Abstract
Adapting to hypoxic stress is pivotal in tumor progression and determining tumor malignancy. The transcriptional factor hypoxia‐inducible factor (HIF) is crucial in modulating tumorous hypoxic responses through altering cell energy metabolism, which includes the modification of glucose and lipid metabolism‐associated gene expression. Stearoyl–CoA desaturase‐1 (SCD1) is the main isoform of SCDs, the rate‐limiting enzymes in the biosynthesis of monounsaturated fatty acids from saturated fatty acids, which is extensively activated in cancer progression. In this study, we found that SCD1 and HIF‐2α were overexpressed in human clear cell renal cell carcinoma (ccRCC) tissues and ccRCC cell lines, and were upregulated in the 786‐0 ccRCC cell line under hypoxia. Knockdown of SCD1 or HIF‐2α impacted the other's expression. Enhancing SCD1 resulted in HIF‐2α upregulation, which could be blocked by inhibiting the PI3K/Akt pathway. Deficiency of SCD1 or HIF‐2α in 768‐0 cells led to apoptosis, less colony formation ability, and decreased cell migration. More obvious effects were observed in 786‐0 cells with double SCD1 and HIF‐2α knockdown. These results indicate a PI3K/Akt‐mediated loop between SCD1 and HIF‐2α that mutually enhances their protein levels. Both SCD1 and HIF‐2α are critical to promoting tumorigenesis by synergistically acting on maintaining cell survival, triggering cell migration, and enhancing the colony formation ability of cancer cells.
The von Hippel‐Lindau (VHL) protein is a tumor suppression protein whose function is to mediate HIFα degradation and whose gene mutation is commonly found in human ccRCCs.1 Aberrations in VHL's functions lead to HIFα accumulation irrespective of hydroxylation and upregulation of a series of hypoxia responsive genes in ccRCC.2 Hypoxia‐inducible factor is a key transcriptional factor mediating cellular adaptation to hypoxic stress in carcinoma cells. It is an α/β heterodimeric DNA binding complex that is regulated through stabilization, dissociation, and degradation of subunit HIFα under normoxia.3 The two best characterized HIFα isoforms are HIF‐1α and HIF‐2α. Although most of their functions are overlapping, the two isoforms have distinct functions in tumor cells.4 Moreover, there is an overexpression trend toward HIF‐2α rather than HIF‐1α in VHL‐defective ccRCC cells,5 which might be due to reciprocal suppression effects between the isoforms.6
Energy metabolism reprogram is a major consequence after HIFα overexpression. Based on unique activities of these HIFα isoforms, HIF‐1α is thought to be primarily involved in glucose metabolism by increasing glucose transporter expression and glycolytic enzymes.7 Many glucose metabolism‐associated genes are regulated by HIF‐1α but not HIF‐2α.7 The latter is supposed to participate in angiogenesis modulation, enhancing tumor growth, and triggering cell cycle progression.6 In another important energy metabolism, lipid metabolism, the two HIFα isoforms showed opposing effects on modulating lipid anabolism in normal cells. Hypoxia‐inducible factor‐1α partially deficient mice showed fewer increases in enzymes controlling lipogenesis in liver, including sterol regulatory element binding protein‐1 and SCD1. The HIF‐1α‐inducing effects of peroxisome proliferator activated receptor‐γ (PPARγ), which in turn activates glycerolipid biosynthesis‐associated genes, are found in ventricle with pathologic cardiac hypertrophy.8 However, the constitutively activated HIF‐2α results in less lipogenesis by decreasing the expression of PPARγ and its target genes (e.g. Hmgcs2, Apoa2, SCD1), impairing mitochondrial fatty acid beta‐oxidation and increasing lipid storage capability in liver cells.9 But whether and, if so, how HIF‐2α impacts lipid metabolism and is further involved in cellular behavior modulation in cancer cells, remains to be clarified.
Stearoyl–CoA desaturases are the rate‐limiting enzymes in the biosynthesis of MUFA from SFA; SCD1 is the main SCD isoform found in all mammalians, with presence in most tissues and cells. This enzyme catalyzes the introduction of the cis double bond in the cis‐Δ9 position of saturated fatty acyl–CoA substrates, mainly converting palmitoyl–CoA to palmitoleoyl–CoA, and stearoyl–CoA to oleoyl–CoA.10 Abnormally high levels of MUFA caused by elevated SCD1 activity is taken as a marker for cancer progression. The substrates SFA for SCD1 in cancer cells mainly come from a shift away from oxidative phosphorylation to enhanced glycolysis under hypoxic stress.11, 12 The alteration in the ratio of unsaturated and saturated fatty acids on cancer cell membrane triggered by extensive activation of SCD1 would contribute to modulations of membrane‐resident signal pathways and impact on cellular behaviors such as cell division, programmed cell death, and cell migration.13, 14 Thus, SCD1 is a one of the central regulators on controlling tumor progression.
In this study, we used the ccRCC line 786‐0, a VHL−/− cell line with only constitutively expressed HIF‐2α, to study the interactions between HIF‐2α and SCD1 expression. We were thereby able to evaluate the contribution of the synergic effects between the two proteins on maintaining cancer cell survival and triggering tumorigenesis.
