Abstract
Heterogeneity of CD20 expression exists in chronic lymphocytic leukemia (CLL), therefore, we explored the prognostic significance of CD20 expression in Chinese patients with CLL. Multiparameter flow cytometry was used to detect the expression of CD20 in CD5+ CD19+ cells. In 172 CLL patients, the median expression percent of CD20 was 97.82% (range, 0–100), and the median mean fluorescence intensity (MFI) of CD20 in CLL cells was 731.45 (range, 0.00–9071.90). The percentage of CD20+ cells in the patient group with mutated variable region of immunoglobulin genes (IGHV) was higher than in the non‐mutant IGHV group (mean, 92.1% vs 80.4%, P < 0.001). There were no differences in the MFI of CD20+ cells in all prognostic factor groups. Representation of the data using a receiver operating characteristic plot reflected separation between the two IGHV groups, with an area under the curve of 0.661 (95% confidence interval, 0.569–0.753). At the cut‐off value of 60.3% for percentage of CD20, the sensitivity and specificity were 90.00% and 38.46%, respectively. Patients whose percentage of CD20 antigen was above 60.3% had longer treatment‐free survival (hazard ratio, 0.452; 95% confidence interval, 0.232–0.884, P = 0.020). Percentage and MFI of CD20 were the variables not associated with treatment‐free survival by multivariate Cox regression analysis (P < 0.05). High level of CD20 expression in de novo CLL appears to be associated with a good prognosis.
Chronic lymphocytic leukemia (CLL), the most frequently occurring leukemia in adults in North America and Europe, is a heterogeneous disease with variable clinical presentation and evolution.1 In recent years, the significant increase in understanding the pathogenesis of CLL has been translated into a biologically oriented assessment of prognosis. In particular, there has been major progress in the identification of both molecular and cellular markers that may predict disease progression.2, 3 Two major molecular subtypes can be distinguished, characterized by a high or low number of somatic hypermutations in the variable region of immunoglobulin genes (IGHV) that features in CLL. However, IGHV gene sequencing remains a demanding technique, so many studies have focused on the identification of alternative markers with similar prognostic significance, and whose expression is easier to investigate, such as CD38 and zeta‐associated protein‐70 (ZAP‐70).4, 5
A great deal of renewed interest in CD20 membrane protein has emerged since 1997, when the chimeric anti‐CD20 mAb rituximab was approved for use in relapsed or refractory low‐grade or follicular B‐cell non‐Hodgkin's lymphoma. Rituximab is commonly incorporated into CD20‐positive B‐cell lymphoma therapy, including CLL, to improve response and prognosis. The CD20 expression level might correlate with treatment outcome in CLL patients. Tam et al.6 showed that CLL patients with trisomy 12 showed strong expression of CD20 and had a high rate of response to rituximab‐based therapy. However, Hsi et al.7 reported that CD20 expression was not significantly correlated with clinical factors, and CD20 expression was also not associated with overall survival (OS). The prognostic significance of CD20 expression in B‐cell precursor acute lymphoblastic leukemia has been extensively studied in children and adults.8, 9, 10 In childhood acute lymphoblastic leukemia (ALL), Borowitz et al.8 found that high CD20 intensity correlated with poorer event‐free survival. Thomas et al.11 also showed that CD20 positivity (20% or greater positive ALL cells) was associated with worse disease‐free survival and OS, an effect especially relevant in patients aged 30 years and younger. Importantly, the ALL patients in these clinical studies were treated with either conventional or intensive frontline chemotherapy regimens that did not include rituximab. Diffuse large B‐cell lymphoma (DLBCL) cells generally express the pan B‐cell marker CD20, which is an important target for treatment. In their study of CD20 levels at onset of disease in patients with DLBCL, Suzuki et al.12 found that CD20 negative status showed significantly inferior OS and progression‐free survival compared to patients who were CD20 normal.
To date, there is limited information on the prognostic value of CD20 in CLL; it is important to shed light on the real function of this B‐lineage differentiation molecule. The aim of our study was to study the correlation between CD20 expression and the clinical–biological characteristics of CLL patients.
Material and Methods
Ethics statement
All patients provided written informed consent before enrolment in this study. The study was approved by the Institutional Review Board of the Nanjing Medical University (Nanjing, China) and carried out according to the Declaration of Helsinki.
