Abstract
Overexpression of the ErbB2/HER2 receptor tyrosine kinase contributes to tumorigenesis. However, mechanisms regulating ErbB2 protein levels remain largely unclear. Here, we identified novel mechanisms of ErbB2 downregulation. ErbB2 constitutively binds to an adaptor protein FRS2β. We found that FRS2β bound to CD2AP and CIN85, which induces endosomal trafficking that targets lysosomes. FRS2β colocalized with CIN85 in the cytoplasm. Expression of wild type FRS2β but not its CIN85 non‐binding mutant, downregulated the ErbB2 protein and inhibited anchorage‐independent cell growth. Moreover, the E3 ubiquitin‐protein ligase Cbl was contained within a complex of FRS2β and CIN85. Knockdown of both CIN85 and CD2AP or of Cbl, or treatment with lysosomal degradation inhibitors diminished FRS2β downregulation of ErbB2. In addition, knockdown of endogenous FRS2β caused upregulation of ErbB2 in primary neural cells. Finally, immunohistochemical analysis showed that human breast cancer tissues that overexpress ErbB2 expressed low levels of FRS2β. Thus, an FRS2β‐CIN85/CD2AP‐Cbl axis for downregulation of ErbB2 may regulate ErbB2 protein levels in physiological and pathological settings. Molecular targeting drugs that can increase or stabilize the ErbB2‐FRS2β‐CIN85/CD2AP‐Cbl axis may have promise for the control of ErbB2‐overexpressing tumors.
The epidermal growth factor receptor (EGFR) and its family member ErbB2 are typical receptor tyrosine kinases (RTK) and are involved in many aspects of biology, including physiological aspects such as development, and pathological aspects such as cancer.1 Epidermal growth factor (EGF) binds to the EGFR, leading to EGFR homo‐dimerization or hetero‐dimerization with its family members, which activates its RTK activity, resulting in the initiation of a variety of cellular signaling pathways that ultimately lead to many cellular processes including cell proliferation, differentiation, migration, survival and adhesion.2 There are also many negative feedback mechanisms to fine‐tune this signaling in a physiological setting.2, 3 In fact, overexpression of the EGFR or ErbB2 contributes to tumorigenesis in many types of cancer, which may be partly due to the fact that negative feedback regulation is insufficient in cells that express too many of these receptors.1 For example, the ErbB2 gene is amplified in approximately 20%–25% of human breast cancers4 It is therefore important to clarify the mechanisms of negative regulation of these receptors in a physiological setting in order to find a way to control pathological processes. Receptor downregulation is a major mechanism for negative regulation of the EGFR. This mechanism involves EGF‐induced receptor endocytosis, which targets the receptor to the lysosome for degradation.5 Although there appears to be no ligand for ErbB2, the extracellular domain of ErbB2 has an extended conformation that mimics the ligand bounded‐extracellular domain of the EGFR, which endows ErbB2 with the capacity to easily homo‐ and hetero‐dimerize and thereby activate cellular signaling pathways.6 However, it is also known that ErbB2 is not degraded as efficiently as the EGFR, and the mechanisms by which ErbB2 is downregulated are still unclear.5
FRS2β belongs to the FRS2 family of docking/scaffolding adaptor proteins.7, 8, 9 This family comprises two members: FRS2α/FRS2 and FRS2β/FRS3. Both members contain N‐terminal myristoylation sites, resulting in their membrane distribution, and a phosphotyrosine‐binding (PTB) domain, which facilitates binding to a limited number of receptor tyrosine kinases (RTKs). FRS2α constitutively binds to the fibroblast growth factor (FGF) receptor via its PTB domain regardless of FGF stimulation and acts as a central mediator in FGF signaling. FRS2α becomes phosphorylated on tyrosine residues in response to FGF, leading to the activation of major signaling pathways including the Ras/ERK pathway.10, 11, 12, 13 FRS2β also constitutively binds to the FGF receptor, becomes tyrosine phosphorylated upon FGF stimulation and may function redundantly with FRS2α.14 However, a growing body of evidence suggests that the main functions of FRS2β are different from those of FRS2α. FRS2β constitutively binds to the EGFR and ErbB2 via its PTB domain regardless of receptor activation, but acts as a feedback inhibitor for EGF signaling.3, 15, 16 FRS2β does not become phosphorylated on tyrosine residues in response to EGF, but does bind to activated ERK thereby inhibiting translocation of ERK to the nucleus, and also inhibits dimerization of the EGFR or ErbB2.16 In contrast to the ubiquitous expression of FRS2α from embryos to adults, FRS2β expression is restricted to only a few tissues: for example, neural tissues and epithelial cells in lung and breast tissues.14, 17 The FRS2β protein colocalizes with intracellular vesicles including lysosomes in neural cells.17 This localization gave rise to the hypothesis that FRS2β functions in intracellular degradation systems.
