Abstract
CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) has been implicated in promoting metastasis of cancer cells through several mechanisms, including the inhibition of anoikis, which is cell death triggered by the loss of extracellular matrix interactions. However, the mechanism inhibiting cell death regulated by CDCP1 remains elusive. Inhibition of CDCP1 expression using small interfering RNA (siRNA) induced the cell death of suspended cancer cells without cleaving caspase‐3, a marker of apoptosis; cell death was not inhibited by a general caspase inhibitor, suggesting that the loss of CDCP1 induces caspase‐independent cell death. In contrast, knockdown of CDCP1 as well as protein kinase Cδ (PKCδ), a downstream effector of CDCP1, in a suspension culture of lung cancer cells resulted in marked induction of membranous microtubule‐associated protein 1 light chain 3 (LC3)‐II protein, a hallmark of autophagy, and caused the formation of an autophagosome structure visualized using green fluorescent protein‐tagged LC3‐II. Expression and phosphorylation of exogenous CDCP1 by Fyn kinase reduced the formation of autophagosomes and inhibited phosphorylation of CDCP1 by PP2, a Src kinase inhibitor or inhibited PKCδ by rottlerin, stimulating autophagosome formation. Moreover, death of suspended lung cancer cells induced by CDCP1 siRNA or by PKCδ siRNA was reduced by the autophagy inhibitor 3‐methyladenine. These results indicate that CDCP1‐PKCδ signaling plays a critical role in inhibiting autophagy, which is responsible for anoikis resistance of lung cancer cells.
CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1), also known as SIMA135 and TRASK,1, 2 is a type I transmembrane protein with three putative CUB domains as extracellular domains and several tyrosine residues that are phosphorylated by Src family kinases (SFK) in the cytoplasmic domain.1, 2, 3, 4, 5, 6 Expression of CDCP1 has been reported in several human malignancies, such as colon and breast cancers.3, 7 CDCP1 expression is strongly associated with progression and poor prognosis of various cancers, including renal cell carcinoma, lung adenocarcinoma and pancreatic cancer.8, 9, 10 CDCP1 was reported to bind directly to protein kinase Cδ (PKCδ) at a unique C2 domain in a phosphorylation‐dependent manner.5 We recently determined the biological significance of this interaction with PKCδ. We found that tyrosine‐phosphorylated CDCP1 regulates cell migration, invasion and anoikis resistance of cancer cells by physically linking between SFK and PKCδ.8, 11
Anoikis is a form of cell death triggered by the loss of cell survival signals based on interactions with the extracellular matrix (ECM).12 Anoikis is considered to be physiologically important for maintaining homeostasis and tissue architecture.13 However, anoikis resistance acquired during carcinogenesis has been described as a core aspect for tumor progression and metastasis of cancer cells.14 Because anoikis resistance is unique to metastatic cancer cells, it might be a good target for developing antimetastatic therapy, which has minimal effects on normal tissue cells. However, it is unknown how CDCP1 signaling confers resistance to anoikis in cancer cells.
In the present study, we found that the cellular response compatible with autophagy is involved in anoikis of lung cancer cells caused by the suppression of CDCP1 signaling. Autophagy is an evolutionally conserved process that is characterized by an increase in the number of autophagosomes surrounding cellular organelles such as the Golgi complex, mitochondria and endoplasmic reticulum.15 High levels of autophagy can lead to cell death.16 In contrast to apoptosis, cell death mediated by autophagy is caspase independent and does not involve classic DNA laddering.17 Accumulating evidence suggests that cancer cells generally show reduced autophagy during cancer progression, supporting the hypothesis that defective autophagy provides resistance to metabolic stress such as hypoxia, acidity and chemotherapeutics, promotes tumor cell survival and plays a role in tumorigenesis.18
Here, we present in vitro evidence that CDCP1‐PKCδ signaling plays an essential role in suppressing autophagy in anchorage‐free lung cancer cells, thus protecting the cells from anoikis during the development of cancer metastasis.
