clinical isolates of A. fumigatus from human patients were typed and resistance profile to azoles was determined as previously described.27 For the initial experiments we tested a set of A. fumigatus strains, including resistant ones, indicated in Table 1 for their sensitivity to PepBiotics. These strains were standardly cultured for 4 days on PDA (Difco) at 37°C. Conidia were harvested from fresh cultures by scraping the surface gently using 10 ml saline solution with 0.002% Tween 20 (Sigma). For the second set of experiments we tested a larger variety of fungal species and strains indicated in Table 2, for their sensitivity for two PepBiotics (CR172 and CR184). These fungi were cultured for one week on oatmeal agar (Quakers and Oxoid) at 25°C except for Malassezia spp., which were cultured on modified Dixon agar (Sigma, Difco, Fluka, Baker, and Oxoid) at 30°C. Conidia or yeast cells were gently removed from the plates with ice-cold 10 mM ACES-buffer, pH 6.8 (N-(2-acetamido)-2-aminoethanesulfonic acid, 0.02% Tween 80) and filtered using glass wool. After centrifugation for 5 minutes at 1811 RCF, cells or conidia were resuspended in 20 ml ice-cold ACES buffer and centrifuged again. Finally, cells or conidia were resuspended in 5 ml ice-cold ACES buffer. The number of isolated yeast cells or conidia was determined using a Bürker-Turk haemocytometer and appropriate dilutions were prepared as indicated in the corresponding experiments. Stock solutions of yeast cells or conidia were stored at −25°C in 30% glycerol in 0.5× saline solutions.
Table 1.
Aspergillus fumigatus strains.
| A. fumigatus | Source | Reference |
|---|---|---|
| DTO 342-B2α | Canine patient with SNA | 27 |
| DTO 327-A82 | Human ICU patient; Resistant to ITC (>16), POS (0.25), VRC (>16) | 27 |
| DTO 326-I1β | Human patient from hematology; Resistant to ITC (>16), POS (1), Intermediate for VRC (2) | 27 |
| CEA10ΔKU80 | Derivative of CEA10; human patient with invasive aspergillosis | 49 |
| DTO 327-D4 (Af293) | Human patient with invasive aspergillosis | 50 |
αDTO is the code used for strains present in the working collection of the Department of Applied and Industrial Mycology (DTO).
βclinical isolates from ICU or hematology patients at Utrecht Medical Center and suspected of invasive aspergillosis were obtained from sputum. MIC concentrations (mg/l) according to EUCAST for the azoles are indicated between brackets. ITC, itraconazole; POS, posaconazole; VRC, voriconazole. More information is available in the indicated reference.
Table 2.
Fungal species.
| Species | DTO number | CBS numberα |
|---|---|---|
| Aspergillus fumigatus | 368-D5 | CBS 113.26 |
| 368-D6 | CBS 113.37 | |
| Candida albicans | 262-G7 | n.d. |
| Candida auris | 384-D4 | CBS 12777 |
| Candida dubliniensis | 384-D3 | CBS 11947 |
| Candida glabrata | 384-C9 | CBS 12544 |
| Pichia kudriavzevii (Candida krusei) | 105-F6 | CBS 573 |
| Candida parapsilosis | 384-D2 | CBS 12541 |
| Candida tropicalis | 384-D1 | CBS 13079 |
| Cryptococcus bacillisporus | 384-D9 | CBS 10081 |
| Cryptococcus deneoformans | 384-D7 | CBS 10511 |
| Cryptococcus deuterogattii | 384-D8 | CBS 10082 |
| Cryptococcus neoformans | 384-D6 | CBS 9172 |
| Cryptococcus tetragattii | 384-E2 | CBS 10101 |
| Fusarium oxysporum | 363-B8 | n.d. |
| Neocosmospora (Fusarium) solani | 321-I6 | n.d. |
| Malassezia furfur | 383-D8 | CBS 1878 |
| 383-D9 | CBS 7019 | |
| Talaromyces marneffei | 299-F6 | CBS 389.87 |
| Meyerozyma guillermondii | 384-D5 | CBS 12072 |
αStrains with CBS code are from the fungal collection of the Westerdijk Fungal Biodiversity Center, Utrecht, The Netherlands; n.d., only present in the working collection of the Department of Applied and Industrial Mycology (DTO).
For metabolic assays, growth or viability experiments Aspergillus-minimal medium (MM) was used containing per liter: 6.0 g NaNO3, 1.5 g KH2PO4, 0.5 g KCl, 0.5 g MgSO4, 200 μl trace element solution (10 g EDTA/l, 4.4 g ZnSO4 · 7H2O/l, 1.01 g MnCl2 ·4H2O/l, 0.32 g CoCl2 ·6H2O/l, 0.315 g ZuSO4 ·5H2O/l, 0.22 g (NH4)6Mo7O24· 4H2O/l, 1.47 g CaCl2 ·2H2O/l and 1.0 g FeSO4 ·7H2O/l), pH 6.0 (NaOH), and 2% w/v glucose for all fungal species (except Malassezia). For metabolic assays and growth/viability assays we used 0.125×MM and 1× MM, respectively. Culturing was performed at 37°C or 25°C as described in the experiments. For Malassezia spp. viability assays MM with 0.4% v/v glycerol (instead of glucose) and supplemented with 1% v/v Tween 60 (Sigma-Aldrich), 0.4% v/v olive oil, (MMM) was used at 30°C. The specific time points corresponding to specific results are indicated in the figures and/or their legends.
