Abstract
The prognostic value of mucin expression has been reported in several studies. We examined the association between mucin expression and other previously reported prognostic factors, including infiltration of CD10+ myeloid cells, transforming growth factor‐β1 (TGF‐β1) expression, and tumor budding at invasion fronts. Immunohistochemical analysis of 206 colorectal samples was carried out to determine whether MUC1, MUC2, MUC4, and MUC5AC expression could predict the survival of colorectal cancer patients. Serial sections were stained for CD10, TGF‐β1, and pan‐cytokeratin in order to detect tumor budding. As per multivariate analyses, MUC1 expression appeared to be the most significant predictor of both recurrence‐free survival and overall survival. MUC4 was only significant to predict recurrence‐free survival, and MUC5AC could be a good marker in stage IV colorectal cancers that require additional chemotherapy. MUC1 (CD227) expression was associated with infiltration of CD10+ myeloid cells, TGF‐β1 expression, and tumor budding grade. These findings suggest that MUC1 is indicative of poor prognoses that may be associated with immunosuppression and epithelial–mesenchymal transition. Furthermore, MUC1 expression appears to be a chemoattractant for CD10+ stromal cells.
Promising curative treatments of advanced cancers almost always depend on adjuvant therapies, which are designed according to primary tumor size, regional lymph node status, and distant metastasis (TNM staging).1 However, the newest revisions in the TNM classification of colorectal cancer (CRC) do not meet our expectations mainly because of the lack of sufficient improvement in their predictive value for prognosis.2 Thus, identification and validation of biomarkers that can predict high‐risk CRCs is urgently needed to identify patients requiring more intensive treatment.
Mucins are high‐molecular‐weight epithelial glycoproteins with a high content of clustered oligosaccharides that are O‐glycosidically linked to tandem repeat peptides rich in threonine, serine, and proline. The two structurally and functionally distinct classes of mucins are secreted gel‐forming mucins (MUC2, MUC5AC, and MUC6), and transmembrane mucins (MUC1, MUC3A, MUC3B, MUC4, MUC12, and MUC17).3 The changes in mucins that occur in CRC may be broadly classified as aberrant mucin gene regulation and glycosylation.4 Of these glycoproteins, MUC1, MUC2, MUC4, and MUC5AC have been studied most comprehensively in association with CRC.4 Expression of MUC1 mucin (CD227), as detected immunohistochemically, increases with colon cancer and correlates with a worse prognosis.5, 6, 7, 8 MUC1 expression has been implicated in several processes involved in tumor invasion and metastasis, such as immune suppression, cell–cell interaction and adhesion, and epithelial–mesenchymal transition (EMT).3, 6, 9, 10, 11 Tumor cells that have undergone EMT are characterized histologically by the presence of tumor budding, defined as single cells or small clusters of de‐differentiated tumor cells at the invasion front in CRC.12 The expression of MUC2, a secreted gel‐forming mucin, is generally decreased in colorectal adenocarcinomas but preserved in mucinous carcinomas.7, 13, 14 Iwase et al.15 reported that reduced MUC2 expression correlated with the development and progression of colorectal neoplasms. A recent study indicated that loss of MUC2 expression is associated with poor outcome in CRC patients.16 Although moderate MUC4 expression levels have been observed in the uninvolved colonic epithelium, these expression levels varied from high to none in CRC. Increased MUC4 expression was associated with poor survival, specifically in patients with early‐stage CRC.17 A product of normal gastric mucosa, MUC5AC is absent in the normal colon but is frequently present in colorectal adenomas and colon cancers. Patients with MUC5AC‐negative tumors have worse survival rates than those with MUC5AC‐positive tumors.18, 19
In the present study, we focused on several prognostic factors, including CD10 expression patterns (tCD10 in tumor cells, sCD10 in stromal fibroblast cells, and iCD10 in infiltrating myeloid cells), transforming growth factor‐β1 (TGF‐β1) expression, and tumor budding grade. We recently reported that the tumor budding associated with iCD10 and TGF‐β1 expression at tumor invasion fronts was of a highly malignant phenotype.20 We also found that iCD10 expression was associated with high‐risk stage II CRC.21 The origin of iCD10+ cells was CD11b+CD15+ cells infiltrating at the invasion fronts of CRCs, which was similar to the myeloid‐derived suppressor cells, and functioned as the source of proteases in the microenvironment of tumor stroma.20, 21 Transforming growth factor‐β (TGF‐β) is one of the most important cytokines aberrantly secreted from tumor cells. It induces immune suppression in the tumor microenvironment and promotes tumor growth and metastasis.22, 23 Hase et al.24 first described tumor budding as a valuable prognostic factor in CRC after evaluating 663 CRC patients without using multivariate analysis. Recently, Wöhlke et al.25 reported tumor budding as an independent prognostic factor in a series of 485 CRC cases. To date, there have been no reports detailing the relationship between mucins, especially MUC1 expression, and iCD10, TGF‐β1 expression, and tumor budding grade.
The present study measured the expression profiles and compared the prognostic values of MUC1, MUC2, MUC4, and MUC5AC. We attempted to determine the highest prognostic significance of mucin expression using multivariate analysis and compared the prognostic significance at different disease stages. Then the correlation between mucin expression and CD10 expression patterns, TGF‐β1 expression, and tumor budding grade was investigated.
