Abstract
Most cancer cells are aneuploid, which could be caused by defects in chromosome segregation machinery. Nucleoporins (Nup) are components of the nuclear pore complex, which is essential for nuclear transport during interphase, but several nucleoporins are also known to be involved in chromosome segregation. Here we report a novel function of Nup188, one of the nucleoporins regulating chromosome segregation. Nup188 localizes to spindle poles during mitosis, through the C‐terminal region of Nup188. In Nup188‐depleted mitotic cells, chromosomes fail to align to the metaphase plate, which causes mitotic arrest due to the spindle assembly checkpoint. Both the middle and the C‐terminal regions were required for chromosome alignment. Robust K‐fibers, microtubule bundles attaching to kinetochores, were hardly formed in Nup188‐depleted cells. Significantly, we found that Nup188 interacts with NuMA, which plays an instrumental role in focusing microtubules at centrosomes, and NuMA localization to spindle poles is perturbed in Nup188‐depleted cells. These data suggest that Nup188 promotes chromosome alignment through K‐fiber formation and recruitment of NuMA to spindle poles.
An abnormal number of chromosomes, which is called aneuploidy, is a hallmark of most cancer cells.1 Aneuploidy is often preceded by chromosomal instability, an elevated rate of gain or loss of chromosomes per cell cycle.2 This pathological condition typically arises from defects in the chromosome segregation machinery. In virtually all animal cells, chromosome segregation is carried out by the spindle, which consists of two separated centrosome pairs at spindle poles and microtubule bundles that connect spindle poles to chromosomes. Microtubules attach to chromosomes by virtue of kinetochores, large protein complexes that reside on the centromeric region of chromosomes. Proteins localizing to the spindle and kinetochores are known to act together to conduct chromosome segregation. The number of these proteins has been increasing, including those that have been characterized to function for cellular processes other than mitosis.3
Nuclear pore complexes (NPC) mediate all transport across the nuclear envelope (NE). Nuclear pore complexes consist of approximately 30 proteins called nucleoporins (Nup), which are organized in subcomplexes within the NPC.4 At the onset of mitosis, NE and NPC disassemble, although Nup often remain associated in subcomplexes, some of which are found associated with mitotic structures, such as kinetochores or the spindle.5 Over the past few years, a growing number of Nup have been linked to mitotic processes like spindle assembly, kinetochore function and the spindle assembly checkpoint (SAC).6, 7 For example, the largest Nup subcomplex called Nup107–160 complex localizes to kinetochores and controls kinetochore–microtubule interaction.8, 9, 10 In contrast, Rae1 and Nup88, other Nup, were found to localize to the spindle and function for spindle formation.11, 12, 13
Intriguingly, several Nup have been linked to cancers.14 Caused by chromosomal translocations, Nup98, Nup214 and Tpr form fusion proteins with various partners, found mainly in leukemias.15, 16, 17, 18 Nup88 was found to be overexpressed in a wide variety of tumors.19, 20, 21 Remarkably, defects in nuclear transport have not been reported in the case of nucleoporin translocations, suggesting that impairment of other functions of Nup contributes to tumorigenesis. In addition, gene knockout of Nup358 (RanBP2) in mice led to increased aneuploidy and development of tumors,22 supporting the idea that deregulation of Nup affects mitotic fidelity and promotes tumor formation.
Nup188 is a component of the Nup93 subcomplex, the second major structural unit of the NPC.23 This subcomplex is known to control the passage of membrane proteins from the outer nuclear membrane to the inner nuclear membrane and is therefore crucial for integrity of nuclear membranes.23 Importantly, the NUP188 gene resides in the region (9q34) that is frequently deleted in adult B‐cell acute lymphoblastic leukemia (B‐ALL).24 However, the pathological significance of this deletion and/or mutation remains unclear. Here, we addressed whether Nup188 has any function other than membrane transport and found that it plays an important role in mitosis. Nup188 appears essential for the formation of K‐fibers (microtubule bundles attaching to kinetochores) and for the recruitment of the spindle‐organizing protein NuMA to spindle poles. These findings identify a novel mitotic function for Nup and might provide the pathological grounds for Nup188 inactivation.
Materials and Methods
Cell culture and transfections
HeLa cells were used unless otherwise indicated. Cells were cultured in DMEM supplemented with 10% fetal bovine serum. For synchronization at the G1/S boundary, cells were cultured in the presence of 2 mM thymidine for 18 h. For synchronization at M phase, cells were cultured in the presence of 2 mM thymidine for 18 h, washed once with PBS, cultured in fresh medium for 3 h and then cultured in the presence of 100 ng/mL nocodazole or 1 μM taxol for 12 h. Transfection of siRNA oligonucleotides was carried out by incubating 100 nM duplexed siRNA with RNAi MAX (Life Technologies, Carlsbad, CA, USA) in antibiotic‐free growth medium. For control transfections, the same annealing reaction was performed using H2O instead of siRNA oligos. Transfection of expression plasmids was performed using FuGENE 6 Transfection Reagent (Promega, Madison, WI, USA). For rescue experiments, siRNA‐treated cells were transfected with the indicated plasmids 36 h later using Lipofectamine 2000 (Life Technologies), according to the manufacturer's protocol. Cells were observed 24 h after transfection of the expression plasmids.