Materials and Methods
Plasmids, antibodies and reagents
Plasmids encoding CDCP1, the CDCP1 rescue mutant that resists treatment of CDCP1 siRNA and the CDCP1 Y734F mutant (Tyr734 to Phe) have been described previously.8, 11 N‐terminus‐truncated CDCP1, which contains the amino acid sequence of CDCP1 from Lys369 till Glu836 with the C‐terminus FLAG tag, was generated by PCR using a KOD‐Plus Mutagenesis kit (Toyobo Co. Ltd., Osaka, Japan). Rabbit anti‐cleaved caspase‐3 (Asp175) antibody (no. 9664) was purchased from Cell Signaling Technology Inc. (Beverly, MA, USA). Rabbit polyclonal antibodies against CDCP1 and tyrosine‐phosphorylated CDCP1 (Tyr734) were prepared as described previously.11 Mouse anti‐α‐tubulin clone B‐5‐1‐2 antibody (T5168), mouse anti‐FLAG M2 peroxidase antibody (A8592) and rabbit anti‐LC3B antibody (L7543) were purchased from Sigma (St Louis, MO, USA). Goat anti‐caspase‐3 p20 (N‐19) antibody (sc‐1226) and rabbit anti‐PKCδ (C‐20) antibody (sc‐937) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The green fluorescent protein (GFP)‐LC3 plasmid was provided by Dr Hirofumi Arakawa (National Cancer Center Research Institute, Tokyo, Japan). The retroviral vectors pQCXIN (Clontech, Mountain View, CA, USA) and pCMSCVbsd were used to express human CDCP1 and Y734F with a FLAG‐tag at the C‐terminus and Fyn, a Src family kinase with a double HA‐tag at the C‐terminus, respectively. pCMSCVbsd contains the blasticidin resistance gene in place of the puromycin resistance gene of pCMSCVpuro (Clontech).19 These retroviruses were converted into the destination vectors with a vector conversion kit (Invitrogen, Carlsbad, CA, USA). The inhibitor, PP2, was purchased from Calbiochem‐Novabiochem (Darmstadt, Germany). Etoposide, rottlerin and 3‐methyladenine (3MA) were purchased from Sigma‐Aldrich (St. Louis, MO, USA). Z‐VAD‐FMK, a general caspase inhibitor, and Z‐FA‐FMK, a negative control for caspase inhibitor, were purchased from BD Bioscience (Franklin Lakes, NJ, USA).
Cell culture and siRNA treatment
Human lung adenocarcinoma cell lines, A549 and H322, were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2. To investigate the effect of PP2, etoposide, rottlerin or 3MA treatment, cells were treated with 10 μm of PP2, 80 μm of etoposide, 4 μm of rottlerin and 20 mm of 3MA, respectively.
Two sets of siRNA for CDCP1 were synthesized as follows (Invitrogen): siCDCP1‐1 (siCD‐1) sense, 5′‐UAAUGUUGCUUUCUCGUGGCAGAGC‐3′, antisense, 5′‐GCUCUGCCACGAGAAAGCAACAUUA‐3′; and siCDCP1‐2 (siCD‐2) sense, 5′‐CAGAGUCCUGAGAAUCACUUUGUCA‐3′; antisense, 5′‐UGACAAAGUGAUUCUCAGGACUCUG‐3′. Two sets of siRNA for PKCδ were synthesized as follows: siPKCδ‐1 sense, 5′‐GGUGCAGAAGAAGCCGACCAUGUAU‐3′, antisense, 5′‐AUACAUGGUCGGCUUCUUCUGCACC‐3′; and siPKCδ‐2 sense, 5′‐CCAAGGUGUUGAUGGUCGGUUCAGUA‐3′, antisense, 5′‐UACUGAACCGACCAUCAACACCUUGG‐3′. Control siRNA (scramble II duplex: 5′‐GCGCGCUUUGUAGGAUUCGdTdT‐3′) was purchased from Dharmacon (Lafayette, CO, USA). siRNA (40 pmol) were incorporated into cells using Lipofectamine 2000 (Life Technologies Corporation, Carlsbad, CA, USA) according to the manufacturer's instructions. The cells were subjected to further experiments 72 h after siRNA treatment.
Western blotting
Cell lysates were prepared using protease inhibitors in PLC lysis buffer (10 mm Tris–HCl [pH 7.5], 5 mm EGTA, 150 mm NaCl, 1% Triton X‐100, 10% glycerol, 10 μg/mL aprotinin, 1 mm sodium orthovanadate [Na3VO4] and 100 μg/mL leupeptin). Protein concentration was measured using the BCA Protein Assay (Pierce, Rockford, IL, USA). Samples were separated using SDS‐PAGE and transferred to a polyvinylidene difluoride membrane (Immobilon‐P, Millipore, Billerica, MA, USA). After blocking the membrane using a blocking buffer (Blocking One, Nakarai Tesque, Kyoto, Japan), the membrane was probed using antibodies. The membrane was further probed with horseradish peroxidase (HRP)‐conjugated anti‐mouse and anti‐rabbit IgG (1:4000) for antibody visualization. Images were acquired using a molecular imager GS‐800 (Bio‐Rad, Hercules, CA, USA).