Peptides
This study includes the use of 2 natural cathelicidins, human LL-37 and chicken L-CATH-2, and 9 cathelicidin-inspired therapeutic peptides denoted as PepBiotics and coded as ‘CR###’. Peptide codes and sequences are listed in Table 3. All peptides used in this study were synthesized by ChinaPeptides Co., Ltd. (Chuanhong Road, Shanghai, China) using classical solid phase peptide synthesis (SPPS) and 9-fluorenylmethoxycarbonyl (Fmoc) as a protective group at the N-terminus.28 After synthesis, the peptides were purified by HPLC with a linear gradient system (gradient: 5–95% B in 6 minutes, flow: 1 ml/min, eluent A: 100% H2O + 0.05% TFA; eluent B: 100% CH3CN + 0.05% (v/v) TFA) using a C18 RP-HPLC column (i.e. C18, 3 μm, 20 × 2 mm) and detection at 220 nm. All peptides used were ≥95% pure as verified by electrospray ionization Mass Spectrometry. To enhance stability and activity, all peptides were amidated at their C-terminus (-NH2) except CR175. After purification, stock solutions of 1.6 mM were prepared by dissolving in sterile H2O and aliquots were stored at −80°C. Working solutions were prepared by diluting stocks with sterile H2O or in buffer solutions as indicated, depending on experimental conditions. A patent application has been filed with regard to the unique and novel core amino acid sequence shared by PepBiotics and use of PepBiotics for a wide variety of infection-related applications (patent pending, PCT/EP2018/060402; “antimicrobial peptides and their use”).
Table 3.
Antimicrobial peptides used in this study.
| Code | Amino acid sequence | Number of amino acids |
|---|---|---|
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES-NH2 | 37 |
| L-CATH-2 | RFGRFLRKIRRFRPKVTITIQGSARF-NH2 | 26 |
| CR170 | RRWVQRWIRRWRPKRIVQRIKDFLRNLV-NH2 | 28 |
| CR171 | RRWGRFLRKIRRFRPKFKRIVQRIKDFLRNLV-NH2 | 32 |
| CR172 | RRWVQRWIRRWRPKVAAARRWVQRWIRRWRPKV-NH2 | 33 |
| CR173 | RRWVQRWIRRWRKVAAARRWVQRWIRRWRPKV-NH2 | 32 |
| CR175 | RRWVQRWIRRWRPKVLQKKGI | 21 |
| CR181 | APKAMRWVQRWIRRWRPKLQKKGI-NH2 | 24 |
| CR182 | APKAMWVQRWIRRWRPLQKKGI-NH2 | 22 |
| CR183 | APKAMRRWVQRWIRRWRPKVLQKNNYL-NH2 | 27 |
| CR184 | APKAMRRWVQRWIRRWRPKVFQVTGSSA-NH2 | 28 |
The patented sequence ‘RRWVQRWIRRWR’ as present in most peptides used in this study is displayed in bold characters (truncated in CR181 and CR182). All peptides were aligned to maximize overlap of the residues present in those peptides that only have part of this sequence present. CR172 and CR173 contain a tandem repeat of the patented sequence, separated by six (CR172) or five (CR173) spacer residues.
Metabolic assay
Minimal medium (MM or dilutions of MM, 50 μl) of the appropriate concentration was mixed with 30 μl milliQ water, 5 μl glucose (final 2%), 5 μl conidia or yeast suspension (106 cfu/ml, 5 × 103 spores or yeasts), 5 μl resazurin stock of 2.1 mM (final is 105 μM) or 210 μM (final 10.5 μM), Sigma, Cat. No. R7017-5 G) and 5 μl AMPs (diluted in water to obtain the final concentration) and were added to a well of a nonbinding ELISA plate (Greiner Bio-One, Cat. No. 655180; 96 well cell culture plate). The final volume was 100 μl and either 105 μM (incubations at 37°C) or 10.5 μM (incubations at 25°C) resazurin was used. Resazurin (blue) is an oxidation-reduction indicator and conversion into resorufin (pink) is a measure of metabolic activity.29,30 It can be further reduced to a colorless dihydroresorufin mainly occurring upon total consumption. This latter compound can be converted back to the pink resorufin requiring atmospheric oxygen.29 Only a limited set of compounds was described that can give false positive reactions.31 Conversion to pink or colorless is regarded as a positive result indicating metabolic activity, but after conversion to the colorless dihydroresorufin no quantification of metabolic activity can be made. We therefore determined for the different culturing conditions the optimal amount of MM and resazurin as well as the incubation time to enable quantification of metabolic activity. During further experiments we always used 1:8 diluted MM (0.125×MM) which provided optimal measure of metabolic activity. The absorbance of the samples was measured at 570 nm using plate readers (Thermo Scientific Multiskan EX or a Spectrostarnano from BMG-Labtech) and routinely corrected for the absorbance of the negative control (no fungal cells). Plates were covered with a lid and were repeatedly measured during 24 hours to max 48 hours for incubations at 37°C and up to 72 hours at 25°C, in the latter case plates were incubated while shaking at 60 rpm. Positive (no AMPs) and negative controls (no fungal cells) were routinely incorporated and the wells at the edges of the plate were filled with water and not with samples to reduce evaporation effects at 37°C. Occasionally, plates were measured at 405 nm to determine cell density.32 No metabolic assays were possible for Malassezia spp. due to false negative results; no conversion of resazurin was observed while growth was detected by microscopic analysis. All fungal species (except Malassezia) used in this study did grow in 1/8 diluted MM at 25°C or in case of Malassezia 1/8 diluted MMM medium at 30°C in absence of antimicrobial peptides.
Viability and growth assay
Fifty μl of a solution of AMPs (diluted in 1×MM to obtain a final concentration which ranged between 0.1 and 5 μM) containing conidia/yeast cells (1500 cfu/ml) in 1×MM or 1×MMM (in case of Malassezia spp.) was mixed with 50 μl sterile water in a well of a 96-well plate (Costar 3879, Corning Incorporated, Corning, NY, USA) and incubated at 25°C or 30°C for Malassezia spp. The amount of cfu/ml was determined after 3, 24, and 48 hours by the addition of Triton X-100 (final concentration 1% v/v) to a well and the complete content was plated and incubated on a malt extract agar (MEA) plate at 25°C or mDixon plate and culturing for 7 days at 30°C (for Malassezia) followed by colony counting. Positive controls lacked AMPs and negative controls lacked fungal cells but contained 0.16 μM AMP. At the start of the experiment an additional sample was directly plated to determine the exact starting amount of cfu/ml at t = 0. The viability assay by direct plating worked only for Malassezia yeast cells but not for filamentous fungi (e.g., Aspergillus, Fusarium, and Talaromyces spp.). In the latter case fungal material was not plated but was inspected with a stereomicroscope (Nikon AZ100) to determine the extent of growth by mycelium formation. In addition, the metabolic assay was used for all filamentous but only for all yeast species (except Malassezia spp.).