Plasmid construction
The cDNA coding for human NUP188 were PCR‐amplified from a human brain cDNA library. The cDNA were subcloned into pcDNA5/FRT (Life Technologies) or pEGFP‐C1 (Clontech, Santa Clara, CA, USA) expression vectors. The expression plasmids for NUP188 deletion mutants were constructed using PCR amplification. The expression plasmids for RNAi‐resistant NUP188 constructs were constructed using the QuikChange Multi Site‐Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). The cDNA coding for human NuMA was kindly gifted from Dr K. Mizuno (Tohoku University). For protein expression using the baculovirus system, the cDNA coding for GST‐tagged NuMA and Nup188 was subcloned into a pFastBac1 vector (Life Technologies).
Immunofluorescence analysis
Cells grown on coverslips were fixed with acetone/methanol (1:1) for 10 min at room temperature or with methanol for 10 min. For Figures 1c, 2c, 3d, 6a and 7b, S1(B) and S2(C), cells were pre‐permeabilized with 0.1% Triton X‐100 for 1 min. For Figures 1a,b, 2e, S1(A) and S2(B), cells were fixed with 4% formaldehyde for 15 min at room temperature and permeabilized with 0.2% Triton X‐100 for 5 min. Cells were incubated with primary antibodies overnight at room temperature, followed by incubation with secondary antibodies for 1 h. The secondary antibodies used in the present study were goat anti‐rabbit IgG Alexa‐Fluor‐488 and 568, goat anti‐mouse Alexa‐Fluor‐488 and 568 and goat anti‐human IgG Alexa‐Fluor‐568 (Life Technologies). For antibody dilutions, 0.01% (v/v) Triton X‐100 in PBS with 1% BSA (w/v) was used. After a 5‐min incubation with 0.1 μg/mL 4', 6‐diamidino‐2‐phenylindole (DAPI), cells were mounted with Fluorescent Mounting Medium (Agilent Technologies). Image stacks were acquired using a Personal DV microscope (Applied Precision, Issaquah, WA, USA) with a x100 1.40 NA Plan Apochromat oil objective lens (Olympus, Tokyo, Japan) and a cooled charge‐coupled device camera (CoolSNAP HQ; Photometrics, Tucson, AZ, USA), driven by softWoRx software (Applied Precision). Z‐series image stacks (which were deblurred using softWoRx deconvolution in Figs 2e, 3a,c,d, 5a,b, and S2C) were presented as maximal intensity projections.
Figure 1.
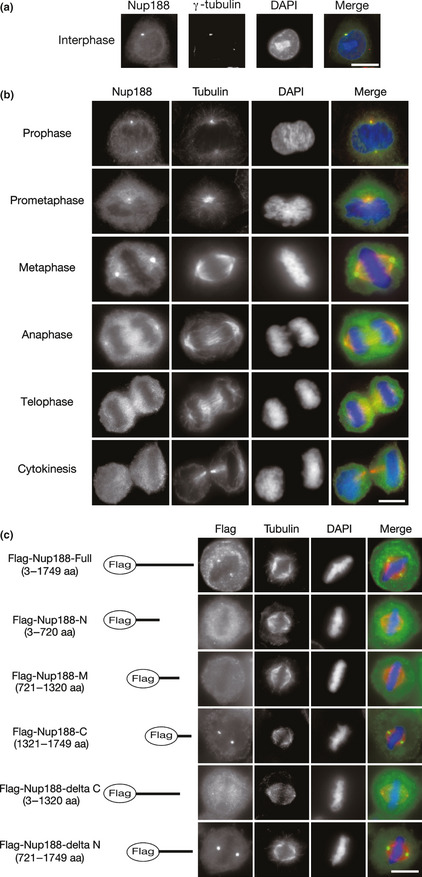
Nup188 localizes to centrosomes in mitosis. (a) Subcellular localization of Nup188 in interphase. HeLa cells were stained with anti‐Nup188 (green) and anti‐γ‐tubulin (red) antibodies. DNA was stained with DAPI (blue). Bar, 10 μm. (b) Subcellular localization of Nup188 during mitosis. HeLa cells were stained with anti‐Nup188 (green) and anti‐α‐tubulin (red) antibodies. DNA was stained with DAPI (blue). Bar, 10 μm. (c) Subcellular localization of Nup188 deletion mutants in metaphase. Localization of Flag‐tagged full‐length Nup188 or deletion mutants expressed in HeLa cells is shown (green). Microtubules were stained with anti‐α‐tubulin antibody (red) and DNA was stained with DAPI (blue). Bar, 10 μm.
Figure 2.
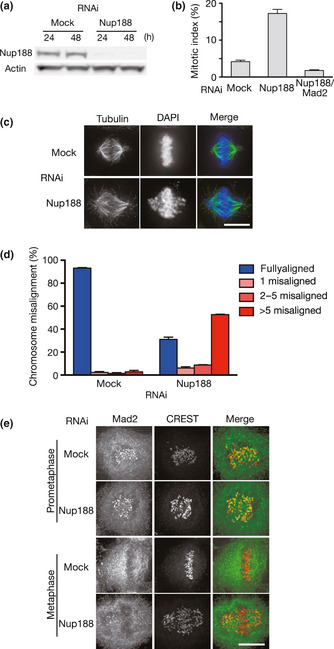
Nup188 depletion causes chromosome misalignment. (a) Depletion of Nup188 with a siRNA. Total cell lysates prepared from HeLa cells treated with mock or Nup188 siRNA for the indicated periods were separated by SDS‐PAGE and probed using western blotting with anti‐Nup188 or anti‐actin antibody. (b) Increased mitotic index in Nup188‐depleted cells. The mitotic index for mock‐, Nup188 siRNA‐ or Nup188/Mad2 siRNA‐treated HeLa cells was measured 48 h after transfection. Error bars represent the SD. (c) Nup188‐depleted cells showed chromosome misalignment. HeLa cells treated with mock or Nup188 siRNA for 48 h were stained with an anti‐tubulin antibody (green). DNA was stained with DAPI (blue). Bar, 10 μm. (d) Quantitative analysis of chromosome misalignment in Nup188‐depleted cells. HeLa cells were treated with mock or Nup188 siRNA for 48 h and with MG132 (10 μM) for the final 2 h. The number of misaligned chromosomes per cell was scored. Error bars represent the SD. (e) Mad2 localizes to kinetochores in Nup188‐depleted cells. HeLa cells treated with mock or Nup188 siRNA for 48 h were stained with an anti‐Mad2 antibody (green) and a CREST serum (red). Bar, 10 μm.