Detection of cell death (trypan blue staining)
Trypan blue staining (0.4%; Gibco, Grand Island, NY, USA) was used to visualize cell death. The cells were transfected with or without siRNA targeting CDCP1 and PKCδ mRNA. After 24 h, the cells were reseeded into a 2‐methacryloyloxyethyl phosphorylcholine (MPC)‐coated plate (Nunc, Penfield, NY, USA) or normal cell culture plates with or without Z‐FA‐FMK, Z‐VAD‐FMK or 3MA treatment. After 48 h, cells were treated with trypan blue stain reagent (Gibco). The number of cells positive or negative for trypan blue staining was determined by counting cells on four slides for two plates (five fields per one slide) at a magnification of ×20 and the percent of trypan blue‐positive cells was calculated as the number of trypan blue‐positive cells/total number of cells. The results are expressed as the mean of three independent experiments.
Measurement of GFP‐LC3 dot‐positive cells
A549 cells and H322 cells were plated 24 h before transfection. Plated cells were transfected with or without siRNA targeting CDCP1 and PKCδ mRNA. After 24 h, cells were reseeded into a MPC‐coated plate or normal cell culture plates and transfected with the GFP‐LC3 plasmid using Lipofectamine 2000 with or without PP2 rottlerin or 3MA treatment. After 48 h, the cells were fixed with 4% paraformaldehyde, washed three times with phosphate‐buffered saline (PBS), mounted using SlowFade Gold antifade reagent (Molecular Probes, Eugene, OR, USA) and analyzed using a fluorescence microscope equipped with a Charge Coupled Device camera at a magnification of ×100. GFP‐LC3 dot‐positive cells were counted as the cells generating more than 15 GFP‐LC3 dots. Approximately 80–130 cells were used to calculate the percent of GFP‐LC3 dot‐positive cells. The mean (SE) of at least three independent determinations was calculated.
Results
Cell death induced by CDCP1 siRNA in suspended lung cancer cells is caspase independent
Our previous report revealed that CDCP1 inhibits anoikis in suspension cultures of lung adenocarcinoma cells;11 however, it was unclear which type of cell death was induced by the loss of CDCP1. Treatment of anchorage‐independent A549 lung adenocarcinoma cells by CDCP1 siRNA induced cell death in suspension conditions, as previously reported.11 However, it did not induce a detectable level of cleaved caspase‐3, a marker of apoptosis, either 48 or 96 h after treatment, while treatment with etoposide, an apoptosis inducer, generated cleaved caspase‐3 in 48 h (Fig. 1a). The total caspase‐3 level decreased following treatment with etoposide, whereas it did not change after treatment with CDCP1 siRNA (Fig. 1a).
Figure 1.
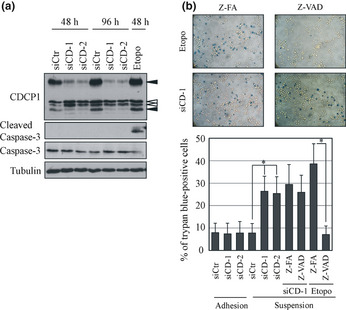
Cell death induced by CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) in lung cancer cells cultured under suspension condition is caspase independent. (a) A549 cells were treated with control (siCtr) or CDCP1 siRNA (siCD‐1 and siCD‐2) and cultured in a low cell‐binding plate (2‐methacryloyloxyethyl phosphorylcholine plate) for 48 or 96 h, respectively. The cell lysate was subjected to western blot analysis with the indicated antibodies. Black arrowheads indicate CDCP1. White arrowheads indicate non‐specific bands. (b) The effect of CDCP1 siRNA, Z‐VAD‐FMK (Z‐VAD) and etoposide (Etopo) on cell death in suspension culture was determined using trypan blue staining and expressed as a percentage. Upper panels represent trypan blue staining of A549 cells treated with siCD‐1 and Z‐FA‐FMK (Z‐FA) or Z‐VAD and treated with etoposide and Z‐FA or Z‐VAD. Data are the means ± SD from three experiments. *P‐values are based on Student's t‐test and were considered significant for P < 0.01.
Approximately 10% of the original A549 cells were positive for trypan blue staining, an indicator of cell death, either in adhesion culture or in suspension culture, which increased to approximately 20–38% after treatment with CDCP1 siRNA only in the suspension culture (Fig. 1b). The inhibitor of general caspase, Z‐VAD‐FMK, reduced cell death of A549 cells treated with etoposide, while it did not affect cell death induced by CDCP1 siRNA (Fig. 1b). Z‐FA‐FMK, a negative control, showed no reduction in cell death of A549 cells (Fig. 1b). These results indicate that anoikis induced in lung cancer cells by CDCP1 depletion is caspase independent.