Materials and Methods
Clinical materials and immunohistochemistry
Human ccRCC and corresponding adjacent non‐tumorous parenchyma specimens from 20 patients were obtained at nephrectomy. The specimens were subjected to analyses on total SCD1 and HIF‐2α levels with immunohistochemistry. The tissues were fixed in formalin, embedded in paraffin and sectioned (3 μm). Then the sections were stained with primary antibodies anti‐human SCD1 (Abcam, Cambridge, UK) and anti‐human HIF‐2α (Novus Biologicals, Littleton, CO, USA), followed by counterstaining with hematoxylin, then visualized under bright field microscope.
Cell culture and treatment
The human ccRCC cell lines 786‐0, A498, and RCC4, and the normal human renal tubular epithelial HK‐2 cell line were obtained from ATCC (Manassas, VA, USA) and maintained in RPMI‐1640 medium supplemented with 10% FCS, 2 mM l‐glutamine, 50 IU/mL penicillin, and 50 μg/mL streptomycin at 37°C in 95% humidity with 5% CO2/21% O2/74% N2 for normoxia and 5% CO2/1% O2/94% N2 in an airtight anaerobic incubator for hypoxia.
Transient transfection
The full‐length human SCD1 was cloned into expression vector pcDNA3 to construct an expressing plasmid SCD1. The plasmids SCD1 or pcDNA3 empty vector (2 μg for each) were transfected into 4 × 105 786‐0 cells using Lipofectamine (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cells with transiently overexpressed SCD1 were available for further analysis 48 h after transfection.
Retroviruses
Oligonucleotides containing the shRNA sequence for HIF‐2 (5′‐GACAAGGTCTGCAAAGGGT‐3′) and SCD1 (5′‐GCGCATCTCTATGGATATC‐3′) were cloned into the pRetroSuper Retroviral vector. To produce retroviral supernatants, each retroviral plasmid was co‐transfected with the retroviral packing plasmid pCL‐Ampho into 293T cells seeded in duplicate in 35‐mm dishes using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. The medium containing retroviruses was collected at 72 h post‐transfection and filtered through a 45‐μm pore size filter. To create stable pools of retrovirally infected cells, 786‐0 cells were incubated overnight in a mixture (1:1) of retroviral supernatant and fresh medium supplemented with polybrene (10 μg/mL). Two days later, cells were selected with puromycin (1 μg/mL) for a further 14 days to establish cell lines with stably expressed shRNA.
Western blot analysis
Equal amounts of total proteins from cell lysate or clinical material extractions were separated using SDS‐PAGE and probed with primary antibodies anti‐human HIF‐2α (Novus Biologicals), anti‐human SCD1 (Abcom), anti‐human Akt and anti‐human Akt pS473 (both from Cell Signaling Technology, Danvers, MA, USA), and anti‐human β‐actin (Sigma, St. Louis, MO, USA) and corresponding secondary antibodies. The results were visualized with ECL (Pierce, Rockford, IL, USA).
Analysis of mRNA
Total RNA was extracted from cells with the RNeasy mini kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. The cDNA was synthesized from 500 ng total RNA using the SuperScript III First‐Strand Synthesis System for RT–PCR (Invitrogen). Real‐time PCR was carried out using the SYBR GreenER qPCR SuperMix (Invitrogen) and the following primers: β‐actin (forward, 5′‐GTCCTCTCCCAAGTCCACAC‐3′; reverse, 5′‐GGGAGACCAAAAGCCTTCAT‐3′); HIF‐2α (forward, 5′‐TCGTGAGAACCTGAGTCTCAAA‐3′; reverse, 5′‐ATCCGGTACTGGCCACTTACTA‐3′); and SCD1 (forward, 5′‐CCGGGAGAATATCCTGGTTT‐3′; reverse, 5′‐GCGGTACTCAACTGGCAGAGT‐3′).
Colony formation assay
Anchorage‐independent growth was determined by the ability of cells to form colonies in soft agar. The 786‐0 cells (50 000 cells/plate) with stably expressed shRNA were trypsinized to single cells and resuspended in 0.4% agar (Sigma), then seeded to a preset 1% bottom agar layer in six‐well plates. Cells were cultured at 37°C in 95% humidity with 5% CO2/21% O2/74% N2 for 14 days. The colonies were counted using micrographs.
Transwell migration assay
The 786‐0 cells with stably expressed shRNA (1 × 105) were suspended in 300 μL medium containing 1% FBS and seeded into Transwell inserts (8‐μm pore; Chemicon, Temecula, CA, USA) precoated with growth factor‐reduced Matrigel (1 μg/mL; BD Biosciences, Franklin Lakes, NJ, USA). Complete medium was added to the lower chamber. After incubation for 24 h, invasive cells on the lower surface of the chamber were fixed with methanol, stained with DAPI, and observed under a fluorescent microscope. Five photographs were taken at fixed positions of the lower surface, top right and left, bottom right and left, and the central field. The total cells in each field were counted and average cell number in each field was calculated.
