Abstract
The c‐myc transcriptional suppressor, far‐upstream element (FUSE)‐binding protein (FBP)‐interacting repressor (FIR), is alternatively spliced in colorectal cancer tissue (Matsushita et al., Cancer Res 2006). Recently, the knockdown of SAP155 pre‐mRNA‐splicing factor, a subunit of SF3b, was reported to disturb FIR pre‐mRNA splicing and yield FIRΔexon2, an exon 2‐spliced variant of FIR, which lacks c‐myc repression activity. In the present study, novel splicing variants of FIR, Δ3 and Δ4, were also generated by SAP155 siRNA, and these variants were found to be activated in human colorectal cancer tissue. Furthermore, the expression levels of FIR variant mRNA were examined in the peripheral blood of colorectal cancer patients and healthy volunteers to assess its potency for tumor detection. As expected, circulating FIR variant mRNA in the peripheral blood of cancer patients were significantly overexpressed compared to that in healthy volunteers. In particular, the area under the receiving operating characteristic curve of FIR, FIRΔexon2 or FIRΔexon2/FIR, was greater than those of conventional carcinoembryonic antigen or carbohydrate antigen 19‐9. In addition, FIRΔexon2 or FIR mRNA expression in the peripheral blood was significantly reduced after operative removal of colorectal tumors. Thus, circulating FIR and FIRΔexon2 mRNA are potential novel screening markers for colorectal cancer testing with conventional carcinoembryonic antigen and or carbohydrate antigen 19‐9. Taken together, our results indicate that overexpression of FIR and its splicing variants in colorectal cancer directs feed‐forward or addicted circuit c‐myc transcriptional activation. Clinical implications for colorectal cancers of novel FIR splicing variants are also discussed in the present paper.
Several non‐invasive biomarkers, including DNA alterations, mRNA and peptides/proteins, are specifically expressed in tumor cells.1, 2 Far‐upstream element (FUSE) is a sequence required for the proper expression of the human c‐myc gene.3 FUSE is located 1.5 kb upstream of the c‐myc promoter P1 and binds the FUSE binding protein (FBP), a transcription factor that stimulates c‐myc expression in a FUSE‐dependent manner.4, 5, 6 Yeast two‐hybrid analysis has revealed that FBP binds to a protein, the FBP interacting repressor (FIR). FIR represses c‐myc transcription by delaying promoter escape.7, 8 FIRΔexon2, an exon2‐splice variant of FIR that lacks c‐myc repression activity, has been frequently identified in human primary colorectal cancers.9 SAP155, a subunit of SF3b, and FIR have been reported to form a complex.10 SAP155 is required for FIR pre‐mRNA splicing. SAP155/FIR (/FIRΔexon2) forms a complex and is mutually upregulated in cancers.11 Despite the splice mutation, FIRΔexon2 retains binding activity to FUSE and is able to displace repression‐competent FIR from FUSE, which thwarts FIR‐mediated transcriptional repression by FUSE.11
Up to 60% of all human genes have at least one alternative splice variant and alternative splicing has been documented to play a significant role in human diseases, including cancer.12 Although the precise mechanism by which alternative splicing is manipulated in cancer remains unclear, cancers are potentially addicted13, 14 to overexpressed SAP155/FIR/FIRΔexon2/c‐myc feed‐forward signaling networks.11 Thus, cancer‐specific alternative splicing can be valuable for the development of cancer therapy targets15 as well as for cancer detection.16
In this context, the present study reveals that SAP155 knockdown yields novel splicing variants of FIR, Δ3 and Δ4, in HeLa and colon cancer cells. Interestingly, Δ3 and Δ4, as well as FIR and FIRΔexon2, were activated in human colon cancer tissues, as detected by quantitative real‐time PCR. Conventional carcinoembryonic antigen (CEA) and carbohydrate antigen (CA)‐19‐9 were compared with FIR and FIRΔexon2 mRNA to evaluate their suitability as molecular markers for colorectal cancer detection in the peripheral blood (PB). FIRΔexon2 and FIR mRNA levels in the PB of colorectal cancer patients were significantly reduced after operative excision of tumors. We show that FIRΔexon2 and FIR mRNA detected in the PB of colorectal cancer patients, but not in that of healthy volunteers, can act as potent novel screening markers of colorectal cancer.
Materials and Methods
Human operatively excised tissue samples and peripheral blood samples of leukemia patients and healthy volunteers
Tissue samples from 36 patients with primary colorectal cancer were surgically excised at Chiba University Hospital. The tumor samples were obtained from the tumor epithelium immediately after operative excision; tumor tissue and the corresponding non‐tumor epithelial samples were 5–10 cm apart. All excised tissues were immediately placed into liquid nitrogen and stored at −80°C until analysis. Paired‐PB (10 mL) was obtained before and after tumor excision from 36 colorectal cancer patients during operation under anesthesia and the day after operation. To avoid skin contamination, the first 20 mL of blood was not used. The same amount of PB from 41 healthy volunteers and five leukemia patients was obtained following the same procedure. Written informed consent was obtained from healthy volunteers and patients.