Live-dead staining with propidium iodide
To investigate viability of A. fumigatus, staining with propidium iodide (PI) was performed. As positive control, 106 conidia of A. fumigatus CEA10 in 100 μl 0.125×MM in an Eppendorf tube were heat-inactivated for 20 min at 90°C. After centrifugation for 60 seconds in an Eppendorf centrifuge at 13.200 g, the conidia were resuspended in 10 μl PI (15 μM final concentration, Sigma-Aldrich). After a 15 minutes incubation at 37°C, conidia were microscopically analyzed by fluorescence microscopy with a TRITC filter and an Axioskop 2 plus fluorescent microscope at 100× magnification. To determine the effect of AMPs on viability of fungal cells using PI, mixtures of conidia in MM were prepared as described above in the absence or presence of AMPs (1 or 5 μM) and incubated at 37°C up to 48 hours, similar as described for the metabolic assays but without resazurin. After various time intervals samples were taken, centrifuged, stained with PI and analyzed using fluorescent microscopy as described above. We initially tested a large set of PepBiotics in duplo experiments to obtain a more qualitative view of the effects on A. fumigatus strains. In the subsequent experiments with a subset of PepBiotics we tested their effects in three independent experiments on a large variety of different fungal species.
Cytotoxicity test
Cytotoxicity of PepBiotics was determined using primary human nasal epithelial cells, isolated and cultivated at UMCU to confluency in Bronchial Epithelical Cell Medium (Sciencell Research Laboratories, Sanbio, Uden, Netherlands).33 Cells were exposed to 5 μM of peptide for 3 hours at 37°C followed by three washings with phosphate-buffered saline (PBS). WST-1 (10% v/v in cell medium) was used for the quantification of cell viability using colorimetric analysis as previously described.34 Controls included no peptide (100% viability) and 70% ethanol (total loss of cell viability). Experiments were performed in triplo. Statistical analysis peptide-treatment versus ‘no peptide’ by two-tailed paired Student t test.
Results
Effects of AMPs on metabolic activity of A. fumigatus
We first compared the effect of L-CATH-2 and a set of PepBiotics on the metabolic activity of A. fumigatus strains Af293, CEA10 and DTO342-B2 at 1 μM. We observed in these initial experiments that a subset of the PepBiotics was markedly more active against these A. fumigatus strains as compared to L-CATH-2 at this concentration. PepBiotics CR170, 171, 172, 173 and 183 inhibited the metabolic activity strongly at 1 μM whereas the other PepBiotics did not and results were comparable to L-CATH-2. (results not shown).
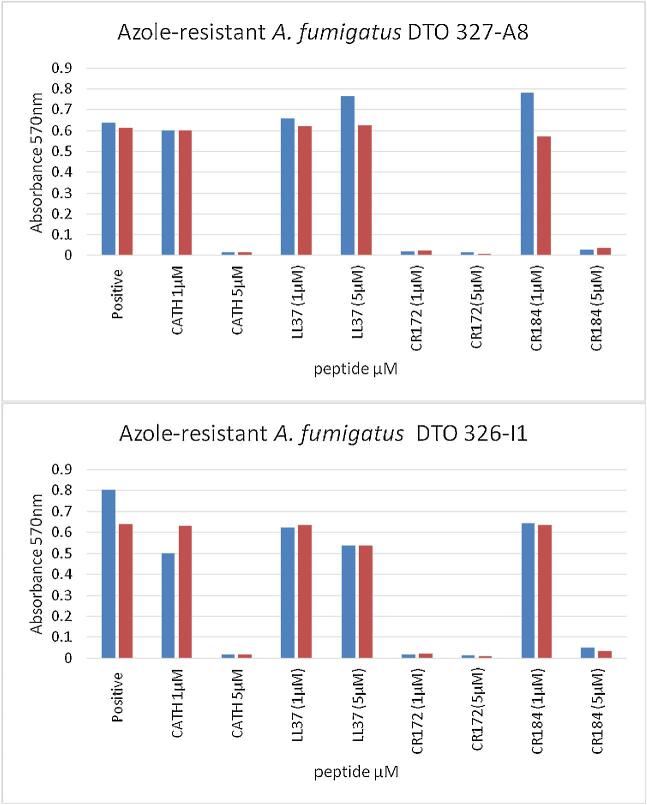
We next tested the activities of AMPs LL-37 and L-CATH-2 with CR172 and CR184 in more detail at 1 μM and 5 μM against two azole-resistant A. fumigatus strains, DTO 327-A8 and 326-I1. Both azole-resistant isolates were sensitive for PepBiotics (Fig. 1). CR172 inhibited metabolic activity strongly at 1 μM whereas CR184 and L-CATH-2 inhibited metabolic activity at 5 μM. The AMP LL-37 did not inhibit metabolic activity under these conditions. Similar results were obtained for Af293 and CEA10 (results not shown).
Figure 1.
Azole-resistant A. fumigatus strains are sensitive for PepBiotics. The indicated azole-resistant strains were used in the metabolic assay using 0.125×MM and 105 μM resazurin at 37°C for 27 h in absence (positive) or presence of either L-CATH-2, LL-37, CR172 or CR184 at 1 or 5 μM final concentration. Results of two experiments are shown (blue and red bar).
Killing potency of PepBiotics against A. fumigatus using PI staining
We next investigated whether the PepBiotics were also able to kill A. fumigatus using the live-dead propidium iodide staining. This stain is used to assess cell viability since it cannot pass the cytoplasmic membrane of living cells. Indeed, more than 95% of the conidia of A. fumigatus were fluorescently labeled when heat-inactivated for 20 minutes at 90°C (Fig. S1F). We subsequently incubated A. fumigatus CEA10ΔKU80 in absence (negative control) or presence of 5 μM L-CATH-2 in 0.125×MM at 37°C and tested the viability of fungal cells by PI staining up to 20 hours. No PI positive staining of conidia, swollen conidia or hyphal forms was detected in the presence of L-CATH-2 (Fig. S1B). We repeated these experiments with the more active PepBiotic CR172 at 5 μM (Fig. S2), but although this concentration is strongly inhibiting metabolic activity, no PI positive staining of fungal cells was observed (Fig. S2D). In addition, no PI-positive staining of CEA10ΔKU80 was observed when we added this peptide to a 23 hour culture followed by an incubation of 30 minutes (Fig. S2F). Remarkably, we did observe killing with two variant PepBiotics on a different A. fumigatus strain. Addition of 1 μM CR173 or CR183 to an 18-hour culture of a different A. fumigatus strain DTO 342-B2 followed by a 15-minute incubation did result in PI-positive staining (Fig. 2). Only hyphal structures were stained whereas dormant and swollen conidia were not PI positive. We observed that germination in the MM was much lower as compared to rich medium and preliminary data indicated that approximately 6% of conidia were germinated in 8 hours, whereas around 60% were swollen. This explains the relative low amount of hyphal structures observed (Figs. S1 and S2). Overall, these results indicate that L-CATH-2 and PepBiotics CR172, CR173, CR183, and CR184 strongly inhibit the metabolic activity or viability of A. fumigatus strains.