Staining with TUNEL
The 786‐0 cells with stably expressed shRNA (1 × 105) were seeded in a 35‐mm dish and cultured in RPMI‐1640 medium supplemented with 10% FCS, 2 mM l‐glutamine, 50 IU/mL penicillin, and 50 μg/mL streptomycin at 37°C in 95% humidity with 5% CO2/21% O2/74% N2 for 72 h. The cells were fixed and subjected to direct TUNEL labeling assay (Roche, Basel, Switzerland) to label the apoptotic cells. The nuclei were visualized by DAPI staining. Then the cells were photographed at randomly selected fields with fluorescence microscopy and at least 300 cells were counted. The percentage of cells with positive TUNEL staining was calculated and taken as the apoptotic rate.
Statistical analysis
All experiments were done in triplicate and analyzed with two‐tailed Student's t‐test. Data are presented as the mean ± SEM. P < 0.05 was considered significantly different.
Results
Upregulation of SCD1 in renal cancer cells in response to HIF‐2α increase
SCD1 is a protein significantly upregulated in various tumors.15 Thus, we first analyzed the SCD1 expressions in pair specimens of ccRCC tumor and adjacent non‐tumorous parenchyma. Compared with the normal parenchyma tissues, SCD1 was increased in all tumor specimens we tested (Fig. 1A). In normal renal tissue, the renal tubules are well organized with constitutively low SCD1 expression, which is indicated by punctate staining. However, in tumor tissues, the renal tubules' structure was disrupted and extensively expressed SCD1 was diffused in the tumor tissue (Fig. 1B). The SCD1 overexpression was not related with the stage of the tumor. As SCD1 is a gene upregulated under hypoxia,16 and HIFα was found overexpressed in many cell lines originated from ccRCC, we analyzed the SCD1 expression in ccRCC cell lines with overexpression of various HIFα isoforms. Compared with HK‐2 cells taken as normal control, SCD1 overexpression was found in all three ccRCC cell lines, A498, RCC4, and 786‐0 (Fig. 1C). These cell lines have constitutive HIF‐2α overexpressions and selective HIF‐1α expressions, namely HIF‐1α and HIF‐2α in RCC4, and only HIF‐2α in A498 and 786‐0.6, 17 That means that HIF‐2α might be more involved in modulating SCD1 overexpression in ccRCC. Furthermore, we found HIF‐2α overexpression in specimens from ccRCC tumors (Fig. 1D). The overexpression of HIF‐2α and SCD1 in ccRCC tumors could probably be further increased under microenvironmental hypoxia. To prove this hypothesis, 786‐0 cells were cultured under normoxia and hypoxia. It was found that SCD1 increased in parallel with HIF‐2α upregulation under hypoxia (Fig. 1E). As 786‐0 is a VHL‐deficient cell line and might not respond to hypoxia correctly, we overexpressed VHL in 786‐0 cells and observed the SCD1 expression under hypoxia. We found that SCD1 was increased under hypoxia in 786‐0 cells with restored VHL expression or in RCC cells (ACHN) with normal VHL expression (Fig. S1), which meant that VHL deficiency did not affect the SCD1 increase in response to hypoxia.
Figure 1.
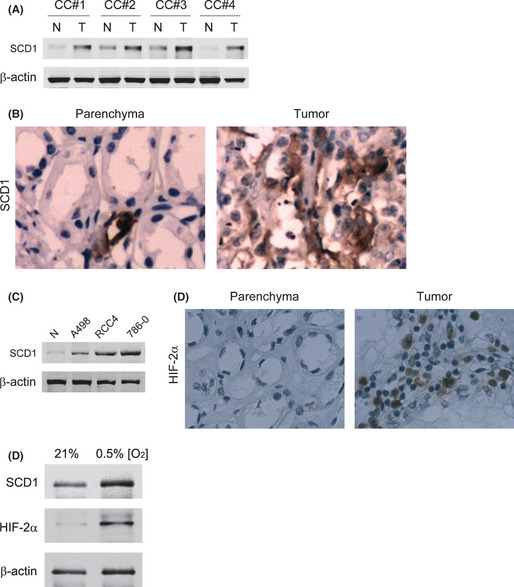
Expression of stearoyl–CoA desaturase‐1 (SCD1) in renal cancer cells and its response to hypoxia‐inducible factor‐2α (HIF‐2α) upregulation. (A) SCD1 levels in four representative specimen pairs (clear cell renal cell carcinoma [ccRCC] tumor [T] and their adjacent non‐tumorous parenchyma [N]) from ccRCC patients were analyzed with Western blot. CC#1–4 indicate specimen numbers. (B) Specimens from ccRCC patients were also subjected to immunohistological staining with an antibody against SCD1. (C) SCD1 expression was assayed in the HK‐2 cell line (taken as normal control [N]) and three ccRCC cell lines, A498, RCC4, and 786‐0, reported to have various HIFα isoform expressions. The HIF‐2α levels in ccRCC tumors were detected with immunohistochemistry. (D) Representative results from four specimen pairs. (E) SCD1 and HIF‐2α levels were analyzed in 786‐0 cells exposed to normoxia (21% O2) or hypoxia (0.5% O2).