Figure 3.
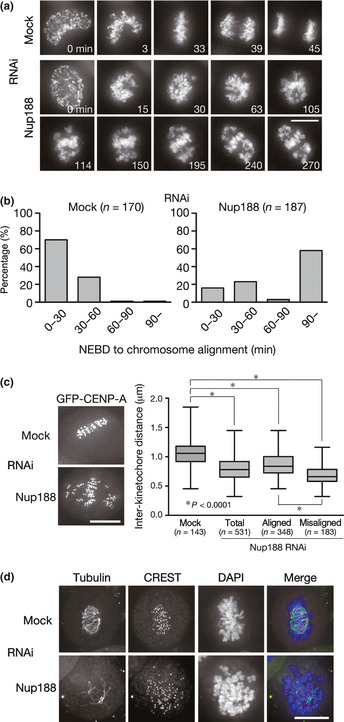
Defective K‐fiber formation in Nup188‐depleted cells. (a) Live cell imaging of HeLa cells expressing histone H2B‐GFP 48 h after mock‐ or Nup188‐siRNA transfection. T = 0 was defined as the time when the nuclear envelope was broken down (NEBD). Bar, 10 μm. Also see Movies S1 and S2. (b) Prolonged mitosis in Nup188‐depleted cells. HeLa cells were treated as in (a). The time from NEBD to chromosome alignment on the metaphase plate was measured in live cell imaging and categorized. The percentages of cells in each category are shown in the graph. (c) Quantitative analysis of interkinetochore distance in Nup188‐depleted cells. HeLa cells expressing GFP‐CENP‐A were treated with mock or Nup188 siRNA for 48 h and the distance between CENP‐A on sister kinetochores was measured. (d) Instability of kinetochore microtubules in Nup188‐depleted cells. HeLa cells were treated as in (c) and then incubated on ice for 10 min before fixation. Cells were stained for kinetochores (CREST, red), tubulin (green) and DNA (blue). Bar, 10 μm.
Figure 6.
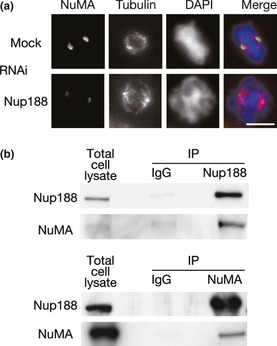
Centrosomal localization of NuMA is affected by Nup188 depletion. (a) Subcellular localization of NuMA in Nup188‐depleted cells. HeLa cells were treated with mock or Nup188 siRNA for 48 h and with MG132 for the final 2 h. Cells were stained for NuMA (green), tubulin (red) and DNA (blue). Bar, 10 μm. (b) Nup188 interacts with NuMA. Total cell lysates prepared from HeLa cells treated with nocodazole were subjected to immunoprecipitation (IP) with anti‐Nup188 (upper) or anti‐NuMA (bottom) antibody. Lysates and IP were separated using SDS‐PAGE and probed using western blotting with anti‐Nup188 or anti‐NuMA antibody.
Figure 7.
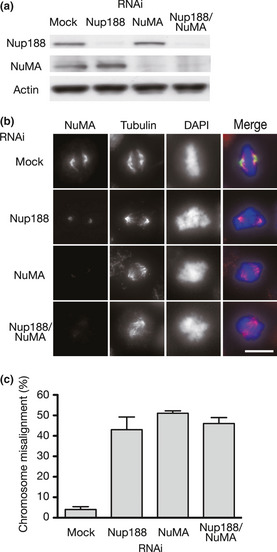
Chromosome misalignment in Nup188‐ and/or NuMA‐depleted cells. (a) Depletion of Nup188 and NuMA with siRNA. Total cell lysates prepared from HeLa cells treated with mock or indicated siRNA for 48 h were separated using SDS‐PAGE and probed using western blotting with indicated antibodies. (b) NuMA localization in Nup188 and/or NuMA‐depleted cells. HeLa cells were treated with mock or indicated siRNA for 48 h and with MG132 for the final 2 h. Cells were stained for NuMA (green), tubulin (red) and DNA (blue). Bar, 10 μm. (c) Quantitative analysis of chromosome misalignment in cells depleted of Nup188/NuMA. HeLa cells were treated as in (b). The percentage of cells with misaligned chromosomes is shown. Error bars represent the SD.
Figure 5.