Patients
One hundred and seventy‐two consecutive Chinese patients with CLL were enrolled from January 2003 and December 2011. Blood samples collected at diagnosis were available for CD20, ZAP‐70, and CD38 analyses. The diagnosis of CLL was based on clinical characteristics, peripheral blood morphologies, immunophenotype, and B‐lymphocytes ≥5.0 × 109/L.13 Clinical stage was determined using the Binet staging system according to the International Workshop on Chronic Lymphocytic Leukemia (IWCLL) criteria. Treatment of CLL was based on the IWCLL criteria. The chemotherapy regimens used included: fludarabine and cyclophosphamide; fludarabine, cyclophosphamide, and rituximab; high‐dose methylprednisolone; rituximab and high‐dose methylprednisolone; cyclophosphamide, vincristine, and prednisone; and chlorambucil.
Cellular immunophenotypic analysis
Flow cytometric analysis was carried out on fresh peripheral blood samples. All flow cytometric analyses were carried out on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The instrument was aligned and calibrated daily with the use of a four‐color mixture of CaliBRITE beads (BD Biosciences) with FACSComp Software according to the manufacturer's instructions. The beads were used to adjust instrument settings, set fluorescence compensation, and check instrument sensitivity in order to monitor instrument performance over time. Whole blood was stained with the following three‐color combination of mAbs, CD19/CD22/CD20, CD19/CD5/CD38, and CD19/Lambda/kappa, directly conjugated with phycoerythrin‐cyanin 5.1 (PC5), phycoerythrin (PE), and FITC, respectively. Murine isotype control (BD Biosciences) conjugated with FITC, PE, and PC5 were applied with each sample to set the negative gate. ZAP‐70‐PE (Caltag Laboratories, Burlingame, CA, USA) was used to determine cytoplasmic ZAP‐70 expression according to the manufacturer's instructions. Multicolor flow cytometry was carried out as previously described.14, 15 Briefly, fresh whole blood samples were incubated with a mixture of fluorescence‐labeled mAbs for 15 min at room temperature in the dark. Lysis of mature red blood cells was done using Tris‐NH4Cl solution. Data analysis was carried out with CellQuest software (BD Biosciences), cell subpopulations of interest were delineated using CD45/side scatter dot plots, and after subgating CD19‐positive tumor cells, CD20 expression positivity and mean fluorescence intensity (MFI) were calculated. At least 10 000 events were measured from each sample. Cut‐off points of 30% and 20% were used to define positivity for CD38 and ZAP‐70, respectively.
Detection of molecular cytogenetic aberrations by FISH
The FISH analysis was carried out for conventional cytogenetic studies. Fluorescent‐labeled probes (Vysis, Downers Grove, IL, USA) were used in interphase cytogenetic analyses to detect prognostically relevant abnormalities of chromosomal regions 6q23 (LSI MYB), 11q22.3 (LSI ATM), 13q14 (LSI D13S319), 14q32 (LSI IGHC/IGHV), 17p13 (LSI p53), and centromere 12 (CEP12). The analysis was carried out as previously described.15 The cut‐off levels for positive values (mean of normal control ± 3 SD), determined from samples of eight cytogenetically normal persons, was 7.5%, 7.7%, 10.3%, 8.9%, 5.2%, and 3.0% for del(6q23), del(11q22.3), del(13q14), 14q32 translocation, del(17p13), and trisomy 12, respectively.
Analysis of p53 mutation and IGHV mutation status
Genomic DNA was isolated from PBMC preparations stored at −80°C. Primers to amplify exons 2/3, 4, 5/6, 7, 8/9, 10, and 11 of the human p53 gene and adjacent intronic sequences (GenBank accession NG_017013.1) were designed using the Primer 5 program (http://www.premierbiosoft.com). The PCR products were purified using standard methods (Invitrogen, Shanghai, China) and directly sequenced with primer p53 ex2‐3‐F, ex4‐R, ex5‐6‐F, ex‐7‐F, ex8‐9‐F, ex‐10‐F, and ex‐11‐F using the ABI3730XL 96‐capillary DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequencing of IGHV was carried out as previously described.16 A germline homology of 98% was used as the cut‐off between IGHV mutated and non‐mutated cases.