Materials and Methods
Patients and specimens
We examined tissue samples from CRC patients who underwent surgical treatments between 1998 and 2005 at the Shiga University of Medical Science Hospital (Otsu, Japan). Of these patients, 206 met our criteria for enrolment in this retrospective study. The following specific criteria were required for inclusion: (i) histologically diagnosed non‐mucinous colorectal adenocarcinomas; (ii) at least 5‐year follow‐up; and (iii) possibility to retrieve maximal tissue size. Baseline clinicopathological features are listed in Table 1. Patients who died from other causes were excluded. At the last follow‐up, 48 of 169 (28.4%) patients at stages I–III had recurrence of cancer after surgery, and 70 of 206 (34.0%) patients in the data pool had died from CRC.
Table 1.
Correlation between mucin expression profile and baseline characteristics in patients with non‐mucinous colorectal adenocarcinomas (n = 206)
| MUC1 | P‐value | MUC2 | P‐value | MUC4 | P‐value | MUC5AC | P‐value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | Low | High | ||||||
| Age | |||||||||||||
| ≤60 years | 73 | 29 | 44 | 0.356 | 52 | 21 | 0.244 | 50 | 23 | 0.429 | 49 | 24 | 0.508 |
| >60 years | 133 | 48 | 85 | 87 | 46 | 88 | 45 | 88 | 45 | ||||
| Gender | |||||||||||||
| Female | 92 | 36 | 56 | 0.373 | 58 | 34 | 0.142 | 63 | 29 | 0.399 | 63 | 29 | 0.162 |
| Male | 114 | 41 | 73 | 81 | 33 | 75 | 39 | 74 | 40 | ||||
| Location | |||||||||||||
| Colon | 114 | 46 | 68 | 0.202 | 69 | 45 | 0.013 | 75 | 39 | 0.399 | 72 | 42 | 0.162 |
| Rectum | 92 | 31 | 61 | 70 | 22 | 63 | 29 | 65 | 27 | ||||
| Histological type | |||||||||||||
| Well | 50 | 19 | 31 | 0.940 | 30 | 20 | 0.418 | 32 | 18 | 0.667 | 34 | 16 | 0.171 |
| Mod | 144 | 53 | 91 | 101 | 43 | 99 | 45 | 98 | 46 | ||||
| Poor | 12 | 5 | 7 | 8 | 4 | 7 | 5 | 5 | 7 | ||||
| Tumor depth | |||||||||||||
| pT1 | 14 | 7 | 7 | 0.477 | 8 | 6 | 0.036 | 10 | 4 | 0.100 | 11 | 3 | 0.283 |
| pT2 | 34 | 15 | 19 | 21 | 13 | 24 | 10 | 17 | 17 | ||||
| pT3 | 92 | 34 | 58 | 58 | 34 | 67 | 25 | 61 | 31 | ||||
| pT4 | 66 | 21 | 45 | 52 | 14 | 37 | 29 | 48 | 18 | ||||
| Lymph node | |||||||||||||
| pN0 | 112 | 48 | 64 | 0.003 | 68 | 44 | 0.008 | 83 | 29 | 0.046 | 74 | 38 | 0.370 |
| pN1 | 61 | 26 | 35 | 43 | 18 | 35 | 26 | 37 | 24 | ||||
| pN2 | 33 | 3 | 30 | 28 | 5 | 20 | 13 | 26 | 7 | ||||
| Metastasis | |||||||||||||
| M0 | 169 | 66 | 103 | 0.192 | 112 | 57 | 0.280 | 119 | 50 | 0.022 | 107 | 62 | 0.027 |
| M1 | 37 | 11 | 26 | 27 | 10 | 19 | 18 | 30 | 7 | ||||
| TNM stage | |||||||||||||
| I | 31 | 17 | 14 | 0.033 | 19 | 12 | 0.067 | 25 | 6 | 0.002 | 21 | 10 | 0.226 |
| II | 71 | 27 | 44 | 42 | 29 | 53 | 18 | 44 | 27 | ||||
| III | 67 | 22 | 45 | 51 | 16 | 41 | 26 | 42 | 25 | ||||
| IV | 37 | 11 | 26 | 27 | 10 | 19 | 18 | 30 | 7 | ||||
| Total | 206 | 77 | 129 | 67 | 139 | 138 | 68 | 137 | 69 | ||||
Values in bold indicate P < 0.05. Mod, moderate.
Cancer tissue specimens were evaluated after informed consent was obtained from the patients in accordance with institutional guidelines. Specimens were fixed in 10% formalin for 24–48 h and embedded in paraffin blocks. Tissue slices were sampled along the maximum tumor diameter and included the deepest site of cancer invasion. Serial 3‐μm sections were prepared from paraffin blocks for immunohistochemical studies.