The CBL‐interacting protein 85 kDa (CIN85) and CD2‐associated protein (CD2AP) belong to the same family of adaptor proteins.18, 19, 20, 21, 22 CIN85 and CD2AP each have three SH3 domains that mediate protein–protein interactions by binding to proline (P)‐rich motifs of a variety of proteins and are involved in many cellular processes including the degradation of EGFRs that are transported in endosomes. Cbl, a E3 ubiquitin ligase, is one such interacting protein and is responsible for directing the lysosomal degradation of these RTKs by their ubiquitination.23
In order to obtain further insights into the specific functions of FRS2β, we screened for novel proteins that interact with FRS2β, but not with FRS2α using a high‐throughput technique that combined affinity purification of interaction partners from cell lysates with subsequent liquid chromatography‐coupled tandem mass spectrometry (LC‐MS/MS). We found that CIN85 and CD2AP were novel binding partners of FRS2β and that the complex between CIN85 or CD2AP with FRS2β contributed to downregulation of the ErbB2 protein.
Materials and Methods
Cell culture
Mouse neural stem/progenitor cell (NSPC) culture and primary culture of embryonic neurons were as previously described.24, 25 Further details are provided in the Supplementary Information.
Identification of FRS2 β binding proteins by LC‐MS/MS
The following constructs were transfected into HEK293T cells to obtain FRS2β binding proteins for LC‐MS/MS analysis: pEF‐BOS‐FLAG, pEF‐BOS‐FRS2β‐FLAG encoding FLAG‐tagged FRS2β and pEF‐BOS‐FRS2α‐FLAG encoding FLAG‐tagged FRS2α. Cell lysates were immunoprecipitated with M2‐FLA‐G‐Agarose (Sigma, St. Louis, MO, USA) for 2 h at 4°C. Further details are provided in the Supplementary Information.
Plasmids and siRNA
The FLAG tag or fluorescent proteins were added to the C‐terminus of FRS2β to avoid disruption of the myristoylation site at the N‐terminus as previously described.17 For lenti‐virus mediated fluorescent‐tagged protein expression, FRS2β‐eGFP was constructed using a standard PCR method and was subcloned into the EcoRI and HpaI digested CSII‐pEF vector.26 mCherry was fused to the N‐terminus of CIN85 using a standard PCR procedure and was subcloned into the EcoRI and XbaI multi‐cloning site in the CSII‐pEF vector. Further details are provided in the Supplementary Information.
Immunoprecipitation and quantitative analysis of Western blotting
Immunoprecipitation and Western blotting were as described previously.16 Western blotting was quantitatively analyzed using the LAS system (Fujifilm, Tokyo, Japan).
Immunocytochemical and imaging analysis
Fluorescence protein‐tagged FRS2β or CIN85 was expressed in cells using lenti‐viral vectors as previously described.24 Further details are provided in the Supplementary Information.
Soft agar colony formation assays
T98G cells expressing FRS2β or its mutant were analyzed in a soft agar colony formation assay as previously described.16 After 3 weeks, cell colonies >100 μm in diameter were counted.
Human tissue specimens and tissue microarrays (TMAs)
Human breast cancer tissue specimens were obtained from surgically resected breast at Kosei Chuo Hospital, from 2009 to 2010. The clinicopathological characteristics are shown in Table S1. The tissue specimens were fixed in 10% formalin solution and embedded in paraffin. TMAs, which were composed of 12 specimens, each of 6‐mm diameter, on one slide, were made by using these fixed tissue specimens.
All patients provided written informed consent. This study was conducted following the Declaration of Helsinki and was approved by the institutional review boards of Kosei Cho Hospital and the Institute of Medical Science, University of Tokyo.
Immunohistochemistry
For immunohistochemical staining of formalin‐fixed paraffin‐embedded (FFPE) sections, we used the Ventana HX System Benchmark (Roche Diagnostic, Penzberg, Germany). Immunohistochemical staining of FRS2β17 and ErbB2/Her2 was assessed semi‐quantitatively by determining the intensity and extent of staining of entire tissue sections as described.27 Expression levels of FRS2β were scored and divided into two groups: − or + (Her2 negative) and ++ or +++ (Her2 positive).
Statistical analysis
Data in the graphs are shown as means ± SD. Statistical significance was assessed using Student's t‐test (two‐tailed). Statistically significant differences are indicated with an asterisk (P < 0.05) in the figures.
Results
Identification of novel FRS2β‐binding proteins by LC‐MS/MS
We screened for novel FRS2β‐binding proteins by LC‐MS/MS analysis of proteins that co‐precipitated with FLAG‐tagged FRS2β from cell lysates. FLAG‐tagged FRS2β or FRS2α was transiently expressed in HEK293T cells. Cells were then stimulated with ligand (final concentration of 100 ng/mL EGF or basic fibroblast growth factor [bFGF], respectively) or were left unstimulated for 10 min. Cell lysates were then precipitated with anti‐FLAG antibody and the precipitates were subjected to LC‐MS/MS analysis. Several molecules were found in common in the FLAG‐precipitates of lysates of bFGF‐stimulated, but not unstimulated, cells expressing either FRS2β or FRS2α. These molecules included the SH2‐containing tyrosine phosphatase Shp2, Grb2 and SOS, which are known to bind to phosphorylated tyrosine residues on either FRS2β or FRS2α.7, 8, 9 As expected from previous reports, ERK was also found in the FLAG‐precipitates derived from cells expressing either FRS2β or FRS2α, following either EGF or bFGF stimulation, but not in the absence of stimulation.16, 28 We then searched for molecules that specifically bound FRS2β but not FRS2α regardless of ligand stimulation. We found that CD2AP protein was among the top 10 proteins in a list of such proteins (%Cov[95] were 33, 26 and 29 in the samples derived from unstimulated cells, EGF‐stimulated cells and bFGF‐stimulated cells, respectively, analyzed by ProteinPilot software (Invitrogen/Applied Biosystems, Carlsbad, CA, USA). This result led us to test the possibility that CD2AP may be a novel FRS2β‐binding protein.