Statistical analysis
All statistical analyses were carried out with spss version 17.0 (SPSS Inc., Chicago, IL, USA). Group‐wise comparisons of CD20 percentages, MFI, and the other biological and clinical variables were carried out, applying two‐tailed Mann–Whitney U‐tests and Kruskal–Wallis tests with Dunn multiple comparisons. Receiver operating characteristic curve (ROC) and area under the ROC curve (AUC) were established to verify the diagnostic value of CD20 in differentiating the mutation or non‐mutation of IGHV. Treatment‐free survival (TFS) was defined as the period from the diagnosis date to either the time of the first CLL‐specific treatment or to the last follow‐up date. Treatment‐free survival was estimated according to the Kaplan–Meier method and compared between groups by means of the log–rank test. Cox's proportional hazards regression models were used to assess the independent effect of covariables on the TFS. A P‐value of <0.05 was considered statistically significant, and all tests were two‐tailed.
Results
Patient characteristics
The baseline characteristics of the study participants are presented in Table 1. One hundred and thirteen patients were male, and 59 were female (male:female ratio, 1.92:1.00). The median age of the group overall was 62 years (range, 35–93 years); 62.79% were aged 60 or older. According to the Binet clinical staging system, 83 patients (48.26%) were classified as Binet stage A, 36 (20.93%) as Binet stage B, and 53 (30.81%) as Binet stage C. Flow cytometry analysis of peripheral blood specimen lymphocytes showed that 119 (70.41%) of 169 patients had <30% of lymphocytes expressing CD38; and 29.59% of patients had more than 30% CD38‐positive lymphocytes. Forty‐four patients (26.35%) had CLL cells that expressed ZAP‐70 by flow cytometry, and 52 patients (33.98%) had CLL cells that expressed non‐mutated IGHV. Mutation of the p53 gene was present in 28 patients (17.95%). Nine patients (5.23%) had del(13q14) as the sole abnormality, 45 patients (26.16) had del(17p13) deletion or del(11q22.3). With a median follow‐up period of 27 months (range, 1–97 months) from CLL diagnosis, eight patients died of CLL‐related causes. Eighty‐one of 172 patients (47.09%) received chemotherapy for their disease according to the IWCLL criteria. Eighteen patients (50%) were treated with rituximab chemotherapy in the treatment‐naïve state. Eighteen patients were also treated with rituximab chemotherapy, but these patients were all in the relapsed state of disease.
Table 1.
Baseline characteristics of 172 Chinese patients with chronic lymphocytic leukemia (n = 172)
| Parameter | Median (range) | Cases (%) |
|---|---|---|
| Gender (n = 172) | ||
| Male | 113 (65.70) | |
| Female | 59 (34.30) | |
| Age | 62 (35–93) | |
| ≥60 years | 108 (62.79) | |
| <60 years | 64 (37.21) | |
| Binet stage (n = 172) | ||
| A | 83 (48.26) | |
| B | 36 (20.93) | |
| C | 53 (30.81) | |
| CD38 (n = 169) | ||
| Positive | 50 (29.59) | |
| Negative | 119 (70.41) | |
| ZAP‐70 (n = 167) | ||
| Positive | 44 (26.35) | |
| Negative | 123 (73.65) | |
| IGHV (n = 153) | ||
| Unmutated (≤2% deviation from a germline) | 52 (33.98) | |
| Mutated (>2% deviation from a germline) | 101 (66.02) | |
| p53 Mutation status (n = 156) | ||
| Presence | 28 (17.95) | |
| Absence | 128 (82.05) | |
| Treatment (n = 172) | ||
| No | 91 (52.91) | |
| Yes | 81 (47.09) | |
| Treatment regime (n = 81) | ||
| Rituximab‐based regimes | 36 (44.44) | |
| Other regimes | 45 (55.56) | |
| Cytogenetics (n = 172) | ||
| del(17p13) or del(11q22.3) | 45 (26.16) | |
| del(13q24) as the sole abnormality | 9 (5.23) | |
| Other | 118 (68.60) | |
| CD20% (n = 172) | 97.82% (0–100) | |
| CD20 MFI (n = 172) | 731.45 (0–9071.90) | |
IGHV, variable region of immunoglobulin genes; MFI, mean fluorescence intensity; ZAP‐70, zeta‐associated protein‐70.