Hypoxia‐inducible factor‐2α controls SCD1 expression in ccRCC
In order to assess whether HIF‐2α affects SCD1 expression in ccRCC, shHIF‐2α was designed and conducted into 786‐0 cells. We analyzed the HIF‐2α and SCD1 levels in 768‐0 cells with scrambled sequence siRNA or shHIF‐2α under hypoxia and found that both HIF‐2α and SCD1 mRNA and protein levels were decreased by shHIF‐2α (Fig. 2). We then proposed that SCD1 upregulation under hypoxic conditions is in response to HIF‐2α increase. However, siRNA against HIF‐2α did not decrease SCD1 levels under hypoxia in cells with normal VHL expression (ACHN) or in 786‐0 cells with restored VHL expression (786‐0/VHL), although slight HIF‐2α decreases were observed in ACHN and 786‐0/VHL cells (Fig. S2).
Figure 2.
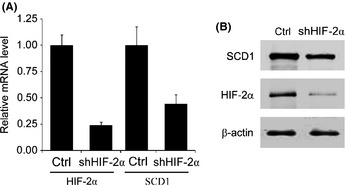
Hypoxia‐inducible factor‐2α (HIF‐2α) affects stearoyl–CoA desaturase‐1 (SCD1) expression in 786‐0 clear cell renal cell carcinoma cells. 786‐0 cells were transfected with siRNA against HIF‐2α (shHIF‐2α) and exposed to hypoxia. Scrambled sequence of shHIF‐2α was taken as control (Ctrl). The HIF‐2α and SCD1 mRNA levels (A) and protein levels (B) were detected.
Stearoyl–CoA desaturase‐1 is required for PI3K/Akt pathway activation
Overexpression of SCD1 resulted in modifications to the membrane‐resident cell signal pathway and was reported to be essential to Akt full activation in cancer cells.12, 18 To investigate whether SCD1 is required for Akt activation in 786‐0 cells, we infected 786‐0 cells with retrovirus‐producing shSCD1 and detected the Akt expression and activation under SCD1 deficiency. The SCD1 mRNA and protein levels and Akt activation, indicated by phosphorylation at Ser473, were decreased efficiently after introduction of shSCD1, but Akt expression was not impacted (Fig. 3A,B). As expected, the SCD1 overexpression in 786‐0 cells by transiently transfecting an expression vector containing the full‐length SCD1 gene enhanced Akt activation but not Akt expression (Fig. 3C).
Figure 3.
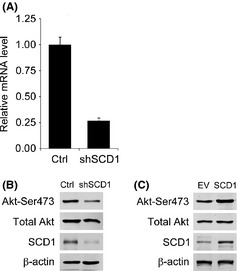
Impact of stearoyl–CoA desaturase‐1 (SCD1) level change on PI3K/Akt pathway activation in 786‐0 renal cancer cells. Levels of SCD1 in 786‐0 cells were knocked down by transfecting siRNA against SCD1 (shSCD1) or increased by transiently transfecting an expression vector containing the full‐length SCD1 gene (SCD1). The scrambled sequence of shSCD1 (Ctrl) or empty vector (EV) was taken as control. Gene silencing effects of shSCD1 were determined by realtime RT‐PCR on SCD1 (A). Total Akt protein levels and Akt activation indicated by phosphorylation at Ser473 were evaluated under SCD1 deficiency (B) and overexpression (C).
Hypoxia‐inducible factor‐2α protein level correlates with SCD1 mediating Akt activation
Expression of HIF‐2α is depended on Akt activation in RCC.19 We then further investigated whether SCD1 levels would affect HIF‐2α expression by enhancing Akt activation. Hypoxia increased both SCD1 and HIF‐2α levels, shown by comparing the cells infected with scrambled sequences as control under normoxia and hypoxia. But these increases on SCD1 and HIF‐2α in response to hypoxia were blocked by shSCD1 (Fig. 4A). We doubted whether HIF‐2α expression was correlated with SCD1 expression. The latter was overexpressed in 786‐0 cells by transfecting SCD1 expression plasmids, which finally resulted in higher HIF‐2α levels in 786‐0 cells (Fig. 4B). The modulating effect of SCD1 on HIF‐2α did not depend on VHL, because similar decreasing effects of shSCD1 on HIF‐2α were observed in RCC cells with or without VHL expression (Fig. S3). Furthermore, these HIF‐2α increases triggered by SCD1 overexpression were attenuated by the presence of PI3K/Akt inhibitor Ly294002. But total Akt was not impacted with or without SCD1 overexpression or PI3K/Akt inhibitor (Fig. 4C). Thus, PI3K/Akt pathway activation mediates SCD1‐regulated HIF‐2α overexpression.
Figure 4.
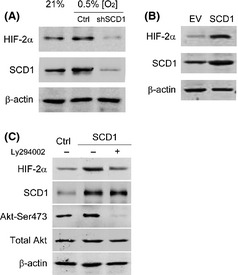
Inhibition on the PI3K/Akt pathway attenuates hypoxia‐inducible factor‐2α (HIF‐2α) upregulation triggered by stearoyl–CoA desaturase‐1 (SCD1) overexpression. (A) Scramble sequence or siRNA against SCD1 (shSCD1) was transfected into 786‐0 renal cancer cells. After puromycin selection (1 μg/mL for 14 days), the cells were exposed to hypoxia for 16 h. The protein levels for SCD1 and HIF‐2α were analyzed with Western blot. Cells transfected with scrambled sequence and cultured under normoxic conditions were taken as control (Ctrl). (B) SCD1‐expressing plasmids (2 μg) were transiently transfected into 786‐0 cells. The protein levels for SCD1 and HIF‐2α were analyzed with Western blot after 48 h. Cells transfected with empty vector (EV; 2 μg) were taken as control. (C) SCD1‐expressing plasmid (2 μg) was transfected into 786‐0 cells. After 24 h, the cells were replaced with medium containing PI3K/Akt pathway inhibitor Ly294002 (25 μM) or vehicle (DMSO) and incubated for a further 24 h. The levels of HIF‐2α, SCD1, total Akt, and phosphorylated Akt were detected by Western blot. Cells transfected with empty vector (2 μg) and incubated with DMSO were taken as control (Ctrl).