We found that CIN85, which belongs to the same protein family as CD2AP, was also expressed in HEK293T cells (Fig. 1A). To confirm FRS2β binding to CD2AP family proteins, we transiently expressed FLAG‐tagged FRS2β or FRS2α in HEK293T cells, immunoprecipitated the cell lysates with an anti‐FLAG antibody and immunoblotted the precipitates with anti‐CIN85 or CD2AP antibodies. As shown in Figure 1(A), FRS2β but not FRS2α bound to CIN85 and CD2AP. Interestingly, we also repeatedly found that the amount of CIN85 protein was decreased in cells expressing FRS2β but not in cells expressing FRS2α. Since it is known that CIN85 and CD2AP are involved in the degradation of RTKs, we determined if overexpression of FRS2β or FRS2α could influence the expression of EGFR and ErbB2 in cells. We found that the ErbB2 protein level was decreased in cells expressing FRS2β but not in cells expressing FRS2α whereas the expression level of the EGFR did not greatly change under these conditions (Fig. 1B).
Figure 1.
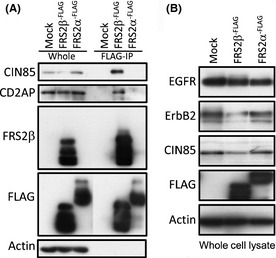
Transient FRS2β expression and its interaction with CIN85 or CD2AP in HEK293T cells. (A) Lysates of HEK293T cells expressing FLAG‐tagged FRS2β or FRS2α or lysates of control vector‐transfected cells were immunoprecipitated with an anti‐FLAG antibody. The immunoprecipitates (FLAG‐IP) and the total cell lysate (whole) were analyzed by Western blotting with the indicated antibodies. (B) Western blotting of whole lysates of cells expressing FLAG‐tagged FRS2β or FRS2α or control vector‐transfected cells (Mock).
FRS2β interacts with CIN85 (or CD2AP) via PxPxxR motifs
Previous reports indicated that the sequence PxPxxR (P, proline; R, arginine; x, any amino acid) can function as binding motif for the SH3 domains of CIN85 or CD2AP.22 We found two PxPxxR motifs in FRS2β (amino acid residues, 246–251 and 287–392), but not in FRS2α (Fig. 2A). The ‘R’ residue has been reported to be the binding core of this motif sequence.22 Therefore, to determine if these motifs meditated FRS2β binding to CIN85 or CD2AP, we replaced the R residue of each, or both of, these motifs in FRS2β with alanine (A) (R251A, R392A, R251/392A) (Fig. 2B). We then transiently expressed FLAG‐tagged wild type or mutant FRS2β into HEK293T cells. When we immunoprecipitated FRS2β from cell lysates with an anti‐FLAG antibody, CIN85 or CD2AP co‐immunoprecipitated with the wild type FRS2β and its R392A mutant (Fig. 2C). However, only weak co‐precipitation of CIN85 or CD2AP with the FRS2β R251A or the R251/392A mutant was observed. We also found that the amount of CIN85 or CD2AP protein in the whole cell lysate was decreased in cells expressing FRS2β, compared with vector‐transfected cells. However, the protein level of CIN85 was not changed in cells expressing the FRS2β R251A or R251/392A mutant and it was moderately decreased in cells expressing the FRS2β R392A mutant. We further examined the amount of ErbB2 protein in these cells and found that it was decreased in cells expressing wild type FRS2β, but not in cells expressing the FRS2β mutants (Fig. 2C).
Figure 2.
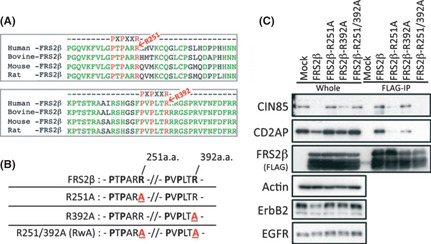
Identification of the binding motif(s) that mediate(s) the FRS2β‐CIN85 or CD2AP interaction. (A) Alignment of the ‘PxPxxR’ CIN85 or CD2AP binding motifs in FRS2β amino acid sequences of different species. (B) Schematic representation of the constructed FRS2β mutants. (C) Immuno‐precipitation of lysates of HEK293T cells expressing FLAG‐tagged FRS2β or its mutants and analysis of the precipitates and the whole cell lysate by Western blotting.
These results suggest that FRS2β interacts with CIN85 or CD2AP via the ‘PxPxxR’ motif(s), mainly via the motif that includes R251. The motif that includes R392 may contribute to this binding to a small extent. The data also suggest that the binding between FRS2β and CIN85 or CD2AP contributes to a reduction in the protein levels of ErbB2.