Expression of CD20 and association with other prognostic factors
In a total of 172 CLL cases, the median expression percent of CD20 was 97.82% (range, 0–100%), and MFI of CD20 on CLL cells was 731.45 (range, 0–9071.90). The possibility of interaction between CD20 and other known prognostic factors was analyzed in our cohort (Table 2). There were no differences in the percentage of CD20‐positive cells in the prognostic factors groups, such as Binet stage, ZAP‐70, CD38 expression, p53 mutation status, and cytogenetic abnormalities (P > 0.05), but expression of CD20 was associated with IGHV mutational status in CLL (P < 0.001). The percentage of CD20‐positive cells in cases of mutated IGHV groups was higher than non‐mutant groups (mean, 92.1% vs 80.4%). There were also no differences in the MFI of CD20‐positive cells in each prognostic factors group (P > 0.05). Compared with non‐mutant groups, MFI of CD20‐positive cells in mutated IGHV groups was higher (mean, 1396 vs 1051.4), however, there was no statistical significance (P = 0.19)
Table 2.
Correlations between CD20 expression and disease features of chronic lymphocytic leukemia
| Parameter | CD20% | CD20 MFI | ||||
|---|---|---|---|---|---|---|
| No. of cases | Median (P5–P95) | P‐value | No. of cases | Median (P5–P95) | P‐value | |
| Gender (n = 172) | ||||||
| Male | 113 | 96.22 (37.26–100) | 0.30 | 113 | 863.44 (88.78–4451.36) | 0.20 |
| Female | 59 | 100 (51.10–100) | 59 | 588.52 (81.05–3466.56) | ||
| Age (n = 172) | ||||||
| ≥60 years | 109 | 97.71 (46.55–100) | 0.88 | 109 | 952.26 (99.05–3933.46) | 0.06 |
| <60 years | 63 | 98.40 (36.84–100) | 63 | 524.15 (53.50–4477.11) | ||
| Binet stage (n = 172) | ||||||
| A | 83 | 97.71 (53.70–100) | 0.52 | 83 | 731.64 (77.23–4630.52) | 0.48 |
| B | 36 | 91.90 (31.87–100) | 36 | 547.28 (84.81–3250.45) | ||
| C | 53 | 100 (27.60–100) | 53 | 833.09 (159.80–4044.24) | ||
| CD38 (n = 169) | ||||||
| Positive | 50 | 98.73 (56.09–100) | 1.00 | 50 | 733.38 (92.45–3933.07) | 0.60 |
| Negative | 119 | 97.71 (37.97–100) | 119 | 731.64 (82.81–3915.94) | ||
| ZAP‐70 (n = 167) | ||||||
| Positive | 44 | 97.08 (52.40–100) | 0.86 | 44 | 598.21 (97.36–3935.30) | 0.84 |
| Negative | 123 | 97.46 (39.00–100) | 123 | 751.56 (96.03–3857.34) | ||
| IGHV (n = 153) | ||||||
| Unmutated | 52 | 90.48 (3.03–100) | <0.001 | 52 | 480.71 (78.74–3569.62) | 0.19 |
| Mutated | 101 | 100 (50.99–100) | 101 | 751.56 (83.86–4666.35) | ||
| p53 mutation status (n = 156) | ||||||
| Presence | 28 | 96.61 (3.97–100) | 0.36 | 28 | 469.89 (145.07–4737.80) | 0.73 |
| Absence | 128 | 98.29 (51.92–100) | 128 | 731.45 (81.84–3933.82) | ||
| Treatment (n = 172) | ||||||
| No | 91 | 100 (56.42–100) | 0.10 | 91 | 930.26 (79.14–4324.41) | 0.30 |
| Yes | 81 | 95.06 (37.56–100) | 81 | 559.89 (107.62–3533.46) | ||
| Cytogenetics (n = 172) | ||||||
| del(17p13) or del(11q22.3) | 45 | 98.96 (3.78–100) | 0.59 | 45 | 548.16 (93.49–6766.42) | 0.21 |
| del(13q14) as the sole abnormality | 9 | 88.83 (65.93–100) | 9 | 313.30 (82.81–3199.19) | ||
| Others | 118 | 97.82 (44.60–100) | 118 | 818.77 (90.59–3930.09) | ||
IGHV, variable region of immunoglobulin genes; MFI, mean fluorescence intensity; ZAP‐70, zeta‐associated protein‐70.