Immunocytochemistry
Colorectal cancer tissues and corresponding normal tissues, as well as leukemia cells, were collected and treated for immunocytochemistry, as previously described.9, 17
RT‐PCR and real‐time quantitative PCR
Total RNA of HeLa cells was extracted using the RNeasy Mini Kit (Qiagen, Tokyo, Japan). Total RNA of PB from patients and healthy volunteers was extracted using the PAX gene Blood RNA kit PreAnalytiX (Qiagen, Tokyo, Japan). cDNA were synthesized from total RNA using the First Strand cDNA Synthesis Kit for RT‐PCR (Roche, Mannheim, Germany). Using the cDNA as a template, FIR cDNA was amplified with suitable primers by RT‐PCR: forward 5′‐GGCCCCATCAAGAGCATC‐3′, reverse 5′‐GGGGCTGGGCCAGGGTCAG‐3′. As a control, β‐actin cDNA was amplified. The amino terminal region of FIR was amplified by RT‐PCR with the following primers: forward 5′‐AGACAGCGGAAGGAGCAAGAGTGG‐3′, reverse 5′‐CTGTGCAGCTTCGGGGACCTCATA‐3′. The PCR product was loaded on a 2.5% agarose gel (Promega, WI, USA), purified using a Gel Extraction Kit (Qiagen, Tokyo, Japan), and cloned with the pGEM‐T Easy vector system (Promega) for DNA sequencing. Real‐time quantitative PCR of c‐myc or FIR cDNA using the LightCycler instrument (Roche, Mannheim, Germany) was carried out in 20 μL of reaction mixture made up of a master mixture (LightCycler‐FastStart DNA Master SYBR Green I) containing FastStart Taq DNA polymerase, dNTP mixture and buffer (LightCycler DNA Master hybridization probes, Roche, Mannheim, Germany), 3.0 mM MgCl2, 0.5 μM each of sense and antisense primers, and 1 μL of template cDNA in a LightCycler capillary. LightCycler software version 3.3 (Roche, Mannheim, Germany) was used for analysis of quantitative RT‐PCR. Primers and PCR Cycle Program Data for FIR (FIR, FIRΔexon2, Δ3 and Δ4) have been listed previously.11
Colorectal cancer and immortalized fibroblast cell lines
The following were purchased from ATCC (Manassas, VA, USA): HeLa cells; human colorectal adenocarcinoma cell lines, SW480 and HCT116; human fibroblast cell lines, MRC5 and WI38; and an African Green Monkey SV40‐transfected kidney fibroblast cell line, COS‐7. Cells were grown at 37°C in 5% CO2 in Iscove's Modified Dulbecco's Medium supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA) and 1% penicillin–streptomycin (Invitrogen).
Protein extraction, immunoblotting and antibodies
Whole cell extract proteins were dissolved in a sample buffer, as described previously.9 The primary mouse monoclonal antibody against FIR C‐terminus (6B4) was prepared by Dr Nozaki.18 In brief, synthetic peptides C+KVVAEVYDQERFDNSDLSA (C+541–559; the numbers indicate the amino acids of PUF60)19 were used as the immunization antigens. Anti‐FIR antibodies (6B4) at a dilution of 1:100 were used in the blocking buffer for western blotting. Anti‐c‐Myc (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti‐SAP155 (MBL, Nagoya, Japan: D221‐3) were mouse monoclonal antibodies, and anti‐FIR rabbit polyclonal antibodies were prepared and purchased from Japan BioService (Saitama, Japan). Antibodies were used in these experiments according to the manufacturer's recommendations.
SAP155 siRNA interference
The target sequence for SAP155 siRNA oligonucleotides has been described previously.11 Transient transfection of siRNA was carried out using Lipofectamine 2000 (Gibco BRL, MA, USA) according to manufacturer's instructions. The transfected cells were cultured for 72 h at 37°C in a CO2 incubator.
Plasmids, Clcn1 and dystrophin minigene
Full‐length FIR, FIRΔexon2, FIRΔ3 and FIRΔ4 cDNA was cloned into the p3xFLAG‐CMV‐14 vector (Sigma, MO, USA) to introduce the FLAG‐tag at the amino terminus.11 Mouse Clcn1 minigene, containing exons 6–7a–7, was cotransfected into HCT116 and COS‐7 cells with FIR or FIRΔexon2 expression vectors to examine splicing activities. MBNL1 expression vector was used as a positive control.20, 21 Alternatively, approximately 1 kb of FIR genome around exon2 was subcloned into minigene plasmid consisting of the area between Dystrophin exons 18 and 20, including part of the adjacent introns.22 In brief, genomic DNA extracted from HeLa cells was amplified around the region of exon2 by PCR with forward primers at intron 1 attached at the Nhe I site and reverse primer at intron 2 attached at the Bam HI site: forward, 5′‐GGGCTAGCCTTCAGGAGAGAACTGGGAC‐3′; reverse, 5′‐GCGGATCCACAGACCAAGACCCACCTGC‐3′. PCR products were loaded onto a 1.0% agarose gel (Promega) and bands were excised and purified, and DNA sequenced. PCR product size was 1099 bp. PCR products were double digested with Nhe I and Bam HI restriction enzymes and ligated to the same sites in H492 cassette vectors,21 which were designed to examine alternative splicing of specific exons, FIR‐exon2 in this study, as minigenes. Plasmids were prepared by CsCl ultra‐centrifugation or using a Endofree Plasmid Maxi Kit (Qiagen, Germantown, MD, USA), and the DNA sequences were verified. Cells were transfected with plasmids containing minigenes by Lipofectamine 2000 reagents (Gibco BRL).