Figure 2.
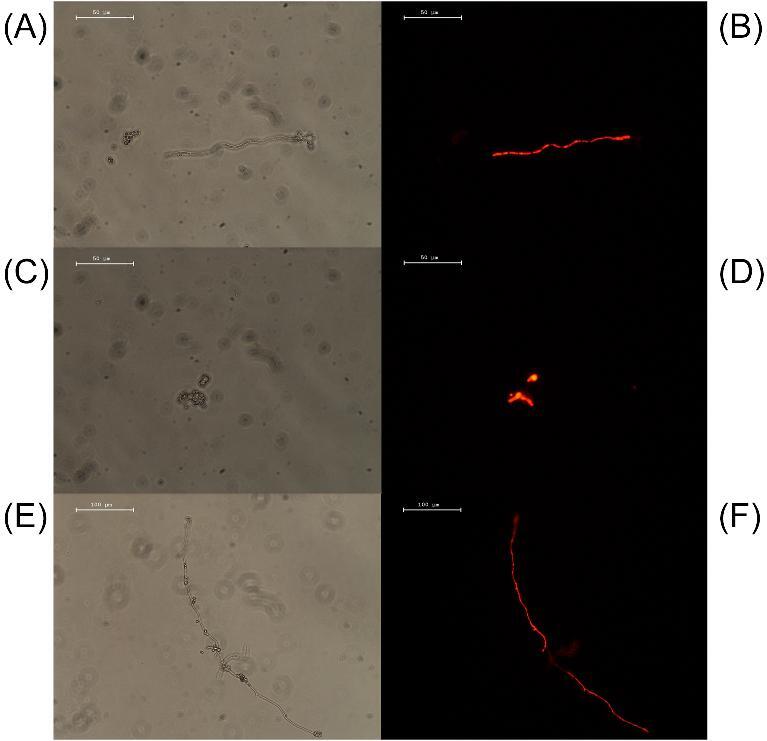
Propidium iodide staining of A. fumigatus with PepBiotics CR173 and CR183. (A) and (C) (normal filter), (B) and (D) (TRITC filter) shows an 18-h culture of A. fumigatus DTO 342-B2 which was subsequently incubated for 15 min with 1μM CR-173, washed and stained with PI (400x). (E) (normal filter) and (F) (TRITC filter): similar strain and culture conditions but a 15-min incubation with 1 μM CR183 (200×). (A–D), size bar 50 μm; (E–F), size bar 100 μm.
Antifungal effects of PepBiotics against clinically relevant fungi
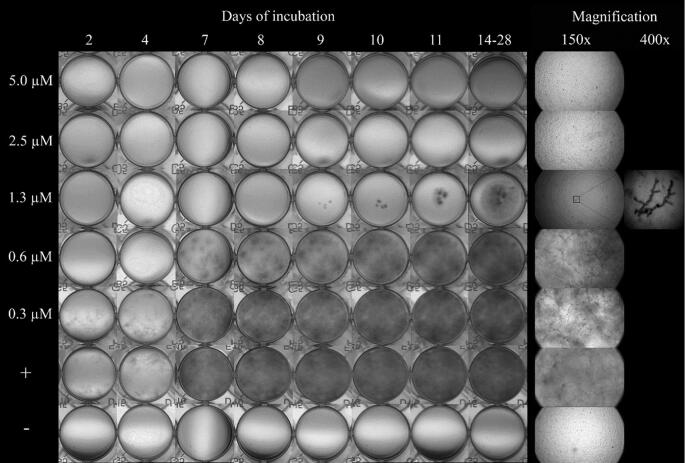
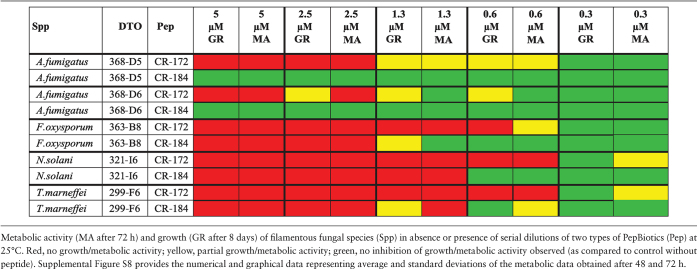
We next investigated whether the PepBiotics CR172 and CR184 are able to inhibit metabolic activity, growth and viability of a large set of clinically relevant fungal species (Table 2) using incubations in MM but at lower growth temperatures. This lower temperature was chosen to reduce the evaporation of medium during prolonged culturing in ELISA plates. All strains indicated in Table 2 were able to grow in the same Aspergillus MM at 25°C, except Malassezia, which required 30°C and addition of the lipidic compounds (1% Tween-60 v/v and 0.4% olive oil v/v) due to its inability to synthesize fatty acids de novo.35 We first optimized the metabolic activity assay using A. fumigatus DTO 368-D5 and D6 for these new conditions in order to be able to follow activity over a prolonged period. The metabolic assay showed a steady increase in absorbance at 570 nm between 25 hours and 72 hours at 25°C using 0.125×MM and 10.5 μM resazurin (results not shown). The viability of Malassezia yeast cells was determined by plating of cells after washing the wells with 1% TX-100 and is expressed as cfu/ml. The growth of yeasts and filamentous fungal species was also followed microscopically (indicated as GR). The viability of filamentous fungal species could not be determined by plating since these fungal species could not be quantitatively removed from the wells by washing with TX-100. Table 4 summarizes the results of three independent experiments of metabolic assays (MA) and growth assays (GR) of a set of filamentous fungal species and representative images can be found in the supplementary Fig. S3-S7. In agreement with our findings at 37°C, A. fumigatus is strongly inhibited by CR172 (Fig. 3) whereas CR184 does not inhibit at 25°C (Fig. S3). Sometimes, a single spore germinated and formed a hyphal structure (see 400× magnification in Fig. 3). Similarly, the metabolic activity and growth of Fusarium and Talaromyces species tested were both more sensitive for PepBiotic CR172 and were inhibited at 0.625 μM (Table 4).