Analyses using ROC and AUC
We carried out the ROC curve and AUC analyses to assess the sensitivity and specificity of the CD20 signature for CLL prognosis risk estimation. Representation of the data using a ROC plot reflected strong separation between these two subgroup of CLL with an AUC of 0.661 (95% confidence interval [CI], 0.569–0.753) for the percentage of CD20 (Fig. 1). At the cut‐off value of 60.3% for percentage of CD20, the sensitivity and specificity were 90.00% and 38.46%, respectively.
Figure 1.
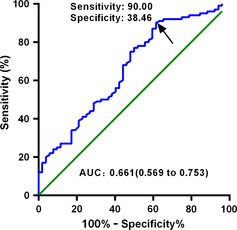
Receiver operating characteristic curve of CD20 expression to predict the mutation status of the variable region of immunoglobulin genes in chronic lymphocytic leukemia. AUC, area under the curve. Arrows point to cutoff value of 60.3% percentage of CD20, the sensitivity and specificity were 90.00% and 38.46%, respectively.
Prognostic impact of CD20 on TFS
According to the set cut‐off value of percentage of CD20, TFS was researched in the CD20 subgroup. Patients whose percentage of CD20 antigen was above 60.3% had a better TFS (hazard ratio [HR], 0.452; 95% CI, 0.232–0.884, P = 0.020) (Fig. 2). Pretreatment characteristics that correlated with longer TFS by univariable analysis are shown in Table 3. Predictors for progression‐free survival were ZAP‐70 (≥20%) (HR = 1.959; 95% CI, 1.23–3.12, P = 0.005) and IGHV mutation (HR = 0.478; 95% CI, 0.303–0.755, P = 0.002). To assess whether CD20 percentage alone represents a prognostic factor, the group of patients who underwent rituximab chemotherapy were analyzed separately from the patients without rituximab. The rituximab treatment affected the prognostic impact of the percentage of CD20 in patients in a naïve state of disease (P = 0.12; P = 0.52) (Fig. S1), however, we cannot draw any conclusions from this data because of the limited number of patients. We researched the co‐expression pattern of ZAP‐70 and CD20 and their impact on TFS (Fig. S2). There were significant statistical differences between the groups (P = 0.01) The low ZAP‐70/high CD20 group had longer TFS when compared with the other groups, with median TFS times being 72 months (low ZAP‐70/high CD20), 15 months (high ZAP‐70/high CD20), 4 months (low ZAP‐70/low CD20), and 3 months (high ZAP‐70/low CD20), respectively. In multivariable analysis, no variable selection technique was used, and all variables remained in the multivariable model. Multivariate Cox proportional hazards models were used to study factors associated with TFS endpoints in CLL by both Enter and Forward conditional methods, pretreatment patient characteristics independently associated with lower risk of disease progression included Binet A, higher CD20 expression, lower ZAP‐70 expression, and IGHV mutation (Table 4). Percentage of CD20 was not associated with TFS by multivariate Cox regression analysis (P = 0.087). In conclusion, a higher level of CD20 expression in de novo CLL appears to be associated with a better prognosis.
Figure 2.
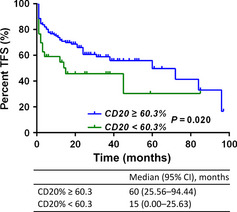
Prognostic impact of CD20 on treatment‐free survival (TFS) in patients with chronic lymphocytic leukemia. Those patients whose percentage of CD20 antigen was above 60.3% had a better TFS (hazard ratio [HR], 0.452; 95% confidence interval [CI], 0.232–0.884, P = 0.020).
Table 3.