Immunohistochemical studies
Immunohistochemical studies were carried out by the polymer method (Histofine MAX PO Multi; Nichirei, Tokyo, Japan). All primary antibodies were incubated for 18 h at 4°C except for pan‐cytokeratin (AE1/AE3; Dako, Glostrup, Denmark) mAb, which was ready‐to‐use, was incubated for 1 h at room temperature. Antigen retrieval was carried out using a high temperature antigen unmasking technique. Immunohistochemical staining was carried out using mouse mAbs for MUC1 (Clone Ma695, 1:100; Leica, Newcastle, UK), MUC2 (Clone Ccp58, 1:100; Leica), MUC4 (Clone 1G8, 1:100; Invitrogen, Carlsbad, CA, USA), MUC5AC (Clone CLH2, 1:100; Leica), CD10 (Clone 56C6, 1:100; Leica), TGF‐β1 (Clone TB21, 1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and pan‐cytokeratin (Clone AE1/AE3, ready‐to‐use; Dako) as the primary antibodies. Peroxidase binding sites were visualized using diaminobenzidine solution (Histofine; Nichirei), and nuclei were lightly counterstained with hematoxylin. Internal positive and negative controls were used for MUC1, MUC2, MUC4, and MUC5AC from adjacent normal mucosa. MUC1 is totally negative in normal mucosa and always has positive specimens in any staining procedure for a parallel control. MUC2 is always positive in goblet cells of normal mucosa. MUC4 is weakly positive in all normal mucosa specimens. The staining of MUC4 and MUC5AC found positivity at the border between normal mucosa and tumor, and was used as the internal positive control. A negative control was prepared without primary antibody. Normal adjacent mucosa from 196 of 206 (95.1%) specimens was used as an internal control for MUC2, MUC4, and MUC5AC expression. A parallel control was used for specimens without normal mucosa.
Pathologic evaluation
All specimens were evaluated by two investigators (KDT, KM). Staining of MUC1 and MUC4 was majorly observed within the cytoplasm and cell membrane. Staining of MUC2 and MUC5AC was observed only within the cytoplasm. Mucin expression was classified into four grades: 0 = negative 0–5%; 1+ = 6–25%; 2+ = 26–50%; and 3+ = >50%. Each grade was analyzed for the lowest P‐value by log–rank test to determine a suitable cut‐off point for each marker. Grades 1+ to 3+ represented high expression for the highest statistical significance. MUC1 and MUC4 expression was of grades 2+ and 3+, respectively. MUC2 expression was also of grade3+. In contrast, MUC5AC was rarely expressed in our study specimens.
The TGF‐β1 expression level was evaluated along the invasive front over the whole section (7–10 view fields per section) and divided into four grades on the basis of the percentage of tumor cells encountered: 1 = negative or weak staining if <5% tumor crypts were encountered; 2 = 1+ if 5–30% tumor crypts were encountered; 3 = 2+ if 30–60% tumor crypts were encountered; and 4 = 3+ if >60% tumor crypts were encountered.
The budding grade of tumors was assessed according to the method of a previous study.12 Immunohistochemical staining of pan‐cytokeratin was carried out for strict detection of tumor budding. Sections were viewed at scanning magnification, and three areas with maximal budding were located. Subsequently, all separate microclusters of tumor cells with five or fewer nuclei or single tumor cells were counted in three maximal budding areas at ×200 magnification using captured microscopic images on a monitor. The final budding grade was calculated as the average of three maximal budding areas.
Expression of iCD10 was graded from 0 to 3+ according to our previous study,20, 21 as follows: 1 = 0, negative or weak expression; 2 = 1+, strong expression in small areas (<1/4 of the area of a ×200 field) with a low density of iCD10+ cells (positive in <50% of infiltrating immune cells); 3 = 2+, strong expression in one to two large areas with a high density of iCD10+ cells (positive in >50% of infiltrating immune cells); and 4 = 3+, strong expression in three or more large areas with a high density of iCD10+ cells or showing diffuse strong positivity (over 1/2 the area of the invasive front).
Statistical analyses
Correlations between the clinicopathological variables and expression patterns of mucins were analyzed using the χ2‐test and Fisher's exact test. Correlations between mucin expression and CD10 expression patterns and TGF‐β1 expression were analyzed using a linear‐by‐linear association test. Correlations between mucin expression and tumor budding grade were analyzed using one‐way anova. The χ2‐test was used for univariate analysis of high risk of disease relapse, and the logistic regression model was used for multivariate analysis. Correlations between the expression levels of these biomarkers with overall survival (OS) and recurrence‐free survival were analyzed using Kaplan–Meier curves, and the differences were estimated using the Mantel–Cox log–rank test. For multivariate analysis of independent factors, we used a Cox regression model to calculate the hazard ratio and 95% confidence interval for prognostic factors. P < 0.05 was considered statistically significant.
Results
Differences in mucin expression in CRC tumor cells
The expression patterns of MUC1, MUC2, MUC4, and MUC5AC are shown in Figure 1. High MUC1 expression was found in 129 (62.6%) of 206 patients. High MUC2 expression was found in 139 (67.5%) of 206 patients. Similar to MUC1 expression, high MUC4 expression was found in 68 (33%) of 206 patients. Compared with the expression of other mucins, MUC5AC expression was rarely observed in the tumor cells. Of the 206 patients, only 39 patients (18.9%) showed 1+ positivity, 17 (8.3%) showed 2+ positivity, and 14 (6.8%) showed 3+ positivity. Therefore, of the 206 patients, 70 (34.0%) were judged as having high MUC5AC expression and 136 (66.0%) as having low MUC5AC expression.
Figure 1.