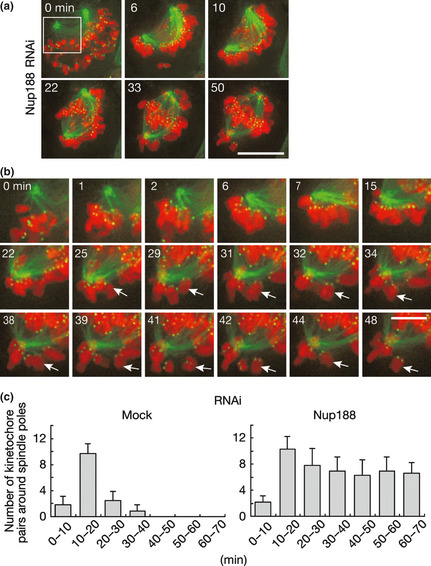
Defective kinetochore–microtubule attachment in Nup188‐depleted cells. (a) Live cell imaging of HeLa cells expressing histone H2B‐mCherry (red), GFP‐CENP‐A (green) and GFP‐α‐tubulin (green). Cells were treated with Nup188 siRNA for 48 h before live cell imaging. T = 0 was set at nuclear envelope breakdown (NEBD). Bar, 10 μm. Also see Movie S3. (b) Magnified image sequence of the field shown by rectangle in (a). Arrows indicate a chromosome showing the back and forth motion. Bar, 5 μm. (c) Chromosomes around spindle poles are continuously seen in Nup188‐depleted cells. Cells used in (a) 48 h after mock‐ or Nup188‐siRNA transfection were observed using live cell imaging. The number of kinetochore pairs around spindle poles was counted and shown in graphs according to the time after NEBD. Error bars represent the SD.
Antibodies
The following polyclonal rabbit antibodies were used: NUP188 (A302‐322A; Bethyl Laboratories, Montgomery, TX, USA); Mad2 (Novus Biologicals, Littleton, CO, USA); Aurora A (ab12875; abcam, Cambridge, UK); and NuMA (A301‐509A; Bethyl Laboratories). The following monoclonal mouse antibodies were used: tubulin (B‐5‐1‐2; Sigma, Saint Louis, MO, USA); GFP (Roche Diagnostics, Mannheim, Germany); Flag (M2; Sigma); γ‐tubulin (GTU‐88; Sigma); Nup93 (CBX00489; Cosmo Bio, Tokyo, Japan); TPX2 (NB100‐74556; Novus Biologicals); and Plk1 (sc‐17783; Santa Cruz Biotechnology, Dallas, TX, USA). We also used a human autoantibody against centromeric antigens (CREST serum)25 and a goat polyclonal anti‐actin antibody (Santa Cruz Biotechnology).
siRNA
The targeted Nup188 sequence was 5′‐AUUUCUAGCAGCAUGGACUGUUCCC‐3′ (Stealth, Life Technologies). The targeted sequences for Mad2 and Nup93 were 5′‐AGAAUUGGUAAUAAACUGUGGUCCC‐3′ and 5′‐AUUCCUUG AGCCCUAUACCGUUCGG‐3′, respectively (Stealth, Life Technologies). The targeted NuMA sequences were 5′‐UUCAGCUUCAGCUCGCAGCUCUUUG‐3′ (Figs 7 and S4A) and 5′‐UAU AAGAGCAGCAUGGUCAUCUUCG‐3′ (#2; Fig. S4B) (Stealth, Life Technologies).
Western blotting
Cells were lysed in immunoprecipitation (IP) buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 20 mM β–glycerophosphate, 5 mM MgCl2, 0.1% NP‐40 and 5% glycerol) supplemented with protease inhibitor cocktails (Complete Mini EDTA‐free; Roche Diagnostics). Protein concentration was measured and adjusted by the Bradford method (Protein Assay system; Bio‐Rad Laboratories, Hercules, CA, USA) and the cell extracts were resolved using SDS‐PAGE and transferred to a PVDF membrane (Immobilon‐P; Merck Millipore, Darmstadt, Germany). Blocking and antibody incubations were performed in 5% non‐fat dry milk. Horseradish peroxidase‐labelled secondary antibodies (Santa Cruz Biotechnology) were visualized using chemiluminescence with luminal and coumaric acid (GE Healthcare, Uppsala, Sweden).
Immunoprecipitation
Cells were lysed in IP buffer supplemented with protease inhibitor cocktails and phosphatase inhibitors (Wako, Osaka, Japan), and cleared by centrifugation for 30 min at 4°C at 16 000g After preclearing with sheep anti‐rabbit IgG conjugated to magnetic beads (Dynabeads M‐280, Life Technologies), the lysates were incubated with 5 μg antibodies for 1 h in the presence of RNase A and DNase I at 4°C, followed by incubation with anti‐rabbit or anti‐mouse IgG conjugated to magnetic beads for 2 h at 4°C. After washing twice with IP buffer and twice with TBS containing 0.1% Tween 20, the beads were suspended in SDS‐PAGE sample buffer and subjected to western blotting.