PxPxxR motifs of FRS2β are essential for co‐localization of FRS2β and CIN85 and for FRS2β inhibition of anchorage‐independent cell growth
We next examined the intracellular localization of FRS2β and CIN85 by introducing eGFP‐fused FRS2β or its R251/392A mutant (RwA) together with mCherry‐fused CIN85 into T98G glioblastoma cells using a lenti‐viral vector. CIN85 exhibited a punctate pattern in the cytoplasm consistent with the localization of CIN85 in endosomes as was previously reported21 (Fig. 3A,D). FRS2β also exhibited a punctate pattern of expression as we previously reported in neural cells17 (Fig. 3B). Merging of these images indicated that these two proteins colocalized in the cytoplasm (Fig. 3C). In contrast, the FRS2β RwA mutant did not show a punctate pattern of expression, nor did it show colocalization with CIN85 (Fig. 3D–F). It is thus the binding between the PxPxxR motif of FRS2β and CIN85 that is required for the co‐localization of these proteins. These results suggest that FRS2β is able to co‐localize with CIN85 in endosomes via its PxPxxR motifs.
Figure 3.
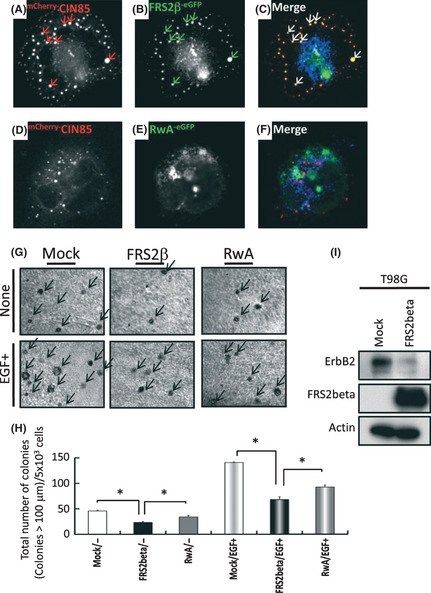
FRS2β‐eGFP and mC herry‐CIN85 co‐localized in T98G cells and expression of FRS2β inhibited anchorage‐independent cell growth. (A–C) mC herry‐CIN85 (A) and FRS2β‐ eGFP (B) showed a punctuate expression pattern and frequent colocalization (C). (D–F) mC herry‐CIN85 showed a punctate expression pattern (D), whereas the RwA‐ eGFP mutant exhibited a diffuse, non‐punctate, intracellular localization (E) and no colocalization with mC herry‐CIN85 (F). (G–I) Soft agar colony formation assay. Photographs were taken after 3 weeks. (H) Quantification of the number of colonies in soft agar after 3 weeks. Arrows indicate the colonies with a diameter >100 μm. (I) Cell lysates were immunoblotted with the indicated antibodies.
We previously reported that FRS2β inhibits anchorage‐independent growth of cells expressing oncogenic ErbB2 carrying mutations in the transmembrane domain.16 We next examined whether FRS2β affects the anchorage‐independent cell growth of T98G cells expressing wild type ErbB2. We found that expression of FRS2β inhibits anchorage‐independent cell growth either in the presence or absence of EGF (Fig. 3G,H). Downregulation of ErbB2 proteins by expressed FRS2β was also confirmed (Fig. 3I).
Knockdown of CIN85 and CD2AP or of Cbl diminished the downregulating effect of FRS2β on the ErbB2 protein
In order to test whether FRS2β downregulates the ErbB2 protein level by interacting with CIN85 or CD2AP, we examined the ErbB2 protein level in HEK293T cells in which CIN85 and/or CD2AP was knocked down by using Cin85 or Cd2ap‐specific siRNA. In addition to the downregulation of the ErbB2 protein that was observed in cells expressing FRS2β, the protein level of CIN85 and CD2AP was also decreased in these cells to some extent (Fig. 4A,B). ErbB2 protein levels were still decreased in cells expressing FRS2β following knockdown of either CIN85 or CD2AP. However, when both CIN85 and CD2AP were simultaneously knocked down, ErbB2 protein levels remained unchanged even when FRS2β was expressed in the cells. These results suggest that expression of CIN85 or CD2AP is required for induction of efficient downregulation of ErbB2 by expression of FRS2β. These data suggest that CIN85 and CD2AP function redundantly for downregulation of the ErbB2 protein.
Figure 4.
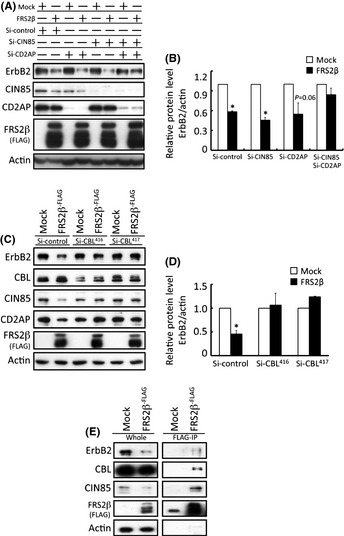
FRS2β downregulates ErbB2 protein levels in HEK293T cells via CIN85, CD2AP and Cbl interaction. (A) Representative results of western blotting analyses of lysates of cells with single knockdown of CIN85 or CD2AP, or dual knockdown of CIN85 and CD2AP. CIN85 and CD2AP dual knockdown diminished ErbB2 downregulation by FRS2β. (B) Relative protein level of ErbB2 in the cells in (A) was calculated from three independent experiments and was determined using the LAS system. ErbB2 protein level was determined relative to that of actin for each cell lysate and the level in Mock‐transfected cells for each experiment was assigned a value of 1. *P < 0.05 versus Mock. Values are means ± SD. (C) Representative results of Western blotting analyses of lysates of Cbl‐knockdown cells. The expression of two different siRNA sequences for Cbl knockdown individually diminished the ErbB2 downregulation mediated by FRS2β. (D) Relative protein level of ErbB2 in the cells in (C) calculated from three independent knockdown experiments was determined as described for (B). *P < 0.05 versus Mock. Values are means ± SD. (E) Immunoprecipitation of FRS2β from lysates of HEK293T cells expressing FLAG‐tagged FRS2β or control vector (Mock) and Western blotting analysis of the precipitates and the whole cell lysate. FRS2β immunoprecipitates included CIN85, Cbl and ErbB2.