Table 4.
Sensitivity of clinically relevant filamentous fungal species for PepBiotics.
|
Figure 3.
Growth of A. fumigatus (DTO 368-D5) in absence or presence of the antifungal peptide CR-172. Growth was evaluated up to 28 days at 25°C. Details of growth after 7 days of incubation as observed with a stereomicroscope at 150× and 400× magnification are shown. Incubation with conidia in absence of CR172 (indicated as +) served as positive control, culture medium in absence of conidia but presence of 0.16 μM CR172 (indicated as −) served as negative control.
In the growth assay, two strains of Malassezia furfur (DTO 383-D8 and DTO 383-D9) showed different sensitivity for the peptides tested. Strain DTO 383-D8, was moderately susceptible for inhibition by CR-184 (1.3 μM) and CR-172 (5 μM) within the 48-hour incubation period (Table 5). In contrast, strain DTO 383-D9 was highly sensitive for both PepBiotics (growth assay, Table 5) and the lowest concentration tested of both CR184 and CR172 (0.3 μM) resulted in undetectable growth. Both strains were also plated after incubations with the PepBiotics to determine the viability expressed as the amount of cfu/ml after 3-, 24-, and 48-hour incubations with the PepBiotics (Fig. S7). M. furfur (strain DTO383-D8) strain lost viability after a 48-hour incubation with 2.5 μM CR172 (Fig. S7A) and after 24-hour incubation with 0.6 μM CR184 (Fig. S7B). In contrast, M. furfur (strain DTO 383-D9) lost viability already after a 3-hour incubation with 0.6 μM CR172 (Fig.S7 panel C) while no viable cells were recovered after a 48-hour incubation with 0.3 μM CR184 (Fig. S7D). These results underscore a remarkable difference in sensitivity for PepBiotics between Malassezia strains.
Table 5.
Effects of PepBiotics on growth of Malassezia furfur.
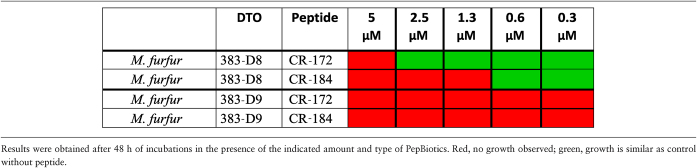
|
Metabolic activity assays with a set of yeasts strains indicated that all tested species were highly sensitive to both PepBiotics. Concentrations of <1  M PepBiotics abolished metabolic activity in most cases as determined after 72 hours (Table 6A,B). These included five Cryptococcus and six Candida strains, among them C. auris (which did not show activity at 0.6 μM, CR-172). Cryptococcus gattii did not grow in the MM used and sensitivity for PepBiotics to this important emerging pathogen could not yet be tested.36
M PepBiotics abolished metabolic activity in most cases as determined after 72 hours (Table 6A,B). These included five Cryptococcus and six Candida strains, among them C. auris (which did not show activity at 0.6 μM, CR-172). Cryptococcus gattii did not grow in the MM used and sensitivity for PepBiotics to this important emerging pathogen could not yet be tested.36
Table 6.
A. Sensitivity of clinically relevant yeast strains for PepBiotics.
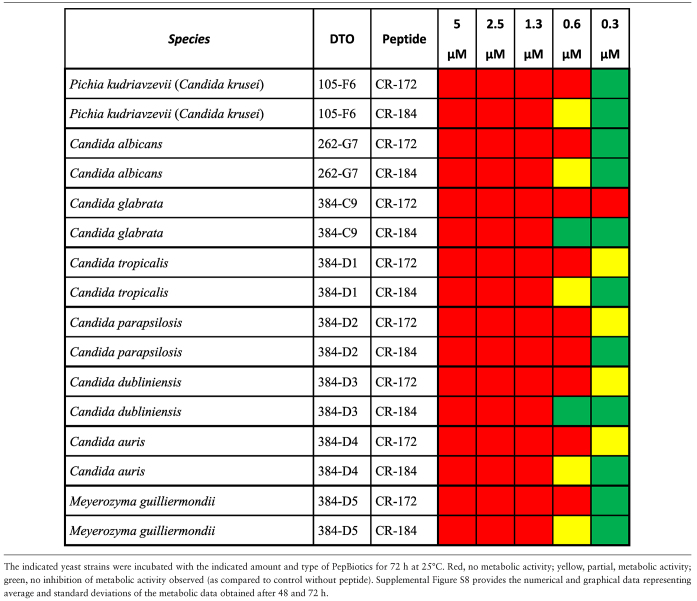
|
Table 6.
B. Sensitivity of Cryptococcus spp. for PepBiotics.
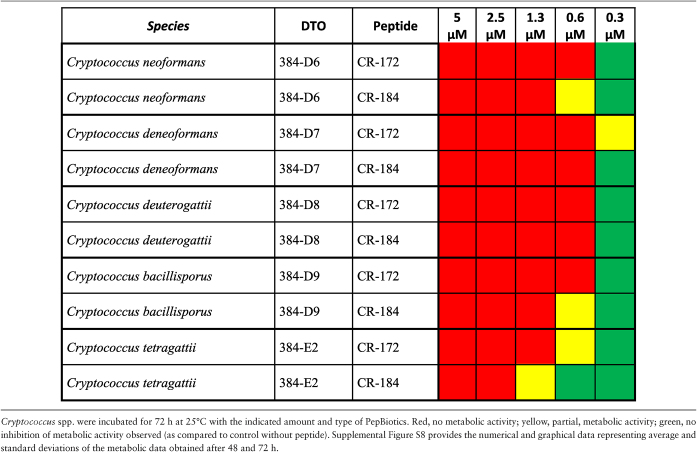
|
Cytotoxicity of 5 μM PepBiotics was determined using primary human nasal epithelial cells and compared to positive (ethanol) and negative control (no peptide) (Table 7). The PepBiotics CR170, CR171, CR172 CR 182, CR184 showed no significant cytotoxic effect (P < 0.05), whereas L-CATH-2 and ethanol did show increased cytotoxic activity to the epithelial cells.