CDCP1 and PKCδ inhibit autophagy in lung cancer cells cultured under suspension conditions
While investigating several caspase‐independent types of cell death, we evaluated whether CDCP1 regulates autophagy, which has recently been proposed as a cause of caspase‐independent cell death.20 Suppression of CDCP1 expression by siRNA induced the expression of LC3‐II, a hallmark of autophagy, in lung cancer cell lines A549 (Fig. 2a), HCC44 and PC14 (data not shown) cultured under suspension conditions. In lung cancer cells transfected with GFP‐LC3, CDCP1 siRNA induced the formation of GFP‐LC3 dots, which are defined as autophagosomes in suspension culture, but these dots were not obvious in the normal adhesion culture (Fig. 2b). In the nutrient depletion conditions, enhancement of GFP‐LC3 dot formation by CDCP1 knockdown was observed in adherent cancer cells while the level of dot formation was very low (Fig. S1). To confirm the association between GFP‐LC3 dot formation and autophagy, we treated A549 cells with 3MA, an inhibitor of autophagy, and found that inhibition of autophagy decreases the formation of GFP‐LC3 dots (Fig. 2b,d). Using electron microscopy, significant induction of vacuoles was observed in A549 cells treated with CDCP1 siRNA (Fig. 2c). The autophagosome formation was also observed with treatment of PKCδ siRNA, a downstream signal molecule of CDCP1 (Fig. 2a,d). These results indicate that CDCP1 and its downstream molecule PKCδ inhibit autophagy in suspension culture. It has been reported that tissue transglutaminase (TG2), which might be under the control of PKCδ, is involved in the inhibition of autophagy21 Therefore we checked the expression of TG2 and found that loss of CDCP1 by siRNA reduces the expression of TG2 protein in several cancer cells during induction of autophagy (Fig. S2), suggesting a possible role of TG2 in CDCP1‐mediated suppression of autophagy.
Figure 2.
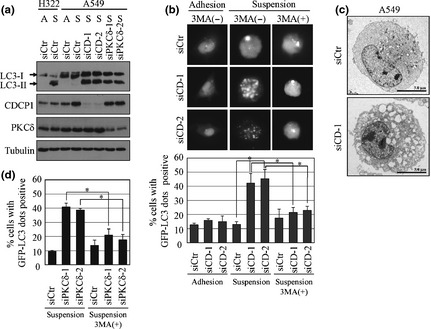
CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) and protein kinase Cδ (PKCδ) inhibit autophagosome formation in lung cancer cells cultured under suspension conditions. (a) A549 cells and H322 cells were treated with control (siCtr), CDCP1 (siCD‐1 and siCD‐2), or PKCδ siRNA (siPKCδ‐1 and siPKCδ‐2) and cultured in a normal cell culture plate or 2‐methacryloyloxyethyl phosphorylcholine plate for 48 h. The cell lysate was subjected to western blot analysis with the indicated antibodies. A, adhesion culture; S, suspension culture. (b) The effect of CDCP1 siRNA with or without 3‐methyladenine (3MA) treatment on autophagosome formation in adhesion or suspension cultures was determined by measuring the green fluorescent protein (GFP)‐LC3 dot‐positive cells as a percentage. The upper panel represents the GFP‐LC3 dot‐positive A549 cells treated with siCD‐1 and siCD‐2 with or without 3MA treatment. The lower panel represents the percentage of GFP‐LC3 dot‐positive cells. (c) Electron micrographs showing the structure of A549 cells treated with control siRNA (siCtr) or CDCP1 siRNA‐1 (siCD‐1). (d) The effect of PKCδ siRNA with or without 3MA treatment on autophagosome formation in suspension culture of A549 cells was determined by measuring GFP‐LC3 dot‐positive cells as a percentage. Data are the means ± SD from three experiments. *P‐values based on Student's t‐test were considered significant for P < 0.01.
Phosphorylation of CDCP1 is required for CDCP1‐mediated inhibition of autophagy in anchorage‐free lung cancer cells
It has been found that phosphorylation of CDCP1 by Src family kinases (SFK) such as Fyn and Yes11 initiates signaling associated with anoikis resistance of cancer cells. Involvement of phosphorylation‐dependent CDCP1 signaling in autophagy was then examined in H322 lung cancer cells. The original H322 cell line was anchorage dependent based on its low CDCP1 expression and low SFK activity, which has been shown to convert to anchorage independent by expression of both CDCP1 and Fyn.11 Suspension of H322 cells caused a remarkable induction of LC3‐II and formation of GFP‐LC3 dots compared with A549 cells (Figs 2a,b 3a). A reduction in the number of GFP‐LC3 dots in suspended H322 cells was observed only when CDCP1 and Fyn were expressed together, while expression of either CDCP1 alone, Fyn alone or a combination of Fyn with Y734F CDCP1, which lacks the tyrosine phosphorylation site of CDCP1, did not affect GFP‐LC3 dot formation (Fig. 3a). Considering the reported phosphorylation status of CDCP1,11 these results clearly demonstrate that expression of phosphorylated CDCP1 is required for suppression of autophagy. Additionally, SFK inhibitor PP2, which reduces phosphorylation of CDCP1, or rottlerin, which might inhibit PKCd, induce formation of GFP‐LC3 dots in A549 cells (Fig. 3b). In contrast, N‐terminus‐truncated CDCP1 (NtCDCP1), which mimics the cleaved CDCP1, could rescue A549 cells from the formation of GFP‐LC3 dots induced by CDCP1 siRNA (Fig. 3c).