Univariable Cox proportional hazards models for treatment‐free survival in Chinese patients with chronic lymphocytic leukemia (n = 172)
| Characteristic | No. of cases | Hazard ratio (95% CI) | P‐value |
|---|---|---|---|
| Age (years), <60 vs ≥60 | 172 | 0.141 (0.894–2.197) | 0.1410 |
| Gender (male vs female) | 172 | 1.326 (0.825–2.133) | 0.2440 |
| Binet stage A (vs C) | 136 | 0.231 (0.131–0.405) | <0.0001 |
| Binet stage B (vs C) | 89 | 0.979 (0.589–1.626) | 0.9340 |
| CD38 ≥ 30% (vs <30%) | 169 | 1.493 (0.948–2.354) | 0.0840 |
| ZAP‐70 ≥ 20% (vs <20%) | 167 | 1.959 (1.230–3.120) | 0.0050 |
| IGHV mutation (vs non‐mutation) | 153 | 0.478 (0.303–0.755) | 0.0020 |
| p53 mutation (vs non‐mutation) | 156 | 1.507 (0.879–2.582) | 0.1360 |
| Unfavorable karyotypea (vs favorablea) | 54 | 1.072 (0.457–2.513) | 0.8730 |
| Others (vs favorablea) | 127 | 0.590 (0.261–1.335) | 0.2050 |
| CD20 ≥ 60.3% (vs <60.3%) | 172 | 0.452 (0.232–0.884) | 0.0200 |
†Unfavorable karyotype, del(17) and del(11q). ‡Favorable karyotype, del(13q24) as the sole abnormality. CI, confidence interval; IGHV, variable region of immunoglobulin genes; MFI, mean fluorescence intensity; ZAP‐70, zeta‐associated protein‐70. Bold values indicate statistically significant.
Table 4.
Multivariate Cox proportional hazards models for treatment‐free survival (TFS) in chronic lymphocytic leukemia
| Characteristic | Hazard ratio (95% CI) | P‐value |
|---|---|---|
| TFS (n = 138) | ||
| Age (years), <60 vs ≥60 | 1.117 (0.661–1.885) | 0.680 |
| Gender (male vs female) | 1.226 (0.726–2.072) | 0.446 |
| Binet stage A (vs C) | 0.339 (0.177–0.647) | 0.001 |
| Binet stage B (vs C) | 1.051 (0.548–2.013) | 0.882 |
| CD38 ≥ 30% (vs <30%) | 0.963 (0.535–1.731) | 0.898 |
| ZAP‐70 ≥ 20% (vs <20%) | 2.428 (1.354–4.355) | 0.003 |
| IGHV mutation (vs non‐mutation) | 0.787 (0.454–1.364) | 0.393 |
| p53 Mutation (vs non‐mutation) | 0.961 (0.529–1.748) | 0.897 |
| Unfavorable karyotypea (vs favorablea) | 1.337 (0.508–3.520) | 0.557 |
| Unfavorable karyotypea (vs others) | 0.980 (0.380–2.530) | 0.967 |
| CD20 ≥ 60.3% (vs <60.3%) | 0.510 (0.236–1.104) | 0.087 |
†Unfavorable karyotype, del(17p13) and del(11q22.3). ‡Favorable karyotype, del(13q14) as the sole abnormality. CI, confidence interval; IGHV, variable region of immunoglobulin genes; ZAP‐70, zeta‐associated protein‐70. Bold values indicate statistically significant.
Discussion
Chronic lymphocytic leukemia is less effectively treated than other B‐cell malignancies with the anti‐CD20 agent rituximab, presumably, at least in part, due to low CD20 expression.17 Expression of CD20 is typically measured by flow cytometry. The aim of our study was to study the correlated CD20 expression with clinical–biological characteristics.
Previously, there are few data on the level of CD20 expression in Chinese CLL patients. Data presented in this report confirm that total levels of CD20 protein are low in CLL B cells. The median expression percent of CD20 was 97.82% (range, 0–100%), and the median MFI of CD20 on CLL was 731.45 (range, 0.00–9071.90). Ginaldi et al.18 first reported that the level of CD20 expression in patients with the leukemic form of B‐cell lymphoma is significantly lowest in CLL and the highest in hairy cell leukemia. The number of CD20 molecules was lower in CLL than in normal B cells and the other B‐cell leukemias (P < 0.05). CD20 is a transmembrane phosphoprotein that functions as a calcium channel, and it has been shown to play an important role in B cell activation and differentiation.19 CD20 expression appears later than other B‐cell markers during normal B lymphocyte development and its membrane density progressively increases during differentiation.20 The low percentage and intensity of CD20 in CLL probably reflects an earlier stage of maturation of the B‐CLL lymphocytes compared with normal blood B lymphocytes and other malignant B cells. The CLL clones use either mutated or non‐mutated IGHV genes and this feature gave rise to the postulate that the two subgroups of CLL originated from distinct cell types. We found that lower CD20 expression was significantly correlated with non‐mutation of IGHV. Low expression of CD20 CLL cells may originate from antigen‐experienced B lymphocytes that have no IGHV gene mutation.