Table 7.
Cytotoxicity tests with a set of PepBiotics.
| Peptide | Average (%) | SD | P-value |
|---|---|---|---|
| No Peptide | 100.0 | ||
| EtOH | 17.5 | 2.9 | .00045 |
| CR-170 | 94.7 | 16.9 | .58 |
| CR-171 | 91.0 | 21.6 | .46 |
| CR-172 | 66.7 | 24.8 | .074 |
| CR-182 | 104.3 | 24.0 | .79 |
| CR-184 | 95.8 | 13.6 | .58 |
| CATH-2 | 45.8 | 9.3 | .010 |
To determine cytotoxic effects of PepBiotics cells were exposed to 5 μM of the indicated peptide. Primary human nasal epithelial cells were used that were isolated and cultivated to confluency in Bronchial Epithelical Cell medium as described in Methods. Controls included no peptide (100% viability) and ethanol (total loss of cell viability). Experiments were performed in triplo.
Discussion
An increase in prevalence of fungal infections is observed worldwide, amongst others due to new medical technologies and possibilities to treat critically ill patients. Especially, the increase in immunocompromised patients is expected to increase the prevalence of invasive and noninvasive fungal infections.36 In addition, this category of patients is also vulnerable to infections by less common fungal species resulting in emerging fungal infections and species of the Mucorales group already belong to that group.37 In addition, increased outbreaks of fungal infections associated with travel or relocation, occupation and contacts in new environmental niches contribute.38 Among the species responsible for such outbreaks are Fusarium sp. causing keratitis, endophthalmitis, and fungemia and Candida auris infections.39,40 The current increase in outbreaks of the multidrug resistant C. auris is very worrying. No established C. auris-specific susceptibility breakpoints have been determined and the currently used breakpoints are based on closely related Candida species.1 The C. auris strain used in this study is the type strain CBS 12777 (also known as CBS10913, DSM21092 or JCM15448)40,41 and antimicrobial resistance testing showed reduced susceptibility to amphotericin B, which is also observed in a large group of C. auris isolates.42 The increased use of antifungals in society and the accompanied increase in antifungal resistance is expected to limit antifungal therapy in the near future even further. Development of new antifungals based on natural and synthetic antifungal peptides has increased accordingly.36 Recent developments in the design of LL-37 based peptides with a minimal core region resulted in various peptides with different biological activities.43
Based on the current knowledge of the antimicrobial properties of the cathelicidin family, a set of cathelicidin-inspired PepBiotics was developed with strong antibacterial activity. These PepBiotics are based on a variety of sequences of cathelicidins and have a unique core sequence (see Table 3). PepBiotics containing the full-length patented sequence either once or twice showed in general antifungal activity at 1 μM. We show that a subset of PepBiotics has broad range antifungal activity against clinically relevant fungal species, including azole-resistant A. fumigatus and C. auris with a reduced susceptibility for mphotericin B. PepBiotics strongly interfere with metabolic activity, growth and/or viability at a submicromolar level, among others of emerging pathogens like Fusarium spp. and Candida auris. All yeasts and filamentous fungi tested in this study were inhibited by two PepBiotics, CR172 and CR184, although 5 μM CR184 was required at 37°C to inhibit A. fumigatus. It is remarkable that the PepBiotics CR172 and CR184 did inhibit metabolic activity of A. fumigatus strains at 37°C more effectively as compared to lower temperature of 25°C. Whether strain-specific differences are involved needs further investigation including the killing potential of different PepBiotics on different A. fumigatus strains. Strain-specific differences for these two PepBiotics were observed for two M. furfur strains and marked differences in the efficiency of killing were observed. It is unclear why strain-specific sensitivities occur, but it is tempting to speculate that binding to target sites but also to other binding sites at the fungal surface might be involved but more research is required to address this. Alternatively, differences in target-site composition might be responsible for observed strain-specific differences. We do not know the targets of the PepBiotics used but it cannot be excluded that intracellular targets like mitochondria are involved as was shown for histatin 5.44 Interestingly, we also observed differences between PepBiotics in their ability to affect viability of A. fumigatus. PepBiotics CR173 and CR183 were able to inactivate hyphae of this fungal species (in this case DTO 342-B2) as observed with PI-staining in contrast to CR172. However, CR172 inhibits growth and metabolic activity. In the presence of the PepBiotics CR172 and CR184 metabolic activity was inhibited but probably not completely or below the detection level of the metabolic assay since germination was still observed. This might be explained by a specific effect of these PepBiotics on mitochondria and possibly comparable to histatin 5,44 but more research is required to address this possibility. Furthermore, the difference in activity between the different PepBiotics (CR173 and CR183 vs CR172) on A. fumigatus might be related to a difference in target sites or efficiency of disrupting the target sites. The metabolic activity assay was not used for M. furfur since no activity was observed despite the observed visible inhibition of growth. We do not have a clear explanation for this observation. Possibly, the mitochondrial activity is already very low of these species in this assay and below the detection level of the assay. An alternative explanation may be that it has been reported that resazurin is trapped in the lipid-rich cell wall layer of Malassezia.45 The PepBiotics CR172 and CR184 affected the viability of M. furfur strains indicating fungicidal activity. Both PepBiotics also affected the growth and metabolic activity of F. oxysporum, F. solani, and T. marneffei, indicative of at least fungistatic activity. The latter species is a thermally dimorphic species causing systemic infections in human immunodeficiency virus (HIV) and non-HIV infected patients and is an emerging pathogen amongst others due to delayed diagnostics.19