In vitro splicing assay
For in vitro splicing assays, HCT116 or COS‐7 cells were plated onto 3.5 or 6‐cm dishes and cultured for 24 h in DMEM plus 10% FBS before plasmid transfection. Cells were grown to 60–80% confluence and then transiently cotransfected with plasmids, 300 ng splicing reporter, and 400 ng MBNL1, FIR‐FLAG or FIRΔexon2‐FLAG. Cells were collected after 48 h, total RNA was extracted using an Rneasy Kit (Qiagen, Valencia, CA), and the samples were analyzed by RT‐PCR. Reverse transcription was performed using Prime‐Script Reverse Transcriptase (TaKaRa, Shiga, Japan). Spliced products were amplified using EGFP primer (forward, CATGGTCCTGCTGGA GTTCGTG; reverse, GTTTCAGGTTCAGGGGGAGGTGTG) and separated by 2% agarose gel.
Adenovirus vector
The FIR and FIRΔexon2 adenovirus vectors were prepared as previously described.11
Detection of FIR mRNA in the culture medium of FIR‐transfected cells
For FIR mRNA detection in the culture medium, HeLa cells were plated in 3.5 or 6‐cm dishes (or 12‐well plates) and cultured for 24 h in improved Eagle's minimum essential medium plus 10% FBS without antibiotics before plasmid transfection. Cells were grown to 60–80% confluence and then transiently cotransfected with 700 or 1400 ng FIR/p3xflag‐CMV14 vector using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The supernatants of the cultured media were collected after 48 h culturing by centrifuging (20 600g for 15 min), and total RNA was xtracted using an ISOGEN (Nippongene, Toyama, Japan). cDNA were synthesized using the First Strand cDNA Synthesis Kit for RT‐PCR (Roche), and the samples were analyzed by RT‐PCR (FIR forward, 5′‐CCATAGCTCTCCAGGTCA‐3′ [exon1–2]; FIR reverse: 5′‐CGTAGACGCGGCACATGA‐3′ [exon6]). PCR conditions were 95°C, 10 min; (95°C, 30 s; 60°C, 30 s; and 72°C, 30 s) × 35 cycles; 72 C, 10 min; DNA polymerase used was AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen).
Conventional carcinoembryonic antigen and carbohydrate antigen detection
CEA and CA 19‐9 were measured by CLEIA using a Lumipulse f (Fujirebio, Tokyo, Japan), with cut‐off values of 5 ng/mL and 37 U/mL, respectively.
Receiver operating characteristic curves of FIR, FIRΔexon2, conventional carcinoembryonic antigen and carbohydrate antigen detection
The overall diagnostic efficiencies of FIRΔexon2, CEA and CA19‐9 were evaluated by comparison of receiver operating characteristic (ROC) curves.23 The area under each ROC curve was calculated, and the statistical significance of the difference between ROC curves was assessed as described previously.24 A P‐value <0.05 was considered to be significant. ROC curves were generated and the area under the curve (AUC) values were calculated using StatFlex software version 5.0.
Statistical analysis
The colorectal cancer tissues and non‐tumor tissues of cancer patients were compared by χ 2‐test. P‐values were calculated using a statistical software program, StatFlex (Artech, Osaka, Japan). Correlation of FIR and SAP155 expression was evaluated using the Pearson product‐moment correlation coefficient.
Results
SAP155 is required for FIR pre‐mRNA splicing in HeLa, RKO, DLD1 and HCT116 cells
SAP155 siRNA treatment produces novel FIR splicing variants in HeLa cells (Fig. 1a). Do these novel FIR splicing variants actually exist in endogenous human colorectal cancer tissues? In addition, if they exist, do these novel FIR splicing variants play a functional role in carcinogenesis or other mechanisms? In the present study, we attempted to detect these novel FIR splicing variant mRNA in colorectal cancer cell lines (Fig. 1a). We successfully confirmed the presence of all four FIR splicing variants by DNA sequencing of PCR products (Fig. 1b). In particular, an increase in Δ3 mRNA by knockdown of SAP155 was most prominent in all colorectal cancer cell lines examined (Fig. 1c).