Live cell imaging
HeLa cells were grown in chambered coverslips (Laboratory‐Tek; Thermo Fisher Scientific, Waltham, MA, USA). One hour before imaging, the medium was changed to pre‐warmed Leibovitz's L‐15 medium (Life Technologies) and the chamber lids were sealed with silicone grease. Recordings were made at 37°C using a temperature‐controlled incubator. Time‐lapse images were collected using a Personal DV microscope. For the experiment in Figure 5(a), cells expressing histone H2B‐mCherry, EGFP (enhanced green fluorescent protein)‐CENP‐A and EGFP‐α‐tubulin were flattened by the pressure of the agar overlay, as previously described.26
In vitro binding assay
GST‐tagged proteins were expressed in Sf21 cells using the Bac‐to‐Bac baculovirus expression system (Life Technologies) according to the manufacturer's instructions. For purification of GST‐Nup188, cells were lysed and sonicated in lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1% NP‐40, 10% glycerol, 1 mM EGTA and 1 mM DTT). Cell lysates were clarified using centrifugation for 10 min at 4°C at 16 000g. The supernatants were incubated with glutathione‐Sepharose 4B (GE Healthcare) for 1 h at 4°C. After washing with lysis buffer, GST‐tagged proteins were eluted using elution buffer (50 mM Tris‐HCl, pH 8.0, 20 mM glutathione, 100 mM NaCl and 1 mM DTT). Eluted protein was applied to a PD‐10 column (GE Healthcare) equilibrated with lysis buffer. For purification of NuMA, GST‐NuMA was expressed in Sf21 cells and purified with glutathione‐Sepharose 4B for 1 h at 4° C. After washing with lysis buffer, the bound GST‐NuMA was treated with prescission protease (GE Healthcare) in a buffer containing 50 mM Tris‐HCl pH 7.5, 150 mM NaCl, 1% NP‐40 and 1 mM DTT and eluted. For the in vitro binding assay, recombinant NuMA was incubated with purified GST or GST‐Nup188 bound to glutathione‐Sepharose 4B for 3 h at 4°C. Precipitates were washed twice with lysis buffer containing 500 mM NaCl, three times with the same buffer containing 150 mM NaCl and then analyzed using SDS‐PAGE and Coomassie Brilliant Blue staining.
Results
Nup188 localizes to centrosomes during mitosis
We first observed the subcellular localization of Nup188 in immunofluorescence staining of HeLa cells. In interphase cells, the Nup188 signal was prominently found at centrosomes, which were defined by the localization of γ–tubulin, in addition to the nuclear membrane, where it serves as a nucleoporin (Figs 1a and S1A). Such centrosomal localization in interphase was not seen for Nup88, another Nup localizing to the spindle in mitosis (Fig. S1A). The centrosomal localization of Nup188 persisted during mitosis from prophase to anaphase (Fig. 1b). A faint Nup188 signal was also detected on the mitotic spindle.
Next, we determined the region of Nup188 responsible for centrosomal localization. Nup188 protein was divided into three fragments and expressed as Flag‐tagged proteins in HeLa cells. Consistent with the finding in immunofluorescence staining, Flag‐tagged full‐length Nup188 was found at centrosomes, as well as the nuclear membrane in interphase cells (Fig. S1B). In mitotic cells, only the C‐terminal fragment was found at centrosomes, while the N‐terminal and the middle regions diffusely localized in the cytoplasm (Fig. 1c). Consistent with these findings, Nup188 failed to localize to centrosomes when it lacked the C‐terminal end. These data suggest that the C‐terminal region of Nup188 is required for the centrosomal localization of Nup188.
Nup188 is involved in chromosome alignment
To study the function of Nup188 during mitosis, we used siRNA to deplete Nup188 in HeLa cells. Western blot analysis estimated that Nup188 expression decreased to <10% of mock‐treated cells 48 h after siRNA transfection (Figs 2a and S2A). Immunofluorescence staining revealed that the centrosomal signal was largely decreased in cells depleted of Nup188 (Fig. S2B). Among Nup188‐depleted cells, 17% of cells were in mitosis, whereas 4% of cells were in a mock‐treated condition (Fig. 2b), suggesting that Nup188 depletion caused mitotic arrest. Observation of mitotic cells after depletion of Nup188 revealed that a significant number of chromosomes seemed not aligned on the metaphase plate (Fig. 2c). This chromosome misalignment phenotype was seen in approximately 70% of cells, where more than five chromosomes were misaligned per cell in most cases (Fig. 2d). We could verify that this phenotype was specific for Nup188 depletion, as chromosome alignment was largely restored by the expression of RNAi‐resistant Nup188 fused with green fluorescent protein (GFP) (see Fig. 4). We also confirmed in a microarray analysis that transcript levels of genes reported to be involved in chromosome alignment were not altered in Nup188‐depleted cells, excluding the possibility of an off‐target effect (data not shown). Furthermore, Nup188 depletion caused chromosome misalignment not only in HeLa cells but also in U2OS osteosarcoma and HCT116 colon adenocarcinoma cell lines (Fig. S2C). These data suggest that Nup188 is required for proper chromosome alignment on the metaphase plate.
Figure 4.
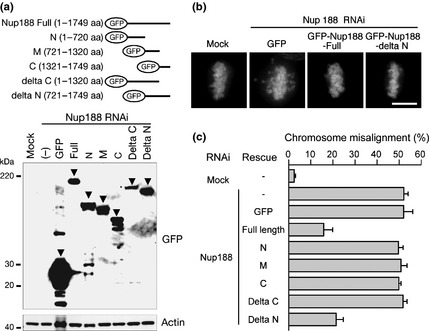
The middle and C‐terminal region of Nup188 are responsible for chromosome alignment. (a) Expression of GFP‐tagged deletion mutants. Lysates were prepared from HeLa cells transfected with RNAi‐resistant GFP‐Nup188 mutants for 24 h, preceded by transfection with Nup188 siRNA for 36 h. Cell lysates were separated using SDS‐PAGE and probed using western blotting with an anti‐GFP or anti‐actin antibody. Arrowheads indicate the bands of corresponding mutants. (b) Representative images of cells expressing GFP, GFP‐Nup188‐Full and GFP‐Nup188‐delta N as well as mock‐treated cells. HeLa cells were transfected with RNAi‐resistant GFP‐Nup188 constructs for 24 h, preceded by transfection with Nup188 siRNA for 36 h. Cells were treated with MG132 for the final 2 h. DNA was stained with DAPI. Bar, 10 μm. (c) The percentage of cells with misaligned chromosomes for cells expressing each construct shown in (a). HeLa cells were treated as in (b). Error bars represent the SD.