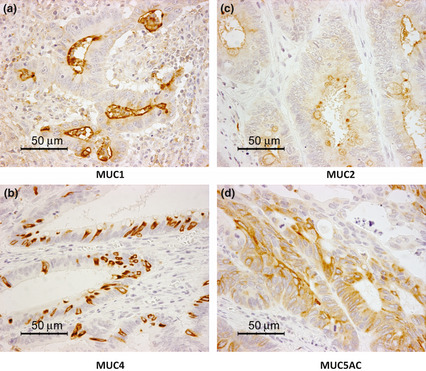
Expression patterns of mucins MUC1, MUC2, MUC4, and MUC5AC in colorectal cancer. MUC1 (a) and MUC4 (c) are expressed mostly on the apical membrane of tumor cells, whereas MUC2 (b) and MUC5AC (d) are expressed in the cytoplasm of tumor cells. Magnification, ×200.
Correlations between mucin expression profiles and baseline characteristics
The correlation between mucin expression profiles and baseline characteristics are detailed in Table 1. Expression of MUC2 in colon cancer was significantly higher than in rectal cancer (P = 0.013, χ2‐test). MUC1, MUC2, and MUC4 expression was associated with lymph node metastasis (P = 0.003, P = 0.008, and P = 0.046, respectively, χ2‐test). MUC4 and MUC5AC expression was associated with distant metastasis (P = 0.022 and P = 0.027, respectively, χ2‐test). MUC1 and MUC4 expression was correlated with TNM staging (P = 0.033 and P = 0.002, respectively, χ2‐test).
Correlations between mucin expression profiles and clinical outcome
The association between mucin expression and recurrence‐free survival (RFS) was assessed in 169 stage I–III patients showing no distant metastases before surgery (Table 2). In univariate analyses, tumor grade, pT, pN, and MUC1, MUC2, and MUC4 expression were significant for predicting RFS. In multivariate analyses, only tumor grade, pN, MUC1, and MUC4 were significant (P = 0.003, P = 0.011, P = 0.026, and P = 0.011, respectively, log–rank test). These results indicate that MUC1 and MUC4 are independent prognostic factors for RFS. Overall survival analysis was carried out for all 206 patients. In univariate analysis, histological type, pN, pT, and expression of all mucins were found to be associated with OS with different P‐values as provided in Table 2. According to multivariate analysis, only MUC1 was an independent prognostic factor (P = 0.007, log–rank test,), same as pT and pN of TNM staging. For predicting both RFS and OS, MUC1 appeared to be superior compared with the other mucins. Kaplan–Meier analysis of expression of all mucins is provided in Figure 2. In this context, the prognostic value of MUC4 appeared to be more significant than that of MUC1. These results suggest that MUC1 is the best marker for predicting RFS and OS independently. Conversely, MUC4 appears to be a useful marker for predicting RFS in CRC.
Table 2.
Univariate and multivariate analyses of recurrence‐free survival in pM0 patients and overall survival in 206 patients with colorectal carcinoma
| Factor (risk indicator) | Recurrence‐free survival (n = 169) | Overall survival (n = 206) | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | HR (95% CI) | P‐value | HR (95% CI) | P‐value | |
| Age (>60 years) | 1.66 (0.86–3.19) | 0.129 | – | – | 0.97 (0.59–1.58) | 0.895 | – | – |
| Gender (male) | 1.40 (0.78–2.51) | 0.260 | – | – | 2.48 (0.91–2.40) | 0.116 | – | – |
| Location (rectum) | 1.22 (0.69–2.15) | 0.488 | – | – | 1.05 (0.66–1.68) | 0.830 | – | – |
| Tumor grade (poor) | 2.66 (1.05–6.73) | 0.039 | 4.62 (1.66–12.84) | 0.003 | 2.09 (0.90–4.83) | 0.085 | 2.58 (1.03–6.45) | 0.042 |
| Tumor depth (pT4) | 1.98 (1.11–3.56) | 0.022 | 1.31 (0.69–2.46) | 0.408 | 2.82 (1.76–4.50) | <0.001 | 2.07 (1.25–3.41) | 0.004 |
| Node (positive) | 3.07 (1.71–5.51) | <0.001 | 2.22 (1.20–4.09) | 0.011 | 3.07 (1.85–5.09) | <0.001 | 2.15 (1.27–3.65) | 0.005 |
| MUC1 (high) | 2.41 (1.23–4.72) | 0.011 | 2.23 (1.10–4.50) | 0.026 | 2.35 (1.35–4.12) | 0.003 | 2.27 (1.25–4.13) | 0.007 |
| MUC2(low) | 1.94 (0.99–3.80) | 0.054 | 1.92 (0.92–3.98) | 0.082 | 1.82 (1.04–3.18) | 0.036 | 1.51 (0.83–2.72) | 0.177 |
| MUC4 (high) | 2.55 (1.45–4.50) | 0.001 | 2.30 (1.21–4.36) | 0.011 | 2.24 (1.40–3.58) | 0.001 | 1.51 (0.91–2.53) | 0.114 |
| MUC5 (negative) | 1.76 (0.93–3.33) | 0.082 | 1.89 (0.95–3.74) | 0.070 | 1.74 (1.01–3.01) | 0.046 | 1.65 (0.93–2.92) | 0.087 |
Values in bold indicate P < 0.05. –, not applicable; CI, confidence interval; HR, hazard ratio.