Figure 1.
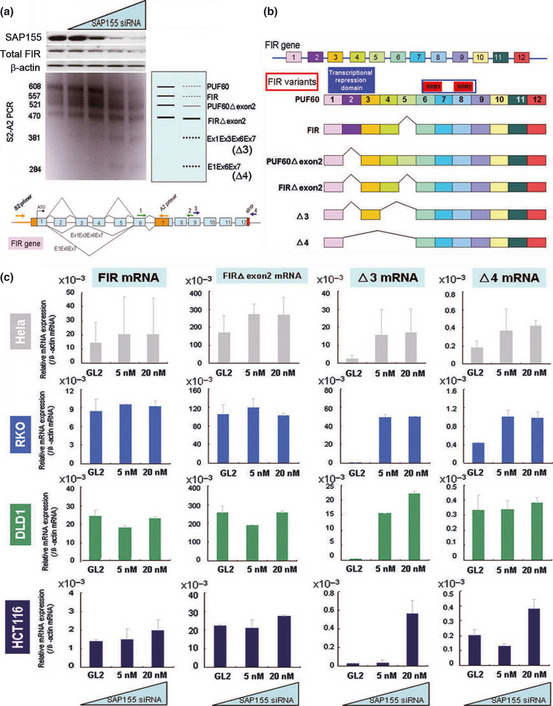
SAP155 siRNA interfered with FIR mRNA splicing. (a) SAP155 siRNA interfered with endogenous FIR pre‐mRNA splicing. (b) Constructs of FIR splicing variants are shown. Each FIR splicing variant was extracted from the DNA gel, subcloned into a suitable plasmid, and DNA sequenced. Scheme of each FIR splicing variant is shown. SAP155 siRNA induced the alternative splicing of FIR‐pre mRNA. (c) FIR splicing variant, Δ3, mRNA was increased by SAP155 siRNA in HeLa, RKO, DLD1 and HCT116 cells.
FIR and its variants are strongly expressed in colorectal cancer tissues. Total FIR protein expression, including the expression of FIR and/or its splicing variants, was significantly increased in colorectal cancer tissues compared to corresponding non‐cancer epithelial glands (upper left panel, Fig. 2a). In addition, mRNA levels of FIR and/or its splicing variants were upregulated in cancer tissues compared to corresponding non‐cancer tissues (Fig. 2b). It must be noted that FIRΔexon2 has been reported to inhibit competitively c‐myc suppression by authentic FIR in vivo and in vitro, and that exon2 of FIR includes the amino terminal suppression domain and is necessary for both endogenous c‐Myc suppression and apoptosis induction.9, 11 Notably, FIRΔexon2 potentially interferes with FIR to bind to FUSE, indicating that FIRΔexon2 disturbs authentic FIR's c‐myc transctiptional suppression.11
Figure 2.
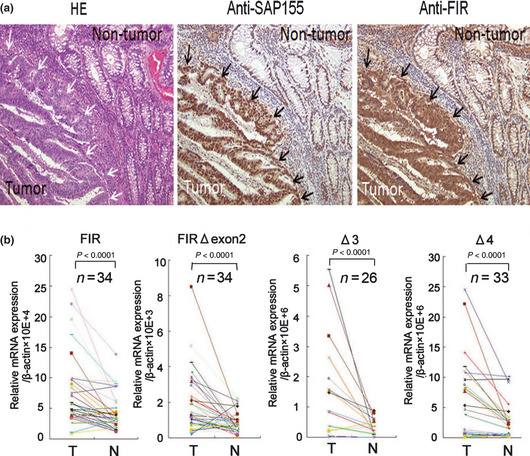
FIR and SAP155 were strongly expressed in colorectal cancer tissues. (a) Total FIR (right panel) and SAP155 (middle panel) were more strongly expressed in colorectal cancer tissues than in normal colonic mucosa. HE staining of colorectal cancer tissues and normal colonic mucosa (left panel) is also shown. (b) Quantitative analysis of FIR and FIR splicing variants, FIRΔexon2, Δ3 and Δ4, by real‐time PCR in colorectal cancer tissues (T). FIR, FIRΔexon2, Δ3 and Δ4 were significantly overexpressed in cancer tissues compared to non‐cancer epithelium (N) (P < 0.0001).
FIR and its splicing variants potentially affect endogenous c‐Myc expression in HeLa cells
FIR‐FLAG, FIRΔexon2FLAG, FIRΔ3‐FLAG and FIRΔ4‐FLAG (Fig. 3a) were transiently transfected to HeLa cells to examine their cellular localization and endogenous c‐Myc expression. Cellular localization of FIR‐FLAG, FIRΔexon2‐FLAG, FIRΔ3‐FLAG and FIRΔ4‐FLAG was mainly in the nucleus (Fig. 3b). Endogenous c‐Myc was suppressed by FIR‐FLAG and increased by FIRΔexon2‐FLAG, as reported previously.9, 11 FIRΔ3‐FLAG and FIRΔ4‐FLAG also enhanced c‐Myc expression (Fig. 3b, arrows). The first RNA recognition motif (RRM1) of FIR (amino acids 112–187, Fig. 1b) binds nucleic acids, and, therefore, it would be helpful to examine whether a conformational change would occur in FIRΔexon2, FIRΔ3 and/or FIRΔ4 because FIR has been shown to dimerize upon nucleic acids binding.25 Further study is required to reveal the significance of dimerization of FIR and/or its splicing variants in terms of FUSE/FBP/c‐Myc pathway and SAP155 interaction in oncogenesis.