Double deficiency in HIF‐2α/SCD1 enhances apoptosis and attenuates colony formation capability
As HIF‐2α and SCD1 mutually enhance their expression in 786‐0 cells, and both proteins are associated with tumor growth, we wanted to know whether this cooperation would affect tumor cell growth and survival. We introduced scrambled sequence or siRNA against SCD1 or HIF‐2α, or both, to 786‐0 cells. Single deficiency of either SCD1 or HIF‐2α reduced the expression levels for both. Double deficiency in SCD1 and HIF‐2α led to lower levels for both proteins compared to single deficiency (Fig. 5A). The apoptosis in infected cells was detected by TUNEL assay. The inhibitions on SCD1 or HIF‐2α induced 786‐0 cell apoptosis, which was indicated by means of DNA fragmentation marked as TUNEL‐positive staining. Double deficiency on SCD1 and HIF‐2α resulted in more enhanced apoptosis compared with single inhibition on SCD1 or HIF‐2α, which was illustrated by increased apoptotic cell percentages in cells with double deficiency (Fig. 5B). The consequences of decreased apoptosis would increase the cell survival and colony formation capability in cancer cells. We observed the attenuated colony formation in cells with reduced SCD1 or HIF‐2α expression. And further blockage on colony formation was found in cells with double deficient SCD1 and HIF‐2α (Fig. 5C). This result indicated synergistic effects between SCD1 and HIF‐2α on reducing 786‐0 cell apoptosis, which might contribute to colony formation.
Figure 5.
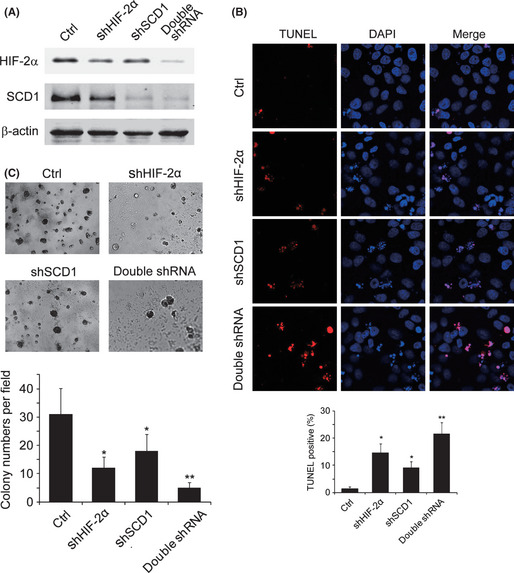
Effects of stearoyl–CoA desaturase‐1 (SCD1) and hypoxia‐inducible factor‐2α (HIF‐2α) on apoptosis and colony formation ability. (A) Renal carcinoma 786‐0 cells were transfected with the following siRNA combinations. After selection, the protein levels for SCD1 and HIF‐2α were analyzed using Western blot. Lane 1, scrambled sequences for SCD1 and HIF‐2α (Ctrl); lane 2, siRNA against HIF‐2α (shHIF‐2α) + scrambled sequences for siRNA against SCD1 (shSCD1); lane 3, shSCD1 + scrambled sequences for shHIF‐2α; lane 4, shSCD1 + shHIF‐2α. (B) 786‐0 cells infected with various siRNA combinations were cultured for 72 h and subjected to TUNEL staining; DAPI was used to indicate the nuclei. At least 300 cells from randomly selected fields were counted and the percentage of TUNEL positively stained cells was calculated. (C) 786‐0 cells infected with siRNA were seeded at 1 × 105 in a 35‐mm dish in soft agar and cultured for 14 days for colony formation. *P < 0.05 for shSCD1 or shHIF‐2α versus Ctrl; **P < 0.05 for double shRNA versus shSCD1 or shHIF‐2α.
Synergistic action of HIF‐2α and SCD1 on cell migration
The metastasis of tumor cells was determined by their migration capacity. The effects of SCD1 and HIF‐2α on 786‐0 cell migration were evaluated using a Transwell assay. Knockdown of either SCD1 or HIF‐2α reduced cell migration through the Transwell membrane. Compared with cells with single protein knockdown, fewer migrated cells were observed in those with double knockdown of SCD1 and HIF‐2α (Fig. 6).
Figure 6.