In summary, our results show that PepBiotics are very promising antifungals. We showed fungicidal activity for the two Mallasezia furfur strains and fungistatic activity for all yeast species tested and, depending on strain and type of PepBiotic also fungicidal or fungistatic activity for the filamentous fungus A. fumigatus. Since T. marneffei is a thermally dimorphic species and experiments were performed on the filamentous form only (at 25°C), further investigations are required to determine if the pathogenic yeast form is also affected by PepBiotics. As a next step, the use of synthetic peptides enables us to design and further develop species-specific antifungals and to improve the growth inhibiting and/or killing properties against fungal cells. It should be mentioned that reducing growth without killing can already be an advantage in medical treatments as the innate or adaptive immune response, or treatment in combination with antifungals, can work in concert. Future research requires additional testing of PepBiotics on fungal biofilms since these are notoriously more resistant to antifungals.46,47 In this respect, it might also45 be worthwhile to investigate combinations of PepBiotics with other known biofilm-reducing agents which could result in synergistic anti-biofilm activity. Nevertheless, fungal cells escape mature biofilms in the process of generating systemic infections and these can be direct targets for PepBiotics as well.48 In addition, natural biofilms are often fungal-bacterial mixtures and developing PepBiotics with both antifungal and antibacterial activity is clearly an advantage. Other studies have demonstrated the strong and broad range potential of PepBiotics against a wide range of gram-negative and gram-positive pathogens, including multiresistant bacterial isolates derived from CF-patients (manuscript in preparation). Resistance development studies have provided preliminary evidence that PepBiotics do not induce bacterial resistance. In addition, several PepBiotics exhibit anti-biofilm activity against preformed Pseudomonas aeroginosa biofilms, an important pathogen that contributes importantly to persistence of chronic lung infections and frequently develops in co-infection with fungal pathogens. Since the antimicrobial activity of PepBiotics is preserved at physiological conditions (0.9% NaCl) and most PepBiotics possess only mild cytotoxic properties, the data presented in this study indicate that PepBiotics have potential to be an important novel class of antimicrobial drugs to fight several clinically important fungal species effectively.
Supplementary Material
Acknowledgements
The authors acknowledge financial support from “Nederlandse Cystic Fibrosis Stichting” and ZonMw (project 95104010).
Footnotes
Contributor Information
Martin van Eijk, Division of Molecular Host Defence, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Stephanie Boerefijn, Division of Molecular Host Defence, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands; Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Lida Cen, Molecular Microbiology, Department of Biology, Faculty of Science, Utrecht University, Utrecht, The Netherlands.
Marisela Rosa, Division of Molecular Host Defence, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Marnix J H Morren, Division of Molecular Host Defence, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Cornelis K van der Ent, Department of Pediatric Pulmonology, Wilhelmina Children's Hospital, University Medical Center, Utrecht, The Netherlands.
Bart Kraak, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Jan Dijksterhuis, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Ivan D Valdes, Molecular Microbiology, Department of Biology, Faculty of Science, Utrecht University, Utrecht, The Netherlands.
Henk P Haagsman, Division of Molecular Host Defence, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Hans de Cock, Molecular Microbiology, Department of Biology, Faculty of Science, Utrecht University, Utrecht, The Netherlands.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
Author contributions
M.v.E., H.H., H.d.C., I.V., J.D., B.K., and C.v.d.E. designed experiments. M.v.E., H.H., H.d.C., and J.D. wrote the article. L.C., I.V., S.B., M.R., M.M., B.K., and C.v.d.E. read the article and approved it. L.C., S.B., M.R., M.M., and B.K, performed the experiments
References
- 1. Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis. 2017; 17: e383–e392. [DOI] [PubMed] [Google Scholar]
- 2. Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018; 360: 739–742. [DOI] [PubMed] [Google Scholar]
- 3. Revie NM, Iyer KR, Robbins N, Cowen LE. Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol. 2018; 45: 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verweij PE, Chowdhary A, Melchers WJG, Meis JF. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis. 2016; 62: 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kombrink A, Tayyrov A, Essig A et al.. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019; 13: 588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kościuczuk E, Lisowski P, Jarczak J et al.. Cathelicidins: family of antimicrobial peptides. A review. Mol Biol Rep. 2012; 39: 10957–10970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Izadpanah A, Gallo RL. Antimicrobial peptides. J Am Acad Dermatol. 2005; 52: 381–390. [DOI] [PubMed] [Google Scholar]
- 8. Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016; 6: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011; 29: 464–472. [DOI] [PubMed] [Google Scholar]
- 10. Malmsten M. Antimicrobial peptides. Ups J Med Sci. 2014; 119: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Harten RM, van Woudenbergh E, van Dijk A, Haagsman HP. Cathelicidins: immunomodulatory antimicrobials. Vaccines. 2018; 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zanetti L, Tomasinsig M. The cathelicidins: structure, function and evolution. Curr Protein Pept Sci. 2005; 6: 23–34. [DOI] [PubMed] [Google Scholar]
- 13. Coorens M, Scheenstra MR, Veldhuizen EJA, Haagsman HP. Interspecies cathelicidin comparison reveals divergence in antimicrobial activity, TLR modulation, chemokine induction and regulation of phagocytosis. Sci Rep. 2017; 7: 40874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bandurska K, Berdowska A, Barczyńska-Felusiak R, Krupa P. Unique features of human cathelicidin LL-37. Biofactors. 2015; 41: 289–300. [DOI] [PubMed] [Google Scholar]
- 15. van Dijk A, Molhoek EM, Bikker FJ, Yu P-L, Veldhuizen EJA, Haagsman HP. Avian cathelicidins: paradigms for the development of anti-infectives. Vet Microbiol. 2011; 153: 27–36. [DOI] [PubMed] [Google Scholar]