Figure 3.
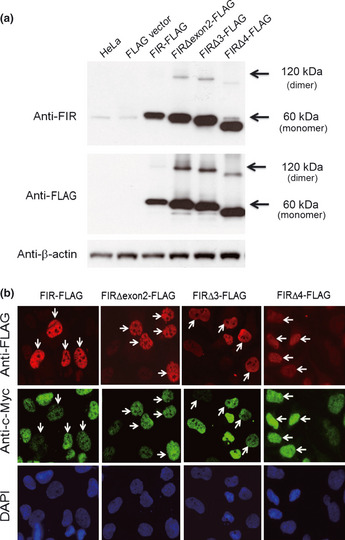
FIR and its splicing variants potentially affect endogenous c‐Myc expression in HeLa cells. (a) FIR‐FLAG, FIRΔexon2‐FLAG, FIRΔ3‐FLAG and FIRΔ4‐FLAG were transiently transfected to HeLa cells, to examine their cellular localization as well as the effect to endogenous c‐Myc expression. (b) Cellular localization of FIR‐FLAG, FIRΔexon2‐FLAG, FIRΔ3‐FLAG and FIRΔ4‐FLAG was examined by immunohistchemistry. Endogenous c‐Myc was suppressed by FIR‐FLAG whereas increased by FIRΔexon2‐FLAG (arrows). FIRΔ3‐FLAG and FIRΔ4‐FLAG also enhanced c‐Myc expression (arrows).
FIR and FIRΔexon2 is involved in both minigene splicing and c‐Myc expression in vitro
Given that enhanced protein expression levels of FIR and its splicing variants play a role in carcinogenesis, these proteins potentially affect alternative splicing or gene expression in cancer cells and, thus, are applicable for cancer screening. Do splicing variants of FIR play a significant role in splicing and c‐myc transcriptional regulation? We previously reported that SAP155 was co‐immunoprecipitated with both FIR and FIRΔexon2, and, thus, disturbed expression of FIR or FIRΔexon2 would affect splicing.11 When the Clcn1 minigene was transfected into HCT116 cells, 45% of the spliced products contained exon7a (Fig. 4a). As expected, cotransfection of FIR or FIRΔexon2 with the Clcn1 minigene increased alternative splicing in HCT116, colorectal adenocarcinoma cells (Fig. 4a). MBNL1, which acts as a splicing promoting factor, was used as a positive control and produced strong repression of exon7a inclusion. Furthermore, to investigate potential differences or disturbances in the splicing mechanism between cell lines, we prepared another minigene plasmid consisting of 1 kb of the FIR gene around exon 2 between dystrophin exons18 and 20, including part of the adjacent introns (Fig. 4b). As expected, exon2 of FIR is more frequently excluded in cancer cell lines, HeLa, HCT116 and SW480, than in non‐cancer cell lines, WI38 and MRC5 (Fig. 4c). However, the ratio of endogenous FIRΔexon2/FIR mRNA among these cells was not consistent with those of minigene, for an undetermined reason (data not shown). These results indicate that FIR, or FIRΔexon2, is potentially involved in splicing, directly or indirectly. In addition, minigene analysis indicates that the splicing mechanism of FIR exon 2 is significantly disturbed in cancer cells. Finally, we found that Ad‐FIR suppressed c‐Myc, whereas Ad‐FIRΔexon2 increased c‐Myc (Fig. 4d). Together, our results indicate that FIR and/or FIRΔexon2 are potentially involved in both transcriptional and splicing regulation.
Figure 4.