Figure 2.
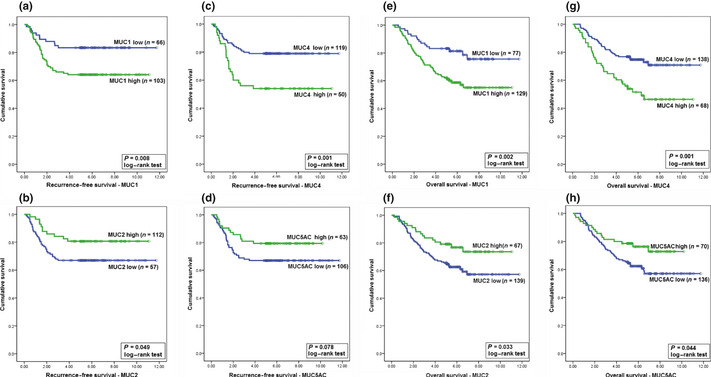
Kaplan–Meier analyses of the prognostic roles of mucins MUC1, MUC2, MUC4, and MUC5AC in predicting recurrence‐free survival and overall survival in colorectal cancer. (a–d) Recurrence‐free survival analyses using MUC1 (a), MUC2 (b), MUC4 (c), and MUC5AC (d) expression as prognostic factors. (e–h) Overall survival analyses using MUC1 (e), MUC2 (f), MUC4 (g), and MUC5AC (h) expression as prognostic factors.
Influence of TNM staging on the prognostic impact of mucin expression
Kaplan–Meier analysis of mucin expression was carried out for each disease stage for RFS and OS. The P‐values of log–rank tests are provided in Table 3. MUC1 expression could predict poor outcomes in stage II for both RFS and OS (P = 0.034 and P = 0.017, respectively). MUC4 expression could predict RFS in stage III (P = 0.003). In stages I–III, low MUC5AC expression was associated with good outcomes but the results were not significant. On the contrary, high MUC5AC expression could predict better OS in stage IV (P = 0.001). These results indicate the influence of TNM staging on the prognostic role of mucin expression.
Table 3.
Comparison of prognostic value of mucin expression between different stages of colorectal cancer by log–rank test
| Factor | Recurrence | Survival | |||||
|---|---|---|---|---|---|---|---|
| Stage I n = 31 | Stage II n = 71 | Stage III n = 67 | Stage I n = 31 | Stage II n = 71 | Stage III n = 67 | Stage IV n = 37 | |
| MUC1 | 0.642 | 0.034 | 0.130 | 0.908 | 0.017 | 0.234 | 0.196 |
| MUC2 | 0.157 | 0.198 | 0.753 | 0.255 | 0.141 | 0.222 | 0.703 |
| MUC4 | 0.386 | 0.184 | 0.003 | 0.484 | 0.285 | 0.070 | 0.179 |
| MUC5AC | 0.187 | 0.299 | 0.256 | 0.288 | 0.533 | 0.116 | 0.001 |
Values in bold indicate P < 0.05.
Association between mucins, CD10 expression patterns, TGF‐β1 expression, and tumor budding grade
We found a correlation between MUC1 expression and sCD10 and iCD10 expression (P = 0.04 and P < 0.001, linear‐by‐linear association; Table 4). Similarly, TGF‐β1 expression by tumor cells at invasion fronts was associated with MUC1 expression (P < 0.001, linear‐by‐linear association). Tumor budding grade was also found to increase significantly following MUC1 expression (P = 0.003, one‐way anova). The association between MUC1 expression and iCD10, TGF‐β1 expression, and tumor budding grade is shown in Figure 3. The expression of other mucins, including MUC2, MUC4, and MUC5AC, was only correlated with tCD10 (P < 0.001, P < 0.001, and P = 0.019, respectively) and not associated with sCD10, iCD10, TGF‐β1 expression, and tumor budding grade. These results suggested that the role of CD10 expression in tumor cells and stromal cells might be different.
Table 4.