In Nup188‐depleted cells, a SAC component Mad2 was detected on kinetochores of misaligned chromosomes, suggesting that the SAC was not satisfied (Fig. 2e). Indeed, when we depleted both Nup188 and Mad2, the increase in the mitotic index was overcome (Fig. 2b). In addition, most of the Nup188‐depleted cells arrested in mitosis in the presence of spindle poison, nocodazole or taxol, in a Mad2‐dependent manner (Fig. S2D). These data indicate that Nup188 is not a component of SAC signaling and dispensable for SAC activity.
Next we determined the Nup188 regions required for chromosome alignment. To do this, we examined chromosome alignment in Nup188‐depleted HeLa cells expressing RNAi‐resistant GFP‐Nup188‐deletion mutants. Comparable expression of each construct was confirmed using western blotting (Fig. 4a). Chromosome misalignment was largely rescued when full‐length Nup188 tagged with GFP was expressed (Fig. 4b,c). The delta N mutant (GFP‐Nup188 encompassing the middle and the C‐terminal region) was the only mutant that could rescue the phenotype among those tested (Fig. 4b,c). The finding that M (middle) and C (C‐terminal region) mutants failed to rescue the misalignment (Fig. 4c) indicates that both the middle and C‐terminal regions of Nup188 are required for successful chromosome alignment.
Defective K‐fiber formation in Nup188‐depleted cells
To examine the mechanism of chromosome misalignment in Nup188‐depleted cells, we performed live cell imaging analysis using HeLa cells expressing histone H2B‐GFP with or without Nup188 depletion. In the vast majority of mock‐treated cells, chromosomes swiftly aligned on the metaphase plate within 1 h after the nuclear envelope breakdown (NEBD) (Fig. 3a,b). In contrast, chromosome congression to the metaphase plate was less efficient in Nup188‐depleted cells and many chromosomes stayed in the vicinity of centrosomes for prolonged periods of time (Fig. 3a,b). Remarkably, we found that the distance between sister kinetochores (interkinetochore distance) was smaller in Nup188‐depleted cells than that in mock‐treated cells for both misaligned and aligned chromosomes (Fig. 3c), indicating that proper tension was not applied on kinetochores, even for the aligned chromosomes, in the absence of Nup188. To examine the stability of kinetochore microtubules, or K‐fiber, mitotic cells were exposed to a low temperature to disassemble as‐yet‐stabilized microtubules. In contrast to mock‐treated cells that showed thick K‐fibers attaching to kinetochores, much fewer cold‐stable K‐fibers were observed in Nup188‐depleted cells (Fig. 3d). These results indicate that K‐fiber formation is defective in Nup188‐depleted cells.
We further monitored kinetochore–microtubule attachment using HeLa cells expressing histone H2B‐mCherry, GFP‐CENP‐A and GFP‐α‐tubulin in live cell imaging analysis. In Nup188‐depleted cells, a significant number of chromosomes stayed close to centrosomes while others moved to the metaphase plate (Fig. 5a). When kinetochores around centrosomes were carefully observed, we found that they repetitively moved back and forth between the centrosome and the metaphase plate (Fig. 5b). In mock‐treated cells, the number of chromosomes around centrosomes decreased during the time course (Fig. 5c). In contrast, the number did not decrease in Nup188‐depleted cells and thick K‐fiber formation was barely detectable.
Nup188 is required for localization of NuMA to centrosomes
These observations led us to surmise that Nup188 contributes to K‐fiber formation through microtubule organization at centrosomes. To address this possibility, we observed the centrosomal localization of proteins that have been implicated in centrosome regulation. In Nup188‐depleted cells, TPX2, Plk1 and Aurora A were detected at centrosomes at similar levels as in the mock‐treated cells (data not shown). However, localization of NuMA at centrosomes was markedly decreased (Fig. 6a). Western blot analysis indicated that the reduction in centrosomal staining of NuMA was not due to a decrease in NuMA expression (Fig. S3A). We then performed IP experiments to investigate if NuMA might interact with Nup188. In nocodazole‐treated mitotic HeLa cell extracts, NuMA was detected in Nup188 IP when TPX2 and Plk1 were not detected (Fig. 6b; data not shown). We could also detect Nup188 in a NuMA IP (Fig. 6b). These co‐immunoprecipitation data suggest that Nup188 is required for the localization of NuMA at centrosomes through its association with NuMA. We further investigated whether the interaction between Nup188 and NuMA is direct or indirect using purified proteins (Fig. S3B,C). NuMA was not detected in an in vitro binding assay with GST‐fused Nup188 (Fig. S3D), suggesting that the interaction is indirect.
NuMA is known to tether spindle microtubules to spindle poles27 and it was recently reported that NuMA depletion causes chromosome misalignment.28 To correlate decreased spindle pole localization of NuMA and chromosome misalignment in Nup188‐depleted cells, we compared the rates of chromosome misalignment in cells depleted of Nup188 and NuMA. Effective depletion of both proteins was confirmed in western blotting (Fig. 7a). The NuMA signal on centrosomes was decreased in cells depleted of Nup188 and/or NuMA (Fig. 7b). Under these conditions, Nup188‐ and NuMA‐depleted cells exhibited similar rates of chromosome misalignment (Figs 7c and S4A). Importantly, when both Nup188 and NuMA were depleted simultaneously, an additive effect was not seen, implying that these proteins are working in the same pathway. These results were confirmed using another siRNA targeted for NuMA (Fig. S4B).