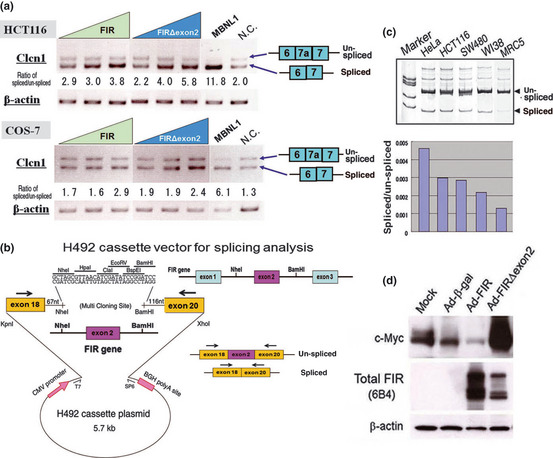
FIR and FIRΔexon2 exhibited direct or indirect splicing activity in vitro. (a) Mouse myotonic dystrophin minigene was cotransfected into HCT116 or COS‐7 cells with FIR or FIRΔexon2 expression vectors to examine splicing activities by PCR analysis. Splicing scores indicate the ratio of PCR product intensity of spliced (exons6–7)/unspliced (exons6–7a–7) bands. N.C., vacant vector as negative control. (b) Construct of H492 minigene cassette vector for splicing analysis. Genomic FIR gene including exon2 was subcloned between human dystrophin exons18 and 20. (c) Endogenous splicing activity for the H492 that contain FIRexon2 minigene in cancer and non‐cancer cells. (d) Ad‐FIR suppressed c‐Myc, whereas Ad‐FIRΔexon2 increased c‐Myc in HeLa cells.
FIR and FIRΔexon2 mRNA expressed in peripheral blood of early‐stage colorectal cancer patients
Before trying to detect circulatory mRNA in the PB, we examined whether FIR or FIRΔexon2 mRNA were detectable in the culture medium of pcDNA3.1‐FIR transfected HeLa cells (Fig. 5a). FIR mRNA were detected in the supernatant of culture medium (Fig. 5b). Next, FIR and/or FIRΔexon2 mRNA were examined as potential molecular markers for cancer screening. This study revealed that the ratios of FIRΔexon2/FIR in the PB of colorectal cancer patients were significantly higher than those in healthy volunteers (Fig. 5c). Ratios of FIRΔexon2/FIR were evaluated against Dukes stages of colorectal cancer invasiveness, and we found that the ratios were increased even in Dukes' A stage (earliest stage) colorectal cancer patients (Fig. 5c, left).
Figure 5.
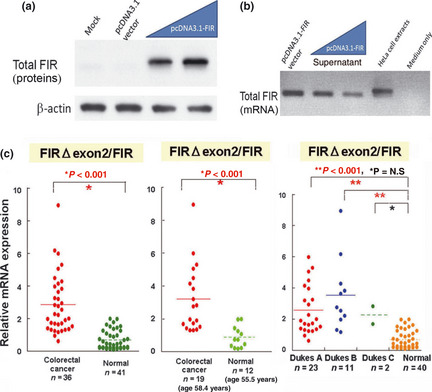
Ratios of FIRΔexon2/FIR mRNA in peripheral blood (PB) of colorectal cancer patients are significantly larger than controls. (a) Total FIR protein was detected in culture medium of pcDNA3.1‐FIR transfected HeLa cells. (b) Total FIR mRNA was detected in culture medium of pcDNA3.1‐FIR transfected HeLa cells. (c) Relative mRNA expression of FIRΔexon2/FIR in the PB is significantly activated (P < 0.001) in colorectal cancer patients (n = 36) compared to healthy volunteers (n = 41) (left). Age‐matched samples showed the same significant results (P < 0.001, middle). Expression levels of both FIRΔexon2 and FIR in the PB of cancer patients are increased at a relatively early stage (P < 0.001, Dukes A stage; no lymph node metastasis) (right).
Receiver‐operating characteristic curves of FIRΔexon2, CEA and CA19‐9 indicating specificity and quantitative capacity as tumor markers
The AUC of FIR (blue, 0.91) and FIRΔexon2/FIR (green, 0.92) was significantly greater than that of CEA (0.63) and CA19‐9(0.63), indicating that the ratio of FIRΔexon2/FIR is a superior tumor marker to CEA or CA19‐9 for screening of colorectal cancer (Fig. 6). Assuming that cancer cells or cell‐free RNA are circulating in the PB, these results suggest that FIR and/or FIRΔexon2 cDNA detection in the PB can be used as a potent molecular marker for early cancer detection. In sum, in colorectal cancers, our results indicate that an imbalance in FIR‐SAP155 expression disturbs FIR pre‐mRNA splicing and generates novel FIR splicing variants.
Figure 6.
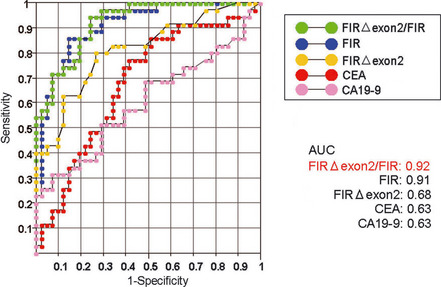
Receiver operating characteristic (ROC) curves. FIRΔexon2/FIR (green line), FIR (blue), FIRΔexon2 (red), conventional carcinoembryonic antigen (CEA) (yellow) and carbohydrate antigen (CA)‐19‐9 (pink) are shown. The area under curve (AUC) of the ratio of FIRΔexon2/FIR (0.92) was significantly greater than that of CEA (0.63) and CA19‐9 (0.63), indicating the ratio of FIRΔexon2/FIR is a candidate screening marker for colorectal cancer.