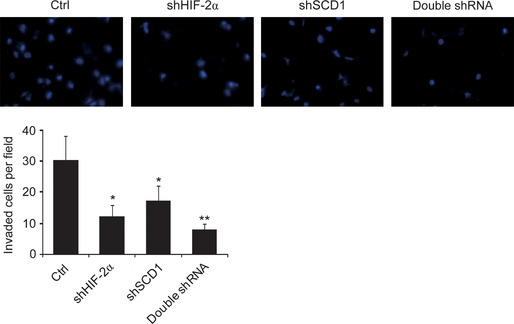
Effects of hypoxia‐inducible factor‐2α (HIF‐2α) and stearoyl–CoA desaturase‐1 (SCD1) on cell migration. Renal carcinoma 786‐0 cells infected with shRNA were seeded at 1 × 105 into a Transwell insert and cultured for 24 h for cell migration. Photographs of the migrated cells were taken from five fields. Representative photographs for each group are shown and the average cell numbers per field were calculated. Panel 1, scrambled sequences for SCD1 and HIF‐2α (Ctrl); panel 2, siRNA against HIF‐2α (shHIF‐2α) + scrambled sequences for siRNA against SCD1 (shSCD1); panel 3, shSCD1 + scrambled sequences for shHIF‐2α; panel 4, shSCD1 + shHIF‐2α. *P < 0.05 for shSCD1 or siRNA against HIF‐2α (shHIF‐2α) versus Ctrl; **P < 0.05 for double shRNA versus shSCD1 or shHIF‐2α.
Discussion
The aberrations of VHL tumor suppressor protein and consequent HIFα overexpression are critical in the development of ccRCC.2 In clinical investigations, according to HIFα expression in tumor cells, the human ccRCCs can be separated into three distinct groups: no HIFα protein detected due to WT VHL expression (approximately 10% of total cases); both HIF‐1α and HIF‐2α detected; and only HIF‐2a detected exclusively. Interestingly, no tumors were found to express only HIF‐1α, suggesting a critical role for HIF‐2α in the development of VHL‐deficient ccRCCs.20 In this study, we investigated the relationship between HIF‐2α and SCD1, an enzyme catalyzing MUFA production from SFA, in ccRCC cells. We found mutual promoting effects on expression between the two proteins which potentiate each other to tumorigenesis by maintaining cell survival and enhancing colony formation and cell migration.
Abnormally elevated SCD1 activity was reported to be associated with several types of cancer. This enzyme was implicated to be crucial in the onset and progression of cancer.21, 22 In animal models, high SCD1 expression in normal cells will bring genetic susceptibility to tumorigenesis.23 Elevated SCD1 activity resulted in abundant MUFA, caused by the shift in the SFA/MUFA ratio. High MUFA levels are correlated with poor prognosis and degree of tumor malignancy in patients.24, 25 Inhibition of SCD1 led to cancer cell death by depletion of MUFA.26 It is supposed that MUFA is associated with Akt pathway activation and AMP‐activated protein kinase (AMPK) pathway inhibition.11, 27 Coincidently, SCD1‐deficient lung cancer cells showed less Akt activation and higher levels of cell death.18 How MUFA accumulation induces PI3K/Akt pathway activation is still not fully understood. Probably the enhanced membrane fluidity caused by more MUFA in the membrane contributes to the modulations of membrane‐resident signal pathways.10 Ultimately, it seems that the SCD1–MUFA–Akt axis mediates SCD1 overexpression, triggering tumor progression. As a modulator of the PI3K/Akt pathway, SCD1 depletion induced caspase‐3 activation and PARP‐cleavage.28 Except for preventing cell apoptosis, SCD1 enhances tumor growth by promoting cell cycle progression.29 These reports give reasonable interpretations why we observed more cell apoptosis and less colony formation ability in SCD1‐depleted ccRCC cells. Furthermore, SCD1 knockdown induced fewer invasions in 786‐0 cells. This might be due to the decreased cell migration and invasion phenotype mediated by PI3K/Akt pathway activation.30
SCD1 is one of the target genes upregulated under hypoxic stress.16 In our study, using the human VHL‐deficient RCC line 786‐0, which has constitutively expressed HLF‐2α and gives a model to investigate the effects of HLF‐2α on its target genes, we found that SCD1 is upregulated under hypoxia and modulated by HLF‐2α. Of the two HIFα isoforms, HLF‐2α has a higher oncogenic potential because it activates pro‐tumorigenic target genes and induces more aggressive cell behaviors, whereas HIF‐1α decreases the tumor growth rate by limiting proliferation and inducing cell apoptosis.6, 31, 32 In our experiments, we observed HIF‐2α knockdown not only impacted on SCD1 expression, but led to higher levels of apoptosis and less colony formation ability in 786‐0 cells. These effects under HIF‐2α deficiency could be mediated by SCD1. Furthermore, synergistic effects between HIF‐2α and SCD1 on modulating cancer cell survival, colony formation ability, and cell migration were observed. These effects are due to a feedback loop between HIF‐2α and SCD1, mediated by PI3K/Akt pathway activation. In this positive feedback loop, increases or decreases in both HIF‐2α and SCD1 would result in stronger or weaker functions, respectively, than those obtained by varying a single protein. It has been reported that HIF‐2α expression was dependent on Akt3 signaling.19 Feedback loop regulation could be a common adjustment to HIFα in modulating tumorigenesis flexibly and efficiently.33 Interestingly, once VHL expression is restored in RCC cells lacking VHL, SCD1 levels change become less sensitive to changes in HIF‐2α levels. But SCD1 can still affect HIF‐2α expression. We cannot exclude the possibility that SCD1 is modulated by other compensatory pathways controlled by VHL once HIF‐2α is absent. The connection between HIF‐2α and SCD1 might be more complex in the presence of VHL; alternative hypoxia signalling pathways have been reported to be both VHL‐dependent and ‐independent manner.34 It is worthwhile investigating the relationship between HIF‐2α and SCD1 in a broader panel of ccRCC cell lines to gain a further understanding of the profiles and about the mutual regulation between the two proteins in ccRCC.