It is thought that CIN85 or CD2AP recruits Cbl to RTKs and that Cbl promotes degradation of the RTKs by enhancing their lysosomal targeting.23 In order to test whether Cbl is involved in the downregulation of ErbB2 by FRS2β‐CIN85 or CD2AP interaction, we knocked down Cbl in these cells by transfection with two different siRNAs against Cbl. ErbB2 protein levels were not significantly changed in Cbl‐knockdown cells, even when FRS2β was expressed in these cells (Fig. 4C,D). When FRS2β was immunoprecipitated with an anti‐FLAG antibody, not only ErbB2 and CIN85, but also Cbl, was co‐immunoprecipitated (Fig. 4E). These results suggest that recruitment of CIN85 or CD2AP and Cbl to FRS2β contributes to the FRS2β‐mediated downregulation of ErbB2.
In order to further examine the mechanisms by which FRS2β downregulates ErbB2, we analyzed the effect of cell treatment with proteasomal or lysosomal degradation inhibitors on FRS2β‐mediated ErbB2 downregulation. Treatment with the proteasomal inhibitors, MG132 or lactacystin, did not affect the downregulation of ErbB2 resulting from FRS2β expression (Fig. 5). In contrast, downregulation of ErbB2 was not observed in cells expressing FRS2β following treatment with the lysosomal inhibitors bafilomycin or chloroquine. This result further supports a role for Cbl in the downregulation of ErbB2 by lysosomal targeting that is mediated by the FRS2β‐CIN85/CD2AP complex.
Figure 5.
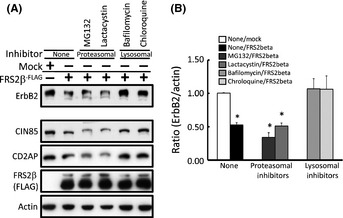
FRS2β downregulates ErbB2 protein levels by inducing its intracellular lysosomal degradation. (A) Identification of the pathway by which FRS2β induces ErbB2 intracellular degradation by treatment of the cells with proteasomal or lysosomal degradation inhibitors. Proteasomal inhibitor treatment (MG132 or Lactacystin) had no effect on ErbB2 downregulation by FRS2β whereas lysosomal inhibitor treatment (bafilomycin or chloroquine) diminished ErbB2 downregulation by FRS2β. (B) The relative protein level of ErbB2 in the cells in (A) was calculated from three independent experiments and was determined using the LASsystem. ErbB2 protein level was determined relative to that of actin in each cell lysate and its level in Mock‐transfected cells for each experiment was assigned a value of 1. *P < 0.05 versus Mock. Values are means ± SD.
Reciprocal expression of ErbB2 and FRS2β in primary cultured neural cells under differentiated and undifferentiated conditions and an increase in ErbB2 protein levels by knockdown of endogenous FRS2β
We next examined whether endogenous FRS2β contributes to downregulation of the ErbB2 protein in a more physiological setting. Although the level of expression of the FRS2β protein was too low for our intended experiments in the commonly used cultured cell lines, this protein is expressed in primary fetal cortical neural cells when cultured in a differentiation medium.17 We therefore cultured primary cortical cells under one of the following two conditions: in a medium that induces neural differentiation or in a medium that is used for the culture of neural stem/progenitor cells (NSPCs). FRS2β and CIN85 were more highly expressed in the differentiated neural cells than in the NSPCs (Fig. 6A). CD2AP was not expressed in these cells at significant levels (Fig. S1). In contrast, ErbB2 was more highly expressed in the NSPCs than in the differentiated neural cells, as previously reported.29 These results indicate that FRS2β/CIN85 and ErbB2 show reciprocal expression in differentiated and non‐differentiated neural cells.
Figure 6.
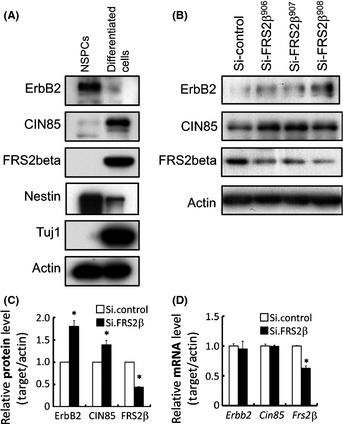
Knockdown of endogenous FRS2β upregulated ErbB2 protein levels in primary cultured embryonic neural cells. (A) Expression of ErbB2, CIN85 and FRS2β in neural stem/progenitor cells (NSPCs) and differentiated neural cells was analyzed by Western blotting. Nestin and Tuj1 were also analyzed as cell type markers. (B‐D) ErbB2 protein levels were upregulated by knockdown of FRS2β in differentiated neural cells. (B) Whole cell lysates were analyzed by Western blotting with the indicated antibodies. (C) The levels of the indicated proteins were quantitatively analyzed in the three independent knockdown studies of (B) using the LAS system. Protein level was normalized to that of actin in the same lysate and is expressed relative to that of the control, which was assigned a value of 1. *P < 0.05 versus control. (D) The mRNA levels of the indicated genes in the cells in (B) were also quantitatively analyzed using quantitative real‐time polymerase chain reaction. *P < 0.05.