From a clinical viewpoint, numerous clinical and preclinical studies on rituximab indicate that treatment with this drug has outgrown its empirical use. We can confirm its efficacy only in patients in whom CD20 expression was determined in CLL cells, but we are not sure whether the level of surface CD20 expression and CD20 MFI could be a predictive factor, identifying the patients in whom treatment with rituximab would be efficient and those in whom the use of this drug would be inefficient. Johnson et al.21 showed that CD20 expression and response to treatment with rituximab are associated in DLBCL patients. Patients with a level of CD20 expression above the cut‐off value had a significantly longer OS and a significantly higher overall response rate than those patients with CD20 expression levels below the cut‐off value. However, we observed no significant difference in TFS between the two groups, although there was a trend toward lower risk for achievement of treatment (P = 0.087). The reason that we did not see any significant P‐values for CD20 expression in either current model may be related to parameter and parameter interaction. As our study was retrospective, both factors contributed to our results, therefore the study might not necessarily be representative.
The results of our study indicate that the level of CD20 expression is an important predictive factor. Datasets compiled by the prospective collection of well‐characterized CLL patients are now surely warranted in order to confirm the clinical significance of CD20 in CLL.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Prognostic impact of CD20 on treatment‐free survival in the group of patients with rituximab chemotherapy and non‐rituximab treatment. Rituximab treatment affected the prognostic impact of the percentage of CD20 in patients in a naïve state of disease (P = 0.12; P = 0.52).
Fig. S2. Co‐expression pattern of ZAP‐70 and CD20 and their impact on treatment‐free survival (TFS). There were significant statistical differences between groups in terms of TFS (P = 0.01). The low ZAP‐70/high CD20 group had a longer TFS compared with other groups; the median TFS was 72 months (low ZAP‐70/high CD20), 15 months (high ZAP‐70/high CD20), 4 months (low ZAP‐70/low CD20) and 3 months (high ZAP‐70/low CD20), respectively.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 30871104, 30971296, 81170485, 81170488), the Natural Science Foundation of Jiangsu Province (Grant No. BK2010584), Key Projects of Health Department of Jiangsu Province (Grant No. K201108), Jiangsu Province's Medical Elite Program (Grant No. RC2011169), the University Doctoral Foundation of the Ministry of Education of China (Grant No. 20093234110010), the Program for Development of Innovative Research Teams in the First Affiliated Hospital of Nanjing Medical University, Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and Project of National Key Clinical Specialty.