Figure 3.
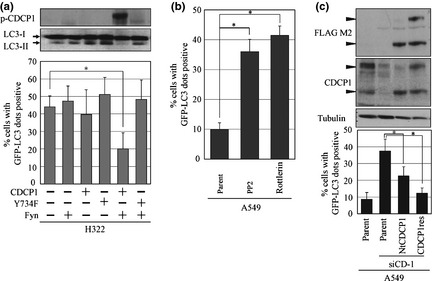
Phosphorylation of CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) inhibits autophagy in suspension culture. (a) H322 cells overexpressing CDCP1 or CDCP1 mutant (Y734F) and Fyn tagged with HA (Fyn) were used to measure autophagosome formation. Note that phosphorylated CDCP1, which is present following overexpression of CDCP1 and Fyn, reduced the number of LC3‐II protein and green fluorescent protein (GFP)‐LC3 dot‐positive cells. (b) A549 cells were treated with or without PP2 or rottlerin in suspension culture and autophagy was detected by GFP‐LC3 dot‐positive cells. (c) A549 cells stably expressed N‐terminus‐truncated CDCP1 (NtCDCP1) or full‐length CDCP1 rescue mutant (CDCP1res) were treated with CDCP1 siRNA and autophagy was detected by the number of GFP‐LC3 dot‐positive cells. Black arrowheads indicate CDCP1. Data are the means ± SD from three experiments. *P‐values were determined using the Student's t‐test and considered significant for P < 0.01.
Inhibition of autophagy by CDCP1‐PKCδ signaling is responsible for anoikis resistance of cancer cells
Because a correlation between the level of cell death and the formation of autophagosome was observed in A549 cells cultured under suspension conditions (Figs 1b,2), we examined whether the suppression of autophagy by CDCP1 signaling contributes to inhibition of anoikis. Increased cell death of A549 cells under suspension conditions treated with each CDCP1 siRNA was rescued after treatment with 3MA, an autophagy inhibitor (Fig. 4a). Reduction of anoikis was observed in parental H322 cells treated with 3MA (Fig. 4b). As expected, H322 cells expressing both CDCP1 and Fyn regained anoikis following treatment with rottlerin or PKCδ siRNA, which was also rescued after treatment with 3MA (Fig. 4b). These results suggest that CDCP1‐PKCδ signaling inhibits anoikis by suppressing autophagy in lung cancer cells.
Figure 4.
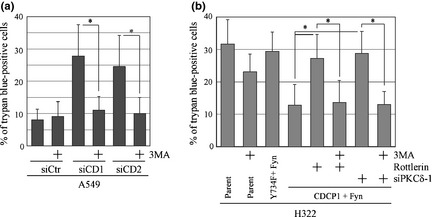
CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1)‐protein kinase Cδ (PKCδ) signaling regulates cancer cell survival by inhibiting autophagy. (a) The effect of the control or CDCP1 siRNA with or without 3‐methyladenine (3MA) treatment on cell death in suspension culture was determined by measuring cells staining positive for trypan blue as a percentage. (b) H322 cells overexpressing CDCP1 or CDCP1 mutant (Y734F) and Fyn tagged with HA (Fyn) were used to measure cell death in suspension culture. The effect of rottlerin and PKCδ siRNA (siPKCδ‐1) with or without 3MA treatment on cell death of H322 cells overexpressing CDCP1 and Fyn in suspension culture was determined by measuring trypan blue‐positive cells as a percentage. Data are the means ± SD from three experiments. *P‐values were determined using Student's t‐test and were considered significant for P < 0.01.
Discussion
Both apoptosis and autophagy are highly regulated forms of programmed cell death and play crucial roles in physiological processes such as the development, homeostasis and elimination of abnormal cells. Our previous study showed that CDCP1 inhibits anoikis in cancer cells.11 Anoikis is an important mechanism for maintaining tissue homeostasis by killing cells that have lost contact with the ECM.22 The ability to survive in the absence of ECM interactions is considered to be a critical feature of metastasis because cancer cells moving through the vasculature or growing to secondary sites are either deprived of matrix or exposed foreign matrix components.23 Indeed, CDCP1 promotes distant metastasis and peritoneal dissemination of cancer cells in mouse models.11, 24 High CDCP1 expression has been observed in various types of human cancers1, 8, 24, 25 and has been associated with poor prognosis of patients in some cases.8, 10 In the present study, we are the first to provide evidence that CDCP1 regulates autophagy in cancer cells and further demonstrate that suppression of autophagy by CDCP1‐PKCδ signaling plays an important role in preventing anoikis of lung cancer cells.