FIRΔexon2 or FIR mRNA expression in peripheral blood is significantly reduced after operative excision of colorectal tumors
If circulating FIR or FIRΔexon2 mRNA are released from cancer cells, these mRNA should be reduced after surgical tumor removal. We measured FIR and FIRΔexon2 mRNA levels in the PB by quantitative real‐time PCR. As expected, both FIR and FIRΔexon2 mRNA levels were significantly reduced after tumor removal (Fig. S1). FIR or FIRΔexon2 proteins are promising biomarker candidates for other cancers (Fig. S2).
Discussion
In this study, first, we identified that authentic FIR and its SAP155‐mediated novel splicing variants of FIRΔexon2, Δ3 and Δ4, mRNA were upregulated in human colon cancer tissues. Second, FIR and FIRΔexon2 were found to play a significant role in FIRexon2‐containing minigene splicing in vitro as well as endogenous c‐Myc expression in HeLa cells. Third, circulating FIR and FIRΔexon2 mRNA in the PB were significantly activated in colorectal cancer patients in comparison to healthy volunteers. Furthermore, FIR and FIRΔexon2 mRNA were detected in the PB of relatively early stage cancer patients (Dukes A stage) (Fig. 5c, right).
Why is FIR activated and alternatively spliced in cancers? Does c‐Myc overexpression play a role in FIR and/or SAP155 activation and FIR pre‐mRNA splicing? One reason might be that FIR is a c‐Myc target. In addition, the tight FIR/FIRΔexon2–SAP155 interaction disables established FIR and SAP155 functions, FIR in c‐myc transcriptional repression and SAP155, a subunit of SF3b, in pre‐mRNA splicing.11 Notably, SAP155 is required for the synthesis of normally spliced FIR mRNA; thus, FIR/FIRΔexon2–SAP155 interaction inhibits ordinal FIR mRNA splicing. The other reason is that FIRΔexon2, FIRΔ3 or FIRΔ4 potently forms a heterodimer with FIR. Thus these heterodimers interfere with FIR to bind to FUSE. These results strongly suggest that FIR splicing variants antagonize FIR in c‐myc transcriptional suppression and simultaneously interfere with SF3b in splicing during tumor progression.
Many such studies have indicated the existence of free DNA/RNA in the PB15, 26, 27 and the importance of spliced variants for screening of early cancers.28, 29, 30 Thus, cancers require activation of the c‐Myc/FIR/SAP155 networks, and this activation is pivotal for carcinogenesis and treatment.11, 31 Furthermore, sensitivity and specificity of FIRΔexon2 mRNA detection by real‐time PCR was superior to conventional tumor markers CEA and CA19‐9. In conclusion, the determination of FIRΔexon2/FIR mRNA ratios in combination with CEA and CA19‐9 improves screening accuracy for colorectal cancers. Consequently, previously reported SAP155‐FIR interactions are pivotal for the mechanism of c‐Myc activation.11 In this study, cancers were found to require SAP155‐FIR interactions to inhibit FIR pre‐mRNA splicing, and, thus, FIR FIRΔexon2, FIRΔ3 and FIRΔ4 mRNA are potent biomarkers for cancer screening.11, 27, 32
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting information
Fig. S1. The expression level of FIR and FIRΔexon2 mRNA in the peripheral blood (PB) was significantly decreased after operation, as detected by quantitative RT‐PCR in colorectal cancer patients.
Fig. S2. Total FIR were activated in gastrointestinal cancers and acute myeloblastic leukemia cells.
Acknowledgments
We thank Dr Shouichi Ishiura (University of Tokyo, Japan) for mouse Clcn1 minigene and MBNL1expression vectors. We also thank Dr Chiaki Nakaseko (Division of Hematology, Chiba University, Japan) for peripheral blood samples of leukemia patients and Dr Yoshitaka Okamoto (Department of Otorhinolaryngology, Chiba University, Japan) for hypopharyngeal cancer tissues.