(Cancer Sci 2013; 104: 996–1001)
References
- 1. Eichhorst B, Dreyling M, Robak T, Montserrat E, Hallek M, Group EGW . Chronic lymphocytic leukemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2011; 22 (Suppl. 6): vi50–4. [DOI] [PubMed] [Google Scholar]
- 2. Hamblin TJ. Searching for surrogates for IGHV mutations in chronic lymphocytic leukemia. Leuk Res 2011; 35: 1432–5. [DOI] [PubMed] [Google Scholar]
- 3. Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50. [DOI] [PubMed] [Google Scholar]
- 4. Malavasi F, Deaglio S, Damle R, Cutrona G, Ferrarini M, Chiorazzi N. CD38 and chronic lymphocytic leukemia: a decade later. Blood 2011; 118: 3470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rassenti LZ, Huynh L, Toy TL et al ZAP‐70 compared with immunoglobulin heavy‐chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med 2004; 351: 893–901. [DOI] [PubMed] [Google Scholar]
- 6. Tam CS, Otero‐Palacios J, Abruzzo LV et al Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in‐situ hybridization in 510 patients. Br J Haematol 2008; 141: 36–40. [DOI] [PubMed] [Google Scholar]
- 7. Hsi ED, Kopecky KJ, Appelbaum FR et al Prognostic significance of CD38 and CD20 expression as assessed by quantitative flow cytometry in chronic lymphocytic leukaemia. Br J Haematol 2003; 120: 1017–25. [DOI] [PubMed] [Google Scholar]
- 8. Borowitz MJ, Shuster J, Carroll AJ et al Prognostic significance of fluorescence intensity of surface marker expression in childhood B‐precursor acute lymphoblastic leukemia. A pediatric oncology group study. Blood 1997; 89: 3960–6. [PubMed] [Google Scholar]
- 9. Jeha S, Behm F, Pei D et al Prognostic significance of CD20 expression in childhood B‐cell precursor acute lymphoblastic leukemia. Blood 2006; 108: 3302–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maury S, Huguet F, Leguay T et al Adverse prognostic significance of CD20 expression in adults with Philadelphia chromosome‐negative B‐cell precursor acute lymphoblastic leukemia. Haematologica 2010; 95: 324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas DA, O'Brien S, Jorgensen JL et al Prognostic significance of CD20 expression in adults with de novo precursor B‐lineage acute lymphoblastic leukemia. Blood 2009; 113: 6330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki Y, Yoshida T, Wang G et al Association of CD20 levels with clinicopathological parameters and its prognostic significance for patients with DLBCL. Ann Hematol 2012; 91: 997–1005. [DOI] [PubMed] [Google Scholar]
- 13. Hallek M, Cheson BD, Catovsky D et al Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute‐Working Group 1996 guidelines. Blood 2008; 111: 5446–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu W, Li JY, Wu YJ et al CD38 as a prognostic factor in Chinese patients with chronic lymphocytic leukaemia. Leuk Res 2009; 33: 237–43. [DOI] [PubMed] [Google Scholar]
- 15. Xu W, Li JY, Wu YJ et al Prognostic significance of ATM and TP53 deletions in Chinese patients with chronic lymphocytic leukemia. Leuk Res 2008; 32: 1071–7. [DOI] [PubMed] [Google Scholar]
- 16. Chen L, Zhang Y, Zheng W et al Distinctive IgVH gene segments usage and mutation status in Chinese patients with chronic lymphocytic leukemia. Leuk Res 2008; 32: 1491–8. [DOI] [PubMed] [Google Scholar]
- 17. Sarro SM, Unruh TL, Zuccolo J et al Quantification of CD20 mRNA and protein levels in chronic lymphocytic leukemia suggests a post‐transcriptional defect. Leuk Res 2010; 34: 1670–3. [DOI] [PubMed] [Google Scholar]
- 18. Ginaldi L, De Martinis M, Matutes E, Farahat N, Morilla R, Catovsky D. Levels of expression of CD19 and CD20 in chronic B cell leukaemias. J Clin Pathol 1998; 51: 364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Musto P, D'Auria F. The clinical and biological role of CD20 membrane antigen modulation under immunotherapy with anti‐CD20 monoclonal antibody rituximab in lymphoprolipherative neoplastic disorders. Expert Opin Biol Ther 2011; 11: 551–7. [DOI] [PubMed] [Google Scholar]
- 20. SH S, E C, NL H et al WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. Lyon, France: IARC Press, 2008. [Google Scholar]
- 21. Johnson NA, Boyle M, Bashashati A et al Diffuse large B‐cell lymphoma: reduced CD20 expression is associated with an inferior survival. Blood 2009; 113: 3773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Prognostic impact of CD20 on treatment‐free survival in the group of patients with rituximab chemotherapy and non‐rituximab treatment. Rituximab treatment affected the prognostic impact of the percentage of CD20 in patients in a naïve state of disease (P = 0.12; P = 0.52).
Fig. S2. Co‐expression pattern of ZAP‐70 and CD20 and their impact on treatment‐free survival (TFS). There were significant statistical differences between groups in terms of TFS (P = 0.01). The low ZAP‐70/high CD20 group had a longer TFS compared with other groups; the median TFS was 72 months (low ZAP‐70/high CD20), 15 months (high ZAP‐70/high CD20), 4 months (low ZAP‐70/low CD20) and 3 months (high ZAP‐70/low CD20), respectively.