Discussion
During mitosis, the NPC disassemble into subcomplexes of Nup.5 It has emerged that a number of Nup play active roles in mitotic processes.6, 7 In the present paper, we report a novel function of Nup188 in mitosis. Nup188 localized to centrosomes and Nup188‐depleted cells arrested in mitosis due to severe chromosome misalignment caused by defective K‐fiber formation. Importantly, we found that Nup188 interacts with NuMA and recruits it to centrosomes. NuMA‐depleted cells showed a similar chromosome misalignment phenotype as Nup188‐depleted cells and an additive defect was not observed when both proteins were depleted simultaneously. Based on these data, it is plausible to predict that Nup188 plays a role in chromosome alignment by facilitating microtubule formation and organization at centrosomes through the recruitment of NuMA.
Human Nup188 is a 1749 aa protein that mainly consists of α‐helical domains, although a detailed structural analysis has not been performed. We found that Nup188 localizes to centrosomes in mitosis, which is dependent on the C‐terminal region. Interestingly, the centrosomal localization is already seen in interphase when the NPC are intact. Localization to the centrosomes in interpahse was not reported for Rae1 and Nup88, nucleoporins known to localize to centrosomes in mitosis.11, 12 Further study is required to clarify whether Nup188 on interphase centrosomes resides in a complex distinct from that in the NPC and plays an interphase‐specific role. In the NPC, Nup188 is known to be in a complex with Nup93.4 Whether Nup93 forms a complex with Nup188 during mitosis and plays a role in chromosome alignment with Nup188 is another important question to be addressed. In addition to its requirement for centrosomal localization of Nup188, mitosis‐specific phsophorylation sites were reported in the C‐terminal region,29, 30 which might be related to a unique function of Nup188 in mitosis. The middle region, in addition to the C‐terminal region, is required for chromosome alignment, which might have a role in microtubule formation and organization on centrosomes. In this regard, it will be particularly interesting to examine the interaction of the middle region with NuMA.
NuMA is known to play a role in focusing microtubules at centrosomes by cross‐linking kinetochore and non‐kinetochore microtubules at spindle poles.31 NuMA localizes in the nucleus in interphase and at NEBD NuMA is phosphorylated so that the phosphorylation promotes its interaction with dynein and therefore its dynein‐dependent spindle pole localization.27 Nup188, which localize to centrosomes before NEBD, might facilitate anchoring of NuMA to spindle poles. Interestingly, Rae1 binds to NuMA and regulates its function to form bipolar spindle.11 Thus, the relationship between Rae1 and Nup188 would be an important question for future study. In Xenopus egg extracts and mice, dissociation of spindle poles from centrosomes is seen after NuMA depletion,31, 32 which was also reported in HeLa cells.28 Such a phenotype was not apparent in the present study, although the spindle was longer in Nup188‐depleted cells than in mock‐treated cells, with thinner and less organized K‐fibers (e.g. Fig. 2c), reflecting defective spindle formation.
In Nup188‐depleted cells, many chromosomes stayed in the vicinity of spindle poles. Close inspection of these chromosomes revealed that a stable kinetochore–microtubule attachment is hardly formed and therefore chromosomes moved back and forth to centrosomes, as a result of repetitive attachment and detachment of micortubules. Defective K‐fiber formation due to reduced tethering of spindle microtubules to centrosomes might be responsible for the phenotype. A similar phenotype is also seen in cells depleted of CENP‐E, a plus end‐directed motor required for chromosome congression.33 However, in CENP‐E‐depleted cells the number of chromosomes distributed around spindle poles gradually decreases, unlike in Nup188‐depleted cells. We checked that spindle and kinetochore localization of CENP‐E is not affected in Nup188‐depleted cells and CENP‐E is not co‐immunoprecipitated with Nup188 in mitotic cell extract (data not shown).
Increasing observation suggest that deregulation of Nup associates with cancer.14 In light of the fact that many cancer cells are aneuploid, mitotic defects might be responsible for some of these cases. It was reported that heterozygous depletion of Nup188 was found in three of 10 B‐ALL patients.24 It is important to investigate if reduced expression of Nup188 or mutations in the remaining allele might affect K‐fiber stability and fidelity of chromosome segregation, leading cells to oncogenic transformation in these patients. Further study is required to elucidate the role of Nup188 on proper chromosome segregation and its relevance to cancer.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Nup188 localizes to centrosomes in interphase cells.
Fig. S2. Verification of the phenotype in Nup188‐depleted cells.
Fig. S3. Relationship between Nup188 and NuMA.
Fig. S4. Chromosome misalignment in Nup188‐ and/or NuMA‐depleted cells.
Movie S1. Live cell imaging of a mock‐treated HeLa cell expressing histone H2B‐GFP (time interval, 3 min; display rate, 5 frames/s).
Movie S2. Live cell imaging of a Nup188‐depleted HeLa cell expressing histone H2B‐GFP (time interval, 3 min; display rate, 5 frames/s).
Movie S3. Live cell imaging of Nup188‐depleted cells expressing histone H2B‐mCherry, GFP‐CENP‐A and GFP‐α‐tubulin (time interval, 1 min; display rate, 5 frames/s).