(Cancer Sci, doi: 10.1111/cas.12058, 2012)
References
- 1. Traverso G, Shuber A, Levin B et al Detection of APC mutations in fecal DNA from patients with colorectal tumors. N Engl J Med 2002; 346: 311–20. [DOI] [PubMed] [Google Scholar]
- 2. Diehl F, Li M, Dressman D et al Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 2005; 102: 16368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avigan MI, Strober B, Levens D. A far upstream element stimulates c‐myc expression in undifferentiated leukemia cells. J Biol Chem 1990; 265: 18538–45. [PubMed] [Google Scholar]
- 4. Duncan R, Bazar L, Michelotti G et al A sequence‐specific, single‐strand binding protein activates the far upstream element of c‐myc and defines a new DNA‐binding motif. Genes Dev 1994; 8: 465–80. [DOI] [PubMed] [Google Scholar]
- 5. Bazar L, Meighen D, Harris V, Duncan R, Levens D, Avigan M. Targeted melting and binding of a DNA regulatory element by a transactivator of c‐myc. J Biol Chem 1995; 270: 8241–8. [DOI] [PubMed] [Google Scholar]
- 6. Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single‐stranded cis elements are associated with activated chromatin of the human c‐myc gene in vivo . Mol Cell Biol 1996; 16: 2656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu J, He L, Collins I et al The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell 2000; 5: 331–41. [DOI] [PubMed] [Google Scholar]
- 8. Liu J, Akoulitchev S, Weber A et al Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 2001; 104: 353–63. [DOI] [PubMed] [Google Scholar]
- 9. Matsushita K, Tomonaga T, Shimada H et al An essential role of alternative splicing of c‐myc suppressor FIR in carcinogenesis. Cancer Res 2006; 66: 1409–17. [DOI] [PubMed] [Google Scholar]
- 10. Corsini L, Hothorn M, Stier G et al Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J Biol Chem 2009; 284: 630–39. [DOI] [PubMed] [Google Scholar]
- 11. Matsushita K, Kajiwara T, Tamura M et al SAP155‐mediated splicing of FUSE‐binding protein‐interacting repressor (FIR) serves as a molecular switch for c‐myc gene expression. Mol Cancer Res 2012; 10: 787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Black DL. Mechanisms of alternative pre‐messenger RNA splicing. Annu Rev Biochem 2003; 72: 291–336. [DOI] [PubMed] [Google Scholar]
- 13. Sekido Y. Genomic abnormalities and signal transduction dysregulation in malignant mesothelioma cells. Cancer Sci 2010; 101: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharma SV, Settleman J. Exploiting the balance between life and death: targeted cancer therapy and “oncogenic shock”. Biochem Pharmacol 2010; 80: 666–73. [DOI] [PubMed] [Google Scholar]
- 15. Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem 2004; 37: 584–94. [DOI] [PubMed] [Google Scholar]
- 16. Kaida D, Schneider‐Poetsch T, Yoshida M. Splicing in oncogenesis and tumor suppression. Cancer Sci 2012; 103: 1611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsushita K, Takenouchi T, Shimada H et al Strong HLA‐DR antigen expression on cancer cells relates to better prognosis of colorectal cancer patients: possible involvement of c‐myc suppression by interferon‐γ in situ . Cancer Sci 2006; 97: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase‐specific phosphorylation of DNA topoisomerase II. J Biol Chem 1996; 271: 21439–45. [DOI] [PubMed] [Google Scholar]
- 19. Page‐McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65‐related splicing activity. RNA 1999; 5: 1548–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet 2004; 13: 495–507. [DOI] [PubMed] [Google Scholar]
- 21. Miller JW, Urbinati CR, Teng‐Umnuay P et al Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J 2000; 19: 4439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishida A, Kataoka N, Takeshima Y et al Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat Commun 2011; 2: 308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zweig MH, Campbell G. Receiver‐operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 1993; 39: 561–77. [PubMed] [Google Scholar]
- 24. Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983; 148: 839–43. [DOI] [PubMed] [Google Scholar]
- 25. Crichlow GV, Zhou H, Hsiao HH et al Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c‐myc inhibition. EMBO J 2008; 27: 277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tong YK, Lo DYM. Diagnostic developments involving cell‐free (circulating) nucleic acids (review). Clin Chim Acta 2006; 363: 187–96. [DOI] [PubMed] [Google Scholar]
- 27. Pathak AK, Bhutani M, Kumar S et al Circulating Cell‐Free DNA in Plasma/Serum of Lung Cancer Patients as a Potential Screening and Prognostic Tool. Clin Chem 2006; 52: 1833–42. [DOI] [PubMed] [Google Scholar]
- 28. Omenn GS, Yocum AK, Menon R. Alternative splice variants, a new class of protein cancer biomarker candidates: findings in pancreatic cancer and breast cancer with systems biology implications. Dis Markers 2010; 28: 241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsushita K, Tomonaga T, Kajiwara T et al c‐myc suppressor FBP‐interacting repressor for cancer diagnosis and therapy. Front Bios 2009; 14: 3401–8. [DOI] [PubMed] [Google Scholar]
- 30. Kaida D, Motoyoshi H, Tashiro E et al Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre‐mRNA. Nat Chem Biol 2007; 3: 576–83. [DOI] [PubMed] [Google Scholar]
- 31. Kitamura A, Matsushita K, Takiguchi Y et al Synergistic effect of non‐transmissible Sendai virus vector encoding the c‐myc suppressor FUSE‐binding protein‐interacting repressor plus cisplatin in the treatment of malignant pleural mesothelioma. Cancer Sci 2011; 102: 1366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J, Chen QM. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene 2012; 1–10. Aug 27. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The expression level of FIR and FIRΔexon2 mRNA in the peripheral blood (PB) was significantly decreased after operation, as detected by quantitative RT‐PCR in colorectal cancer patients.
Fig. S2. Total FIR were activated in gastrointestinal cancers and acute myeloblastic leukemia cells.