Correlation between mucin expression profiles and CD10, transforming growth factor‐β1 (TGF‐β1), and tumor budding grade at invasion fronts in colorectal cancer
| Factor | n | tCD10 | sCD10 | iCD10 | TGF‐β1 expression | Budding grade | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1+ | 2+ | 3+ | 0 | 1+ | 2+ | 3+ | 0 | 1+ | 2+ | 3+ | 0 | 1+ | 2+ | 3+ | Mean (95% CI) | ||
| MUC1 | ||||||||||||||||||
| 0 | 34 | 18 | 5 | 3 | 8 | 13 | 14 | 2 | 5 | 18 | 15 | 1 | 0 | 11 | 13 | 5 | 5 | 15. 9 (11.5–20.5) |
| 1+ | 43 | 24 | 6 | 3 | 10 | 14 | 12 | 10 | 7 | 22 | 11 | 4 | 6 | 8 | 13 | 17 | 5 | 19.3 (15.9–22.6) |
| 2+ | 68 | 27 | 10 | 11 | 20 | 17 | 31 | 16 | 4 | 28 | 21 | 10 | 9 | 13 | 17 | 24 | 14 | 19.3 (16.8–21.8) |
| 3+ | 61 | 35 | 9 | 9 | 8 | 16 | 15 | 15 | 15 | 20 | 14 | 13 | 14 | 7 | 10 | 21 | 23 | 25. 6 (21.2–29.9) |
| P = 0.684 | P = 0.040 | P < 0.001 | P < 0.001 | P = 0.003 | ||||||||||||||
| MUC2 | ||||||||||||||||||
| 0 | 92 | 39 | 12 | 13 | 28 | 27 | 32 | 16 | 17 | 36 | 30 | 11 | 15 | 12 | 24 | 31 | 25 | 22.0 (19.4–24.7) |
| 1+ | 47 | 20 | 10 | 6 | 11 | 13 | 17 | 11 | 6 | 23 | 12 | 8 | 4 | 12 | 8 | 17 | 10 | 20.0 (16.4–23.5) |
| 2+ | 39 | 23 | 6 | 3 | 7 | 12 | 12 | 11 | 4 | 14 | 12 | 8 | 5 | 7 | 10 | 14 | 8 | 20.7 (15.1–26.2) |
| 3+ | 28 | 22 | 2 | 4 | 0 | 8 | 11 | 5 | 4 | 15 | 7 | 1 | 5 | 8 | 11 | 5 | 4 | 16.8 (12.4–21.3) |
| P < 0.001 | P = 0.678 | P = 0.588 | P = 0.239 | P = 0.337 | ||||||||||||||
| MUC4 | ||||||||||||||||||
| 0 | 100 | 45 | 13 | 15 | 27 | 32 | 37 | 20 | 11 | 46 | 27 | 13 | 14 | 16 | 28 | 36 | 20 | 21.6 (18.9–24.3) |
| 1+ | 38 | 13 | 7 | 2 | 16 | 12 | 12 | 9 | 5 | 18 | 13 | 4 | 3 | 8 | 10 | 8 | 12 | 20.1 (15.3–25.0) |
| 2+ | 38 | 22 | 8 | 5 | 3 | 9 | 13 | 8 | 8 | 13 | 10 | 7 | 8 | 7 | 10 | 11 | 10 | 17.9 (13.6–22.3) |
| 3+ | 30 | 24 | 2 | 4 | 0 | 7 | 10 | 6 | 7 | 11 | 11 | 4 | 4 | 8 | 5 | 12 | 5 | 21.3 (16.9–25.6) |
| P < 0.001 | P = 0.057 | P = 0.320 | P = 0.683 | P = 0.537 | ||||||||||||||
| MUC5 | ||||||||||||||||||
| 0 | 136 | 60 | 25 | 16 | 35 | 43 | 50 | 26 | 17 | 58 | 35 | 20 | 23 | 22 | 35 | 46 | 33 | 20.7 (18.8–22.6) |
| 1+ | 39 | 24 | 1 | 5 | 9 | 8 | 14 | 10 | 7 | 17 | 11 | 5 | 6 | 8 | 10 | 13 | 8 | 20.6 (14.7–26.5) |
| 2+ | 17 | 10 | 2 | 4 | 1 | 2 | 7 | 4 | 4 | 5 | 9 | 3 | 0 | 3 | 5 | 6 | 3 | 24.3 (14.6–34.1) |
| 3+ | 14 | 10 | 2 | 1 | 1 | 7 | 1 | 3 | 3 | 8 | 6 | 0 | 0 | 6 | 3 | 2 | 3 | 14.9 (9.2–20.6) |
| P = 0.019 | P = 0.207 | P = 0.052 | P = 0.087 | P = 0.284 | ||||||||||||||
Values in bold indicate P < 0.05. CI, confidence interval; iCD10, CD10 expression in infiltrating myeloid cells; sCD10, CD10 expression in stromal fibroblast cells; tCD10, CD10 expression in tumor cells.
Figure 3.
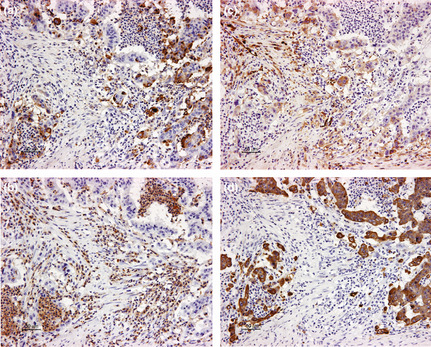
(a) Association between mucin MUC1 expression and iCD10, transforming growth factor‐β1 expression, and tumor budding at invasion fronts on serial sections of colorectal cancer. High MUC1 expression in tumor cells, (b) high‐grade infiltration of CD10+ immune cells (iCD10), (c) high transforming growth factor‐β1 expression in tumor cells, and (d) high tumor budding grade (pan‐cytokeratin) in the same area. Magnification, ×200.