First, using two different approaches, we showed that anoikis induced by CDCP1 siRNA in A549 lung cancer cells is caspase independent. Cell death caused by CDCP1 inhibition did not produce cleaved caspase‐3, one of the key molecules involved in apoptosis (Fig. 1a). Activation of caspase‐3 requires proteolytic processing of its inactive zymogen into activated fragments such as cleaved caspase‐3.26 Furthermore, a general caspase inhibitor did not affect anoikis following treatment with CDCP1 siRNA (Fig. 1b). Thus, our data demonstrate that cell death induced by CDCP1 siRNA in lung cancer cells differs from typical caspase‐dependent apoptosis. We examined cell autophagy and its involvement in this type of cell death.
Autophagy is characterized by an increase in the number of autophagosomes, vesicles that surround such cellular organelles as the Golgi complex, mitochondria, polyribosomes and endoplasmic reticulum.15 The role of autophagy in cell death remains controversial and in some contexts autophagy might even promote cell survival through a number of mechanisms, such as generating nutrients and energy to sustain starving cells or stressed cells, degrading toxic proteins and protecting cells from oxidative stress.27, 28, 29 Most of these protective functions of autophagy are responsive and temporary. In contrast, cancer cells generally tend to undergo a lower level of autophagy than their normal counterparts, which supports the concept that defects in autophagic cell death play a role in the process of tumorigenesis.18 Several autophagy regulators are downregulated in human cancers. In fact, the importance of autophagy in tumor suppression has been supported by studies of tumors in Beclin 1 ± mice.30, 31 It was also reported that Beclin‐1 is monoallelically deleted in 40–75% of human breast, prostate and ovarian tumors.32 Previous studies have demonstrated that autophagy induces cancer cell death.21, 33, 34
Multiple models have been proposed for the progression from autophagy towards cell death; one of the models is that autophagosomes merge with lysosomes and digest organelles, leading to cell death (autophagic cell death).16 In contrast to apoptosis, autophagic cell death is caspase independent.17 Our data show that a reduction of CDCP1 led to autophagy by inducing the LC3‐II protein and GFP‐LC3 dots in lung cancer cells cultured under suspension conditions (Fig. 2). Electron microscopy revealed that massive formation of large vacuoles is induced by the loss of CDCP1 in suspension, while typical autophagosome‐like bilayer structures were not clearly observed (Fig. 2c). Because autophagic markers such as LC3‐II were observed under these conditions, it might be possible that the autophagosome structure was destroyed during the process of the formation of prominent large vacuoles. Expression of phosphorylated CDCP1 reduced autophagic markers; in contrast, an SFK inhibitor induced LC3‐II protein (data not shown) and GFP‐LC3 dots (Fig. 3b), which supports previous reports regarding the effects of Src inhibitors on autophagy.33 Because CDCP1 is a major substrate of SFK in suspended cancer cells,11, 24 CDCP1 might be a key regulator of autophagy controlled by SFK. It has been reported that induction of autophagy was observed in T lymphocytes by the selective knockdown of Fyn.35 In addition, the SFK inhibitor induces autophagy in cancer cells.33, 36, 37 These reports might also intensify the potential role of CDCP1, which is a main substrate of SFK as a general regulator of autophagy.
Recent reports suggest that cleavage of CDCP1 is critical for activation of CDCP1 signaling.38, 39, 40, 41 Cleaved CDCP1 was observed in A549 cells (Fig. 1a) and autophagy induced by CDCP1 siRNA was rescued by NtCDCP1, which mimics the cleaved form of CDCP1 (Fig. 3c). Results of the current study support that cleaved CDCP1 might also have an essential role in suppression of autophagy. It has been revealed that tyrosine phosphorylation of CDCP1 is required for binding to the unique C2 domain of PKCδ.5 The association between phosphorylated CDCP1 and PKCδ causes enzymatic activation of PKCδ,8 resulting in multiple malignant phenotypes in cancer cells including anoikis resistance, cell migration and matrix degradation.6, 8, 11 By examining autophagy induced by suspension using siRNA and a rottlerin, we found that PKCδ can act as a downstream effector of CDCP1 also for autophagy, which is supported by a previous report showing that PKCδ constitutively suppresses autophagy and inhibits autophagic cell death in cancer cells.21 As one possible mechanism, expression of TG2, which is reported to suppress autophagy, was reduced by knockdown of CDCP1 during induction of autophagy in several cancer cells (Fig. S2). Therefore, it is possible that induction of TG2 by CDCP1‐PKCδ signaling might have a role in the suppression of autophagy in a wide range of cancers. The results of the current study and our previous study demonstrate that CDCP1‐PKCδ signaling is essential for both suppression of autophagy and cell death in suspension cultures of cancer cells. Finally, we revealed that anoikis induced by suppression of either CDCP1 or PKCδ in cancer cells can be prevented by inhibiting autophagy with 3MA (Fig. 4), showing that autophagy causes anoikis following the loss of CDCP1. Our data show that CDCP1‐PKCδ signaling might contribute to tumor metastasis by protecting matrix‐detached cancer cells from autophagic cell death.