When endogenous expression of FRS2β was decreased by transfection of siRNA against Frs2β in these differentiated neural cells, the expression levels of ErbB2 and CIN85 proteins were upregulated (Fig. 6B,C) although the expression levels of ErbB2 and CIN85 transcripts were not significantly changed in these cells (Fig. 6D). These data suggest that FRS2β does not affect the transcription levels of Erbb2 or Cin85, but instead, that FRS2β downregulates ErbB2 at the protein level.
Low levels of FRS2β expression in breast cancer tissues overexpressing ErbB2
Since it is well known that overexpression of ErbB2 contributes to tumorigenesis of human breast cancer, we next examined the expression levels of FRS2β in breast cancer tissues from patients. We immunohistochemically analyzed breast tissue microarrays (TMAs), whose clinicopathological characteristics are shown in Table S1 (Fig. 7A–D). Positive staining for ErbB2 was observed in the cytoplasmic membrane within these specimens (Fig. 7A). We also detected positive cytoplasmic and membrane staining for FRS2β (Fig. 7D). We divided the breast cancer specimens into two groups based on the common criteria, Her2 positive (n = 15) and negative (n = 15). All Her2‐positive breast cancer tissues had low levels of FRS2β expression (15/15), whereas Her2‐negative tissues had either low (10/15) or high levels (5/15) of FRS2β expression (Fig. 7E). This result indicates that Her2‐positive breast cancer tissues expressed significantly less FRS2β protein than Her2‐negative tissues. There was an inverse correlation between expression levels of Her2 and FRS2β (r = −0.44; P = 0.013, Spearman's correlation coefficient by rank test). This result is consistent with the notion that FRS2β may contribute to downregulation of the ErbB2 protein in human breast cancer and that a low amount of FRS2β is associated with overexpression of ErbB2.
Figure 7.
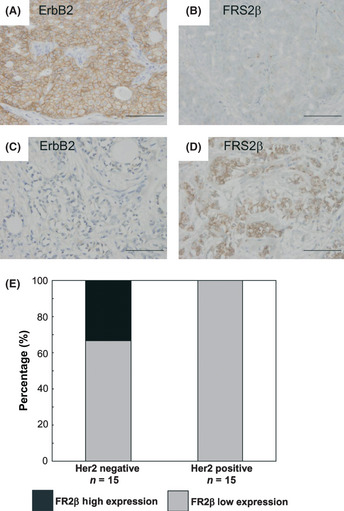
High levels of FRS2β expression are associated with weak ErbB2 expression in breast cancer tissues. (A–D) Representative images of immunohistochemical analysis of breast tissue specimens using anti‐ ErbB2 or anti‐ FRS2β antibodies. A and B or C and D are representative images of adjacent sections from the same specimen. Scale bar = 100 μm. (E) Quantification of staining scores.
Finally we examined expression of CD2AP and CIN85 in various cancer cell lines. We found that both CD2AP and CIN85 were expressed in many cancer cell lines, including breast cancer cells (MCF‐7), glioma cells (A172, T98G and U118) and neuroblastoma cells (SKNMC) (Fig. S1). In addition, we stably expressed FRS2β in MCF‐7 cells. As shown in Fig. S2, expression levels of ErB2 and CD2AP but not EGFR were reduced as compared with mock cells.
Discussion
In this study, using LC‐MS/MS analysis we identified CIN85 and CD2AP as novel FRS2β binding proteins, and demonstrated that an FRS2β‐CIN85/CD2AP complex downregulates the ErbB2 protein, probably by inducing its lysosomal degradation.
The mechanism of EGFR downregulation and that of ErbB2 downregulation that we describe here have many differences. EGF binding to the EGFR accelerates receptor internalization, which is followed by efficient lysosomal targeting of the internalized receptors and results in receptor downregulation. Upon EGF stimulation, the EGFR directly or indirectly binds to Cbl via tyrosine phosphorylated residues on the EGFR. Cbl binds to the SH3 domains of CIN85, and then CIN85 interacts with multiple proteins that are involved in endocytosis, in sorting into early and late endosomes or multi‐vesicular bodies, and in targeting to lysosomes.18, 19, 23 Cbl also ubiquitinates the EGFR and other proteins including CIN85 and it is thought that these ubiquitinated proteins are targeted to lysosomes and do not recycle back to the plasma membrane. We demonstrated that FRS2β can recruit the CIN85/CD2AP‐Cbl degradation complex to ErbB2 regardless of ligand stimulation of ErbB family members. Since there is no ligand for ErbB2, this mechanism may play important roles in the negative regulation of ErbB2 signaling in a physiological setting.