Discussion
Mucins are the major secreted glycoproteins of the gastrointestinal tract. They play a role in the neoplastic progression and metastasis of colon cancer cells. Mucin expression has been studied in various types of cancers, especially CRC.3, 6, 7 The transmembrane mucins, MUC1 and MUC4, were shown to be highly expressed in patients with unfavorable outcomes.6, 17 In contrast, patients with better prognoses expressed the secreted gel‐forming mucins, MUC2 and MUC5AC.3 The focus of the present study was on determining the prognostic value of expression of four common mucins, including MUC1, MUC2, MUC4, and MUC5AC in CRC. Multivariate analysis defined MUC1 as the best prognostic factor for both RFS and OS in CRC. Previous studies also indicated that MUC1, but not MUC2 or MUC3, could be an independent prognostic factor in CRC.6, 7, 14 In the present study, we found that MUC2 was only significant for predicting OS according to univariate analysis. Studies reporting the relationship between MUC4 and prognoses have been rare, with only one recent study showing MUC4 high expression as a predictor of poor survival in CRC.17 In our study, MUC4 expression was also found to significantly predict both RFS and OS according to univariate analysis and only RFS according to multivariate analysis. Absence of MUC5AC expression in tumors can be a prognostic factor for more aggressive CRC.18 Kocer et al.18 found that MUC5AC expression in CRC is rare, with only 3% high expression, 24.2% weak expression, and 72.7% negative expression. In our study, MUC5AC expression was also rare (66.5% low expression or negative expression). Interestingly, we found that MUC5AC expression in stages I–III was not significant to predict outcome; however, high MUC5AC expression in stage IV was an indication of poor prognoses for OS. Because all primary tumors of stage IV in our study were resectable and additional chemotherapy was provided to these patients after surgery, the influence of MUC5AC on stage IV CRC involved chemotherapy. Fluorouracil (5‐FU) was the main chemotherapy agent used during our study. Leteurtre et al.26 reported that the colon cancer cell line HT 29‐5M21, which highly expresses MUC5AC, was 5‐FU‐resistant. In contrast, cells with low MUC5AC expression (HT 29‐5M12, HT 29‐5F7, and HT 29‐5M21) were 5‐FU‐sensitive. These findings suggest that MUC5AC expression might be influenced by additional chemotherapy. However, further studies are required to evaluate the association between MUC5AC expression and 5‐FU sensitivity in vitro and in vivo.
Within the mucin family, MUC1 has recently emerged as a highly attractive target for the development of vaccines.27 Rahn et al.28 reported that MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM‐1. This mechanism could explain the correlation between MUC1 expression and the growth of metastases in distant vital organs. In addition, TGF‐β1 is a well‐known tumor‐derived immunosuppressive factor23 that converts the tumor immunoreaction type 1 into type 2. We found MUC1 and TGF‐β1 expression to be strongly associated in this study. The present study is the first to describe this association and the possibility of the immunosuppressive function of MUC1. Furthermore, a recent study reported that MUC1 enhances invasiveness of pancreatic cancer cells by inducing EMT.11 It is still controversial whether tumor budding in CRC could be a process similar to EMT. Depending on the researcher, names other than tumor budding have been assigned to this phenomenon or to closely related findings in in vitro or experimental systems, such as focal de‐differentiation, tumor cell dissociation, or EMT.29 Baldus et al.8 found that MUC1 and β‐catenin expression at the invasion front were significantly correlated and that these two proteins were coexpressed. As well as interaction with the ECM component ICAM‐1, stimulating MMP‐13 expression to contribute to esophageal carcinoma metastasis, MUC1 exerts anti‐adhesive effects on β‐catenin by reducing its binding to E‐cadherin.28, 29, 30, 31, 32 We also found a strong association between MUC1 expression and tumor budding. Moreover, MUC1 regulates the development of myeloid progenitors into CD11b+Gr1+ myeloid‐derived suppressor cells.10 There was a correlation between MUC1 expression and CD10+ stromal cells at invasion fronts including sCD10+ and iCD10+ cells. The sCD10+ cells almost showed stromal fibroblast cells and iCD10+ cells were also characterized as CD11b+CD15+ myeloid‐derived cells in our previous studies.20, 21 These findings suggested that tumor cells expressing MUC1 might interact with stromal cells to enhance tumor budding or the EMT process in CRC.
Taken together, MUC1 expression appears to be the most useful independent prognostic factor for CRC. The mechanisms of biological behaviors of CRC involving MUC1 expression could be associated with CD10 expression in stromal cells, TGF‐β1 expression, and tumor budding grade.
Disclosure Statement
The authors have no conflict of interest.