It is still under investigation whether anoikis is generally associated with autophagy. Our results show that the induction of LC3‐II and aggregation of GFP‐LC3 in cell death is induced in a suspension culture of anchorage‐dependent cancer cells (Figs 3a,4b), indicating the possibility that autophagy is widely involved in suspension‐induced cell death. Further studies are required to determine the exact mechanism through which autophagy induces anoikis in various types of cells. Our results might suggest that induction of autophagy can act as protective force for cancer metastasis through anoikis sensitization.
Our recent studies demonstrated that CDCP1 is required for various malignant aspects of cancer cells during the progression of human cancers and is a promising therapeutic target of these cancers. Detection of autophagy might be a good indicator of therapeutic effects when targeting the CDCP1‐PKCδ pathway.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Phosphorylated CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) regulates autophagy in adherent cancer cells during nutrient starvation conditions.
Fig. S2. Expression of tissue transglutaminase (TG2) is attenuated by treatment of CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) siRNA in lung, pancreatic and gastric cancer cells.
Acknowledgments
This work was supported by a Grant‐in‐Aid for Cancer Research and Grant‐in Aid for Scientific Research from the Ministry of Education, Culture, Science and Technology of Japan, and in part by a Grant‐in‐Aid from the Ministry of Health, Labour and Welfare of Japan for the third‐term Comprehensive 10‐Year Strategy for Cancer Control. This work was also supported in part by the National Cancer Center Research and Development Fund (23‐B‐24).
(Cancer Sci 2013; 104: 865–870)
References
- 1. Hooper JD, Zijlstra A, Aimes RT et al Subtractive immunization using highly metastatic human tumor cells identifies SIMA135/CDCP1, a 135 kDa cell surface phosphorylated glycoprotein antigen. Oncogene 2003; 22: 1783–94. [DOI] [PubMed] [Google Scholar]
- 2. Bhatt AS, Erdjument‐Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene 2005; 24: 5333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scherl‐Mostageer M, Sommergruber W, Abseher R, Hauptmann R, Ambros P, Schweifer N. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene 2001; 20: 4402–8. [DOI] [PubMed] [Google Scholar]
- 4. Brown TA, Yang TM, Zaitsevskaia T et al Adhesion or plasmin regulates tyrosine phosphorylation of a novel membrane glycoprotein p80/gp140/CUB domain‐containing protein 1 in epithelia. J Biol Chem 2004; 279: 14772–83. [DOI] [PubMed] [Google Scholar]
- 5. Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCδ is a phosphotyrosine binding domain. Cell 2005; 121: 271–80. [DOI] [PubMed] [Google Scholar]
- 6. Uekita T, Sakai R. Roles of CUB domain‐containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci 2011; 102: 1943–8. [DOI] [PubMed] [Google Scholar]
- 7. Bühring HJ, Kuçi S, Conze T et al CDCP1 identifies a broad spectrum of normal and malignant stem/progenitor cell subsets of hematopoietic and nonhematopoietic origin. Stem Cells 2004; 22: 334–43. [DOI] [PubMed] [Google Scholar]
- 8. Miyazawa Y, Uekita T, Hiraoka N et al CUB domain‐containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res 2010; 70: 5136–46. [DOI] [PubMed] [Google Scholar]
- 9. Awakura Y, Nakamura E, Takahashi T et al Microarray‐based identification of CUB‐domain containing protein 1 as a potential prognostic marker in conventional renal cell carcinoma. J Cancer Res Clin Oncol 2008; 134: 1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ikeda J, Oda T, Inoue M et al Expression of CUB domain containing protein (CDCP1) is correlated with prognosis and survival of patients with adenocarcinoma of lung. Cancer Sci 2009; 100: 429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uekita T, Jia L, Narisawa‐Saito M, Yokota J, Kiyono T, Sakai R. CUB domain‐containing protein 1 is a novel regulator of anoikis resistance lung adenocarcinoma. Mol Cell Biol 2007; 27: 7649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frish SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol 2001; 13: 555–62. [DOI] [PubMed] [Google Scholar]
- 13. Michel JB. Anoikis in the carviovascular system: known and unknown extracellular mediators. Arterioscler Thromb Vasc Biol 2003; 23: 2146–54. [DOI] [PubMed] [Google Scholar]
- 14. Hanahan D, Weiberg RA. The hallmarks of cancer. Cell 2000; 100: 57–70. [DOI] [PubMed] [Google Scholar]
- 15. Kondo Y, Kanazawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005; 5: 726–34. [DOI] [PubMed] [Google Scholar]