- 16. Veldhuizen EJA, Brouwer EC, Schneider VAF, Fluit AC. Chicken cathelicidins display antimicrobial activity against multiresistant bacteria without inducing strong resistance. PLoS One. 2013; 8: e61964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ordonez SR, Amarullah IH, Wubbolts RW, Veldhuizen EJA, Haagsman HP. Fungicidal mechanisms of cathelicidins LL-37 and CATH-2 revealed by live-cell imaging. Antimicrob Agents Chemother. 2014; 58: 2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kohler JR, Casadevall A, Perfect J. The spectrum of fungi that infects humans. Cold Spring Harb Perspect Med. 2015; 5: a019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan JFW, Lau SKP, Yuen K, Woo PCY. Talaromyces (penicillium) marneffei infection in non-HIV-infected patients. Emerg Microbes Infect. 2016; 5: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J. Human fungal pathogens: why should we learn? J Microbiol. 2016; 54: 145–148. [DOI] [PubMed] [Google Scholar]
- 21. Howard SJ, Cerar D, Anderson MJ et al.. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis. 2009; 15: 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lestrade PPA, Meis JF, Melchers WJG, Verweij PE. Triazole resistance in Aspergillus fumigatus: recent insights and challenges for patient management. Clin Microbiol Infect. 2019; 25: 799–806. [DOI] [PubMed] [Google Scholar]
- 23. Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. 2001; 39: 2431–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buil JB, Snelders E, Denardi LB, Melchers WJG, Verweij PE. Trends in azole resistance in Aspergillus fumigatus, The Netherlands, 1994–2016. Emerging Infect Dis. 2019; 25: 176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gago S, Denning DW, Bowyer P. Pathophysiological aspects of Aspergillus colonization in disease. Med Mycol. 2019; 57: S219–S227. [DOI] [PubMed] [Google Scholar]
- 26. Engel TGP, Slabbers L, de Jong C et al.. Prevalence and diversity of filamentous fungi in the airways of cystic fibrosis patients: a Dutch, multicentre study. J Cyst Fibros. 2019; 18: 221–226. [DOI] [PubMed] [Google Scholar]
- 27. Valdes Ivan D, van den Berg Joris, Haagsman Annika et al.. Comparative genotyping and phenotyping of Aspergillus fumigatus isolates from humans, dogs and the environment. BMC Microbiol. 2018; 18: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen PR, Oddo A. Fmoc solid-phase peptide synthesis. Methods Mol Biol. 2015; 1348: 33. [DOI] [PubMed] [Google Scholar]
- 29. González-Pinzón R, Haggerty R, Myrold DD. Measuring aerobic respiration in stream ecosystems using the resazurin-resorufin system. J Geophys Res: Biogeosci. 2012; 117 doi: 10.1029/2012JG001965. [Google Scholar]
- 30. O'Brien J, Wilson I, Orton T, Pognan F. Investigation of the alamar blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000; 267: 5421–5426. [DOI] [PubMed] [Google Scholar]
- 31. Guerin TF, Mondido M, McClenn B, Peasley B. Application of resazurin for estimating abundance of contaminant-degrading micro-organisms. Lett Appl Microbiol. 2001; 32: 340–345. [DOI] [PubMed] [Google Scholar]
- 32. Meletiadis J, Meis JF, Mouton JW, Verweij PE. Analysis of growth characteristics of filamentous fungi in different nutrient media. J Clin Microbiol. 2001; 39: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. MAv Meegen, SWJ Terheggen-Lagro, Ent Cornelis K. van der, Beekman JM. CFTR expression analysis in human nasal epithelial cells by flow cytometry. PLoS One. 2011; 6: e27658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Dijk A, Molhoek EM, Veldhuizen EJA et al.. Identification of chicken cathelicidin-2 core elements involved in antibacterial and immunomodulatory activities. Mol Immunol. 2009; 46: 2465–2473. [DOI] [PubMed] [Google Scholar]
- 35. Wu G, Zhao H, Li C et al.. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLos Genet. 2015; 11: e1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bondaryk M, Staniszewska M, Zielińska P, Urbańczyk-Lipkowska Z. Natural antimicrobial peptides as inspiration for design of a new generation antifungal compounds. J Fungi (Basel). 2017; 3: E46 doi: 10.3390/jof3030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dannaoui E. Antifungal resistance in mucorales. Int J Antimicrob Agents. 2017; 50: 617–621. [DOI] [PubMed] [Google Scholar]
- 38. Benedict K, Richardson M, Vallabhaneni S, Jackson BR, Chiller T. Emerging issues, challenges, and changing epidemiology of fungal disease outbreaks. Lancet Infect Dis. 2017; 17: e403–e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spivak ES, Hanson KE. Candida auris: an emerging fungal pathogen. J Clin Microbiol. 2018; 56: e01588–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith DM, Dunstone NJ, Scaife AA, Fiedler EK, Copsey D, Hardiman SC. Corrigendum. J Clim. 2018; 31: 4963–4964. [Google Scholar]
- 41. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009; 53: 41–44. [DOI] [PubMed] [Google Scholar]
- 42. Larkin E, Hager C, Chandra J et al.. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother. 2017; 61: e02396–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang G, Narayana JL, Mishra B et al.. Design of antimicrobial peptides: progress made with human cathelicidin LL-37. Adv Exp Med Biol. 2019; 1117: 215–240. [DOI] [PubMed] [Google Scholar]
- 44. Puri S, Edgerton M. How does it kill?: Understanding the candidacidal mechanism of salivary histatin 5. Eukaryotic Cell. 2014; 13: 958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomas DS, Ingham E, Bojar RA, Holland KT. In vitro modulation of human keratinocyte pro- and anti-inflammatory cytokine production by the capsule of Malassezia species. FEMS Immunol Med Microbiol. 2008; 54: 203–214. [DOI] [PubMed] [Google Scholar]
- 46. Delattin N, Brucker KD, Cremer KD, Cammue BPA, Thevissen K. Antimicrobial peptides as a strategy to combat fungal biofilms. Curr Top Med Chem. 2017; 17: 604–612. [DOI] [PubMed] [Google Scholar]
- 47. Scarsini M, Tomasinsig L, Arzese A, D'Este F, Oro D, Skerlavaj B. Antifungal activity of cathelicidin peptides against planktonic and biofilm cultures of candida species isolated from vaginal infections. Peptides. 2015; 71: 211–221. [DOI] [PubMed] [Google Scholar]
- 48. Reichhardt C, Stevens DA, Cegelski L. Fungal biofilm composition and opportunities in drug discovery. Future Med Chem. 2016; 8: 1455–1468. [DOI] [PubMed] [Google Scholar]
- 49. Ferreira da Silva, Eliana Márcia, Kress Marcia R. V. Z., Savoldi M, et al. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryotic Cell. 2006; 5: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nierman WC, Pain A, Anderson MJ et al.. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005; 438: 1151–1156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.