In ccRCCs, HIF‐2α overexpression contributes to cellular adaptation to microenvironmental hypoxic stress. Upregulation of SCD1 in response to HIF‐2α not only removes the detrimental endogenous SFA originating from enhanced glycolysis in cancer cells, and supplies enough substrate for lipid biosynthesis required for fast cell duplication, but also enhances HIF‐ 2α expression in a feedback loop pattern and cooperates with HIF‐2α in promoting tumorigenesis. Thus, therapy targeting the inhibition of both HIF‐2α and SCD1 could be effective in the treatment of ccRCC.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- HIF
hypoxia‐inducible factor
- MUFA
monounsaturated fatty acids
- RCC
renal cell carcinoma
- SCD1
stearoyl–CoA desaturase‐1
- SFA
saturated fatty acids
- shHIF‐2α
siRNA against HIF‐2α
- shSCD1
siRNA against SCD1
- VHL
von Hippel‐Lindau
Supporting information
Fig. S1. Expression of stearoyl–CoA desaturase‐1 (SCD1) under normoxia or hypoxia in cells with or without von Hippel‐Lindau (VHL) protein expression.
Fig. S2. Effects of hypoxia‐inducible factor‐2α (HIF‐2α) deficiency on stearoyl–CoA desaturase‐1 (SCD1) expression in cells with or without von Hippel‐Lindau (VHL) protein expression.
Fig. S3. Impact of stearoyl–CoA desaturase‐1 (SCD1) knockdown on hypoxia‐inducible factor‐2α (HIF‐2α) expression in cells with or without von Hippel‐Lindau (VHL) protein expression.
Acknowledgments
This work was sponsored by the National Natural Science Foundation of China (Grant No. 81272841), by the Municipal Hospital Level Project for Emerging and Frontier Technology of Shanghai (Grant No. SHDC 12010104), and the Shanghai Natural Science Foundation (Grant No. 10JC1409600).
(Cancer Sci 2013; 104: 416–422)
References
- 1. Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 2006; 441: 437–43. [DOI] [PubMed] [Google Scholar]
- 2. Arjumand W, Sultana S. Role of VHL gene mutation in human renal cell carcinoma. Tumour Biol 2012; 33: 9–16. [DOI] [PubMed] [Google Scholar]
- 3. Majmundar AJ, Wong WJ, Simon MC. Hypoxia‐inducible factors and the response to hypoxic stress. Mol Cell 2010; 40: 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 2012; 12(1): 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krieg M, Haas R, Brauch H et al Up‐regulation of hypoxia‐inducible factors HIF‐1alpha and HIF‐2alpha under normoxic conditions in renal carcinoma cells by von Hippel‐Lindau tumor suppressor gene loss of function. Oncogene 2000; 19: 5435–43. [DOI] [PubMed] [Google Scholar]
- 6. Raval RR, Lau KW, Tran MG et al Contrasting properties of hypoxia‐inducible factor 1 (HIF‐1) and HIF‐2 in von Hippel‐Lindau‐associated renal cell carcinoma. Mol Cell Biol 2005; 25: 5675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang V, Davis DA, Haque M et al Differential gene up‐regulation by hypoxia‐inducible factor‐1alpha and hypoxia‐inducible factor‐2alpha in HEK293T cells. Cancer Res 2005; 65: 3299–306. [DOI] [PubMed] [Google Scholar]
- 8. Krishnan J, Suter M, Windak R et al Activation of a HIF1alpha‐PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab 2009; 9: 512–24. [DOI] [PubMed] [Google Scholar]
- 9. Rankin EB, Rha J, Selak MA et al Hypoxia‐inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol 2009; 29: 4527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim YC, Ntambi JM. Regulation of stearoyl‐CoA desaturase genes: role in cellular metabolism and preadipocyte differentiation. Biochem Biophys Res Commun 1999; 266: 1–4. [DOI] [PubMed] [Google Scholar]
- 11. Scaglia N, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase‐1 inactivates acetyl‐CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS ONE 2009; 4: e6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fritz V, Benfodda Z, Rodier G et al Abrogation of de novo lipogenesis by stearoyl‐CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol Cancer Ther 2010; 9: 1740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vacaresse N, Lajoie‐Mazenc I, Augé N et al Activation of epithelial growth factor receptor pathway by unsaturated fatty acids. Circ Res 1999; 85: 892–9. [DOI] [PubMed] [Google Scholar]
- 14. Zeisig R, Koklic T, Wiesner B et al Increase in fluidity in the membrane of MT3 breast cancer cells correlates with enhanced cell adhesion in vitro and increased lung metastasis in NOD/SCID mice. Arch Biochem Biophys 2007; 459(1): 98–106. [DOI] [PubMed] [Google Scholar]
- 15. Igal RA. Stearoyl‐CoA desaturase‐1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis 2010; 31: 1509–15. [DOI] [PubMed] [Google Scholar]