- 16. Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med 2003; 8: 65–76. [PMC free article] [PubMed] [Google Scholar]
- 17. Kirisato T, Baba M, Ishikawa N et al Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147: 435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell 2003; 4: 422–4. [DOI] [PubMed] [Google Scholar]
- 19. Kyo S, Nakamura M, Kiyono T et al Successful immortalization of endometrial glandular cells with normal structural and functional characteristics. Am J Pathol 2003; 163: 2259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer 2005; 5: 886–97. [DOI] [PubMed] [Google Scholar]
- 21. Akar U, Ozpolat B, Mehta K et al Tissue transglutaminase inhibits autophagy in pancreatic cancer cells. Mol Cancer Res 2007; 5: 241–9. [DOI] [PubMed] [Google Scholar]
- 22. Gilmore AP. Anoikis. Cell Death Differ 2005; 12: 1473–7. [DOI] [PubMed] [Google Scholar]
- 23. Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002; 2: 563–72. [DOI] [PubMed] [Google Scholar]
- 24. Uekita T, Tanaka M, Takigahira M et al CUB‐domain‐containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol 2008; 172: 1729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu H, Ong SE, Badu‐Nkansah K, Schindler J, White FM, Hynes RO. CUB‐domain‐containing protein 1(CDCP1) activates Src to promote melanoma metastasis. Proc Natl Acad Sci USA 2011; 108: 1379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cohen GM. Caspases: the executioners of apoptosis. Biochem J 1997; 326: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy 2005; 1: e10–8. [DOI] [PubMed] [Google Scholar]
- 28. Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest 2005; 115: 2679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 2008; 19: 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qu X, Yu J, Bhagat G et al Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112: 1809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 2003; 100: 15077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang XH, Jackson S, Seaman M et al Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402: 672–6. [DOI] [PubMed] [Google Scholar]
- 33. Le XF, Mao W, Lu Z, Carter BZ, Bast RC. Dasatinib induces autophagic cell death in human ovarian cancer. Cancer 2010; 116: 4980–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petrovski G, Zahuczky G, Katona K et al Clearance of dying autophagic cells of different origin by professional and non‐professional phagocytosis. Cell Death Differ 2007; 14: 1117–28. [DOI] [PubMed] [Google Scholar]
- 35. Harr MW, McColl KS, Zhong F, Molitoris JK, Distelhorst CW. Glucocorticoids downregulate Fyn and inhibit IP3‐mediated calcium signaling to promote autophagy in T lymphocytes. Autophagy 2010; 6: 912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park MA, Reinehr R, Haussinger D et al Sorafenib activates CD95 and promotes autophagy and cell death via src family kinases in gastrointestinal tumor cells. Mol Cancer Ther 2010; 9: 2220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Milano V, Piao Y, Lafortune T, De Groot J. Dasatinib‐induced autophagy is enhanced in combination with temozolomide in glioma. Mol Cancer Ther 2010; 8: 394–406. [DOI] [PubMed] [Google Scholar]
- 38. Spassov DS, Ahuja D, Wong CH, Moasser MM. The structural features of Trask that mediate its anti‐adhesive function. PLoS ONE 2011; 6: e19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Casar B, He Y, Iconomou M, Hopper JD, Quigley JP, Deryugina EI. Blocking of CDCP1 cleavage in vivo prevents Akt‐dependent survival and inhibits metastatic colonization through PARP1‐mediated apoptosis of cancer cells. Oncogene 2012; 31: 3924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Law ME, Corsino PE, Jahn SC et al Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal‐like breast cancer cells through novel mechanisms. Oncogene 2013; 32: 1316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Casar B, Rimann I, Kato H, Shattil SJ, Quigley JP, Deryugina EI. In vivo cleaved CDCP1 promotes early tumor dissemination via complexing with activated ß1 integrin and induction of FAK/PI3K/Akt motility signaling. Oncogene 2012. doi: 10.1038/onc.2012.547 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phosphorylated CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) regulates autophagy in adherent cancer cells during nutrient starvation conditions.
Fig. S2. Expression of tissue transglutaminase (TG2) is attenuated by treatment of CUB (C1r/C1s, urchin embryonic growth factor, BMP1) domain‐containing protein 1 (CDCP1) siRNA in lung, pancreatic and gastric cancer cells.