Acknowledgments
The authors thank K. Mizuno for NuMA cDNA and A. Harata for technical assistance. This work was supported by a Grant‐in‐Aid for Scientific Research from the Japanese Society of Promotion of Science; a Grant‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and grants from the Mitsubishi Foundation, the Naito Foundation, Takeda Science Foundation, Princess Takamatsu Cancer Research Fund (10‐24210), Daiichi‐Sankyo Foundation of Life Science and Gonryo Medical Foundation.
(Cancer Sci 2013; 104: 871–879)
References
- 1. Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep 2012; 13: 501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanaka K, Hirota T. Chromosome segregation machinery and cancer. Cancer Sci 2009; 100: 1158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanaka K. Regulatory mechanisms of kinetochore‐microtubule interaction in mitosis. Cell Mol Life Sci 2013; 70: 559–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure 2009; 17: 1156–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 2009; 10: 178–91. [DOI] [PubMed] [Google Scholar]
- 6. Chatel G, Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal 2011; 23: 1555–62. [DOI] [PubMed] [Google Scholar]
- 7. Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci 2010; 67: 2215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zuccolo M, Alves A, Galy V et al The human Nup107‐160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J 2007; 26: 1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107‐160 complex and gamma‐TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 2010; 12: 164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Platani M, Santarella‐Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW. The Nup107‐160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell 2009; 20: 5260–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong RW, Blobel G, Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc Natl Acad Sci U S A 2006; 103: 19783–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hashizume C, Nakano H, Yoshida K, Wong RW. Characterization of the role of the tumor marker Nup88 in mitosis. Mol Cancer 2010; 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blower MD, Nachury M, Heald R, Weis K. A Rae1‐containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 2005; 121: 223–34. [DOI] [PubMed] [Google Scholar]
- 14. Xu S, Powers MA. Nuclear pore proteins and cancer. Semin Cell Dev Biol 2009; 20: 620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura T, Largaespada DA, Lee MP et al Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet 1996; 12: 154–8. [DOI] [PubMed] [Google Scholar]
- 16. Borrow J, Shearman AM, Stanton VP Jr et al The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet 1996; 12: 159–67. [DOI] [PubMed] [Google Scholar]
- 17. von Lindern M, Fornerod M, van Baal S et al The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia‐specific dek‐can mRNA. Mol Cell Biol 1992; 12: 1687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cooper CS, Park M, Blair DG et al Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984; 311: 29–33. [DOI] [PubMed] [Google Scholar]
- 19. Gould VE, Martinez N, Orucevic A, Schneider J, Alonso A. A novel, nuclear pore‐associated, widely distributed molecule overexpressed in oncogenesis and development. Am J Pathol 2000; 157: 1605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gould VE, Orucevic A, Zentgraf H, Gattuso P, Martinez N, Alonso A. Nup88 (karyoporin) in human malignant neoplasms and dysplasias: correlations of immunostaining of tissue sections, cytologic smears, and immunoblot analysis. Hum Pathol 2002; 33: 536–44. [DOI] [PubMed] [Google Scholar]
- 21. Martinez N, Alonso A, Moragues MD, Ponton J, Schneider J. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer Res 1999; 59: 5408–11. [PubMed] [Google Scholar]
- 22. Dawlaty MM, Malureanu L, Jeganathan KB et al Resolution of sister centromeres requires RanBP2‐mediated SUMOylation of topoisomerase IIalpha. Cell 2008; 133: 103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 2010; 189: 1129–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nowak NJ, Sait SN, Zeidan A et al Recurrent deletion of 9q34 in adult normal karyotype precursor B‐cell acute lymphoblastic leukemia. Cancer Genet Cytogenet 2010; 199: 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci 2007; 120: 4188–96. [DOI] [PubMed] [Google Scholar]
- 26. Itoh G, Kanno S, Uchida KS et al CAMP (C13orf8, ZNF828) is a novel regulator of kinetochore‐microtubule attachment. EMBO J 2011; 30: 130–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Radulescu AE, Cleveland DW. NuMA after 30 years: the matrix revisited. Trends Cell Biol 2010; 20: 214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haren L, Gnadt N, Wright M, Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res Notes 2009; 2: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dephoure N, Zhou C, Villen J et al A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 2008; 105: 10762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res 2009; 8: 4553–63. [DOI] [PubMed] [Google Scholar]
- 31. Silk AD, Holland AJ, Cleveland DW. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J Cell Biol 2009; 184: 677–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 1996; 87: 447–58. [DOI] [PubMed] [Google Scholar]
- 33. Kapoor TM, Lampson MA, Hergert P et al Chromosomes can congress to the metaphase plate before biorientation. Science 2006; 311: 388–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Nup188 localizes to centrosomes in interphase cells.
Fig. S2. Verification of the phenotype in Nup188‐depleted cells.
Fig. S3. Relationship between Nup188 and NuMA.
Fig. S4. Chromosome misalignment in Nup188‐ and/or NuMA‐depleted cells.
Movie S1. Live cell imaging of a mock‐treated HeLa cell expressing histone H2B‐GFP (time interval, 3 min; display rate, 5 frames/s).
Movie S2. Live cell imaging of a Nup188‐depleted HeLa cell expressing histone H2B‐GFP (time interval, 3 min; display rate, 5 frames/s).
Movie S3. Live cell imaging of Nup188‐depleted cells expressing histone H2B‐mCherry, GFP‐CENP‐A and GFP‐α‐tubulin (time interval, 1 min; display rate, 5 frames/s).