Expression of FRS2β is limited to a few tissues. During development and in adults, FRS2β is expressed in the specific area of neural tissues in which differentiated neural cells are localized as well as in several epithelial cells in the lung, gastrointestinal tract and kidney.14, 17 We detected reciprocal expression of FRS2β and ErbB2 in primary neural cells cultured under two different conditions. We also previously reported that FRS2β is expressed in Tuj1‐positive differentiated neuronal cells.17 In contrast, it has been reported that ErbB2 is expressed in the ventricular zone in which NSPCs are localized.29 Our findings raise the intriguing possibility that ErbB2 signaling might be strictly controlled by FRS2β in vivo, by FRS2β‐mediated downregulation of ErbB2 protein levels.
The EGFR protein does not appear to be downregulated by FRS2β in the absence of ligand stimulation by a mechanism similar to that of ErbB2 downregulation. In fact, we could not co‐immunoprecipitate Cbl and the EGFR with FRS2β without ligand stimulation (data not shown). It is possible that the EGFR‐FRS2β‐CIN85 complex may not efficiently form a complex with Cbl due to a conformational problem, although it is difficult to prove such a hypothesis. However, since the EGFR protein is efficiently downregulated upon EGF stimulation of many common cell lines that do not express FRS2β, it appears that FRS2β does not play an important role in downregulation of the EGFR protein.
The CIN85 adaptor protein interacts with a number of proteins and is involved in a number of biological processes including receptor endocytosis.19 A recent screen, using an LC‐MS/MS screening approach, identified approximately 100 proteins that can interact with CIN85 via its SH3 domains.30 However, this list did not include FRS2β, probably because the screen was performed using lysates of cells of the commonly used HeLa cell line, which do not express FRS2β. The site on FRS2β for CIN85/CD2AP binding that includes R251 is also used for FRS2β binding to activated ERK.16 It is possible that CIN85/CD2AP and ERK do not bind to the same FRS2β protein. We previously reported that ERK colocalizes with FRS2β in the plasma membrane.16 It is thus reasonable to assume that the FRS2β‐CIN85/CD2AP complex and the FRS2β‐ERK complex are separately localized in the endosome and plasma membrane, respectively.
Although FRS2β is weakly, or not at all, expressed in many cell lines, it was striking that FRS2β was expressed in many breast cancer tissues at various levels. These findings emphasize the notion that although expression of FRS2β may be disadvantageous to cell growth and/or survival in vitro, FRS2β does have in vivo roles. FRS2β expression levels tended to be low in breast cancer tissues with high ErbB2 expression levels. Thus, FRS2β may have a role in controlling ErbB2 protein levels even in pathological conditions including cancer.
Recently, it was reported that increased levels of CIN85 contributes to breast cancer malignancy.31 The authors examined functions of CIN85 in breast cancer cell lines including MCF‐7 cells in which expression levels of FRS2β are very low.16 Since CIN85 is able to bind many other proteins including Arf GTPase and N‐WASP, it appears to activate the pathways in which these proteins are involved. It is thus likely that functions of CIN85 are changed dependent on the main binding partners in cells.
Dsiclosure Statement
The authors have no conflict of interest.
Supporting information
Doc S1. Supporting information Materials and Methods.
Fig. S1. Expression of CD2AP and CIN85 in various cells.
Fig. S2. Overexpression of FRS2β downregulates expression of ErbB2 and CD2AP but not EGFR in MCF‐7 cells.
Table S1. Clinicopathological characteristics of the breast cancer patients.
Acknowledgments
We are grateful to Drs A. Sawano and Y. Miyawaki for helping us to develop immunofluorescence imaging. The mCherry plasmid was kindly provided by Dr R. Y. Tsien. We thank Dr H. Miyoshi for providing the lenti‐viral vectors. We thank Mr M. Kameoka (Nikon Corporation) for kind instructions regarding the use of the confocal microscope. We thank Dr T. Hamakubo for the continuous supply of the anti‐FRS2β monoclonal antibody. We are grateful to Dr T. Nakazawa for his constructive instruction and discussion. This work was supported by a grant from the Ministry of Health, Labour and Welfare of Japan for the 3rd‐term Comprehensive 10‐year Strategy for Cancer Control, by a Grant‐in‐Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) for Scientific Research on Innovative Areas, and a Grant‐in‐Aid from Japan Society for the Promotion of Science (JSPS) for Scientific Research to NG. This work was also supported by a Research Fellow Grant of the JSPS to YM (JSPS Research Fellow DC2).