- 16. Igwe EI, Essler S, Al‐Furoukh N et al Hypoxic transcription gene profiles under the modulation of nitric oxide in nuclear run on‐microarray and proteomics. BMC Genomics 2009; 10: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shinojima T, Oya M, Takayanagi A et al Renal cancer cells lacking hypoxia inducible factor (HIF)‐1alpha expression maintain vascular endothelial growth factor expression through HIF‐2alpha. Carcinogenesis 2007; 28: 529–36. [DOI] [PubMed] [Google Scholar]
- 18. Scaglia N, Igal RA. Inhibition of Stearoyl‐CoA Desaturase 1 expression in human lung adenocarcinoma cells impairs tumorigenesis. Int J Oncol 2008; 33: 839–50. [PubMed] [Google Scholar]
- 19. Toschi A, Lee E, Gadir N et al Differential dependence of hypoxia‐inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem 2008; 283: 34495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gordan JD, Lal P, Dondeti VR et al HIF‐alpha effects on c‐Myc distinguish two subtypes of sporadic VHL‐deficient clear cell renal carcinoma. Cancer Cell 2008; 14: 435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Ding SF, Habib NA et al Partial characterization of a cDNA for human stearoyl‐CoA desaturase and changes in its mRNA expression in some normal and malignant tissues. Int J Cancer 1994; 57: 348–52. [DOI] [PubMed] [Google Scholar]
- 22. Yahagi N, Shimano H, Hasegawa K et al Co‐ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur J Cancer 2005; 41: 1316–22. [DOI] [PubMed] [Google Scholar]
- 23. Falvella FS, Pascale RM, Gariboldi M et al Stearoyl‐CoA desaturase 1 (Scd1) gene overexpression is associated with genetic predisposition to hepatocarcinogenesis in mice and rats. Carcinogenesis 2002; 23: 1933–6. [DOI] [PubMed] [Google Scholar]
- 24. Chajès V, Joulin V, Clavel‐Chapelon F. The fatty acid desaturation index of blood lipids, as a biomarker of hepatic stearoyl‐CoA desaturase expression, is a predictive factor of breast cancer risk. Curr Opin Lipidol 2011; 22: 6–10. [DOI] [PubMed] [Google Scholar]
- 25. Lane J, Mansel RE, Jiang WG. Expression of human delta‐6‐desaturase is associated with aggressiveness of human breast cancer. Int J Mol Med 2003; 12: 253–7. [PubMed] [Google Scholar]
- 26. Mason P, Liang B, Li L et al SCD1 inhibition causes cancer cell death by depleting mono‐unsaturated fatty acids. PLoS ONE 2012; 7: e33823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hardy S, Langelier Y, Prentki M. Oleate activates phosphatidylinositol 3‐kinase and promotes proliferation and reduces apoptosis of MDA‐MB‐231 breast cancer cells, whereas palmitate has opposite effects. Cancer Res 2000; 60: 6353–8. [PubMed] [Google Scholar]
- 28. Minville‐Walz M, Pierre AS, Pichon L et al Inhibition of stearoyl‐CoA desaturase 1 expression induces CHOP‐dependent cell death in human cancer cells. PLoS ONE 2010; 5: e14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hess D, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase activity blocks cell cycle progression and induces programmed cell death in lung cancer cells. PLoS ONE 2010; 5: e11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stambolic V, Woodgett JR. Functional distinctions of protein kinase B/Akt isoforms defined by their influence on cell migration. Trends Cell Biol 2006; 16: 461–6. [DOI] [PubMed] [Google Scholar]
- 31. Biswas S, Troy H, Leek R et al Effects of HIF‐1alpha and HIF2alpha on growth and metabolism of clear‐cell renal cell carcinoma 786‐0 xenografts. J Oncol 2010; 2010: 757908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu J, Wang B, Xu Y et al Epigenetic regulation of HIF‐1alpha in renal cancer cells involves HIF‐1alpha/2alpha binding to a reverse hypoxia‐response element. Oncogene 2012; 31: 1065–72. [DOI] [PubMed] [Google Scholar]
- 33. Henze AT, Acker T. Feedback regulators of hypoxia‐inducible factors and their role in cancer biology. Cell Cycle 2010; 9: 2749–63. [DOI] [PubMed] [Google Scholar]
- 34. Jiang Y et al Gene expression profiling in a renal cell carcinoma cell line: dissecting VHL and hypoxia‐dependent pathways. Mol Cancer Res 2003; 1: 453–62. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of stearoyl–CoA desaturase‐1 (SCD1) under normoxia or hypoxia in cells with or without von Hippel‐Lindau (VHL) protein expression.
Fig. S2. Effects of hypoxia‐inducible factor‐2α (HIF‐2α) deficiency on stearoyl–CoA desaturase‐1 (SCD1) expression in cells with or without von Hippel‐Lindau (VHL) protein expression.
Fig. S3. Impact of stearoyl–CoA desaturase‐1 (SCD1) knockdown on hypoxia‐inducible factor‐2α (HIF‐2α) expression in cells with or without von Hippel‐Lindau (VHL) protein expression.