(Cancer Sci 2013; 104: 958–964)
References
- 1. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual, 7th edn: USA: Springer, 2010. [Google Scholar]
- 2. Ueno H, Mochizuki H, Akagi Y et al Optimal colorectal cancer staging criteria in TNM classification. J Clin Oncol 2012; 30: 1519–26. [DOI] [PubMed] [Google Scholar]
- 3. Byrd JC, Bresalier RS. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev 2004; 23: 77–99. [DOI] [PubMed] [Google Scholar]
- 4. Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep 2010; 12: 319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Niv Y. MUC1 and colorectal cancer pathophysiology considerations. World J Gastroenterol 2008; 14: 2139–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duncan TJ, Watson NF, Al‐Attar AH, Scholefield JH, Durrant LG. The role of MUC1 and MUC3 in the biology and prognosis of colorectal cancer. World J Surg Oncol 2007; 5: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldus SE, Mönig SP, Hanisch FG et al Comparative evaluation of the prognostic value of MUC1, MUC2, sialyl‐Lewis(a) and sialyl‐Lewis(x) antigens in colorectal adenocarcinoma. Histopathology 2002; 40: 440–9. [DOI] [PubMed] [Google Scholar]
- 8. Baldus SE, Mönig SP, Huxel S et al MUC1 and nuclear beta‐catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clin Cancer Res 2004; 10: 2790–6. [DOI] [PubMed] [Google Scholar]
- 9. Agrawal B, Krantz MJ, Reddish MA, Longenecker BM. Cancer‐associated MUC1 mucin inhibits human T‐cell proliferation, which is reversible by IL‐2. Nat Med 1998; 4: 43–9. [DOI] [PubMed] [Google Scholar]
- 10. Poh TW, Bradley JM, Mukherjee P, Gendler SJ. Lack of Muc1‐regulated beta‐catenin stability results in aberrant expansion of CD11b+Gr1 + myeloid‐derived suppressor cells from the bone marrow. Cancer Res 2009; 69: 3554–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roy LD, Sahraei M, Subramani DB et al MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene 2011; 30: 1449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prall F, Nizze H, Barten M. Tumour budding as prognostic factor in stage I/II colorectal carcinoma. Histopathology 2005; 47: 17–24. [DOI] [PubMed] [Google Scholar]
- 13. Ajioka Y, Allison LJ, Jass JR. Significance of MUC1 and MUC2 mucin expression in colorectal cancer. J Clin Pathol 1996; 49: 560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manne U, Weiss HL, Grizzle WE. Racial differences in the prognostic usefulness of MUC1 and MUC2 in colorectal adenocarcinomas. Clin Cancer Res 2000; 6: 4017–25. [PubMed] [Google Scholar]
- 15. Iwase T, Kushima R, Mukaisho K, Mitsufuji S, Okanoue T, Hattori T. Overexpression of CD10 and reduced MUC2 expression correlate with the development and progression of colorectal neoplasms. Pathol Res Pract 2005; 201: 83–91. [DOI] [PubMed] [Google Scholar]
- 16. Kang H, Min BS, Lee KY et al Loss of E‐cadherin and MUC2 expressions correlated with poor survival in patients with stages II and III colorectal carcinoma. Ann Surg Oncol 2011; 18: 711–9. [DOI] [PubMed] [Google Scholar]
- 17. Shanmugam C, Jhala NC, Katkoori VR et al Prognostic value of mucin 4 expression in colorectal adenocarcinomas. Cancer 2010; 116: 3577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kocer B, Soran A, Erdogan S et al Expression of MUC5AC in colorectal carcinoma and relationship with prognosis. Pathol Int 2002; 52: 470–7. [DOI] [PubMed] [Google Scholar]
- 19. Bu XD, Li N, Tian XQ et al Altered expression of MUC2 and MUC5AC in progression of colorectal carcinoma. World J Gastroenterol 2010; 16: 4089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khanh DT, Mekata E, Mukaisho K et al Prognostic role of CD10+ myeloid cells in association with tumor budding at the invasion front of colorectal cancer. Cancer Sci 2011; 102: 1724–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khanh DT, Mekata E, Mukaisho K et al Myeloid cells positive for CD10 at invasion front can predict poor outcome in stage II colorectal cancer. Int J Clin Oncol 2012; 17: 240–9. [DOI] [PubMed] [Google Scholar]
- 22. Robson H, Anderson E, James RD, Schofield PF. Transforming growth factor beta 1 expression in human colorectal tumours: an independent prognostic marker in a subgroup of poor prognosis patients. Br J Cancer 1996; 74: 753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flavell RA, Sanjabi S, Wrzesinski SH, Licona‐Limón P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol 2010; 10: 554–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hase K, Shatney C, Johnson D, Trollope M, Vierra M. Prognostic value of tumor “budding” in patients with colorectal cancer. Dis Colon Rectum 1993; 36: 627–35. [DOI] [PubMed] [Google Scholar]
- 25. Wöhlke M, Schiffmann L, Prall F. Aggressive colorectal carcinoma phenotypes of invasion can be assessed reproducibly and effectively predict poor survival: interobserver study and multivariate survival analysis of a prospectively collected series of 299 patients after potentially curative resections with long‐term follow‐up. Histopathology 2011; 59: 857–66. [DOI] [PubMed] [Google Scholar]
- 26. Leteurtre E, Gouyer V, Rousseau K et al Differential mucin expression in colon carcinoma HT‐29 clones with variable resistance to 5‐fluorouracil and methotrexate. Biol Cell 2004; 96: 145–51. [DOI] [PubMed] [Google Scholar]
- 27. Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 2009; 9: 874–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rahn JJ, Chow JW, Horne GJ et al MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM‐1. Clin Exp Metastasis 2005; 22: 475–83. [DOI] [PubMed] [Google Scholar]
- 29. Prall F. Tumour budding in colorectal carcinoma. Histopathology 2007; 50: 151–62. [DOI] [PubMed] [Google Scholar]
- 30. Wesseling J, van der Valk SW, Vos HL, Sonnenberg A, Hilkens J. Episialin (MUC1) overexpression inhibits integrin‐mediated cell adhesion to extracellular matrix components. J Cell Biol 1995; 129: 255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ye Q, Yan Z, Liao X et al MUC1induces metastasis in esophageal squamous cell carcinoma by upregulating matrix metalloproteinase 13. Lab Invest 2011; 91: 778–87. [DOI] [PubMed] [Google Scholar]
- 32. Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma‐associated antigen and beta‐catenin. Mol Cell Biol 1998; 18: 7216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]