(Cancer Sci, doi: 10.1111/cas.12086, 2013)
References
- 1. Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2: 127–37. [DOI] [PubMed] [Google Scholar]
- 2. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2010; 141: 1117–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gotoh N. Feedback inhibitors of the epidermal growth factor receptor signaling pathways. Int J Biochem Cell Biol 2009; 41: 511–5. [DOI] [PubMed] [Google Scholar]
- 4. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER‐2/neu oncogene. Science 1987; 235: 177–82. [DOI] [PubMed] [Google Scholar]
- 5. Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res 2008; 314: 3093–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Citri A, Yarden Y. EGF‐ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006; 7: 505–16. [DOI] [PubMed] [Google Scholar]
- 7. Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci 2008; 99: 1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sato T, Gotoh N. The FRS2 family of docking/scaffolding adaptor proteins as therapeutic targets of cancer treatment. Expert Opin Ther Targets 2009; 13: 689–700. [DOI] [PubMed] [Google Scholar]
- 9. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005; 16: 139–49. [DOI] [PubMed] [Google Scholar]
- 10. Gotoh N, Manova K, Tanaka S et al The docking protein FRS2alpha is an essential component of multiple fibroblast growth factor responses during early mouse development. Mol Cell Biol 2005; 25: 4105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hadari YR, Gotoh N, Kouhara H, Lax I, Schlessinger J. Critical role for the docking‐protein FRS2 alpha in FGF receptor‐mediated signal transduction pathways. Proc Natl Acad Sci USA 2001; 98: 8578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamamoto S, Yoshino I, Shimazaki T et al Essential role of Shp2‐binding sites on FRS2alpha for corticogenesis and for FGF2‐dependent proliferation of neural progenitor cells. Proc Natl Acad Sci USA 2005; 102: 15983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gotoh N, Ito M, Yamamoto S et al Tyrosine phosphorylation sites on FRS2alpha responsible for Shp2 recruitment are critical for induction of lens and retina. Proc Natl Acad Sci USA 2004; 101: 17144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gotoh N, Laks S, Nakashima M, Lax I, Schlessinger J. FRS2 family docking proteins with overlapping roles in activation of MAP kinase have distinct spatial‐temporal patterns of expression of their transcripts. FEBS Lett 2004; 564: 14–8. [DOI] [PubMed] [Google Scholar]
- 15. Huang L, Watanabe M, Chikamori M et al Unique role of SNT‐2/FRS2beta/FRS3 docking/adaptor protein for negative regulation in EGF receptor tyrosine kinase signaling pathways. Oncogene 2006; 25: 6457–66. [DOI] [PubMed] [Google Scholar]
- 16. Iejima D, Minegishi Y, Takenaka K et al FRS2beta, a potential prognostic gene for non‐small cell lung cancer, encodes a feedback inhibitor of EGF receptor family members by ERK binding. Oncogene 2010; 29: 3087–99. [DOI] [PubMed] [Google Scholar]
- 17. Minegishi Y, Iwanari H, Mochizuki Y et al Prominent expression of FRS2beta protein in neural cells and its association with intracellular vesicles. FEBS Lett 2009; 583: 807–14. [DOI] [PubMed] [Google Scholar]
- 18. Dikic I. CIN85/CMS family of adaptor molecules. FEBS Lett 2002; 529: 110–5. [DOI] [PubMed] [Google Scholar]
- 19. Havrylov S, Redowicz MJ, Buchman VL. Emerging roles of Ruk/CIN85 in vesicle‐mediated transport, adhesion, migration and malignancy. Traffic 2010; 11: 721–31. [DOI] [PubMed] [Google Scholar]
- 20. Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 2003; 5: 461–6. [DOI] [PubMed] [Google Scholar]
- 21. Schroeder B, Weller SG, Chen J, Billadeau D, McNiven MA. A Dyn2‐CIN85 complex mediates degradative traffic of the EGFR by regulation of late endosomal budding. EMBO J 2010; 29: 3039–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurakin AV, Wu S, Bredesen DE. Atypical recognition consensus of CIN85/SETA/Ruk SH3 domains revealed by target‐assisted iterative screening. J Biol Chem 2003; 278: 34102–9. [DOI] [PubMed] [Google Scholar]
- 23. Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol 2005; 6: 907–18. [DOI] [PubMed] [Google Scholar]
- 24. Sato T, Shimazaki T, Naka H et al FRS2alpha regulates Erk levels to control a self‐renewal target Hes1 and proliferation of FGF‐responsive neural stem/progenitor cells. Stem Cells 2010; 28: 1661–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakamura Y, Sakakibara S. Miyata et al. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci 2000; 20: 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakaue‐Sawano A, Kurokawa H. Morimura et al. Visualizing spatiotemporal dynamics of multicellular cell‐cycle progression. Cell 2008; 132: 487–98. [DOI] [PubMed] [Google Scholar]
- 27. Acs G, Zhang PJ, McGrath CM et al Hypoxia‐inducible erythropoietin signaling in squamous dysplasia and squamous cell carcinoma of the uterine cervix and its potential role in cervical carcinogenesis and tumor progression. Am J Pathol 2003; 162: 1789–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lax I, Wong A, Lamothe B et al The docking protein FRS2alpha controls a MAP kinase‐mediated negative feedback mechanism for signaling by FGF receptors. Mol Cell 2002; 10: 709–19. [DOI] [PubMed] [Google Scholar]
- 29. Fox IJ, Kornblum HI. Developmental profile of ErbB receptors in murine central nervous system: implications for functional interactions. J Neurosci Res 2005; 79: 584–97. [DOI] [PubMed] [Google Scholar]
- 30. Havrylov S, Rzhepetskyy Y, Malinowska A, Drobot L, Redowicz MJ. Proteins recruited by SH3 domains of Ruk/CIN85 adaptor identified by LC‐MS/MS. Proteome Sci 2009; 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samoylenko A, Vynnytska‐Myronovska B, Byts N et al Increased levels of the HER1 adaptor protein Ruk/CIN85 contributes to breast cancer malignancy. Carcinogenesis 2012; 33: 1976–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Doc S1. Supporting information Materials and Methods.
Fig. S1. Expression of CD2AP and CIN85 in various cells.
Fig. S2. Overexpression of FRS2β downregulates expression of ErbB2 and CD2AP but not EGFR in MCF‐7 cells.
Table S1. Clinicopathological characteristics of the breast cancer patients.


